Significance
Bacterial “sex,” or conjugation, is a central mechanism underlying the proliferation of antibiotic resistance. Despite the discovery of conjugation by F plasmids in Escherichia coli over seven decades ago, we have only now visualized the F-encoded transfer channel and F pilus-associated platforms in the E. coli cell envelope by cryoelectron tomography. The channel supports plasmid transfer or assembly of F pili, which remarkably upon synthesis are deposited onto alternative basal structures around the cell surface. The F plasmid transfer system is a paradigm for the bacterial type IV secretion system (T4SS) superfamily. Consequently, the F-encoded structures are broadly informative of mechanisms underlying the biogenesis and function of type IV secretion machines and associated conjugative pili.
Keywords: cryoelectron tomography, DNA conjugation, type IV secretion, pilus, protein transport
Abstract
Bacterial conjugation systems are members of the large type IV secretion system (T4SS) superfamily. Conjugative transfer of F plasmids residing in the Enterobacteriaceae was first reported in the 1940s, yet the architecture of F plasmid-encoded transfer channel and its physical relationship with the F pilus remain unknown. We visualized F-encoded structures in the native bacterial cell envelope by in situ cryoelectron tomography (CryoET). Remarkably, F plasmids encode four distinct structures, not just the translocation channel or channel-pilus complex predicted by prevailing models. The F1 structure is composed of distinct outer and inner membrane complexes and a connecting cylinder that together house the envelope-spanning translocation channel. The F2 structure is essentially the F1 complex with the F pilus attached at the outer membrane (OM). Remarkably, the F3 structure consists of the F pilus attached to a thin, cell envelope-spanning stalk, whereas the F4 structure consists of the pilus docked to the OM without an associated periplasmic density. The traffic ATPase TraC is configured as a hexamer of dimers at the cytoplasmic faces of the F1 and F2 structures, where it respectively regulates substrate transfer and F pilus biogenesis. Together, our findings present architectural renderings of the DNA conjugation or “mating” channel, the channel–pilus connection, and unprecedented pilus basal structures. These structural snapshots support a model for biogenesis of the F transfer system and allow for detailed comparisons with other structurally characterized T4SSs.
In the 1940s, Lederberg and Tatum ushered in the era of bacterial genetics with reports of sexual “mating” orchestrated by the Escherichia coli F “fertility” factor (1). Bacterial sex, now termed conjugation, is widely recognized as a predominant mechanism underlying the rapid and widespread transmission of antibiotic resistance genes and the evolution of untreatable “superbugs” (2). Over the past seven decades, researchers have described many of the underlying regulatory and mechanistic features associated with horizontal gene transfer (3, 4). Only recently, however, have revolutionary advances in high-resolution microscopy enabled visualization of conjugation “nanomachines” to present a structural basis for the proliferation of antibiotic resistance.
Stable subassemblies of conjugation systems have been isolated from the bacterial cell envelope for analysis by single-particle electron microscopy. These include 1.1 megadalton (MDa) outer membrane complexes (OMCs), which span the periplasms and outer membranes of gram-negative species (5–8), and a larger 3-MDa complex consisting of an OMC joined by a thin flexible stalk to an asymmetric inner membrane complex (IMC) (9). This OMC/IMC subassembly was termed the VirB3–10 complex to reflect the fact it is composed of homologs of VirB subunits from the archetypal Agrobacterium tumefaciens VirB/VirD4 system (SI Appendix, Fig. S1) (5–8). Recently, dimers of the VirD4 substrate receptor also were shown to dock with the VirB3–10 complex (10). The resulting VirB3–10/VirD4 substructure represents a major advance, but the absence of a central channel leaves unanswered the question of how DNA substrates are conveyed through the T4SS to recipient cells.
Conjugation systems are grouped together with ancestrally related “effector translocator” systems, which are deployed by pathogenic bacteria to deliver effector proteins to eukaryotic cell targets, as the type IV secretion system (T4SS) superfamily (11). Although intact T4SSs have been refractory to isolation for in vitro structural analysis, recently, by use of cryoelectron tomography (CryoET), we presented an in situ structure of the Dot/Icm “effector translocator” system. This T4SS is used by Legionella pneumophila to deliver several hundred effector proteins into human macrophages for establishment of a replicative vacuole (12). The Dot/Icm system is considerably larger—both compositionally and structurally—than the VirB3–10/VirD4 complex but retains the overall OMC/IMC architecture. Most strikingly, the Dot/Icm machine has a central channel that spans the entire cell envelope, presenting a view of the substrate translocation pathway for a T4SS (SI Appendix, Fig. S1).
In gram-negative bacteria, conjugation machines elaborate not only the mating channel but also an attachment organelle termed the conjugative pilus (11). The best-characterized of these organelles are F pili elaborated by the F “fertility” factors, now known as F plasmids. In a “mate-seeking” mode, the F pilus dynamically extends and retracts to find and then draw a potential recipient cell into direct contact with the donor cell (13). Upon establishment of the stable “mating junction,” the F system transitions to the “mating” or DNA transfer mode (14). Production of the F mating channel and F pilus requires nearly the same set of Tra subunits, and, consequently, the channel and pilus are commonly depicted as a structural unit (SI Appendix, Fig. S2B). Presently, however, the physical and functional relationships of the mating channel and F pilus are undefined. Here, we addressed this deficiency by solving structures of the F plasmid-encoded channel as well as several distinct F pilus basal platforms in the native context of the E. coli cell envelope by in situ CryoET.
Results
The F Transfer Channel Revealed by In Situ CryoET.
We enhanced detection of F-encoded machines by use of pED208, an F plasmid whose derepressed tra operon confers abundant plasmid transfer (≥1 transconjugants per donor; Tc’s/D) and production of up to 20 pili per cell (SI Appendix, Fig. S2) (15). We also exploited an E. coli mreB minC mutant, which grows as unusually thin cells (mreB phenotype) that asymmetrically divide (minC phenotype), resulting in the formation of small (<300 nm in diameter) minicells (SI Appendix, Table S1) (16). Minicells are ideal for structural analyses of bacterial nanomachines because they lack chromosomal DNA but possess the cell envelopes and cytoplasmic constituents, including resident plasmids, of their parent cells. Importantly, E. coli minicells are fully metabolically active in their capacity to produce F pili and transfer F plasmids to viable recipient cells (Fig. 1A and SI Appendix, Fig. S2) (17).
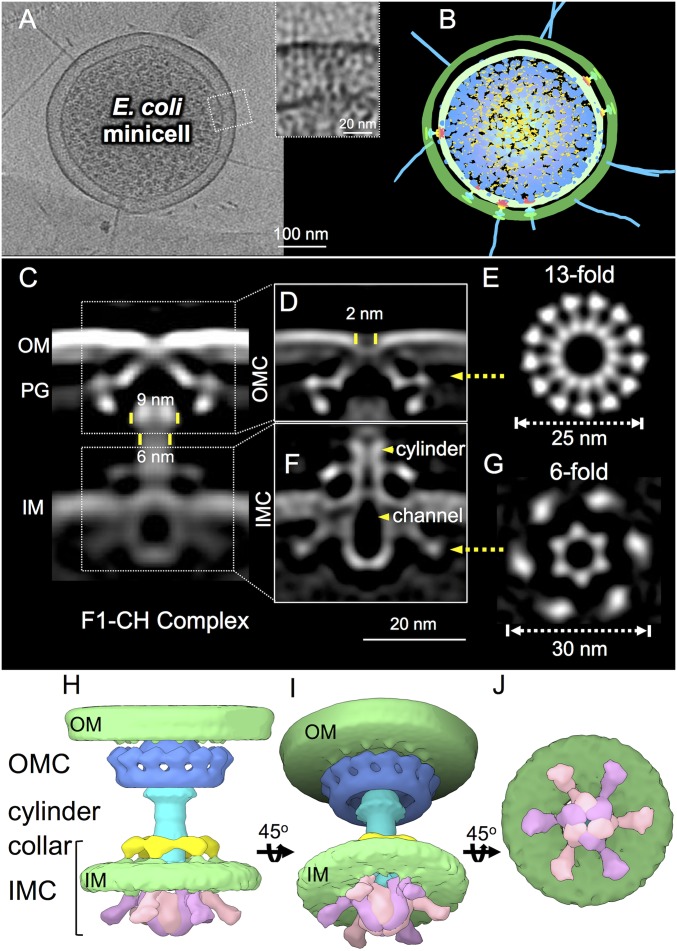
Fig. 1.
In situ structure of the E. coli F-plasmid type IV secretion machine revealed by CryoET and subtomogram averaging. (A) A tomographic slice from a representative E. coli minicell showing multiple T4SS machines embedded in the cell envelope; the boxed region was magnified to show an F1-CH complex. (B) A 3D surface view of the E. coli minicell in A showing T4SS machines and F pili. (C) A central slice of the averaged structure of the F1-CH complex in the cell envelope. Diameters of the flare and cylinder are shown. (D) After refinement, details of the OMC are visible, including a 2-nm gap in the OM. (E) A cross-section view of the region in D marked by a yellow arrow shows 13-fold symmetry of the OMC. (F) After refinement, details of the IMC are visible, including a periplasmic collar, cylinder and central channel (yellow arrowheads), and cytoplasmic inverted “V” structures. (G) A cross-section view of the region in F marked by a yellow arrow shows sixfold symmetry of the IMC. (H–J) Three-dimensional surface renderings of the F1-CH complex shown in different views.
We used a high-throughput CryoET pipeline to visualize thousands of E. coli minicells (SI Appendix, Table S2). Three-dimensional reconstructions revealed the presence of one or more F-encoded structures per minicell, some with and others without an associated F pilus (Fig. 1 A and B and Movie S1). First, we focused on the cell envelope-spanning structure lacking an associated F pilus. From 1,490 machine subtomograms (400 × 400 × 400 voxels) extracted from 511 tomographic reconstructions, an in situ structure was generated at a resolution of ∼2.3 nm (Fig. 1 C and D and SI Appendix, Fig. S3). The envelope-spanning nanomachine is clearly visible, as are densities corresponding to the outer membrane (OM), peptidoglycan (PG), and inner membrane (IM).
The F-encoded channel, here designated the F1 channel or F1-CH complex, is composed of large OMC and IMC subassemblies joined by a central cylinder. The OMC spans and distorts the OM inward, generating a 2-nm gap across the outer leaflet of the OM (Fig. 1D). At the periplasmic face of the OM, the OMC adopts a disk or “flying-saucer” structure with a width of 25 nm and a height of 11.5 nm that extends across the PG layer. In top view, the OMC presents as 13 knobs joined by spokes to a central ring or hub of 13 nm in diameter (Fig. 1E). Three-dimensional classifications revealed the 13-fold symmetric features of the OMC, whose overall architecture was resolved further by imposing a 13-fold symmetry during refinement (SI Appendix, Fig. S3A). The “flying-saucer” architecture and 13-fold symmetry of the F1-CH OMC are features also found in the Dot/Icm system, in contrast to the barrel shape and 14-fold symmetry of the VirB3–10 OMC (Fig. 1 D and E and SI Appendix, Fig. S1).
The F1-CH OMC encircles a central, “flared” cylinder that extends across the periplasm to the IMC (Fig. 1 C, D, and F). The flare and cylinder are 9 and 6 nm wide, respectively, with a central channel of ∼2 nm. The channel extends from the OMC to and through the IMC (Fig. 1 D and F). The cylinder is surrounded by a hexameric collar near the IM, which conforms to the upper boundary of the IMC (Fig. 1F). The IMC extends across the IM, where its cytoplasmic densities consist of two prominent side-by-side inverted “V” structures. The apices of the V structures attach to the IM, the outer arms splay outward, and the inner arms bend inward and connect to close off the entrance to the central channel. In bottom view, the IMC is composed of six concentrically arranged V structures (Fig. 1G). The tips of the inner arms form a central hexameric ring of 10 nm in width, while the outer arms form a knobbed ring of 30 nm. The sixfold symmetric features of the IMC were visible in the initial 3D classifications, and the architecture of the IMC was further resolved by imposing a sixfold symmetry during refinement (SI Appendix, Fig. S3B). Structural details of the entire F1-CH complex are displayed clearly in 3D reconstructions, presented as side and 45° angle views of the intact structure and a bottom view of the hexameric IMC (Fig. 1 H–J). The OMC and IMC structures have been deposited in The Electron Microscopy Data Bank (EMDB) as entries EMD-9344 (18) and EMD-9347 (19), respectively.
The TraC ATPase Forms a Central Hexamer of Dimers at the Channel Entrance.
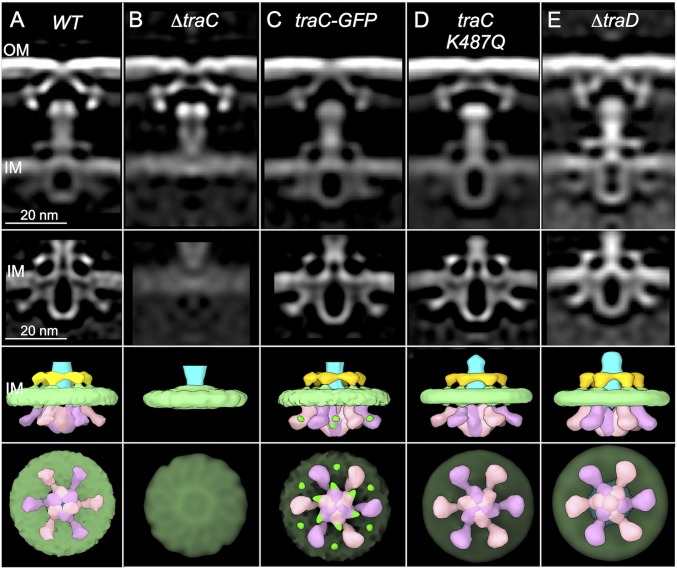
Most T4SSs functioning in gram-negative species require three ATPases (VirB4, VirB11, VirD4) for substrate translocation (SI Appendix, Fig. S1) (11, 20, 21). Two (VirB4, VirB11) also function in pilus biogenesis. In the VirB3–10 structure, the IMC is dominated by two side-by-side hexamers of the VirB4 ATPase (9), whereas in the Dot/Icm system, VirB4-like DotO assembles as six V-shaped structures that are concentrically arranged at the base of the central channel (SI Appendix, Fig. S1) (12). This latter arrangement closely resembles that observed at the base of the F1-CH complex (Fig. 1 F and G). Strikingly, mutant F1-CH complexes from a strain deleted of VirB4-like TraC are devoid of the V-shaped densities, as well as the hexameric collar at the periplasmic face of the IMC (Fig. 2 A and B).
Fig. 2.
Architectures of mutant F1-CH machines. The top row shows the 3D reconstructions of F1-CH complexes visualized in WT, ΔtraC, traC-GFP, traC.K487Q, and ΔtraD mutant cells. The second row shows refinements of the IMCs, and the third and fourth rows show surface renderings from side and bottom views, respectively. (A) Native F1-CH complex (WT) for comparisons. (B) The ΔtraC mutant machine lacks the periplasmic collar and cytoplasmic inverted “V” structures. (C) The mutant machine with TraC-GFP in place of native TraC shows the presence of GFP densities (green dots shown in surface renderings) associated with the inner and outer arms of the inverted “V” structures. (D and E) Mutant machines with TraC.K487Q in place of native TraC or deleted of TraD align well with the WT F1-CH structure.
To confirm that TraC comprises the V densities, we fused a traceable GFP tag to TraC’s C terminus, which yielded a stable and fully functional fusion protein (SI Appendix, Fig. S4). In the solved F1-CH complex from the TraC-GFP-producing strain, the V-shaped densities reappear, but extra densities corresponding to the mass of GFP are associated with both the inner and outer arms of the V structures (Fig. 2C and SI Appendix, Fig. S5). These findings establish unequivocally that the base of the F1-CH complex is composed of a hexamer of TraC dimers at the channel entrance.
VirB4 ATPases require intact Walker A nucleoside triphosphate binding domains for function, but the contributions of ATP binding or hydrolysis to VirB4 oligomerization or association with the transfer channel, or to the overall architecture of the transfer channel, have not been evaluated (22, 23). A TraC mutant with Gln substituted for the conserved K487 residue in the Walker A nucleoside triphosphate binding motif is stably produced but nonfunctional. Production of the K487Q mutant protein in an otherwise WT background also attenuates plasmid transfer and pilus production, suggestive of an interaction with and poisoning effect on the translocation channel (SI Appendix, Fig. S4). The F1-CH complex visualized in a strain producing the K487Q variant aligns well with that from a strain producing native TraC and, most notably, possesses the cytoplasmic V-shaped densities (Fig. 2D). Assembly of the entire F1-CH structure and of TraC as a hexamer of dimers at the channel entrance thus proceed independently of TraC’s capacity to bind or hydrolyze ATP.
In the Dot/Icm system, VirB4-like DotO is a docking platform for VirB11-like DotB, which associates transiently with the T4SS channel (SI Appendix, Fig. S1) (12). VirD4-like DotL, the third ATPase associated with this system, also appears to associate dynamically with the T4SS, as deduced by a lack of effect of the ΔdotL mutation on Dot/Icm machine architecture (12). F systems lack VirB11-like ATPase subunits, but reminiscent of the Dot/Icm system, the WT F1-CH complex aligns well with that visualized in a mutant deleted of VirD4-like TraD (Fig. 2 A and E). TraD and other VirD4-like ATPases are known to bind and deliver substrates to cognate T4SS channels (3, 24, 25), yet our findings for both the F and Dot/Icm systems suggest that these coupling factors might engage only transiently with T4SSs, perhaps as a function of substrate docking.
F Pili Associate with Distinct Basal Platforms.
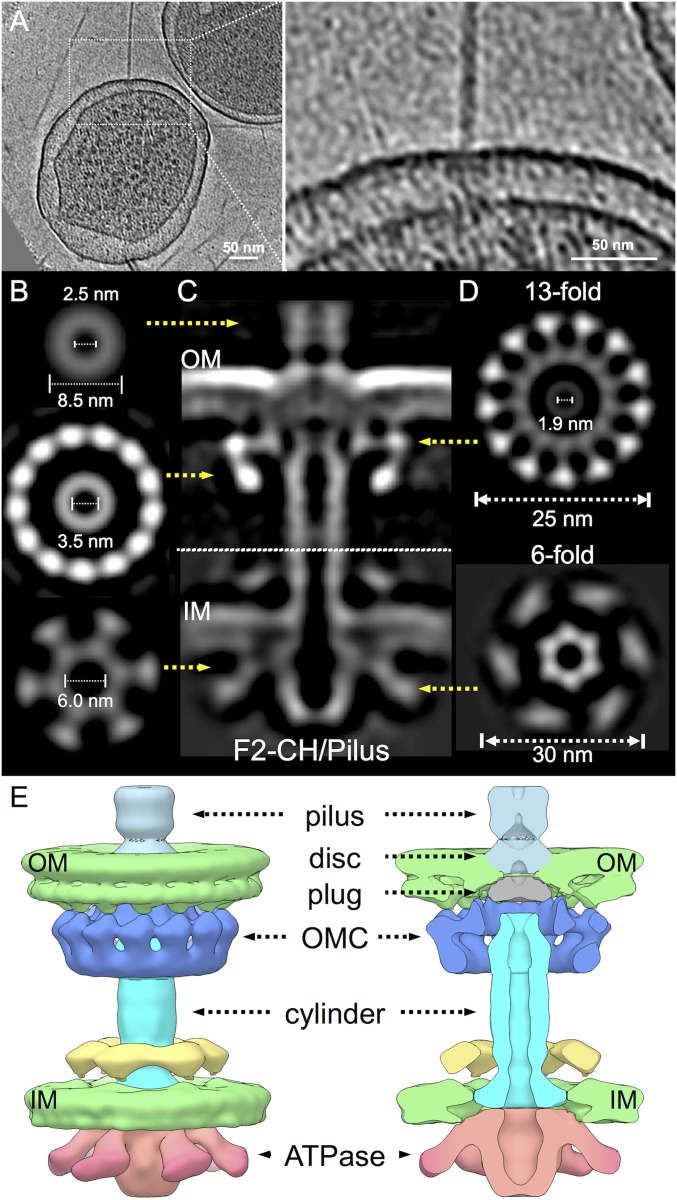
The F1-CH complexes represented ∼30% of all F-encoded structures detectable on the E. coli cell surface. The remaining complexes were associated with the F pilus. One such pilus platform, best described as the F1-CH complex attached to the F pilus, is termed the F2-CH/Pilus complex (Fig. 3A). In the solved structure, the IMC possesses collar and V-shaped densities that align well with those in the F1-CH complex (Figs. 1 F and G and 3 B–D). However, major structural differences exist in the central cylinder and OMC. At the cell surface, the F pilus has dimensions similar to those reported for a high-resolution structure of the F pilus solved by cryoelectron microscopy (Fig. 3B) (26). Intriguingly, at the junction of the pilus with the OMC, the pilus walls narrow to a very thin density (Fig. 3C). This thin density connects to a disk domain, whose location in the OMC eliminates the inward distortion of the OM observed in the F1-CH complex (Figs. 1D and 3C). The disk has a channel approximating that of the pilus lumen, but an adjacent plug domain closes off this channel at the inner leaflet of the OM (Fig. 3 C and E).
Fig. 3.
In situ structure of the F-encoded F2-CH/Pilus complex. (A) A tomographic slice from a representative E. coli minicell and a high magnification view showing the F2-CH/Pilus complex with associated F pilus. (B) Cross-section views show differences in widths of the pilus, cylinder, and inner membrane complex at positions in C indicated by yellow arrows. (C) A central slice of the averaged structure of the F2-CH/Pilus complex across the cell envelope. (D) The OMC shows a 13-fold symmetry in a cross view, and the IMC shows sixfold symmetry in a cross view. (E) Three-dimensional surface rendering of the F2-CH/Pilus and cutaway view with architectural features indicated.
In the F1-CH complex, the crest of the “flying saucer”-shaped OMC forms a ∼30° angle as it joins the OM (Fig. 1C). The corresponding region of the F2-CH/Pilus complex forms a sharp 90° angle (Fig. 3C), creating a more “open” configuration at the OMC–OM interface. This structural transition appears to be necessary to accommodate extension of the central cylinder through the base of the OMC to the inner leaflet of the OM. At this junction, however, the cylinder pinches together, nearly closing off the central channel (Fig. 3C). The observed conformational changes accompanying the F1-CH to F2-CH/Pilus transition (Movie S2) are of postulated importance for F pilus biogenesis (Discussion).
The F pilus is arranged as an 8.5-nm helical fiber composed of 12.8 TraA pilin subunits per turn. Each pilin forms extensive networks of contacts with adjacent pilins and monomers in the upper and lower helical stacks, giving rise to a tightly packed fiber with constant inner lumen, pilus wall, and outer dimensions (26). In this structural context, it is evident that the mature pilus does not span the OMC of the F2-CH/Pilus complex, nor does it extend through the periplasm (Fig. 3C). To evaluate the structural contribution, if any, of the F pilus or TraA pilin to the visualized F-encoded complexes, we solved the structures of mutant machines elaborated by a ΔtraA mutant strain. As expected, the ΔtraA mutant does not elaborate pili or the F2-CH/Pilus complex. However, in the F1-CH complex, the OMC and central cylinder align well with the corresponding substructures from the WT complex (SI Appendix, Fig. S6A), establishing that they are not composed of pilin subunits. Taken together, our findings strongly support a conclusion that the F pilus nucleates assembly at the OM, not at the IM or in the periplasm. The biogenesis pathway for F pili thus differs strikingly from pathways mediating assembly of type IV pili (which are not related to conjugative pili elaborated by T4SSs) or type III secretion system needles, both of which polymerize from IM platforms (SI Appendix, Fig. S6B) (27, 28).
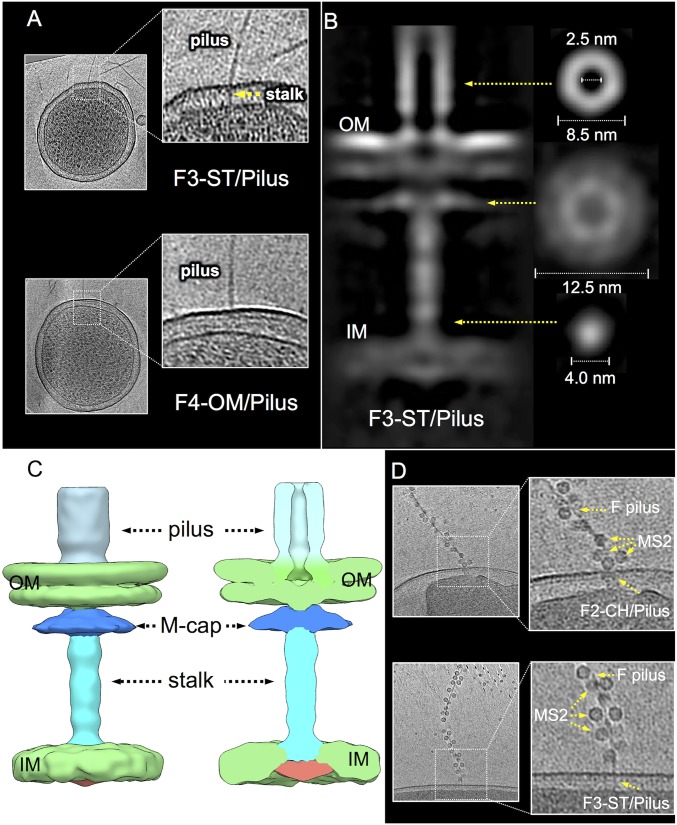
The F2-CH/Pilus complex has the general architecture long predicted for conjugation machines (SI Appendix, Fig. S2B), but, surprisingly, corresponded to only a minor fraction (∼3–5%) of the visualized F pilus-associated structures. Another complex, representing ∼90% of these structures, has never been detected previously and consists of the F pilus attached to a thin, periplasmic stalk (Fig. 4A). In this complex, designated the F3-ST/Pilus, the walls of the F pilus narrow at the OM junction and the pilus appears to connect to OM-spanning disk/plug domains. This is reminiscent of the F2-CH/Pilus complex, although in the F3 structure, the boundaries of the disk/plug domain and the OM are less defined (Figs. 3C and 4 B and C). Remarkably, the F3-ST/Pilus completely lacks the OMC and central cylinder. Instead, a “mushroom-capped” stalk spans the periplasm. The mushroom cap has a diameter of ∼12 nm, is positioned in the PG layer, and lacks a central channel. The stalk is ∼4 nm wide and devoid of structural detail, but also clearly lacks a channel (Fig. 4 B and C). A small density protrudes from the cytoplasmic face of the IM, suggesting that the stalk might span the IM.
Fig. 4.
In situ structures of F-encoded F3-ST/Pilus and F4-OM/Pilus complexes. (A) Tomographic slices from representative E. coli minicells showing the F pilus associated with an envelope-spanning stalk (F3-ST/Pilus) or the OM in the absence of an associated periplasmic density (F4-OM/Pilus). (B) A central slice of the averaged structure of the F3-ST/Pilus complex and cross views of the pilus, “mushroom-cap,” and stalk showing the absence of a central channel; cross-sections views and dimensions are presented for the various structures at positions indicated by yellow arrows. (C) Three-dimensional surface rendering of the F3-ST/Pilus complex and cutaway view with architectural features indicated. (D) MS2 bacteriophage binds to the sides of pOX38-encoded F pili docked onto F2 and F3 basal structures elaborated by intact E. coli cells.
The icosahedryl bacteriophage MS2 binds poorly to pED208 pili, but efficiently to F pili elaborated by another F plasmid, pOX38 (14, 29). pOX38 elaborates only 1–3 pili per cell and was not well-suited for our structural studies, yet its use enabled visualization of MS2 phage-bound F pili extending from intact E. coli MC4100 cells. Importantly, MS2 binds efficiently to F pili associated with F2-CH/Pilus and F3-ST/Pilus basal platforms elaborated by pOX38 (Fig. 4D). These findings confirm that the pili associated with both basal platforms are F pili and further establish that production of both types of pilus-associated complexes is a general property of F plasmids regardless of whether their tra operons are derepressed (abundant pili) or repressed (few pili). Moreover, the data show that F2-CH/Pilus and F3-ST/Pilus structures are elaborated by intact MC4100 cells and are not unique to minicells, a concept underscored by our ability to detect all four F-encoded structures on the surfaces of intact E. coli UU2834 cells (SI Appendix, Fig. S7A).
Finally, we also detected other F pilus platforms. One is a variant of the F2-CH/Pilus complex in which the F pilus binds asymmetrically at the outer edges of the OMC rather than centrally (SI Appendix, Fig. S7B). A second, designated as the F4-OM/Pilus, consists of the F pilus attached to the OM without any associated periplasmic density (Fig. 4A and SI Appendix, Fig. S7A). These latter F4 complexes corresponded to ∼5% of the visualized pilus-associated structures. We also observed spatial clustering of F1-CH–like complexes with F3 or F4 complexes (SI Appendix, Fig. S7B). These complexes were scarce and structurally variable, which prevented detailed structural analysis by in situ CryoET. Nevertheless, their existence offers insights into the F pilus biogenesis pathway (Discussion).
Discussion
A Structure-Driven Model for Biogenesis of the F Transfer System.
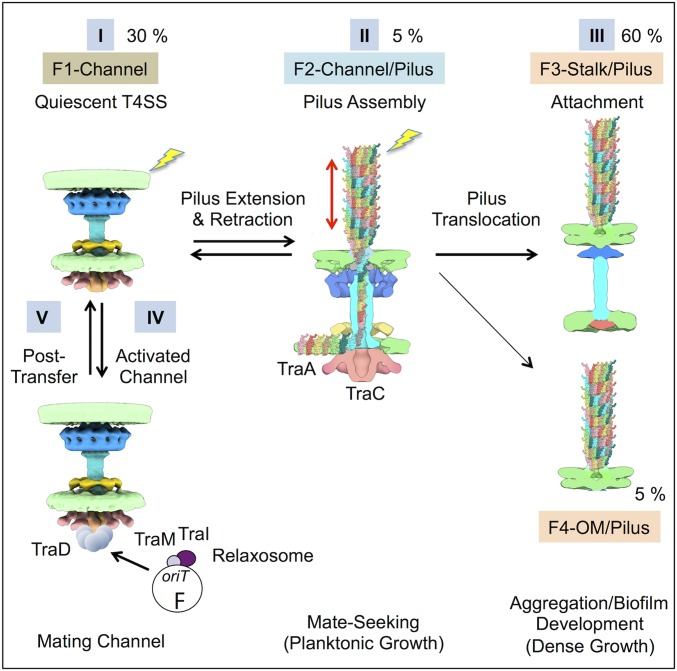
Our detection of at least four morphologically distinct F plasmid-encoded structures raises exciting questions about their biogenesis and biological functions. In a structure-driven model depicted in Fig. 5, we propose that the F-encoded Tra subunits assemble initially as a quiescent F1-CH complex. In the absence of a “mating” signal propagated by recipient cell contact, the F1-CH complex defaults to the “mate-seeking” mode by transitioning to the F2-CH/Pilus complex. The F2-CH/Pilus complex functions as a pilus assembly factory by orchestrating rounds of F pilus extension and retraction but can alternatively deposit assembled F pili onto distinct basal platforms to yield the F3-ST/Pilus and F4-OM/Pilus structures. Upon receipt of a recipient cell signal, the F1-CH complex transitions to the “mating” mode by recruitment of the TraD receptor and the F plasmid substrate to initiate translocation.
Fig. 5.
A structure-driven assembly pathway for the F transfer system. Step I: Tra subunits assemble initially as a quiescent F1-Channel complex. Step II: The F1 complex transitions to the F2-Channel/Pilus for dynamic extension and retraction of the F pilus in a “mate-seeking” mode. Dynamic F pilus extension and retraction by the F2 structure favors formation of distant cell-cell contacts during low cell density, e.g., planktonic, growth. Step III: Assembled F pili are alternatively deposited onto distinct platforms, yielding the F3-Stalk/Pilus or F4-OM/Pilus structures. The presumptively static F3 and F4 structures promote nonspecific aggregation and biofilm development; in polymicrobial settings, these dense growth conditions favor formation of mating junctions. Step IV: Recipient cell contact (lightning bolts) triggers F pilus retraction and recruitment of the TraD receptor and F plasmid substrate to activate the DNA transfer or “mating” mode. Step V: Following plasmid transfer, donor and recipient cells disengage and the “mating” channel reverts to the F1 quiescent complex. The relative percentages of each complex visualized on E. coli minicells by in situ CryoET are shown.
F2-CH/Pilus Complex: The F Pilus Assembly Factory.
The proposed transition from quiescent F1-CH complex to the F2-CH/Pilus for pilus assembly is consistent with previous work showing that nearly identical sets of Tra subunits are required to build both the conjugation channel and pilus (30, 31). The F1 to F2 transition involves major structural changes, particularly in the OMC and central cylinder (Movie S2). Most notably, the transition is marked by elongation of the central cylinder through the OMC to the OM, resulting in formation of a channel that extends from the IM to the OM. The central cylinder, however, is structurally distinct from the F pilus and built from a subunit(s) other than the TraA pilin, and its 3.5-nm central channel cannot accommodate the 8.5-nm pilus fiber. The solved F2-CH/Pilus complex thus constitutes strong evidence that polymerization of the mature F pilus initiates at the OMC as opposed to an IMC platform.
Early steps of F pilus biogenesis are well-defined or inferred from work on related systems: (i) newly synthesized TraA propilins integrate in the IM and undergo proteolytic cleavage and N-terminal acetylation to generate a membrane pool (32), (ii) upon receipt of an activating signal, TraA monomers are extracted from the IM by a mechanism dependent on the TraC ATPase (33), and (iii) during extraction, each monomer of TraA associates with a molecule of IM phospholipid (PL) (26). Once extracted from the IM, how are the potentially thousands of hydrophobic pilin-PL protomers shunted to the OM for F pilus assembly? A dedicated chaperone might shuttle the hydrophobic pilin complexes through the periplasm, reminiscent of the chaperone-usher pathway that directs assembly of Pap/type I pili (34). However, we think this model is unlikely because pilin-PL protomers must move bidirectionally across the cell envelope during the reversible process of F pilus extension and retraction. Instead, we propose a two-stage nucleation model in which the membrane-extracted protomers are recruited to the IMC of the F2-CH/Pilus complex, where they assemble as a thin protofilament (Fig. 5). Through sequential addition of protomers to the base of the protofilament, the protofilament extends through the channel of the visualized central cylinder, a process that ultimately triggers the conformational changes accompanying the F1 to F2 transition (Movie S2). In a stage II nucleation reaction, as the protofilament protrudes across the OM, it packs into the helical array visualized for the mature pilus (26). Stage II nucleation proceeds until receipt of a signal for retraction, whereupon the entire process reverses and pilin-PL molecules are returned to the IM. This two-stage model is compatible with the visualized 3-nm channel of the F2-CH/Pilus complex, a width sufficient to accommodate a thin protofilament, but not the mature pilus. The model is also compatible with evidence that pilin subunits interact with IMC subunits during pilus biogenesis (35) and an early finding that F pili grow by addition of pilin monomers to the base (36). Conceivably, the thinning of the pilus walls at the pilus–OMC junction observed in the F2-CH/Pilus complex corresponds to the protofilament–pilus structural transition (Fig. 3C).
In our model, the F2-CH/Pilus is not competent for substrate transfer when the F pilus is extended. However, evidence exists for F plasmid transfer through F pili that connect distant donor and recipient cells (35). This suggests that under certain conditions, the F2-CH/Pilus complex might be activated to deliver the DNA substrate through an extended pilus. Such translocation events, if they occur, must be rare given the reported low frequency of plasmid transfer at a distance (37) and extensive biochemical and ultrastructural evidence establishing that stable, shear-resistant mating junctions are required for efficient conjugation (14, 38).
Deposition of F Pili onto Alternative Basal Platforms.
The highly dynamic F2-CH/Pilus structures are difficult to detect in our structural snapshots, whereas the F3-ST/Pilus complexes abundantly decorate the E. coli cell surface. The F3 platform lacks densities corresponding to subunits or subassemblies, e.g., TraC ATPase, OMC, and IMC, that are required for F plasmid transfer and F pilus biogenesis. F pili associated with F3 platforms must therefore arise by partial disassembly of the F2-CH/Pilus complex or by movement of the pilus away from the F2 platform. Several observations favor the latter model. First, OMCs are intrinsically highly stable due to an extensive network of interfacial contacts among the constituent subunits (6, 8, 9), which argues against machine disassembly. Second, we observed instances in which F pili are docked asymmetrically onto the F2 basal complex, or F3-ST/Pilus or F4-OM/Pilus complexes are located next to pilus-free, F2-like basal platforms (SI Appendix, Fig. S7B). We propose that these structural variants correspond to assembly intermediates associated with the F2 to F3 or F4 transitions. Finally, we observed that the F pilus associates with OM-spanning disk/plug domains in both the F2-CH/Pilus and F3-ST/pilus complexes (Figs. 3 C and E and 4 B and C). Conceivably, through lateral movement away from the F2-CH/Pilus complex, the disk/plug domains might relocate anchored F pili onto alternative basal platforms for distribution around the cell surface. In our working model, deposition of F pili onto F3 or F4 platforms is a dead-end pathway for the F pilus, although it remains possible that these pili are capable of retraction by an undefined ATP-independent mechanism or by reattachment onto the F2 platform.
In sum, results of our structural studies suggest that F transfer systems have evolved two strategies for enhancing the probability of plasmid transfer. First, they elaborate F2-CH/Pilus complexes, which function as previously envisioned for conjugation machines by stochastically building and retracting pili in a “mate-seeking” mode. This dynamic activity is ideal for establishment of distant contacts with recipient cells under conditions of low cell density, e.g., planktonic growth. Second, assembled pili can be deposited onto alternative F3 or F4 platforms, either through disassembly of the F2 basal structure or active translocation on the OM. Decoration of the cell surface with these presumptively static pili would enhance the probability of intercellular contacts, aggregation, and biofilm development. In polymicrobial communities typically found in nature, such encounters set the stage for ample mating-pair formation and efficient intraspecies and interspecies horizontal gene transfer (39).
Materials and Methods
Bacterial Strains and Plasmids.
Bacterial strains, plasmids, and oligonucleotides used in this study are listed in SI Appendix, Table S1.
Preparation of Frozen-Hydrated Specimens.
Bacterial cultures were grown overnight on lysogeny broth (LB) plates at 37 °C. The bacteria were scratched from the plates and suspended in physiologically buffered saline, then mixed with 10-nm colloidal gold particles (used as fiducial markers in image alignment) and deposited onto freshly glow-discharged, holey carbon grids for 1 min. The grids were blotted with filter paper and rapidly frozen in liquid ethane, using a gravity-driven plunger apparatus as described previously (28, 40).
Cryo-ET Data Collection, Image Classification, and 3D Visualization.
Frozen-hydrated specimens were imaged and data were processed using our previously established protocols (12, 28, 41), with minor modifications as presented in the SI Appendix.
Data Availability.
Density maps and coordinate data that support the F-encoded channel structures determined by cyroelectron tomography have been deposited in The Electron Microscopy Data Bank (EMDB) as EMD-9344 (18) and EMD-9347 (19).
Supplementary Material
Acknowledgments
B.H. was supported in part by McGovern Medical School startup funds and Welch Foundation Grant AU-1953-20180324. P.J.C. was supported by NIH Grant R01GM48476. B.H. and P.J.C. were supported in part by NIH Grant R21 AI 142378. We are grateful to Sandy Parkinson for deletion of the fla gene cluster from the minC mreB E. coli mutant to yield strain UU2834. We are grateful to Craig Roy, Jun Liu, and Tim Cover for insightful suggestions and critiques of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Density maps and coordinate data that support the F-encoded channel structures determined by cryoelectron tomography have been deposited in The Electron Microscopy Data Bank (EMDB), https://www.ebi.ac.uk/pdbe/emdb (entry nos. EMD-9344 and EMD-9347).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904428116/-/DCSupplemental.
References
- 1.Lederberg J., Tatum E. L., Gene recombination in Escherichia coli. Nature 158, 558 (1946). [DOI] [PubMed] [Google Scholar]
- 2.Koraimann G., Spread and persistence of virulence and antibiotic resistance genes: A ride on the F plasmid conjugation module. Ecosal Plus 8, 10.1128/ecosalplus.ESP-0003-2018 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Cabezón E., Ripoll-Rozada J., Peña A., de la Cruz F., Arechaga I., Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 39, 81–95 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Frost L. S., Koraimann G., Regulation of bacterial conjugation: Balancing opportunity with adversity. Future Microbiol. 5, 1057–1071 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Fronzes R., et al. , Structure of a type IV secretion system core complex. Science 323, 266–268 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandran V., et al. , Structure of the outer membrane complex of a type IV secretion system. Nature 462, 1011–1015 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon J. E., et al. , Use of chimeric type IV secretion systems to define contributions of outer membrane subassemblies for contact-dependent translocation. Mol. Microbiol. 105, 273–293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sgro G. G., et al. , Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri. Nat. Microbiol. 3, 1429–1440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Low H. H., et al. , Structure of a type IV secretion system. Nature 508, 550–553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redzej A., et al. , Structure of a VirD4 coupling protein bound to a VirB type IV secretion machinery. EMBO J. 36, 3080–3095 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grohmann E., Christie P. J., Waksman G., Backert S., Type IV secretion in Gram-negative and Gram-positive bacteria. Mol. Microbiol. 107, 455–471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chetrit D., Hu B., Christie P. J., Roy C. R., Liu J., A unique cytoplasmic ATPase complex defines the Legionella pneumophila type IV secretion channel. Nat. Microbiol. 3, 678–686 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke M., Maddera L., Harris R. L., Silverman P. M., F-pili dynamics by live-cell imaging. Proc. Natl. Acad. Sci. U.S.A. 105, 17978–17981 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arutyunov D., Frost L. S., F conjugation: Back to the beginning. Plasmid 70, 18–32 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Lu J., et al. , Analysis and characterization of the IncFV plasmid pED208 transfer region. Plasmid 48, 24–37 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Farley M. M., Hu B., Margolin W., Liu J., Minicells, back in fashion. J. Bacteriol. 198, 1186–1195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kass L. R., Yarmolinsky M. B., Segregation of functional sex factor into minicells. Proc. Natl. Acad. Sci. U.S.A. 66, 815–822 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu B., Khara P., Christie P. J., Type IV outer membrane complex (F plasmid). Protein Data Bank. http://www.ebi.ac.uk/pdbe/entry/EMD-9344/visualization. Deposited 14 November 2018.
- 19.Hu B., Khara P., Christie P. J., Type IV inner membrane complex (F plasmid). Protein Data Bank. http://www.ebi.ac.uk/pdbe/entry/EMD-9347/visualization. Deposited 14 November 2018.
- 20.Atmakuri K., Cascales E., Christie P. J., Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 54, 1199–1211 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cascales E., Christie P. J., Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304, 1170–1173 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger B. R., Christie P. J., The Agrobacterium tumefaciens virB4 gene product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J. Bacteriol. 175, 1723–1734 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabel C., Grahn A. M., Lurz R., Lanka E., The VirB4 family of proposed traffic nucleoside triphosphatases: Common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption. J. Bacteriol. 185, 1045–1058 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zechner E. L., Lang S., Schildbach J. F., Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1073–1087 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larrea D., de Paz H. D., Arechaga I., de la Cruz F., Llosa M., Structural independence of conjugative coupling protein TrwB from its type IV secretion machinery. Plasmid 70, 146–153 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Costa T. R., et al. , Structure of the bacterial sex F pilus reveals an assembly of a stoichiometric protein-phospholipid complex. Cell 166, 1436–1444.e10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang Y. W., et al. , Architecture of the type IVa pilus machine. Science 351, aad2001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu B., Lara-Tejero M., Kong Q., Galan J. E., Liu J., In situ molecular architecture of the Salmonella type III secretion machine. Cell 168, 1065–1074.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caro L. G., Schnös M., The attachment of the male-specific bacteriophage F1 to sensitive strains of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 56, 126–132 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger B. R., Christie P. J., Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176, 3646–3660 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawley T. D., Klimke W. A., Gubbins M. J., Frost L. S., F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224, 1–15 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Paiva W. D., Grossman T., Silverman P. M., Characterization of F-pilin as an inner membrane component of Escherichia coli K12. J. Biol. Chem. 267, 26191–26197 (1992). [PubMed] [Google Scholar]
- 33.Kerr J. E., Christie P. J., Evidence for VirB4-mediated dislocation of membrane-integrated VirB2 pilin during biogenesis of the Agrobacterium VirB/VirD4 type IV secretion system. J. Bacteriol. 192, 4923–4934 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Werneburg G. T., Thanassi D. G., Pili assembled by the chaperone/usher pathway in Escherichia coli and Salmonella. Ecosal Plus 8, 10.1128/ecosalplus.ESP-0007-2017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan Q., et al. , Identification of the VirB4-VirB8-VirB5-VirB2 pilus assembly sequence of type IV secretion systems. J. Biol. Chem. 280, 26349–26359 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Maher D., Sherburne R., Taylor D. E., H-pilus assembly kinetics determined by electron microscopy. J. Bacteriol. 175, 2175–2183 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babic A., Lindner A. B., Vulic M., Stewart E. J., Radman M., Direct visualization of horizontal gene transfer. Science 319, 1533–1536 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Samuels A. L., Lanka E., Davies J. E., Conjugative junctions in RP4-mediated mating of Escherichia coli. J. Bacteriol. 182, 2709–2715 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghigo J. M., Natural conjugative plasmids induce bacterial biofilm development. Nature 412, 442–445 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Morado D. R., Hu B., Liu J., Using tomoauto: A protocol for high-throughput automated cryo-electron tomography. J. Vis. Exp., e53608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu B., et al. , Visualization of the type III secretion sorting platform of Shigella flexneri. Proc. Natl. Acad. Sci. U.S.A. 112, 1047–1052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Density maps and coordinate data that support the F-encoded channel structures determined by cyroelectron tomography have been deposited in The Electron Microscopy Data Bank (EMDB) as EMD-9344 (18) and EMD-9347 (19).