Significance
Despite being a major drug target of this century, kinases are challenging modalities to inhibit because of promiscuity and the consequent adverse effects of compounds targeting their conserved active site. Here we deliver a proof-of-principle approach to overcome these obstacles using allosteric modulation. Taking advantage of the naturally built allosteric network that evolved to be different among kinases, we develop several extremely selective and affine allosteric binders using monobodies (small proteins), causing both strong inhibition and activation. These monobodies allow “dialing-in” of any desired activity, revealing 3 major advantages of targeting allosteric sites: extreme selectivity, ability to activate and inhibit, and avoidance of competing with high cellular ATP. This approach provides a general, powerful path toward rational drug design.
Keywords: kinase, allostery, monobody, Aurora A, allosteric drugs
Abstract
Despite being the subject of intense effort and scrutiny, kinases have proven to be consistently challenging targets in inhibitor drug design. A key obstacle has been promiscuity and consequent adverse effects of drugs targeting the ATP binding site. Here we introduce an approach to controlling kinase activity by using monobodies that bind to the highly specific regulatory allosteric pocket of the oncoprotein Aurora A (AurA) kinase, thereby offering the potential for more specific kinase modulators. Strikingly, we identify a series of highly specific monobodies acting either as strong kinase inhibitors or activators via differential recognition of structural motifs in the allosteric pocket. X-ray crystal structures comparing AurA bound to activating vs inhibiting monobodies reveal the atomistic mechanism underlying allosteric modulation. The results reveal 3 major advantages of targeting allosteric vs orthosteric sites: extreme selectivity, ability to inhibit as well as activate, and avoidance of competing with ATP that is present at high concentrations in the cells. We envision that exploiting allosteric networks for inhibition or activation will provide a general, powerful pathway toward rational drug design.
Aurora A (AurA) has garnered much interest in the last 2 decades as a major oncotarget, as its overexpression is linked to a multitude of cancers (1). AurA coordinates mitotic division not only through regulating centrosome maturation and duplication but also through directly affecting spindle microtubule formation in later stages of mitosis (2, 3). To localize to the spindle microtubules and coordinate proper progression of mitosis, AurA must bind to, and be allosterically activated by, the microtubule-associated protein TPX2 (Targeting Protein for Xklp2) (4, 5). Therefore, disrupting this protein–protein interaction has a double inhibitory effect: loss of kinase activation and disruption of AurA localization to the spindles, which leads to defective mitosis (6) and could trigger attenuated cell viability (7).
Because the interaction surface between AurA and TPX2 is extensive and lacks the classic, small, confined binding pocket that is preferred when designing small molecule inhibitors, it has been challenging to identify potent disruptors of this interaction despite significant efforts from various groups (8–12). Initial efforts focused around fragment screening identified compounds with micromolar affinities to AurA [Kd = 3.6–12 µM range (10–12)]. Later on, screening a library of Wobbegong shark antibodies led to the identification of vNAR-D01 (Kd = 2 µM) (9). This antibody inhibited AurA; however, the authors stress the need for better allosteric molecules because no interaction between AurA and vNAR-D01 in Xenopus egg extracts could be observed, likely as a result of too weak binding or low specificity (9). The authors further reason that the disulfide-containing antibodies may not be used for intracellular targeting (9).
Here we describe an approach using monobodies that addresses both the affinity and disulfide bond problems. Monobodies are synthetic binding proteins developed from highly tailored combinatorial libraries constructed on a fibronectin type III domain scaffold that is small and Cys-free (13). Monobodies as binders with high specificity and affinity to diverse targets have been developed, some of which employ quite small interaction epitopes (14, 15). We select a series of monobodies that bind tightly to the naturally occurring allosteric activation pocket of AurA, and importantly, elicit a range of kinase activity from strong inhibition to strong activation. Quantitative characterization of the monobody–AurA interactions and enzyme activity changes, together with high-resolution structures of inhibiting and activating complexes, reveal the detailed molecular mechanism of allosteric modulation of AurA. Furthermore, the monobodies are extremely specific for AurA, with no detectable binding, even to the closest homolog AurB.
Results and Discussion
Selection of Monobodies That Bind to the Allosteric Hydrophobic Pocket of AurA.
AurA is allosterically activated through TPX2 anchoring to a hydrophobic pocket in the N-terminal lobe of AurA catalytic domain (5) that is widely used in the protein kinase superfamily for allosteric modulation (5, 16). We wanted to explore the concept of developing monobodies in an unbiased way that modulates AurA activity by binding to this pocket, thereby shifting the equilibrium between active and inactive states of the kinase. Obtaining a range of allosteric activators and inhibitors would reveal how AurA is allosterically controlled, and that basic understanding could open opportunities to find a novel kind of very specific kinase drugs.
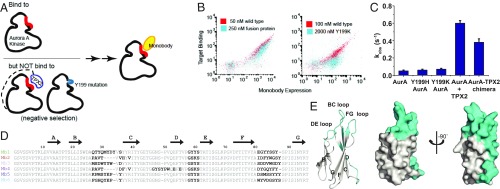
To generate monobodies that specifically bind to this hydrophobic pocket, a scheme that involves both positive and negative selection is designed. Monobodies are selected for binding to wild-type (WT) AurA and against binding to AurA fused to a TPX2-derived peptide, AurA-TPX2 chimera (Fig. 1 A and B and SI Appendix, Fig. S1A). The linker between AurA and TPX2 in AurA-TPX2 chimera fully mimics the linker connecting the C-lobe with the C-terminal activation segment of PKA. We reason that in the AurA-TPX2 chimera, the TPX2-binding site will be occupied by the intramolecular interaction between the kinase and the fused TPX2 peptide. This strategy is used to enrich for monobodies that specifically bind to the hydrophobic pocket and not to other regions of the kinase. Examining binding profiles of the resulting monobody pools for AurA versus AurA-TPX2 chimera prompts us to add an additional negative selection step by designing AurA mutants in the hydrophobic pocket (Y199H or Y199K) with impaired TPX2 binding (SI Appendix, Fig. S1B) but unaltered AurA activity (Fig. 1C). Indeed, this strategy delivers a pool of monobodies that bind much more strongly to WT than to AurA mutants (Fig. 1B), suggesting successful enrichment for monobodies binding to the intended interface (Fig. 1 D and E).
Fig. 1.
Generation of monobodies that bind to the allosteric hydrophobic pocket of AurA. (A) Schematic of monobody selection design. Monobodies were chosen for their ability to bind to AurA but not Y199H or Y199K AurA mutants or AurA-TPX2 chimeric protein. (B) Representative monobody populations after negative selection with AurA-TPX2 chimera or Y199K AurA mutants, as tested using yeast display. (C) Mutation of AurA Y199 to either H or K does not change the activity of the mutant AurA proteins. Activities were measured by the ADP/NADH coupled assay at 25 °C with 1 μM AurA, 1 mM Lats2 peptide and saturating concentrations of ATP and MgCl2. AurA-TPX2 chimera closely mimics the activity of WT protein in the presence of 5 µM TPX2, ideally suiting this decoy protein to be used in monobody counter selection screens. (D) Amino acid sequences of monobody clones. Residues at diversified positions in the combinatorial libraries are shown in bold. Amino acid numbering is shown for Mb1. Although subsequent monobody sequences are shown with gaps for purposes of visual alignment among each other, we do not count gaps in sequence numbering of these monobodies. (E) Mapping of diversified positions (cyan) onto the structure of a prototypical monobody (PDB ID code 1FNF). Errors in C are SD from triplicates.
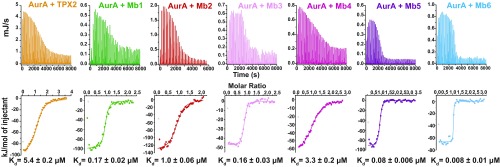
A total of 84 clones are tested for binding to the WT, and Y199H and Y199K AurA and 6 monobodies are chosen for further characterization based on high specificity to WT AurA over the mutants (Fig. 1 D and E). Binding measurements using purified proteins show that these monobodies have a large affinity range, with Kd values ranging from low nanomolar to low micromolar (Fig. 2). Tight binding is shown to be specific for AurA, as no binding could be detected for monobodies Mb1 and Mb2 to the closely related AurB kinase even at kinase:monobodies ratios aimed at measuring high micromolar binding affinities (SI Appendix, Fig. S2).
Fig. 2.
Monobodies bind to AurA with high affinity. Binding of TPX2 and Mb1-6 to AurA at 25 °C as measured by ITC. Fitted dissociation constants, Kd, are shown with the SE for the estimate of Kd, which is a measure of the goodness of fit of the data.
Monobodies Differentially Activate or Inhibit AurA.
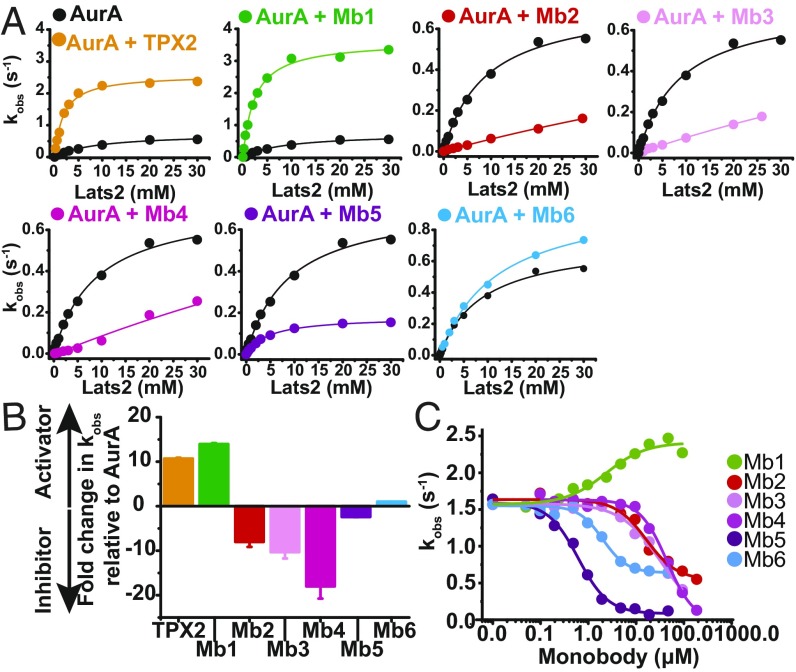
How do the different monobodies affect AurA’s kinase activity? AurA phosphorylation of a peptide fragment of Lats2, a physiological substrate of AurA (17), is followed under saturating concentrations of ATP (SI Appendix, Fig. S3A) and MgCl2, using the ADP/NADH coupled assay (Fig. 3A). High-performance liquid chromatography–based assays are run, detecting phosphorylated Lats2 as control, ensuring that the ADP/NADH-coupled assay indeed reflects AurA kinase kinetics (SI Appendix, Fig. S3 B and C). Assays are run at timescales (up to 900 s) and AurA concentrations (1 µM) at which the kinase remains fully dephosphorylated (SI Appendix, Fig. S3D and ref. 5), as the goal is to target the correct AurA state, the dephosphorylated state found at the cell spindles (18). Strikingly, these monobodies are capable of either inhibiting or activating AurA kinase activity (Fig. 3). In fact, they span a large range of allosteric modulation, starting with strong activators (Mb1) to strong inhibitors (Mb2, Mb3, Mb4, Mb5; Fig. 3 A and B). Inhibitory monobodies prevent AurA’s activation by TPX2 in vitro in a manner that is proportional to their relative AurA’s binding affinities, with, for instance, the tighter binder Mb5 inhibiting AurA more effectively than the weaker binder Mb2 (Fig. 3C). Interestingly, Mb6, the tightest binder of the series, does not affect AurA’s activity. This range of monobody function is in harmony with the original idea of selecting allosteric modulators, as it directly reflects differential binding to active or inactive conformations of the kinase, thereby shifting the allosteric equilibrium. According to this model, activators should have a higher affinity to the active relative to the inactive conformation, and inhibitors the opposite behavior, whereas for Mb6, high affinity to both states is predicted, leading to no significant change in the inactive/active equilibrium relative to apo AurA.
Fig. 3.
Monobodies as activators and inhibitors of AurA activity. (A) The ADP/NADH coupled assay was used to monitor Lats2 peptide phosphorylation by AurA in the absence (black) or presence of TPX2 or monobodies Mb1 to Mb6. Fits to Michaelis Menten kinetics, normalized by the enzyme concentration (kobs), show that the monobodies affect both the kcat and KM of AurA for Lats2. Assays were carried out at 25 °C in the presence of 1 or 0.25 μM AurA and saturating concentrations of monobody or TPX2, ATP, and MgCl2. (B) Comparison of kobs values of AurA at 1 mM Lats2 showing that Mb1 activates AurA 15-fold, a value comparable to allosteric activation by TPX2, whereas monobodies Mb2-Mb5 inhibit AurA between 2-fold and 20-fold, with Mb6 not significantly affecting AurA kinetics. (C) Monobodies can outcompete TPX2 from AurA in a manner consistent with their relative AurA affinities and reflective of their inhibitory/neutral (Mb2-Mb6) or activating (Mb1) propensity. Experiments were conducted in the presence of 1 μM AurA, 5 μM TPX2, 3 mM Lats2, using the same coupled assay as described earlier. Errors in B were determined from jackknifing of data in A.
Crystal Structures of AurA-Mb1 and AurA-Mb2.
The discovery of both activating and inhibiting monobodies offers a unique opportunity to shed light into the molecular mechanism of allosteric modulation of the kinase. X-ray crystal structures of AurA-Mb1 and AurA-Mb2 as representative activating and inhibitory AurA-monobody complexes, respectively, were obtained (Table 1).
Table 1.
X-ray structures data collection and refinement statistics
| Data/statistics | AurA-Mb1-AMPPCP | AurA-Mb2-AMPPCP |
| Data collection | ||
| Space group | I222 | P21 21 21 |
| Cell dimensions | ||
| a, b, c (Å) | 75.34, 91.36, 143.72 | 63.05, 69.92, 173.72 |
| α, β, γ (°) | 90.00 90.00 90.00 | 90.00 90.00 90.00 |
| Resolution (Å) | 2.06 | 2.55 |
| Rmerge | 0.126 (1.214)* | 0.053 (0.528)* |
| I/σI | 10.3 (1.8) | 13.09 (1.03) |
| Completeness (%) | 100.0 (99.5) | 98.2 (83.8) |
| Redundancy | 7.2 (7.2) | 4.9 (2.3) |
| Refinement | ||
| Resolution (Å) | 77.10–2.06 (2.12–2.06) | 46.82–2.55 (2.64–2.55) |
| No. reflections | 29755 | 25450 (2507) |
| Rwork/Rfree | 0.23/0.27 (0.43/0.41) | 0.26/0.32 (0.36/0.39) |
| No. atoms | ||
| Protein | 2890 | 4875 |
| Ligand/Ion | 52 | 62 |
| Water | 145 | N/A |
| B-factors | ||
| Protein | 38.16 | 73.50 |
| Ligand/Ion | 46.11 | 93.26 |
| Water | 33.30 | N/A |
| R.m.s. deviations | ||
| Bond length (Å) | 0.016 | 0.003 |
| Bond angles (°) | 1.916 | 0.90 |
| PDB ID | 5G15 | 6C83 |
Values in parenthesis correspond to the highest-resolution shell.
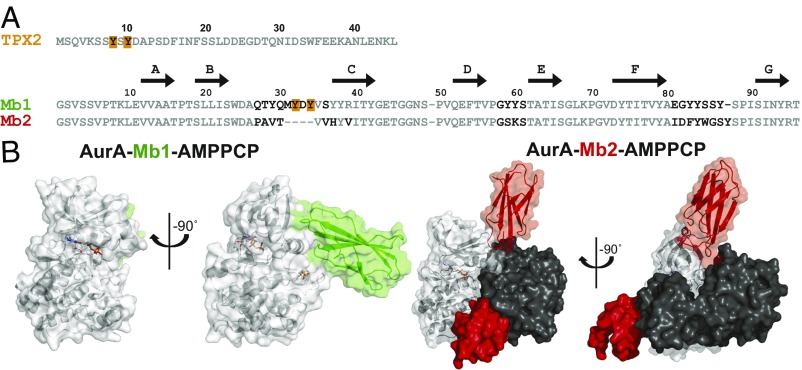
A sequence alignment of all monobodies in comparison with TPX2 shows the presence of a motif containing 2 Tyr separated by a small amino acid in both the allosteric activator TPX2 and the activating monobody Mb1, whereas this YxY motif is absent in the inhibiting monobodies, including Mb2 (Figs. 1D and 4A). Both Mb1 and Mb2 monobodies indeed bind to the hydrophobic pocket, as designed (Fig. 4B), but with different molecular features, detailed here. Primary interface contacts of Mb1 to AurA are the BC and DE loops of the monobody, whereas Mb2 uses an entirely different region, the FG loop, in addition to the BC loop (Fig. 1D). The AurA-Mb2 complex is seen as an AurA2-Mb22 dimer in the crystal, in contrast to a monomeric AurA-Mb1 complex (Fig. 4B and SI Appendix, Fig. S4 A–E). Analytical ultracentrifugation experiments show that the AurA-Mb2 complex is indeed a predominant dimer in solution and persists even at low AurA concentrations (SI Appendix, Fig. S4 F and G). All other monobodies show only a small population of AurA2-Mb2 dimer (SI Appendix, Fig. S4H), and together with the concentration dependence of inhibition by Mb2, can rule out dimerization as the primary source of inhibition by the monobodies (SI Appendix, Fig. S4 F–H).
Fig. 4.
Structures of AurA bound to activating or inhibiting monobody. (A) Sequences of AurA activators, TPX2 and Mb1, and inhibitor, Mb2. The 2 Tyr critical for TPX2-dependent AurA activation (Y8 and Y10; shown in bold, orange), and the structurally equivalent residues in Mb1 (also bold and orange), as well as sequence difference between Mb1 and Mb2 (black bold), are highlighted. Residue numbering is based on a previously published system (16–19). (B) X-ray crystal structures of AurA (gray) bound to AMPPCP and either Mb1 (green, 2.06 Å, PDB ID code 5G15) or Mb2 (red, 2.55Å, PDB ID code 6C83), exposing extensive but different contacts between AurA and Mb1/Mb2.
Molecular Mechanism of Activation and Inhibition by Mb1 and Mb2.
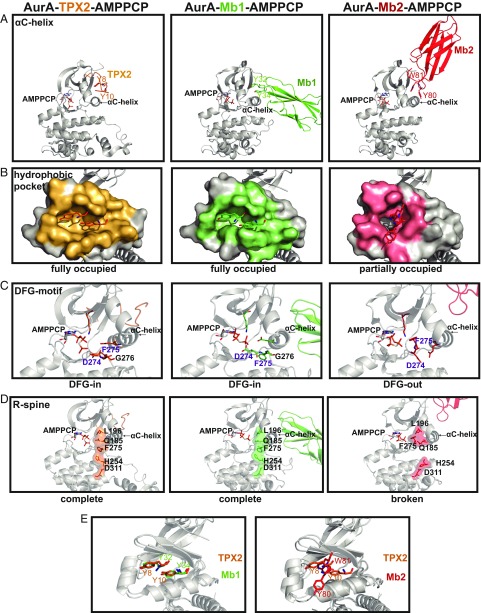
Four major structural features that differ between the inactivating and activating monobody–AurA structures are identified. These attributes characterize the molecular determinants responsible for monobodies’ opposing modulation of AurA’s activity (Fig. 5). First, Mb1 anchors above the αC-helix, at the hydrophobic AurA pocket, stabilizing it in an active conformation reminiscent of TPX2. Contrarily, Mb2 slides differently into this pocket, sandwiching the catalytically crucial αC-helix in a restrictive inactive conformation (Fig. 5A).
Fig. 5.
Monobodies stabilize either an active or inactive conformation of AurA. (A) Structures of AurA bound to TPX2 (PDB ID code 4C3P), Mb1 (PDB ID code 5G15), or Mb2 (PDB ID code 6C83) highlighting (A) differential engagement of AurA’s αC-helix by 2 aromatic residues within TPX2 or Mb1/Mb2, (B) AurA residues interfacing with TPX2 (orange), Mb1 (green) or Mb2 (pink) were calculated using PISA (40), (C) Hallmarks of an active kinase: DFG-in motif, K162-E181 salt bridge, and (D) completion of the R-spine are present in the AurA-TPX2 or AurA-Mb1 complexes and absent in the AurA-Mb2 structure. (E) Y32 and Y34 of Mb1 are superimposable with Y8 and Y10 of TPX2 (Left), whereas Y80 and W81 of Mb2 interact very differently with AurA (Right); see also SI Appendix, Fig. S5.
Second, whereas AurA’s interface residues are abundant and nearly identical among the AurA-TPX2 and AurA-activating monobody Mb1 structures, they are scarcer in the AurA-inhibitory monobody Mb2 complex (Fig. 5B and SI Appendix, Figs. S5 and S6).
Third, the conformation of the DFG motif is different between the inactivating and activating monobody–AurA complexes (Fig. 5C). DFG-in refers to the positioning of the regulatory F275 and the crucial D274 active site residue to coordinate Mg2+, which in turn coordinates to the γ-phosphate of ATP (19), thus precisely positioning the kinase for catalysis. The DFG-out orientation disfavors these interactions and is therefore thought to report on the inactive state of the kinase. The active DFG-in state is indeed seen both in the structure of AurA bound to TPX2 or Mb1.
Fourth, to complete the assessment of the state (active/inactive) of AurA, we examined the R-spine, an additional marker of an active kinase (Fig. 5D) (20). As expected, AurA-Mb1 displays a completed R-spine identical to the one in AurA-TPX2. In contrast, the R-spine is interrupted in the AurA-Mb2 structure, leaving a gap between the N- and C-lobes (Fig. 5D).
Remarkably, Mb1 and TPX2 use the precise superposition of the YxY motif, a well-characterized hot-spot (21, 22) of the AurA-TPX2 interaction, for contacting AurA (Fig. 5E). This suggests that Mb1 and TPX2 share a common mode of allosteric activation despite their vast differences in sequence and structure. In contrast, Mb2 uses a different mode of action: it inserts aromatic residue Trp81 into the hydrophobic pocket and positions Tyr80 on the other side of the αC-helix, thereby shifting it in an inactive conformation. We noticed that the spatial orientation of Trp81 was similar to that of W91 in vNAR-D01 or that of AurkinA (8, 9, 12). This suggests that the space occupied by monobody molecules could be a central hotspot in rationally designing hydrophobic-pocket based allosteric inhibitors of AurA.
In summary, Mb1 and Mb2 exert their opposing effects on AurA activity by differentially interacting with the hydrophobic pocket and its surrounding region. This leads to the modulation of an allosteric network spanning the N-terminal hydrophobic pocket via the active site to the C-terminal lobe, resulting in shifting the equilibrium toward the active (as in the case with Mb1) or the inactive (as is the case with Mb2) state of AurA, as we had hypothesized on the basis of the activity modulation by a classic allosteric model.
Conclusions
With the presence of drug promiscuity, ensuing adverse effects of active site inhibitors, and particularly with the prevalence of drug-resistant mutations, there is an immediate, unfulfilled need to search for more effective, more creative approaches to inhibitor design. Guided by nature’s extensive use of allosteric modulation, an obvious first choice would be to target allosteric sites (8–12, 16, 23–27), thereby taking full advantage of the naturally evolved allosteric networks. Such a strategy offers several advantages. First, inhibition as well as activation can be designed, with the latter being a particularly new and powerful therapeutic avenue that cannot be achieved by active-site binders. Second, targeting of allosteric hotspots promises greater specificity because of the fact that active sites are highly conserved whereas modulatory sites have evolved for differential regulation. Third, scaffolding function would be disrupted in many systems.
Our results using monobodies provide the proof of principle for these concepts, using synthetic protein-based strong allosteric modulators (15-fold activation to 20-fold inhibition) with very high specificity and affinity, thereby opening opportunities for rational drug design. The monobody selection method is very efficient because it is based on highly refined library designs and molecular display technologies enabling the construction of large libraries and efficient directed evolution. The resulting monobodies themselves could be used directly as treatment, if in vivo delivery methods progress to a level suitable for pharmaceutical delivery (28–30). Our structures of AurA bound to monobodies provide the mechanistic atomic-level view for allosteric modulation that can then be directly used in rational small molecule design for mimicking these interactions, including using the partially unoccupied space of the pocket in the Mb2/AurA complex (Fig. 5B and SI Appendix, Fig. S6).
Recent reports on targeting the TPX2 pocket by small molecules and proteomimetics or antibody-based scaffolds (8–12) underscore the emerging high interest in allosteric inhibition. Our results differ in that the monobodies are extremely specific for AurA, whereas such specificity was not measured for the other reported inhibitors (8–12). Second, the affinities of several monobodies described here are much tighter than the reported inhibitors. Third, the series of monobodies delivers allosteric modulation ranging from strong inhibition to strong activation. Fourth, the monobodies described here have the advantage over the reported antibodies that they do not contain disulfide bonds, a feature that prohibits the antibodies to be used for intracellular targets such as AurA kinase.
Targeting this regulatory pocket for allosteric modulation has been reported for other kinases (Pdk1 and PKCζ) by small molecules having micromolar binding capacities (24–26, 31). Although some molecules are activators (24–26), inhibitors have also been identified (24, 31). The unique potential of allosteric inhibitors in cancer treatment has been elegantly demonstrated by the development of the allosteric inhibitor GNF-5 for the kinase Bcr-Abl and SHP099 for the phosphatase SHP2 (27, 32). In addition to allosteric inhibitors, there have been a number of reports of inhibitors that disrupt protein–protein interfaces (23, 33–35).
Our demonstration of allosteric activators and inhibitors of AurA kinase activity goes beyond just disrupting the AurA/TPX2 interface. The immediate advantage of monobody selection for allosteric modulation is apparent and provides a complementary approach to small molecule synthesis. Monobodies as modulators of kinases are not a commonly explored path in drug discovery. In fact, there are currently only 3 reported cases of monobodies binding to Abl kinase, and in all cases, they bind to the SH2 domain rather than the catalytic domains (15, 36). Another synthetic binding protein system, DARPins, has been used to develop inhibitors of ERK, but they bind to the activation loop (37). The present work establishes the feasibility of allosterically activating and inactivating kinases by directing monobodies to the kinase domain itself, specifically the naturally built-in allosteric site away from the ATP binding site.
The concept of exploiting allosteric networks for tuning catalytic activities up or down bears the potential to elevate rational drug design to a new level. For this goal, an atomistic understanding of the allosteric networks and how to manipulate them with man-made drugs is essential. As conceptualized via different monobodies for AurA, monobody selection, together with the corresponding structures when complexed with its targets, provides an exciting experimental approach for intelligent allosteric drug design. This approach of controlling activity of a target protein by monobodies is immediately transferrable to many other targets, and not just targets from the kinome (5, 38, 39). Both the technology and the concept are general, and therefore open opportunities to apply them in other systems to study cellular signaling pathways, and for designing inhibitors or activators in drug design.
Materials and Methods
Expanded methods are available within SI Appendix. In brief, for monobody selection, affinity measurement, expression, and purification we used methods previously described in refs. 13 and 15 with minor modifications. The AurA-TPX2 chimera is composed of AurA122-403+PKA linker (FIPKFKGPGDTSNFDDYEEEEIRVSINEKCGKE) +TPX21-45. For Aurora A and TPX2 expression and purification, as well as biochemical (in vitro kinase assays) and biophysical and structural studies (ITC, X-ray crystallography, analytical ultracentrifugation), we adapted methods previously described in ref. 5. In vitro kinase assays were measured using Lats2 peptide (ATLARRDSLQKPGLE) (17) as substrate via a coupled enzymatic assay that detects ADP production through coupling it with pyruvate kinase and lactate dehydrogenase monitoring oxydation of NADH to NAD+. Kinase assays were performed at 25 °C in assay buffer (20 mM Tri⋅sHCl, 200 mM NaCl, 20 mM MgCl2, 5 mM TCEP, 10% glycerol at pH 7.5).
Supplementary Material
Acknowledgments
We thank R. Padua for helpful guidance with crystallography. We thank the Advanced Light Source, Berkeley, California, for access to beamline BL8.2.2. The Berkeley Center for Structural Biology is supported in part by the National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. We thank the Stanford Synchrotron Radiation Lightsource for access to beamline 7-1. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the U.S. Department of Energy Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). This work was supported by the Howard Hughes Medical Institute, the Office of Basic Energy Sciences, Catalysis Science Program, U.S. Dept. of Energy, award DE-FG02-05ER15699 and NIH (GM100966 to D.K.) and R01-GM090324 and R01-CA194864 (to S. Koide) and P30CA014599 (to The University of Chicago Comprehensive Cancer Center).
Footnotes
Conflict of interest statement: D.K. and A.Z. are the inventors on pending patents applied for by Brandeis University that describe compositions and methods for modulating kinase activity (US20180334510A1 and US20190038582A1). A.K. and S. Koide are listed as inventors on issued and pending patents on the monobody technology filed by The University of Chicago (US Patent 9512199 B2 and related pending applications).
This article is a PNAS Direct Submission.
Data deposition: Macromolecular structural data have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5G15 and 6C83).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906024116/-/DCSupplemental.
References
- 1.Zhou H., et al. , Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 20, 189–193 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Glover D. M., Leibowitz M. H., McLean D. A., Parry H., Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 81, 95–105 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Asteriti I. A., Giubettini M., Lavia P., Guarguaglini G., Aurora-A inactivation causes mitotic spindle pole fragmentation by unbalancing microtubule-generated forces. Mol. Cancer 10, 131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kufer T. A., et al. , Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 158, 617–623 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zorba A., et al. , Molecular mechanism of Aurora A kinase autophosphorylation and its allosteric activation by TPX2. eLife 3, e02667 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird A. W., Hyman A. A., Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A. J. Cell Biol. 182, 289–300 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganem N. J., Godinho S. A., Pellman D., A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayliss R., Burgess S. G., McIntyre P. J., Switching Aurora-A kinase on and off at an allosteric site. FEBS J. 284, 2947–2954 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Burgess S. G., et al. , Allosteric inhibition of Aurora-A kinase by a synthetic vNAR domain. Open Biol. 6, 160089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asteriti I. A., et al. , Identification of small molecule inhibitors of the Aurora-A/TPX2 complex. Oncotarget 8, 32117–32133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McIntyre P. J., et al. , Characterization of three druggable hot-spots in the aurora-A/TPX2 interaction using biochemical, biophysical, and fragment-based approaches. ACS Chem. Biol. 12, 2906–2914 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Janeček M., et al. , Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2. Sci. Rep. 6, 28528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koide A., Wojcik J., Gilbreth R. N., Hoey R. J., Koide S., Teaching an old scaffold new tricks: Monobodies constructed using alternative surfaces of the FN3 scaffold. J. Mol. Biol. 415, 393–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbreth R. N., Koide S., Structural insights for engineering binding proteins based on non-antibody scaffolds. Curr. Opin. Struct. Biol. 22, 413–420 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wojcik J., et al. , A potent and highly specific FN3 monobody inhibitor of the Abl SH2 domain. Nat. Struct. Mol. Biol. 17, 519–527 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gold M. G., Barford D., Komander D., Lining the pockets of kinases and phosphatases. Curr. Opin. Struct. Biol. 16, 693–701 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Yabuta N., Mukai S., Okada N., Aylon Y., Nojima H., The tumor suppressor Lats2 is pivotal in Aurora A and Aurora B signaling during mitosis. Cell Cycle 10, 2724–2736 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Toya M., Terasawa M., Nagata K., Iida Y., Sugimoto A., A kinase-independent role for Aurora A in the assembly of mitotic spindle microtubules in Caenorhabditis elegans embryos. Nat. Cell Biol. 13, 708–714 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Treiber D. K., Shah N. P., Ins and outs of kinase DFG motifs. Chem. Biol. 20, 745–746 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Kornev A. P., Taylor S. S., Ten Eyck L. F., A helix scaffold for the assembly of active protein kinases. Proc. Natl. Acad. Sci. U.S.A. 105, 14377–14382 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayliss R., Sardon T., Vernos I., Conti E., Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell 12, 851–862 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Eyers P. A., Erikson E., Chen L. G., Maller J. L., A novel mechanism for activation of the protein kinase Aurora A. Curr. Biol. 13, 691–697 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Wells J. A., McClendon C. L., Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 450, 1001–1009 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Sadowsky J. D., et al. , Turning a protein kinase on or off from a single allosteric site via disulfide trapping. Proc. Natl. Acad. Sci. U.S.A. 108, 6056–6061 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rettenmaier T. J., et al. , A small-molecule mimic of a peptide docking motif inhibits the protein kinase PDK1. Proc. Natl. Acad. Sci. U.S.A. 111, 18590–18595 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engel M., et al. , Allosteric activation of the protein kinase PDK1 with low molecular weight compounds. EMBO J. 25, 5469–5480 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J., et al. , Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature 463, 501–506 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan M., et al. , A novel intracellular protein delivery platform based on single-protein nanocapsules. Nat. Nanotechnol. 5, 48–53 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Tao P., et al. , In vitro and in vivo delivery of genes and proteins using the bacteriophage T4 DNA packaging machine. Proc. Natl. Acad. Sci. U.S.A. 110, 5846–5851 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaczmarczyk S. J., Sitaraman K., Young H. A., Hughes S. H., Chatterjee D. K., Protein delivery using engineered virus-like particles. Proc. Natl. Acad. Sci. U.S.A. 108, 16998–17003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez-Garcia L. A., et al. , Allosteric regulation of protein kinase PKCζ by the N-terminal C1 domain and small compounds to the PIF-pocket. Chem. Biol. 18, 1463–1473 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Chen Y. N., et al. , Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature 535, 148–152 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Wendt M. D., et al. , Discovery of a novel small molecule binding site of human survivin. Bioorg. Med. Chem. Lett. 17, 3122–3129 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Nero T. L., Morton C. J., Holien J. K., Wielens J., Parker M. W., Oncogenic protein interfaces: Small molecules, big challenges. Nat. Rev. Cancer 14, 248–262 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Fischer G., Rossmann M., Hyvönen M., Alternative modulation of protein-protein interactions by small molecules. Curr. Opin. Biotechnol. 35, 78–85 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grebien F., et al. , Targeting the SH2-kinase interface in Bcr-Abl inhibits leukemogenesis. Cell 147, 306–319 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kummer L., et al. , Structural and functional analysis of phosphorylation-specific binders of the kinase ERK from designed ankyrin repeat protein libraries. Proc. Natl. Acad. Sci. U.S.A. 109, E2248–E2257 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calejo A. I., Taskén K., Targeting protein-protein interactions in complexes organized by A kinase anchoring proteins. Front. Pharmacol. 6, 192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge B., et al. , MAPKK-independent activation of p38alpha mediated by TAB1-dependent autophosphorylation of p38alpha. Science 295, 1291–1294 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.