Abstract
Untargeted metabolomic measurements using mass spectrometry are a powerful tool for uncovering new small molecules with environmental and biological importance. The small molecule identification step, however, still remains an enormous challenge due to fragmentation difficulties or unspecific fragment ion information. Current methods to address this challenge are often dependent on databases or require the use of nuclear magnetic resonance (NMR), which have their own difficulties. The use of the gas-phase collision cross section (CCS) values obtained from ion mobility spectrometry (IMS) measurements were recently demonstrated to reduce the number of false positive metabolite identifications. While promising, the amount of empirical CCS information currently available is limited, thus predictive CCS methods need to be developed. In this article, we expand upon current experimental IMS capabilities by predicting the CCS values using a deep learning algorithm. We successfully developed and trained a prediction model for CCS values requiring only information about a compound’s SMILES notation and ion type. The use of data from five different laboratories using different instruments allowed the algorithm to be trained and tested on more than 2400 molecules. The resulting CCS predictions were found to achieve a coefficient of determination of 0.97 and median relative error of 2.7% for a wide range of molecules. Furthermore, the method requires only a small amount of processing power to predict CCS values. Considering the performance, time, and resources necessary, as well as its applicability to a variety of molecules, this model was able to outperform all currently available CCS prediction algorithms.
Graphical Abstract

Mass spectrometry (MS) is widely used for biomarker discovery and to explore prevailing metabolomic processes. Untargeted MS measurements coupled with high-performance liquid chromatography (LC) allow for the detection of thousands of ions in a matter of minutes. However, even though the resolution, mass accuracy, and sensitivity of mass spectrometers continue to improve, the identification of small molecules is still challenging due to their limited mass range and number of possible isomers.1–3 To date, most methods for metabolite identification are based on mass spectra database comparison. In these comparisons, spectra obtained experimentally are matched to the database containing a list of known molecular masses and fragmentation patterns. However, the vast majority of features in an MS experiment cannot be identified due to limited entries in current databases and/or insufficient fragmentation coverage. Furthermore, even if the chemical formula of a particular species is convincingly identified, structural identification remains a challenge due to the number of isomer species which can exist for any given chemical formula.4
To deal with these challenges, the Metabolomics Standards Initiative has published guidelines for metabolite identification,5,6 which give identification confidence levels depending on the amount of information discerned for the molecule. On the basis of these recommendations, the best way to confidently identify a metabolite is to use two independent and orthogonal data types for each authentic compound analyzed, such as GC or LC retention time, molecular mass, and tandem mass spectra. However, this step can be costly, as numerous authenticated chemical standards are required to attain the highest confidence level (i.e., a level 1 identification). Additionally, chemical standards are oftentimes unavailable for true unknown molecules (novel compounds), thus requiring several orthogonal analytical measurements and/or custom synthesis to support an identification.
Recently, the use of ion mobility spectrometry coupled with mass spectrometry (IM-MS) has become very promising for adding a structural dimension to MS analysis based on collision cross sections (CCSs) in support of metabolomic studies.7 In contrast to other properties, such as retention time, CCS is an ion parameter which can be measured with relative standard deviation (RSD) ranging from 0.29 to 6.2% when using different instrumental platforms8 and 3% to better than 0.5%, when using a standardized method,9–11 making it a valuable property for metabolite identification that is reproducible between different laboratories. The use of CCS can reduce the number of possible identifications and the number of false positive identifications in untargeted metabolomic studies.12–14 Additionally, when reference CCS values are not available in a database, it is possible to compute theoretical CCS values on structures obtained from molecular simulations. Currently this approach requires choosing the proper theory and approach for converting candidate structures to CCS, many of which are computationally expensive without being as precise as an experimental measurement.12,15 A more efficient process to produce CCS values for small molecules is through machine learning approaches.16 By using a training set, the algorithm attempts to identify the relation between an input (usually a set of molecular descriptors) and the CCS values. If the learning step is successful, the function can be applied to new inputs to obtain accurate and efficient predictions. The performance of the model is verified using validation and testing sets that were not used during the training step.
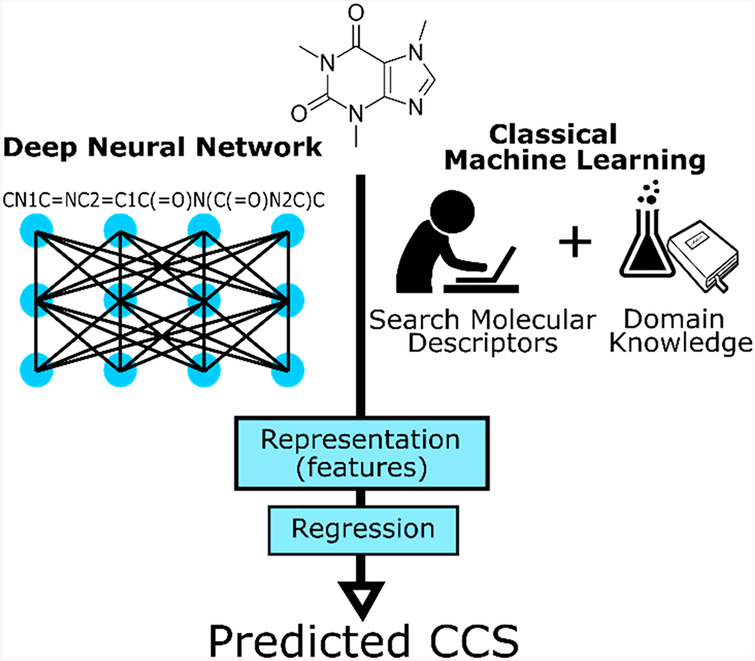
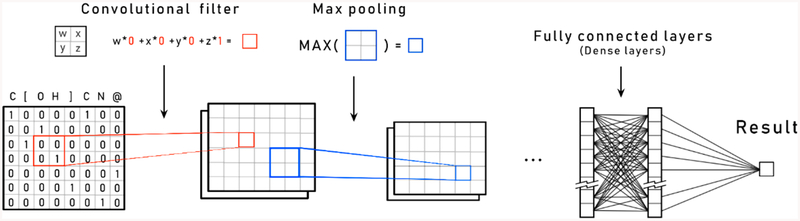
Deep neural networks (DNN), a type of machine learning algorithm, are now commonly used in multiple domains, such as self-driving cars, medical diagnosis, drug optimization, and speech recognition.17–19 Compared with other machine learning algorithms, such as support vector machines and random forests that learn models directly from a set of user-provided features, deep learning algorithms are composed of a cascade of layers which extract increasingly complex features (i.e., combinations of the original features) from the initial input (Figure 1). The DNN models are trained to build a representation which is then used to perform a prediction task. Convolutional neural networks (CNN), a subtype of DNN, are widely used in image recognition due to their capability to resist translation and transformation of features present in the input.20 CNN structures can be separated in two components (Figures 1 and 2). The first is the feature-learning component, which is constituted of multiple successions of convolution filter layers and maximum pooling layers (Figure 2). The output of the feature-learning component is a hidden internal representation of the input constructed by the neural-network. The second component, known as the predictive section, performs classification or regression depending on the task at hand, through a series of fully connected layers using the internal representation as input. DNN and CNN have already been used successfully in chemoinformatics for predicting molecular properties and protein-ligand interactions.21
Figure 1.
Comparison between DNN and classical machine learning for CCS prediction. Blue sections are purely computational, making DNN an almost completely computational approach. Classical machine learning requires an input of well-defined and comprehensive molecular descriptors, which can be adversely influenced by domain knowledge (e.g., CCS is correlated with the m/z value), reducing its accuracy.
Figure 2.
Schematic representation of the different operations performed by a convolutional neural network.
CCS prediction using machine learning has been addressed on multiple occasions in prior studies.12,2,23 Although results from these previous works were published, the prediction models and the code needed to reconstruct the models are unavailable. Moreover, previously published prediction models might not generalize well to new data from multiple laboratories because these models were mostly trained on data sets produced in a single laboratory and on a single instrument, making them highly specific to a certain context. Furthermore, most if not all prediction models to date use a set of molecular descriptors as the input for predictions. This transfers the problem of predicting CCS to the issue of finding or computing the values for a set of molecular descriptors (e.g., polar surface area, molar refractivity, etc.), which is not straightforward and thus is prone to user error. Simplified molecular-input line-entry systems (SMILESs) are structurally descriptive notations which can be readily assigned to any compound with a known structure, and they are already used as input by different methods to compute molecular descriptors.24 In this work, we utilized a chemical SMILES, a chain of characters easily found in chemical compound databases, as the input of a CNN model to predict CCS for different types of molecules. We generated a neural network structure based on CNN for CCS prediction and measured the performances of the generated models on different testing sets. We also evaluated the reusability of the SMILES internal representation learned by the model on a multitask learning problem. Finally, we offer a simple command line tool to use the generated model for CCS predictions.
■ EXPERIMENTAL SECTION
Data Sets.
Five data sets containing CCS and mass information were collected from multiple sources, including measurements from drift tube ion mobility (DTIM) and traveling wave ion mobility (TWIM) instruments, in order to constitute a learning database that includes a large panel of molecules. These sources included the following data sets.
MetCCS.25
The 779 CCS values were measured on an Agilent 6560 DTIM-QTOF-MS instrument (Agilent Technologies, Santa Clara, CA). This data set is already separated between a training (n = 648) and testing (n = 131) set.
Astarita.11
The 205 CCS values were measured on a Waters Synapt G2 Q-TWIM-TOF-MS instrument (Waters Corporation, Manchester, UK). The positive and negative ion mode data sets were used as an independent testing sets for comparison with MetCCS web server predictions.
Baker.26
The 857 CCS values were measured using the Agilent 6560 DTIM-QTOF-MS instrument customized for increased precision and reproducibility. This data set contains multiple types of small molecules.
McLean.27
The 211 CCS values were measured using the Agilent 6560 DTIM-QTOF-MS instrument. This diverse data set contains CCS values for amino acids, lipids, metabolites, and peptides.
CBM2018.22
The 357 CCS values were measured using a Waters Vion TWIM-QTOF-MS instrument. This data set contains pharmaceuticals, drugs of abuse, and their metabolites.
Data Preparation.
For each molecular entry, the SMILES notation was retrieved from PubChem except for amino acids sequences for which the SMILESs were generated using the Python RdKit module. The data sets were filtered to keep only SMILES with less than 250 characters/chemical symbols. This removed only a few entries and allowed the network input to be kept at a reasonable size. The only ions considered in this evaluation were (M + H)+, (M + Na)+, (M − H)−, and (M − 2H)2− in order to have at least 50 examples per ion type.
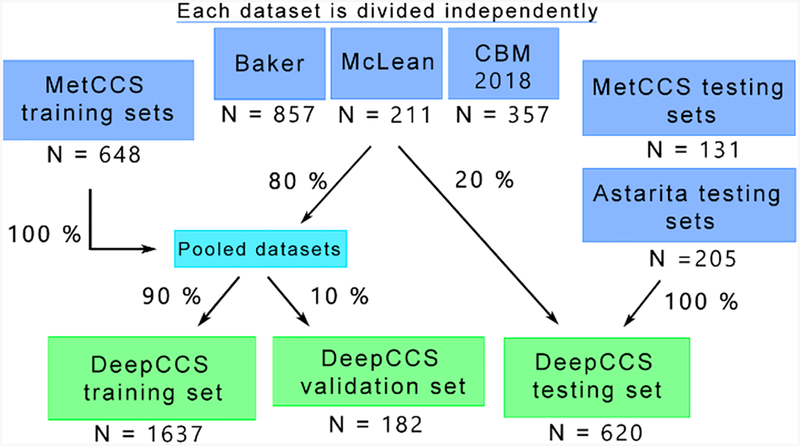
The different data sets were split into a training, validation, and testing set following the schema in Figure 3. The Astarita data sets were all included in the DeepCCS testing set to allow for a valid comparison to MetCCS predictors, while 20% of the Baker, McLean, and CBM2018 data sets were included in the testing set to better evaluate the generalization of the models.
Figure 3.
Partitioning of the different source data sets between the training, validation, and testing set of DeepCCS.
To feed the neural network, SMILESs and ions were encoded using one-hot vector encoding and padded to a length of 250 characters. This resulted in binary matrices of 250 × 36 for SMILESs and binary vectors of length 4 for the ions.
Neural Network Structure Optimization and Training.
The decision to use a CNN was justified by the intuitive way this type of DNN learns. The CNN looks for features in the input, and in the case of image recognition, these features can be lines, points, or a combination of these, leading to more complex objects (e.g, eyes or wheels). In our case, these features can be chemical groups and substructures. Since these groups of atoms can be anywhere in the SMILES representation, the translation resistance characteristic of CNN was a good fit. Furthermore, it was shown that using CNN with SMILES as input can give results equivalent or better than the state-of-the-art in many chemo-informatics applications.28
A CNN structure is modulated by a set of hyperparameters that affect the number and width of layers, convolution filter size, and maximum pooling window size, among others. Different hyperparameters have a different impact on the capability of a model to learn and generalize. In this work, hyperparameter optimization was performed by a 5-fold cross validation using a random search approach. Implementation and experiments were performed in Python. The CNN was built using the Keras library with the Tensorflow backend. After training, the model giving the best score on the validation set at the end of the different epochs was retained (Figure 4). The training of a model using the standard data set partitioning (Figure 3) took around 25 min on a Nvidia Tesla P100 GPU. Prediction of 100 CCS values using the DeepCCS command line tool took approximately 3 s on a standard desktop computer without the use of a GPU. Additional information about model optimization and construction are available in the Supporting Information. All the codes needed to train the network and to reproduce the results on the different testing sets presented in this article are available at github.com/plpla/DeepCCS/.
Figure 4.
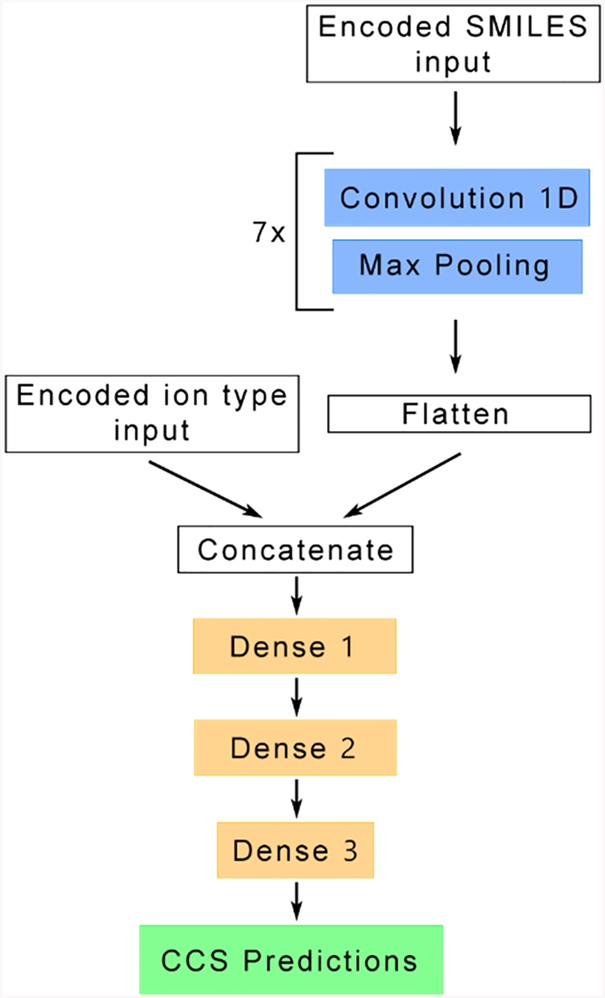
DeepCCS neural network structure. The upper part, in blue, performs a series of convolution and maximum pooling steps to learn an internal representation of the encoded SMILES input. This representation is flattened and concatenated with the ion type to be processed by the lower part of the network, in orange, to perform CCS prediction through a series of dense layers. The detailed network structure is available in Table S3.
Evaluation of the Internal Representation Reusability.
In order to evaluate the reusability potential of the internal representation learned by a CNN using SMILES as input, a multitask experiment was performed. The SMILES and molecular properties of every compound in the Human Metabolome Database (HMDB)29 were extracted. Only compounds with a valid SMILES and valid values for polar surface area, logS, refractivity, polarizability, logP (ALOGPS), and logP (Chemaxon) were retained. This allowed for the extraction of 71 232 compounds. This data set was randomly separated between the training, validation, and testing set using 72, 8, and 20% of the complete data set. The SMILES encoder previously learned was adjusted to include new chemical symbols not seen in the CCS data sets. A new CNN based on the DeepCCS structure (Figure 4) was built with the following changes. The second input (ion type) was removed; the concatenation layer was removed; and six different dense sections, one per property, replaced the single dense section. The resulting multitask CNN structure can be consulted in Table S4. The task of the network was to predict the different properties using a common internal representation. This allowed the network to learn a general internal presentation of a SMILES. After the first training phase, a DeepCCS model was reconstructed using the convolution and maximum pooling layers that were trained on the multitask problem. The weights of the feature learning part layers were locked to prevent further learning. The new half-trained network was retrained for 150 epochs using the CCS data after encoding using the updated SMILES encoder and the exact same data set split.
■ RESULTS AND DISCUSSION
DeepCCS Network Structure.
Convolutional neural networks are known to learn an internal representation of the input through a series of convolution and maximum pooling steps. This internal representation is then used as the input for a multilayer perceptron to perform predictions. The network structure of DeepCCS that was obtained after optimization uses the same principle, transformation of the SMILES provided an input to an internal representation and prediction using this representation and the ion type for which the CCS prediction must be made. The use of a second input allows DeepCCS to separate the internal representation of the input and the ion type to let the network focus only on the molecular structure in the feature-learning part. The final structure of the DeepCCS neural network is presented in Figure 4, and details can be found in Table S3 and in the source code. It contains a series of 7 convolution and maximum pooling layers to study the molecular structure of the SMILES provided in input. It is worth mentioning, that the downscaling factor (strides parameter) was increased to a value of 2 on the last maximum pooling layer to significantly reduce the number of trainable weights in the network without impacting the prediction accuracy.
CCS Prediction.
To evaluate the robustness of the training step, two different experiments were performed (Table 1). First, ten different models were trained on a single data set partition. Since the initialization of the trainable weights are different for each model, the resulting model explored different paths to finally converge to a final state that produced similar predictions. The second experiment consisted of training ten different models using ten different data splits generated randomly. This allowed to study the impact of the training and testing set composition on the performances of the model. For example, if a set of molecules exhibiting a molecular substructure that the network could not interpret properly because of a lack of example was exclusively in the training set from the first experiment, it would have erroneously resulted in good results. As shown in Table 1, the results are very similar between the two experiments showing that data set splitting, and the network initial weights values have a minimal impact on the performances of the various trained models.
Table 1.
Average Coefficient of Determination (R2) and Median Relative Error over Ten Different Models Trained Using Either a Single Dataset Split or Different Dataset Splitsa
| data set | single split | different splits | ||
|---|---|---|---|---|
| R2 | median relative error (%) | R2 | median relative error (%) | |
| Global | 0.976 (0.001) | 2.67 (0.18) | 0.979 (0.004) | 2.37 (0.27) |
| MetCCS test pos. | 0.960 (0.005) | 2.02 (0.24) | 0.964 (0.007) | 1.93 (0.35) |
| MetCCS test neg. | 0.969 (0.005) | 3.11 (0.49) | 0.967 (0.007) | 3.15 (0.63) |
| Astarita pos. | 0.901 (0.013) | 4.86 (0.30) | 0.897 (0.011) | 4.77 (0.44) |
| Astarita neg. | 0.955 (0.006) | 3.13 (0.48) | 0.949 (0.008) | 3.36 (0.37) |
| Baker | 0.954 (0.006) | 2.43 (0.11) | 0.967 (0.010) | 2.02 (0.17) |
| McLean | 0.995 (0.001) | 1.49 (0.14) | 0.996 (0.001) | 1.15 (0.28) |
| CBM 2018 | 0.930 (0.010) | 2.26 (0.28) | 0.969 (0.010) | 1.26 (0.47) |
Only data from the different testing set partitions were used. Standard deviation values are in parentheses.
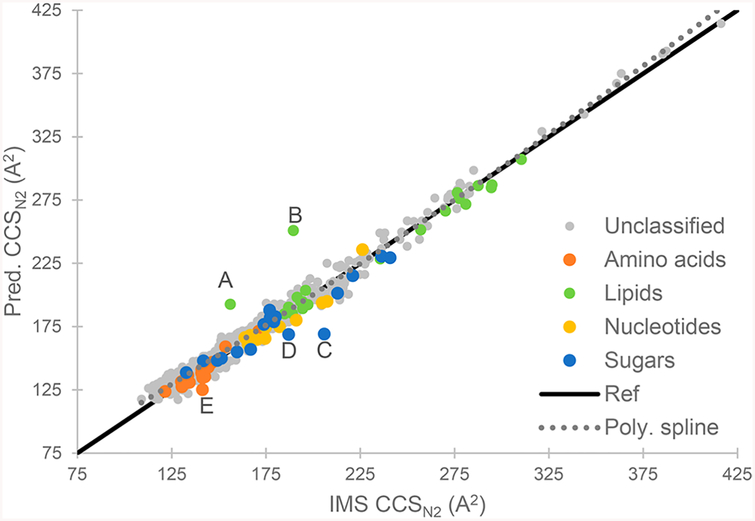
When all testing sets were merged into a single, global testing set, the coefficient of determination (R2) was greater than 0.97, and the absolute median relative error (MRE) was below 2.6%, indicating an excellent accuracy of prediction when compared to experimentally measured values. Considering that this global testing set was not used during the training step and that it contains data originating from five different laboratory and multiple instruments, one can conclude that the model achieved a state of generalization where it can be applied to new molecules. Furthermore, since the reported deviation for ion mobility CCS measurement can be as high as 6.2%,8 these results appear acceptable. Similar results were obtained when removing the test sets of the SMILES–ion combinations that are present in the training set with different CCS values, thus making sure no similar examples were already seen by the model before generating predictions (Table S5). Using the compounds classification provided in the Baker data set, Figure 5 shows that the model performs well for different types of compounds, such as amino acids, fatty acids, and lipids; hence, the model correctly discriminates the differences between multiple types of chemical structures and can predict the CCS value properly. With the exception of a few outliers, the concordance between measured and predicted CCS values is close to the reference line which indicates overall good predictions (Figure 5).
Figure 5.
Comparison of IMS measured and predicted CCS values for all compound from the testing set. Compound classes are from the Baker data set, others are unclassified. The solid line (ref) represents a reference line of perfect fit (a slope of 1). The dotted line indicates a 2nd-order polynomial spline fit to the data to show overall tendency. Letters A–E indicate outliers, respectively, methyl behenate, 1,2-diacyl-sn-glycero 3-phosphocholine, d-maltose, sophorose, and l-threonine (see the Supporting Information).
Although global performances of the algorithm were satisfactory, the Astarita data sets showed poorer correlation compared to the other data sets, with an average R2 lower than 0.9 and a MRE close to 5% for positive ions. Since the performances on the other testing data sets were better, we hypothesized that either the measurement accuracy of the Astarita data sets were lower than the other data sets or that a bias in measurement between data sets is present. To investigate this further, CCS values for identical SMILES and ion type were compared, which allows for the variation between data sets to be evaluated (Table 2). The average difference between the Astarita positive data set and Baker (n = 57) or McLean (n = 14) data sets was approximately 5% for overlapping measurements, which is significantly higher than the differences observed between other data sets. This seems to indicate that a CCS measurement bias is present in these data sets, which serves to decrease the performance of the model during the testing step. In fact, previous studies have demonstrated that large (>5%) differences in CCS can exist when comparing measurements obtained from drift tube (e.g., Baker and McLean data sets) and traveling wave instruments (e.g., Astarita data sets).8 Since both Astarita data sets were not included in the training step, this bias does not affect the model directly. The comparison of CCS measurement in multiple laboratories using different experimental conditions also allows us to get a better idea of the real variation that can be expected when comparing experimental CCS values from different studies.
Table 2.
Comparison of CCS Measurement for Identical Molecules and Ion Type between the Different Datasetsa
| MetCCS train pos. |
MetCCS train neg. |
MetCCS test pos. |
MetCCS test neg. |
Astarita pos |
Astarita neg |
Baker | McLean | CBM 2018 |
|
|---|---|---|---|---|---|---|---|---|---|
| MetCCS train pos. | 0.00 | 0.37 | 4.36 | −1.77 | −0.03 | 1.14 | |||
| MetCCS train neg. | 0.00 | −3.74 | −1.92 | ||||||
| MetCCS test pos. | −0.37 | 0.00 | 2.91 | −2.40 | −1.20 | ||||
| MetCCS test neg. | 0.00 | −0.23 | −4.02 | −3.21 | |||||
| Astarita pos. | −4.36 | −2.91 | 0.00 | −5.54 | −4.88 | ||||
| Astarita neg. | 0.23 | 0.00 | −4.99 | −2.82 | |||||
| Baker | 1.77 | 3.74 | 2.40 | 4.02 | 5.54 | 4.99 | 0.00 | 0.26 | 3.62 |
| McLean | 0.03 | 1.92 | 1.20 | 3.21 | 4.88 | 2.82 | −0.26 | 0.00 | 0.38 |
| CBM 2018 | −1.14 | −3.62 | −0.38 | 0.00 |
The value is the non-absolute mean percent difference relative to the average CCS measured by two datasets.
On the basis of our results, CCS measurements can be reproducible well below the reference 2% values (Baker vs McLean, n = 54), but it can also be as high as 5% (Astarita vs Baker, n = 57). These results corroborate what was observed by Schmitz et al. when comparing CCS values obtained from different IM instrumentation and techniques.8 The differences between TWIMS (i.e., Astarita, CBM2018) and DTIMS (i.e., MetCCS, Baker, McLean) are not systematic for most molecule types, but some show appreciable differences.8 Once again, an appropriate calibration for TWIMS is critical to obtain a high quality measurement using this technique. These results also put emphasis on the importance of using data from different contexts, such as instruments (DTIMS and TWIMS) and laboratories, to obtain a predictor that can generalize to every context when insufficient training data from a specific context are available. As more data sets are published and included in the training step of DeepCCS, the real variation of CCS measurements will become more precise, and the DeepCCS model will improve such that it should be able to predict values closer to the average CCS, therefore increasing its performance. The addition of the IMS measurement technique could also be included in the model as more data become available, making the model adaptable to the different contexts.
Outliers Detection for Database Validation.
Outliers in Figure 5 (points A–E) were further investigated. These data points, respectively, correspond to 1,2-diacyl-sn-glycero 3-phosphocholine, methyl behenate, d-maltose, sophorose, and l-threonine. All outliers, except sophorose, were confirmed as measurement error by remeasuring the CCS value of the compounds (Supporting Information and Figure S1). For sophorose, we hypothesize that the error is similar to the one for d-maltose. Carbohydrates are prone to aggregation, and these multimers readily dissociate between the IM and the MS stage, resulting in ion signals at higher CCS values (Figure S1C). Since the measured value is, like for d-maltose, higher than the predicted value, we hypothesis that it might also be a case of aggregation–dissociation of sugar molecules leading to an erroneous measurement.
Outlier investigation detected four confirmed and one unconfirmed but highly probable erroneous measurements. These results highlight on another potential utility of CCS prediction tools, database validation. By comparing predicted and measured CCS values, one can easily detect suspect measurements and further investigate their validity. The ease of use and good performances of DeepCCS make it ideal for this task.
Comparison to Existing Tools.
The DeepCCS model uses SMILES notation as input, which is easy to obtain for most small molecules. When performing metabolite identification using MS data, a popular approach is to compare the empirically measured spectra to reference spectra from a database. This reference database necessarily contains the structure of the compounds, and therefore, the SMILES notation is either already present or easily computed. In contrast, other CCS predictors use molecular descriptors as input, which are not always available in databases and can require licensing commercial software to compute them.
The MetCCS web server is a CCS prediction tool based on Support Vector Regression and uses 14 common molecular descriptors available in the HMDB database. MetCCS has been used to generate over 176 000 CCS values for over 35 000 small molecule metabolites from the HMDB.12 Although MetCCS does not allow CCS prediction for molecules other than metabolites, a separate tool, LipidCCS, has been developed by the authors for lipids and fatty acids.30 In contrast, CCS prediction for all molecule types can be done directly in DeepCCS. Results from the DeepCCS model were compared with those obtained using the MetCCS server in order to evaluate the performance of each machine learning approach (Figure 6). The MetCCS testing data sets and the Baker data set were used, as MetCCS does not work for all molecule types and requires HMDB identifiers to collect the associated molecular descriptors. The MetCCS prediction server produces the most extreme values on most data sets. This could be explained by the sub representation of certain molecule types in the MetCCS training set that is much smaller. Overall, both predictors perform similarly with most predictions within a 5% window, but we can discern the impact of the different training sets of the predictions. Both models were trained almost exclusively with data from DTIMS (DeepCCS used a fraction of the CBM2018), but MetCCS was trained with data from a single laboratory. The Baker and McLean CCS values are far more distant than the MetCCS training set value to Astarita values.
Figure 6.
Comparison of the error distribution on five different testing sets between DeepCCS and MetCCS. Previously detected database errors were removed from the comparison.
When considering the MRE and R2 (Table 3), the performances were found to be similar when using the MetCCS testing set, but MetCCS performed better when the Astarita data sets were used. This might be explained by the close proximity of MetCCS and Astarita CCS values (Table 2). Although, when evaluating the performances using the Baker data set, DeepCCS performed better and could predict the CCS for more molecules (171 instead of 134). This shows that the model improves on current methods through better generalization and by providing accurate predictions on multiple compound types.
Table 3.
Comparison of DeepCCS and MetCCS Predictive Performances Using Different CCS Testing Sets
| R2 | median relative error (%) | |||
|---|---|---|---|---|
| DeepCCS | MetCCS | DeepCCS | MetCCS | |
| MetCCS test positive | 0.97 | 0.95 | 1.63 | 1.74 |
| MetCCS test negative | 0.98 | 0.97 | 2.30 | 1.54 |
| Astarita positive | 0.93 | 0.93 | 4.22 | 2.96 |
| Astarita negative | 0.97 | 0.97 | 2.21 | 1.47 |
| Baker testing set (n = 171 and 134) |
0.95 | 0.9 | 2.50 | 3.00 |
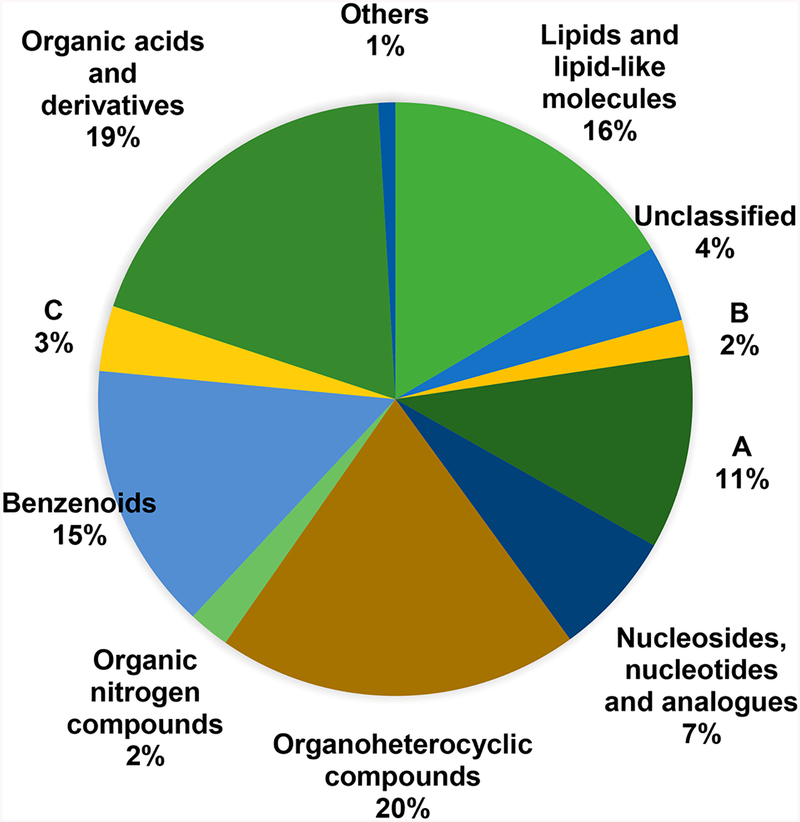
Even with these advances, DeepCCS has its own limitations. It can only perform predictions using features that have already been observed. For example, only chemical symbols of atoms contained within the data set are available for encoding, therefore only SMILES with these symbols can be used for predictions. This limitation is in place to ensure predictions are based on features that are recognized by the CNN; that is, the model cannot perform predictions using items it has never seen. Similar limitations apply to molecule types, thus while DeepCCS can perform CCS prediction on any compound, the predictions might be less accurate for molecule types and substructures not seen during the initial training stage. Figure 7 shows molecule distribution at the superclass level from the ClassyFire taxonomy.31 Even though the training set contains multiple examples, it clearly does not cover all possible molecules. In all cases, these limitations can be solved by generating sufficiently large and diverse CCS data sets to train a new model.
Figure 7.
Classification at the superclass level of the molecules from DeepCCS data sets using the ClassyFire taxonomy. The “Others” group contains the following classes, organohalogen compounds, organic polymers, organosulfur compounds, and homogeneous nonmetal compounds. (A) Organic oxygen compounds. (B) Alkaloids and derivatives. (C) Phenylpropanoids and polyketides. Classification tables at the subclass and class levels are available in Tables S6 and S7.
Generalization of the Internal Representation.
The SMILES input contains all of the structural information on the molecule, and as such, the CNN internal representation should be generalizable to predicting other molecular properties. This assumption was evaluated by performing molecular property predictions on a multitask problem. Six different chemical properties available in the HMDB (polar surface area, logS, refractivity, polarizability, logP (ALOGPS), and logP (Chemaxon)) were predicted with the objective to learn an internal representation. The resulting model predicted these chemical properties with very good accuracy (R2 > 0.98) and a median relative error below 0.7% (Table S2).
This generalized internal representation incorporating the data set from the single split experiment was subsequently used for CCS prediction. Performances similar to what was obtained in previous experiments were also obtained (Table 4). When predicting CCS, this new model performed with a global R2 of 0.968 and a median relative error of 2.6%. The results of this experiment showed that the internal representation learned by a CNN using SMILES can be reused to predict different molecular properties beyond CCS, and that significantly increasing the number of SMILES used to learn the internal representation does not have an impact of the accuracy of CCS predictions. Therefore, we hypothesize that, to further increase CCS prediction accuracy, the predictive part of the network would need additional data for a better understanding of the link between the network internal representation and the CCS value. The other possibility, and probably the best way to increase prediction accuracy, would be to decrease the variations between CCS measurement in the different data sets used for training the model. A large CCS database (n > 3800) exhibiting high measurement precision has recently been developed by the authors and will include further CCS measurement as they will be published.14 It will be used in future work to further improve the predictive capabilities of DeepCCS.
Table 4.
Model Performances on CCS Prediction after Training the Feature Learning Section of the Network on a Multi-Output Problema
| data set | R2 | median relative error (%) |
|---|---|---|
| global | 0.968 | 2.55 |
| MetCCS test pos. | 0.928 | 2.43 |
| MetCCS test neg. | 0.941 | 2.37 |
| Astarita pos. | 0.878 | 4.27 |
| Astarita neg. | 0.945 | 2.89 |
| Baker | 0.950 | 2.15 |
| McLean | 0.986 | 1.50 |
The dataset split is identical to the single split previously used.
■ CONCLUSION
CCS prediction using machine learning is necessary to populate the numerous possible small molecule CCS values with high speed and accuracy. The DeepCCS prediction algorithm uses SMILES notation as an input instead of a more traditional set of molecular descriptors, which allows DeepCCS to be fast (100 predictions in ~3 s) and, due to the CNN structure used, is also generalizable to a large number of different molecule types. Additionally, the DeepCCS command line tool provides an easy way to train a new model using newly generated data or to simply predict CCS values using the provided model. The precision of empirical CCS measurements used as a training set was found to have a significant impact on the overall prediction accuracy of the model. In this case, the wide variations observed (2% to more than 5%) in measured CCS are certainly a limiting factor on the capability of the model to predict CCS with less than 3% error. The performance of machine learning models, such as DeepCCS, will improve as more high-quality CCS measurements are made available.
Supplementary Material
■ ACKNOWLEDGMENTS
The authors acknowledge Alexandre Drouin and Prudencio Tossou for valuable comments and discussions. This work was supported, in part, by a Fond de recherche québécois en santé (FRQS) Doctoral award to P.-L.P. and the Canada Research Chair in Medical Genomics (J.C.). J.C., P.-L.P. and E.F.F. also acknowledge funding from Mitacs (MC00006). J.C.M and J.A.M acknowledge funding from the National Institutes of Health (NIGMS R01GM092218 and NCI 1R03CA222452–01). E.B. would also like to acknowledge funding support from the National Institute of Environmental Health Sciences of the NIH (R01 ES022190 and P42 ES027704). We gratefully acknowledge the support of NVIDIA Corporation for the donation of the Titan Xp GPU used for this research. Part of the computations were performed on the Compute Canada supercomputer infrastructure.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.anal-chem.8b05821.
Outliers description, IM spectra and CCS values for repeated measurement, neural network structures and hyper-parameters values used during cross-validation, HMDB chemical properties prediction results, effect of repeated SMILES-ion combination on model performances, and ClassyFire classification of the compounds (PDF)
Notes
The authors declare no competing financial interest.
■ REFERENCES
- (1).Lynn K-S; Cheng M-L; Chen Y-R; Hsu C; Chen A; Lih TM; Chang H-Y; Huang C; Shiao M-S; Pan W-H; Sung T-Y; Hsu W-L Anal. Chem 2015, 87 (4), 2143–2151. [DOI] [PubMed] [Google Scholar]
- (2).Nguyen DH; Nguyen CH; Mamitsuka H Briefings Bioinf. 2018, 2018, 066. [Google Scholar]
- (3).Blaženović I; Shen T; Mehta SS; Kind T; Ji J; Piparo M; Cacciola F; Mondello L; Fiehn O Anal. Chem 2018, 90 (18), 10758–10764. [DOI] [PubMed] [Google Scholar]
- (4).May JC; McLean JA Annu. Rev. Anal. Chem 2016, 9 (1), 387–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Sumner LW; Amberg A; Barrett D; Beale MH; Beger R; Daykin CA; Fan TW-M; Fiehn O; Goodacre R; Griffin JL; Hankemeier T; Hardy N; Harnly J; Higashi R; Kopka J; Lane AN; Lindon JC; Marriott P; Nicholls AW; Reily MD; Thaden JJ; Viant MR Metabolomics 2007, 3 (3), 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Rochat BJ Am. Soc. Mass Spectrom 2017, 28 (4), 709–723. [DOI] [PubMed] [Google Scholar]
- (7).Schrimpe-Rutledge AC; Codreanu SG; Sherrod SD; McLean JA J. Am. Soc. Mass Spectrom 2016, 27 (12), 1897–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hinnenkamp V; Klein J; Meckelmann SW; Balsaa P; Schmidt TC; Schmitz OJ Anal. Chem 2018, 90 (20), 12042. [DOI] [PubMed] [Google Scholar]
- (9).Stow SM; Causon TJ; Zheng X; Kurulugama RT; Mairinger T; May JC; Rennie EE; Baker ES; Smith RD; McLean JA; Hann S; Fjeldsted JC Anal. Chem 2017, 89 (17), 9048–9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Paglia G; Angel P; Williams JP; Richardson K; Olivos HJ; Thompson JW; Menikarachchi L; Lai S; Walsh C; Moseley A; Plumb RS; Grant DF; Palsson BO; Langridge J; Geromanos S; Astarita G Anal. Chem 2015, 87 (2), 1137–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Paglia G; Williams JP; Menikarachchi L; Thompson JW; Tyldesley-Worster R; Halldórsson S; Rolfsson O; Moseley A; Grant D; Langridge J; Palsson BO; Astarita G Anal. Chem 2013, 86 (8), 3985–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Zhou Z; Shen X; Tu J; Zhu Z-J Anal. Chem 2016, 88 (22), 11084–11091. [DOI] [PubMed] [Google Scholar]
- (13).Nichols CM; Dodds JN; Rose BS; Picache JA; Morris CB; Codreanu SG; May JC; Sherrod SD; McLean JA Anal. Chem 2018, 90 (24), 14484–14492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Picache JA; Rose BS; Balinski A; Leaptrot KL; Sherrod SD; May JC; Mclean JA Chem. Sci 2019, 10, 983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Colby SM; Thomas DG; Nunez JR; Baxter DJ; Glaesemann KR; Brown JM; Pirrung MA; Govind N; Teeguarden JG; Metz TO; Renslow RS ISiCLE: A molecular collision cross section calculation pipeline for establishing large in silico reference libraries for compound identification. 2018, https://arxiv.org/abs/1809.08378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Zhou Z; Tu J; Zhu Z-J Curr. Opin. Chem. Biol 2018, 42, 34–41. [DOI] [PubMed] [Google Scholar]
- (17).LeCun Y; Bengio Y; Hinton G Nature 2015, 521 (7553), 436–444. [DOI] [PubMed] [Google Scholar]
- (18).Ehteshami Bejnordi B; Veta M; Johannes van Diest P; van Ginneken B; Karssemeijer N; Litjens G; van der Laak JAWM; Hermsen M; Manson QF; Balkenhol M; Geessink O; Stathonikos N; van Dijk MC; Bult P; Beca F; Beck AH; Wang D; Khosla A; Gargeya R; Irshad H; Zhong A; Dou Q; Li Q; Chen H; Lin H-J; Heng P-A; Haß C; Bruni E; Wong Q; Halici U; Öner MÜ; Cetin-Atalay R; Berseth M; Khvatkov V; Vylegzhanin A; Kraus O; Shaban M; Rajpoot N; Awan R; Sirinukunwattana K; Qaiser T; Tsang Y-W; Tellez D; Annuscheit J; Hufnagl P; Valkonen M; Kartasalo K; Latonen L; Ruusuvuori P; Liimatainen K; Albarqouni S; Mungal B; George A; Demirci S; Navab N; Watanabe S; Seno S; Takenaka Y; Matsuda H; Phoulady HA; Kovalev V; Kalinovsky A; Liauchuk V; Bueno G; Fernandez-Carrobles MM; Serrano I; Deniz O; Racoceanu D; Venâncio R JAMA 2017, 318 (22), 2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhao K; So H-C Methods Mol. Biol. (N. Y. NY;U. S.) 2019, 1903, 219–237. [DOI] [PubMed] [Google Scholar]
- (20).LeCun Y; Kavukcuoglu K; Farabet C Proceedings of 2010 IEEE International Symposium on Circuits and Systems 2010, 253–256. [Google Scholar]
- (21).Chen H; Engkvist O; Wang Y; Olivecrona M; Blaschke T Drug Discovery Today 2018, 23 (6), 1241– 1250. [DOI] [PubMed] [Google Scholar]
- (22).Mollerup CB; Mardal M; Dalsgaard PW; Linnet K; Barron LP J. Chromatogr. A 2018, 1542, 82–88. [DOI] [PubMed] [Google Scholar]
- (23).Bijlsma L; Bade R; Celma A; Mullin L; Cleland G; Stead S; Hernandez F; Sancho JV Anal. Chem 2017, 89 (12), 6583–6589. [DOI] [PubMed] [Google Scholar]
- (24).Moriwaki H; Tian Y-S; Kawashita N; Takagi TJ Cheminf. 2018, 10 (1), 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zhou Z; Xiong X; Zhu Z-J Bioinformatics 2017, 33 (14), 2235–2237. [DOI] [PubMed] [Google Scholar]
- (26).Zheng X; Aly NA; Zhou Y; Dupuis KT; Bilbao A; Paurus VL; Orton DJ; Wilson R; Payne SH; Smith RD; Baker ES Chem. Sci 2017, 8 (11), 7724–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).May JC; Goodwin CR; Lareau NM; Leaptrot KL; Morris CB; Kurulugama RT; Mordehai A; Klein C; Barry W; Darland E; Overney G; Imatani K; Stafford GC; Fjeldsted JC; McLean JA Anal. Chem 2013, 86 (4), 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Kwon S; Yoon S IEEE/ACM Trans. Comput. Biol. Bioinf 2018, 1. [Google Scholar]
- (29).Wishart DS; Feunang YD; Marcu A; Guo AC; Liang K; et al. Nucleic Acids Res. 2018, 46 (D1), D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Zhou Z; Tu J; Xiong X; Shen X; Zhu Z-J Anal. Chem 2017, 89 (17), 9559–9566. [DOI] [PubMed] [Google Scholar]
- (31).Djoumbou Feunang Y; Eisner R; Knox C; Chepelev L; Hastings J; Owen G; Fahy E; Steinbeck C; Subramanian S; Bolton E; Greiner R; Wishart DS J. Cheminf 2016, 8 (1), 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.