Evidence of microbial communities in the urinary tract (the urinary microbiome or urobiome) surfaced less than a decade ago. Since then, multiple investigators have confirmed this finding 1. More recent studies are now focused on characterizing the urobiome and its associations with human health and disease. We anticipate technological advances that will provide improved information about the status of the urobiome to clinicians; these changes will inform the care of multiple common urinary conditions linked to changes in the urobiome.
Discovery and confirmation of the urobiome was facilitated primarily by two complementary assays — high-throughput 16S ribosomal RNA (rRNA) gene sequencing that detected DNA evidence of bacteria and enhanced urine culture protocols (such as expanded quantitative urine culture (EQUC) that provided evidence that microbes detected by 16S rRNA sequencing and other culture-independent techniques are alive. EQUC, described in 2014, is available for clinical laboratory use and is recommended when the standard clinical urine culture is negative and urinary symptoms persist without explanation 2.
The urine specimen collection technique influences interpretation. In women, voided urine specimens should be described as genitourinary specimens, based on clear evidence of vulvovaginal microbial contribution. By contrast, urine specimens collected by suprapubic aspiration or transurethral catheterization can be considered bladder specimens 3. Consensus conferences are needed to standardize specimen collection and preservation techniques, as well as analytical approaches.
A few teams have begun to characterize the female bladder urobiome. The urobiome has a low biomass relative to the vaginal microbiome and is dominated by species of a few genera, most often Lactobacillus, Gardnerella, and Streptococcus. Our understanding of ‘normal’ urobiome has been limited by the lack of a bladder health definition; one has recenty been proposed by The Prevention of Lower Urinary Tract Symptoms Consortium 4. This consortium define bladder health as “A complete state of physical, mental, and social well-being related to bladder function and not merely the absence of LUTS. Healthy bladder function permits daily activities, adapts to short-term physical or environmental stressors, and allows optimal well-being (travel, exercise, social, occupational, or other activities).” Earlier investigations using symptom absence as an indicator of normalcy have reported findings that reveal associations between the female bladder urobiome and postinstrumentation UTIs, response to overactive bladder treatment, and urgency urinary incontinence (UUI) 1. This proposed bladder health definition may improve clinical phenotyping and related urobiome states.
The NIH-NICHD-funded Pelvic Floor Disorders Network recently reported results from a multicentre cross-sectional study that used catheterized urine samples and 16S rRNA sequencing to describe urobiome characteristics of well-characterized asymptomatic women (n = 84) compared with women with mixed urinary incontinence (MUI) (n = 123) 5. Participants (mean age 53 years) were dichotomized at age 51, the median age for menopause. Using Dirichlet Multinomial Mixture modelling, the investigators detected six community states on the basis of >50% microbial dominance. Although the proportion of women with Lactobacillus predominance did not differ between adult women with MUI and age-matched asymptomatic women, the investigators confirmed previous reports suggesting that some members of the genus Lactobacillus might be associated with urinary symptoms, including urgency urinary incontinence. These authors also suggested that non-Lactobacillus bacterial taxa in the urobiome might further influence symptoms. This study also supported previous reports that both BMI and older age were independently associated with MUI 5.
Fok et al. 6 reported 16S sequencing results from 126 adult women who had catheterized urine samples and vaginal and perineal swabs taken immediately before urogynecological surgery. Lactobacillus was the most abundant genus in bladder urine (30%) and vagina (26%); however this report highlights a relationship between urinary symptoms (overactive bladder) and two fastidious anaerobic Gram-positive microbes from the phylum Actinobacteria (Atopobium vaginae and Finegoldia magna) that were not influenced by either age or hormonal status. These emerging uropathogens are unfamiliar to most clinicians and whether all strains of A. vaginae have the ability to cause symptoms is unknown, especially as A. vaginae is commonly found in the vagina of women without bacterial vaginosis. This study presents the possibility of using a preoperative assessment of the urinary microbiome to reduce bothersome urinary symptoms following surgery and reduce risk of perioperative UTI.
Enhanced culture methods enable investigators to build a reference set of bladder-specific isolates that can be used to understand microbial contributions to bladder health and disease. Thomas-White and co-workers 7 described the phylogenetic diversity of bacteria isolated by EQUC from the bladders of women with and without LUTS by sequencing 149 genomes from 79 different bacterial species. They then compared the protein domains encoded in 64 bladder genomes from women without LUTS with the encoded domains of 92 vaginal and 152 gastrointestinal strains cultivated from asymptomatic women. This analysis revealed some overlap in functions within the bladder and vaginal strains, but a clear distinction from the gastrointestinal microbiota. This study revealed that the the bacteria of the bladder are distinct from those that inhabit the gut — even isolates of the same species differ substantially; to a lesser degree, the same is true for vaginal bacteria. This study also reveraled that public databases (such as GenBank) are inadequate for studies of the urobiome and its relationship to bladder health and disease because these databases lack urobiome-specific genomes.
Compelling evidence associates the female bladder urobiome with lower urinary tract symptoms (LUTS), but a similar association in men has not been established. Thus, Bajic et al. 8 stratified 28 men undergoing benign prostate enlargement (BPE)–LUTS surgery and 21 patients undergoing non-BPE–LUTS surgery by their International Prostate Symptom Score (IPSS). They collected paired voided and catheterized urine specimens, analysing them by EQUC and 16S rRNA sequencing. Overall, 39% of catheterized and 98% of voided specimens contained microbiota, but the catheterized and voided microbiota differed substantially. Thus, the authors concluded that voided urine does not adequately characterize the male bladder urobiome, and that catherized urine should be used instead. Analysing catheterized urine from men with and without BPE, the investigators determined that IPSS severity was associated with detectable bacteria in bladder urine, as microbes were detected in 22.2%, 30.0%, and 57.1% of men with mild, moderate, and severe LUTS, respectively (P = 0.024). The authors concluded that men with more severe urinary symptoms are more likely to have detectable bladder bacteria than those with less severe or no symptoms. Additional studies are needed to identify specific bladder bacteria associated with LUTS in men with BPE, owing to the small sample size of this study 8.
Few studies have investigated the role of the urobiome in urological malignancies. One study used 16S rRNA sequencing to compare voided urine from six healthy individuals with samples from eight patients with bladder cancer and found enrichment of the genus Streptococcus in patients with bladder cancer 9. A more recent study 10 used 16S sequencing to compare voided urine samples from 12 men diagnosed with bladder cancer and 11 healthy, age-matched participants. Although the authors found no significant differences between the two cohorts in terms of microbial diversity or overall microbiome composition, they did identify several taxa that were significantly over-represented in bladder cancer or healthy subgroup. The most common bacteria in healthy individuals were members of the genera Veillonella, Streptococcus, and Corynebacterium. By contrast, several taxa were enriched in the bladder cancer group, including members of the genus Fusobacterium, which has been associated with colorectal cancer. To validate this observation, the authors assessed an independent set of bladder cancer tissues using PCR and detected Fusobacterium nucleatum sequences in 11 of 42 samples. Again, additional studies are needed to determine if the urobiome is associated with bladder cancer, owing to the small sample size of this study 10.
Since the discovery and confirmation of the human urobiome, investigators have engaged in highly influential studies to characterize this microbial community and understand how it relates to human health and disease. Consensus is needed for terminology, specimen collection, storage techniques, and analytical approaches. We anticipate that the coming years will be marked by robust research in this field that will lead to clinically meaningful improvements in treatment of a wide variety of urinary tract disorders.
Fig. 1:
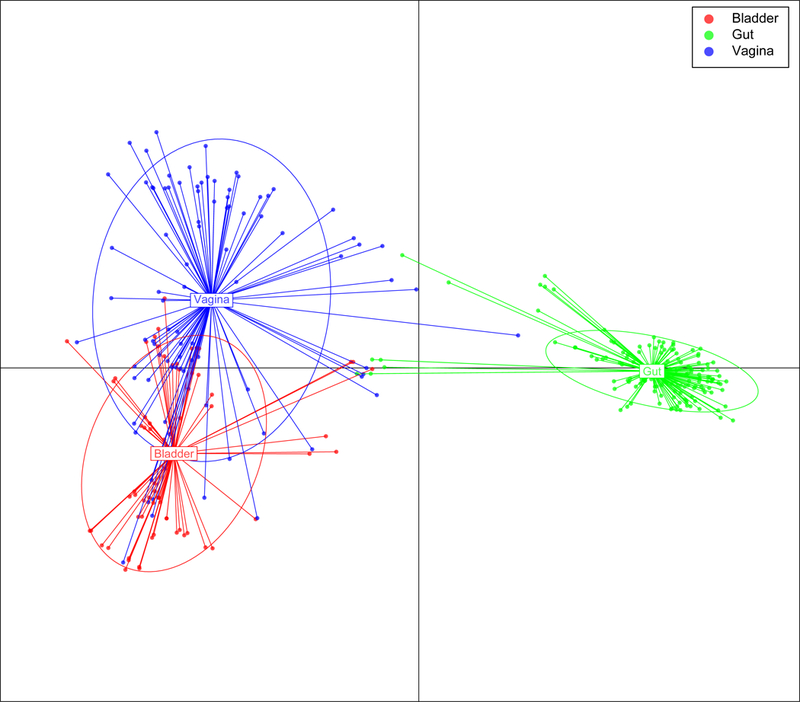
Functional diversity between genomes of bacterial strains isolated from the bladder, vagina and intestine.
Key advances.
Lactobacillus predominance does not differ between adult women with mixed urinary incontinence and age-matched asymptomatic women, but some members of the genus Lactobacillus might be associated with urinary symptoms 5.
Preoperative assessment of the urinary microbiome could reduce bothersome urinary symptoms following surgery and reduce risk of perioperative urinary tract infection 6.
Public databases are inadequate for studies of the urobiome and its relationship to bladder health and disease because these databases lack urobiome-specific genomes 7.
Men with more severe urinary symptoms are more likely to have detectable bladder bacteria than those with less severe or no symptoms. Voided urine does not adequately characterize the male bladder urobiome, and catherized urine should be used instead 8.
The urobiome differs between individuals with bladder cancer and those without 10.
Pull quotes
High-throughput 16S rRNA sequencing and enhanced urine culturing facilitated urobiome research
A proposed bladder health definition may improve clinical phenotyping and related urobiome states.
The bacteria of the bladder are distinct from those that inhabit the gut
Consensus is needed for terminology, specimen collection, storage techniques, and analytical approaches
Acknowlegements
This work was supported by NIH grant 5R01DK104718-03 to A.J.W. and L.B.
Footnotes
Competing interests
A.J.W. discloses Investigator Initiated Study funds from Astellas Scientific and Medical Affairs and Kimberly Clark Corporation. L.B. discloses editorial stipends from JAMA, Female Pelvic Medicine and Reconstructive Surgery and UpToDate.
References:
- 1.Mueller ER, Wolfe AJ & Brubaker L Female urinary microbiota. Curr Opin Urol 27, 282–286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Price TK et al. The Clinical Urine Culture: Enhanced Techniques Improve Detection of Clinically Relevant Microorganisms. J Clin Microbiol (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolfe AJ et al. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol 50, 1376–1383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lukacz ES et al. A Novel Research Definition of Bladder Health in Women and Girls: Implications for Research and Public Health Promotion. J Womens Health (Larchmt) 27, 974–981 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komesu YM et al. The urinary microbiome in women with mixed urinary incontinence compared to similarly aged controls. Int Urogynecol J (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fok CS et al. Urinary symptoms are associated with certain urinary microbes in urogynecologic surgical patients. Int Urogynecol J (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas-White K et al. Culturing of female bladder bacteria reveals an interconnected urogenital microbiota. Nat Commun 9, 1557 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bajic P et al. Male Bladder Microbiome Relates to Lower Urinary Tract Symptoms. Eur Urol Focus (2018). [DOI] [PubMed] [Google Scholar]
- 9.Xu W et al. Mini-review: perspective of the microbiome in the pathogenesis of urothelial carcinoma. Am J Clin Exp Urol 2, 57–61 (2014). [PMC free article] [PubMed] [Google Scholar]
- 10.Popović VB et al. The urinary microbiome associated with bladder cancer. Sci Rep. 8, 12157 ( 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]


