Abstract
The CYP26s are responsible for metabolizing retinoic acid and play an important role in maintaining homeostatic levels of retinoic acid. Given the ability of CYP2C8 to metabolize retinoic acid, we evaluated the potential for CYP2C8 inhibitors to also inhibit CYP26. In vitro assays were used to evaluate the inhibition potencies of CYP2C8 inhibitors against CYP26A1 and CYP26B1. Using tazarotenic acid as a substrate for CYP26, IC50 values for 17 inhibitors of CYP2C8 were determined for CYP26A1 and CYP26B1, ranging from approximately 20 nM to 100 μM, with a positive correlation observed between IC50s for CYP2C8 and CYP26A1. An evaluation of IC50’s versus in vivo Cmax values suggests that inhibitors such as clotrimazole or fluconazole may interact with CYP26 at clinically relevant concentrations and may alter levels of retinoic acid. The findings provide insight into drug interactions resulting in elevated retinoic acid concentrations and expand upon the pharmacophore of CYP26 inhibition.
Keywords: retinoic acid, CYP26, CYP2C8, homology model, tazarotenic acid
Introduction
Endogenous retinoic acid concentrations are highly regulated owing to their importance in cellular development with altered concentrations of retinoic acid known to have pharmacological and toxicological implications (1–6). In humans, retinoic acid binds to the retinoic acid and retinoid X receptors and plays a key role in the regulation of genes that affect the extent of cellular proliferation and differentiation as well as apoptosis (7–11). The regulation of circulating retinoic acid concentrations can occur through modulation of its synthesis, which involves multiple enzymatic steps in the conversion of retinol to all-trans-retinoic acid (at-RA) or through its clearance, which is primarily mediated by cytochrome P450-catalyzed oxidation to 4-hydroxy-at-retinoic acid (12, 13). Within the cytochrome P450 superfamily of xenobiotic metabolizing enzymes, the CYP26 subfamily (CYP26A1, CYP26B1 and CYP26C1) are the primary enzymes involved in retinoic acid metabolism (14–18). While hepatic CYP26 content is primarily a function of CYP26A1 expression, CYP26A1 and CYP26B1 mRNA are ubiquitously expressed, with sites of expression including the skin, lungs, testes and brain (15, 18–24). Less information is available about the expression patterns and functional relevance of CYP26C1.
A significant amount of catalytic overlap is observed for CYP26A1 and CYP26B1 in regard to their metabolism of at-RA, though the sequence homology between the two isozymes is only 42% (19, 21, 25). Perhaps owing to their homeostatic role in the regulation of retinoic acid concentrations and the subsequent pharmacological or toxicological outcomes, the pursuit of selective chemical inhibitors of CYP26A1 or CYP26B1 has received interest in both the inflammation and oncology therapeutic areas (26–34). Many of the compounds designed to inhibit CYP26 activity, also known as retinoic acid metabolism blocking agents (RAMBAs), share a similar pharmacophore, with an extended hydrophobic region that bridges a hydrophobic or aromatic ring system on one end of the molecule to a hydrogen bond accepting group on the opposite end (35, 36). In many cases, the aromatic group described above is an azole-containing ring system, designed to coordinate to the porphyrin iron of CYP26A1 or CYP26B1 and thus inhibit the enzyme (19, 29, 30, 37–39). Liarazole currently represents the most studied example of a RAMBA in clinical use (30, 40).
Approximately 5% to 8% of xenobiotic metabolism has been attributed to CYP2C8, with highly characterized substrates including amodiaquine, repaglinide, rosiglitazone, cerivastatin, paclitaxel and montelukast (41–44). CYP2C8 has also been shown to metabolize at-RA, 9-cis-retinoic acid and 13-cis-retinoic acid (45–49). Potent in vitro inhibitors of CYP2C8-catalyzed metabolism include montelukast, candesartan cilexetil, zafirlukast, clotrimazole and fluconazole (44, 50, 51). Clinically relevant drug interactions attributed to CYP2C8 inhibition have been noted for rosiglitazone, repaglinide and cerivastatin when co-administered with the CYP2C8 inhibitor gemfibrozil (52–55). The active site properties of CYP2C8 which contribute to its substrate and inhibitor profiles are fairly well understood, with the crystal structure of CYP2C8 having been solved with various ligands bound in the active site, including 9-cis-retinoic acid (56). The active site volume of CYP2C8 is relatively large (1438 Å3) and it has been described as having a bifurcated Y-shaped geometry (56, 57). Similar to the ligand profile of CYP26, early pharmacophore models of CYP2C8 ligands suggested the need for a hydrophobic or aromatic group proximal to the site of oxidation, an extended hydrophobic chain distal to the site of oxidation and multiple hydrogen binding sites, properties which are often displayed by various retinoid or retinoid-like compounds (42, 57–59).
Owing to the similar pharmacophore features described for CYP2C8 and CYP26 and as CYP2C8 has also been shown to catalyze the formation of 4-hydroxyretinoic acid from at-RA, the potential exists for the inhibitor binding profile of CYP26A1 and CYP26B1 to overlap with that of CYP2C8. As such, the primary aim of this work was to evaluate the potential for known inhibitors of CYP2C8 to inhibit CYP26A1 or CYP26B1 activity. In vitro inhibition assays were used to determine IC50 values for a set of known CYP2C8 inhibitors against CYP26A1 and CYP26B1. In the process, the use of tazarotenic acid, which has recently been shown to be metabolized by CYP26A1 and CYP26B1, as a probe substrate for CYP26 inhibition assays was evaluated. The mechanism of active site binding and inhibition of CYP26A1 and CYP26B1 was then characterized for compounds with azole moieties as well as those hypothesized to not inhibit the enzymes through type II binding interactions. Finally, in vitro inhibition parameters were compared to reported skin or plasma concentrations following clinically relevant doses of CYP2C8 inhibitors in an attempt to estimate the magnitude of the potential clinical interaction of known CYP2C8 inhibitors on CYP26 activity in vivo.
Materials and Methods
Materials
CYP26A1 and CYP26B1 were generous gifts from Dr. Nina Isoherranen (University of Washington). Recombinant CYP2C8 Supersomes® and purified human cytochrome P450 reductase were obtained from Corning Life Sciences (Tewksbury, MA). Tazarotenic acid, MM11253, liarazole, EC23, AM80 and candesartan were purchased from Tocris Chemicals (Bristol, United Kingdom). Montelukast, pioglitazone, rosiglitazone and zafirlukast were from Cayman Chemical (Ann Arbor, MI). Talarazole was purchased from MedChem Express (Monmouth Junction, NJ). Rapid equilibrium dialysis (RED) device kits were obtained from ThermoFisher Scientific (Waltham, MA). All other chemicals were from Sigma-Aldrich (St. Louis, MO) and were of the highest grade available.
Homology Modeling and Computational Docking Simulations
CYP26A1 and CYP26B1 homology models based on the crystal structure of CYP120 (pdb 2VE3) were designed using Prime modeling software (Schrodinger LLC, New York) as previously described (60). In brief, CYP120 was chosen as a template based on its 33% sequence identity and 53% positive sequence coverage with CYP26A1 and 34% sequence identity and 54% positive sequence coverage with CYP26B1. Ligation of a heme prosthetic group utilized Cys442 (CYP26A1) or Cys441 (CYP26B1), with the entire protein structure subsequently subject to energy minimization with the OPLS_2005 force field constraints in the MacroModel algorithm (Schrodinger LLC, New York). Validation of the models was accomplished using PSIPred (University College London, UK) and SSPro (Schrodinger), through visualization of the Ramachandran plots and through docking simulations with all trans-retinoic acid. The crystal structure of CYP2C8 was obtained from the RCSB Protein Data Bank (pdb 1PQ2). The CYP26A1 or CYP26B1 homology models were superimposed on the CYP2C8 crystal structure using the Super script within Pymol (Schrodinger LLC, New York; http://www.pymolwiki.org/index.php/Super). Structural similarity was determined by calculating the root mean square deviation (RMSD) between the protein structures. Amino acid residues 494 – 512 from the CYP26B1 homology model were not included in the RMSD calculation. Computational docking of clotrimazole (CYP26A1 and CYP26B1), zafirlukast (CYP26A1) or candesartan cilexetil (CYP26B1) was accomplished using an induced fit docking algorithm which incorporated decreased van der Waals radii, spatial repositioning of non-rigid protein side chains and additional energy minimization functions post-ligand docking (61, 62). Compounds for docking simulations were chosen based on inhibition potency as well as their potential (or lack thereof) for type II azole-heme interactions. Docking parameters required the center of mass of the inhibitors to be positioned within a 1728 Å3 grid which was designed to be approximately 3 Å above the protoporphyrin ring system using Glide (Schrodinger LLC, New York). The OPLS_2005 force field constraints used by LigPrep (Schrodinger LLC, New York) were used to prepare the energy minimized structures of clotrimazole (hypothesized to bind through type II ligand interactions) and candesartan cilexetil and zafirlukast (two compounds not hypothesized to interaction through type II binding) prior to docking. Binding orientations obtained from the computational docking experiments were evaluated and scored using GlideScore and eModel, which incorporates aspects of the GlideScore, ligand score and grid score into the final assessment of the plausibility of the docking results.
In Vitro Inhibition Assays
CYP2C8 in vitro IC50 values were obtained from previously reported literature sources (44, 50, 51). An initial single point inhibition screen (n = 3) was then used to estimate the inhibition potency of the set of known CYP2C8 inhibitors against CYP26A1 or CYP26B1. Inhibitors of CYP26 activity were included in the screening set as positive controls. In vitro screening conditions consisted of 10 μM inhibitor, 5 nM CYP26A1 or CYP26B1, 25 nM purified human cytochrome P450 reductase, and 200 nM tazarotenic acid, a compound which has recently been shown to be a substrate of CYP26 (60). The final volume of the incubation was 50 μL. Screening incubations were performed in triplicate and were pre-warmed at 37°C for 3 minutes prior to addition of 1 mM NADPH (final concentration). Incubations were terminated after 10 minutes with three volumes (v/v) of 100 nM tolbutamide in acetonitrile and centrifuged for 20 minutes at 1240 x g. A portion of the resulting supernatant was transferred for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. IC50 values were then determined for compounds exhibiting greater than 50% inhibition in the screening assay for at least one of the CYP26 isoforms. Incubations conditions (n = 3) were similar to those used in the screening assay except for the inhibition concentrations, which ranged from 0 – 100 μM. CYP26A1 and CYP26B1 IC50 values were estimated using a three parameter inhibition model as shown in Equation 1, where Activitymax represents the observed probe substrate activity with no inhibitor, Activitymin is the probe substrate activity at the maximum inhibitor concentration and [I] is the concentration of inhibitor in the incubation. IC50 incubations were performed in triplicate. The organic content of each incubation was kept to less than 1% of the total volume and product formation under the conditions described above had previously been determined to be linear with respect to incubation time and protein content.
| Equation 1 |
Spectral Binding Determination
Spectral binding characterizations (n = 3) were carried out to determine the binding orientation of the most potent azole-containing compound (clotrimazole) for CYP26A1 and CYP26B1, as well as zafirlukast (CYP26A1) and candesartan cilexetil (CYP26B1). The binding of clotrimazole to CYP2C8 was also explored. Ligand concentrations ranged from 0 – 20 μM. A protein concentration of 500 nM was used in spectral binding assays. Following each addition of ligand, cuvettes (1 cm path length) were inverted multiple times and allowed to settle for 1 minute prior to measuring the difference spectra from 350 – 550 nm using a Cary 4000 UV-Vis spectrophotometer (Agilent Technologies, Santa Clara, CA). Spectral binding constants (Ks) were estimated using nonlinear regression of the absorbance difference (ΔAbs) for each enzyme (CYP26A1, λ430nm – λ413nm; CYP26B1, λ430nm – λ400nm; CYP2C8, λ430nm – λ390nm) as shown in Equation 2.
| Equation 2 |
Assessment of In Vitro Free Fraction
In order to determine the unbound fraction of clotrimazole in the IC50 and spectral binding assays, equilibrium dialysis was conducted under relevant conditions. Experiments were performed in triplicate using the Rapid Equilibrium Dialysis Device (Thermo Fisher Scientific, Waltham, MA) which was prepared according to the manufacturer’s recommendations. In brief, 1 μM of clotrimazole was added to 5 nM or 500 nM CYP26A1 or CYP26B1 in potassium phosphate buffer (100 μL, pH 7.4) and was dialyzed for 12 hours at 37 °C against 300 μL of control potassium phosphate buffer. The plate was agitated using an orbital shaker set to 200 rpm. Upon completion of the incubation period, a 50 μL aliquot was removed from each side of the equilibrium dialysis membrane and added to 50 μL of control enzyme or buffer to normalize for potential matrix effects. Protein precipitation was achieved by adding three volumes of 100 nM tolbutamide in ice cold acetonitrile and centrifuging the samples for 20 minutes at 1240 x g. A portion of the resulting supernatant was transferred for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. The unbound fraction was determined as shown in Equation 3.
| Equation 3 |
In Vitro Stability of Candesartan Cilexetil
Candesartan is the pharmacologically active form of the prodrug candesartan cilexetil, which is hydrolyzed by intestinal esterases following oral administration (63). In order to determine whether the observed inhibition potency of candesartan cilexetil was due to the prodrug or to the hydrolysis product, the in vitro stability of candesartan cilexetil was determined using CYP26A1, CYP26B1 and CYP2C8. Briefly, 1 μM candesartan cilexetil was added to incubations containing 5 nM CYP26A1, CYP26B1 or CYP2C8 and 25 nM purified human cytochrome P450 reductase in 100 mM potassium phosphate buffer (pH 7.4; n = 3). Incubations were performed at 37 °C and initiated through addition of 1 mM NADPH (final concentration) in order to mirror the conditions of the IC50 assay. Aliquots were removed at 0, 1, 5 and 10 minutes and immediately placed into ice cold acetonitrile containing 100 nM tolbutamide as an internal standard. Samples were vortex-mixed and centrifuged for 20 minutes at 1240 x g. A portion of the supernatant was transferred for LC-MS/MS analysis of candesartan cilexetil degradation and candesartan formation in the incubations.
Calculation of Cmax,u / IC50
Previously reported Cmax and unbound fraction values in plasma were obtained for 17 known inhibitors of CYP2C8 (benzbromarone, candesartan, candesartan cilexetil, clotrimazole, 17α-ethynylestradiol, fluconazole, itraconazole, mometasone furoate, montelukast, pioglitazone, quercetin, raloxifene, repaglinide, ritonavir, rosiglitazone, tamoxifen and zafirlukast) at clinically relevant doses (64–73). As no reported plasma concentrations of clotrimazole after oral administration were available, skin concentrations following a topical administration were used. The ratio of the unbound Cmax values to the in vitro IC50 values was calculated using Equation 4.
| Equation 4 |
Liquid Chromatography – Mass Spectrometry Analysis
Tazarotenic acid sulfoxide, clotrimazole, candesartan and candesartan cilexetil was monitored using LC-MS/MS. The mass spectrometer incorporated electrospray ionization coupled to an Applied Biosystems 4000 QTrap (Applied Biosystems, Foster City, CA). Samples were injected (10 μL) using a LEAP CTC HTS PAL autosampler (CTC Analytics, Carrboro, NC) and introduced to the mass spectrometer using two LC-20AD binary pumps with an in-line DGU-20A5 solvent degasser (Shimadzu, Columbia, MD). A rapid gradient using 0.1% formic acid (v/v) in water (A) and 0.1% formic acid in methanol:acetonitrile (1:1; B) with a Synergi 2.5 μm Hydro RP 100 Å (50 × 2.0 mm) column (Phenomenex, Torrance, CA) was utilized. Gradient conditions were as follows: 2.0% B (0 – 0.2 minutes), 2.0% B – 95% B (from 0.2 – 1.0 minutes), 95% B (from 1.0 – 1.5 minutes) and re-equilibration at 2.0% B for 0.3 minutes. Tazarotenic acid sulfoxide (positive ion, 340.3 / 280.3) clotrimazole (positive ion, 345.4 / 277.0), candesartan (positive ion, 441.0 / 263.1), candesartan cilexetil (positive ion, 611.1 / 567.2) and the internal standard tolbutamide (positive ion, 271.2 / 91.1; negative ion, 268.9 / 169.7) were detected under MRM (multiple reaction monitoring) conditions. Additional parameters that were used in the analytical method included the source temperature (500 °C), curtain gas (12 arbitrary units), ion spray voltage (5000 V), CAD gas (medium), and ion source gas 1 and gas 2 (30 arbitrary units, each).
Results
Evaluation of tazarotenic acid sulfoxide formation as a probe substrate of CYP26
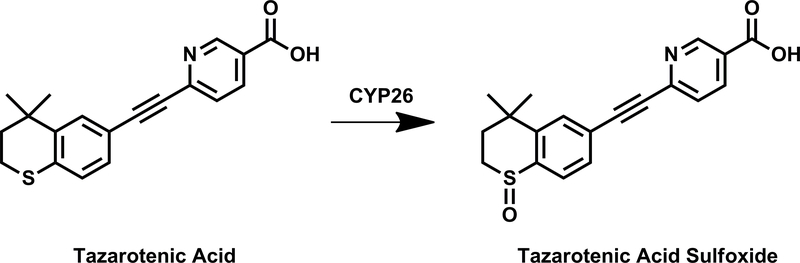
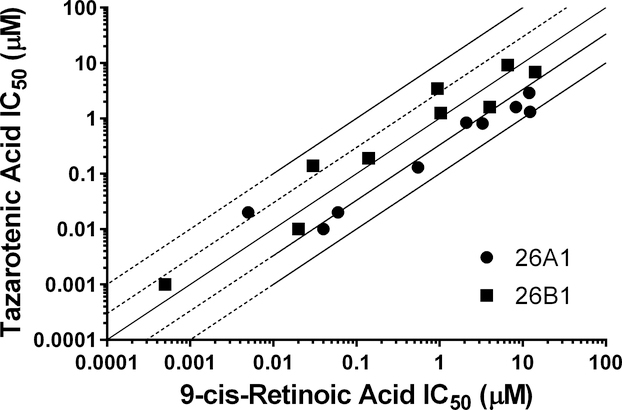
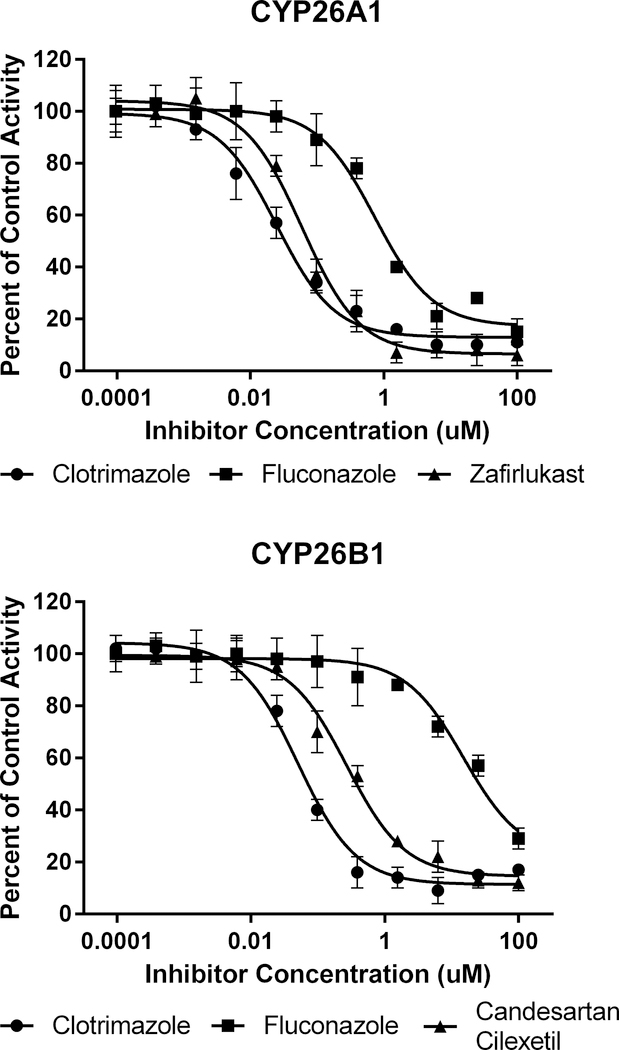
Tazarotenic acid (Figure 1) has recently been identified as a xenobiotic substrate of CYP26A1 and CYP26B1 (60). Prior to utilizing the tazarotenic acid assay to screen new compounds for inhibition of CYP26A1 and CYP26B1, IC50 values were generated for a test set of known CYP26 inhibitors (Figure 2) using tazarotenic acid as the probe substrate and compared to previously published results obtained when 9-cis-retinoic acid was the probe substrate (Table 1) (39, 74). Inhibitor potency rankings were generally the same and a statistically significant correlation was observed between the IC50 values obtained using the two assays. Correlation coefficients (r2) for the IC50 values obtained using tazarotenic acid assay and the 9-cis-retinoic acid assay were 0.78 and 0.62 for CYP26A1 and CYP26B1, respectively, suggesting that formation of tazarotenic acid sulfoxide is an appropriate probe substrate for determining inhibition of CYP26 activity (Figure 2).
Figure 1.
CYP26-catalyzed metabolism of tazarotenic acid to tazarotenic acid sulfoxide.
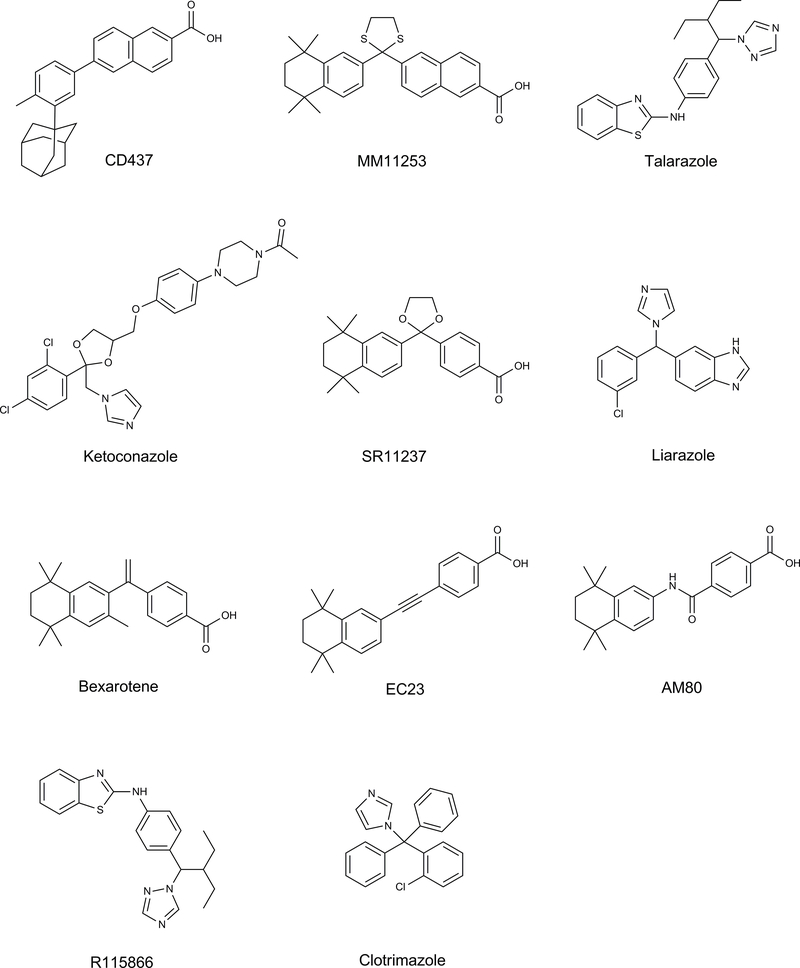
Figure 2.
Structures of key compounds used to compare inhibition of tazarotenic acid sulfoxidation to retinoic acid 4-hydroxylation as noted in Table 1 or in docking simulations.
Table 1.
Inhibition of tazarotenic acid sulfoxide formation in recombinant CYP26 enzymes by known inhibitors of retinoic acid hydroxylation (39, 74). IC50 values are shown ± standard error (calculated or reported; N.R. = Not Reported).
| IC50 (μM) |
||||
|---|---|---|---|---|
| CYP26A1 | CYP26B1 | |||
| Inhibitor | Tazarotenic Acid | 9-cis-Retinoic Acid | Tazarotenic Acid | 9-cis-Retinoic Acid |
| CD437 | 0.01 ± 0.01 | 0.04 (N.R.) | 0.14 ± 0.05 | 0.03 (N.R.) |
| MM11253 | 0.02 ± 0.01 | 0.06 (N.R.) | 1.25 ± 0.44 | 1.03 (N.R.) |
| Talarazole | 0.02 ± 0.01 | 0.005 ± 0.0008 | 0.001 ± 0.001 | 0.0005 ± 0.0003 |
| Ketoconazole | 0.13 ± 0.10 | 0.55 ± 1.3 | 0.19 ± 0.14 | 0.14 ± 0.21 |
| SR11237 | 0.81 ± 0.34 | 3.3 (N.R.) | 6.86 ± 0.46 | 14.2 (N.R.) |
| Liarazole | 0.84 ± 0.87 | 2.1 ± 1.1 | 0.01 ± 0.01 | 0.02 ± 0.01 |
| Bexarotene | 1.31 ± 0.48 | 12.3 (N.R.) | 1.60 ± 0.27 | 4.0 (N.R.) |
| EC23 | 1.60 ± 0.74 | 8.3 ± 5.23 | 3.45 ± 2.80 | 0.94 ± 0.63 |
| AM80 | 2.89 ± 0.24 | 12 ± 3.98 | 9.21 ± 0.83 | 6.6 ± 6.7 |
In Vitro Inhibition Screening and IC50 Determination
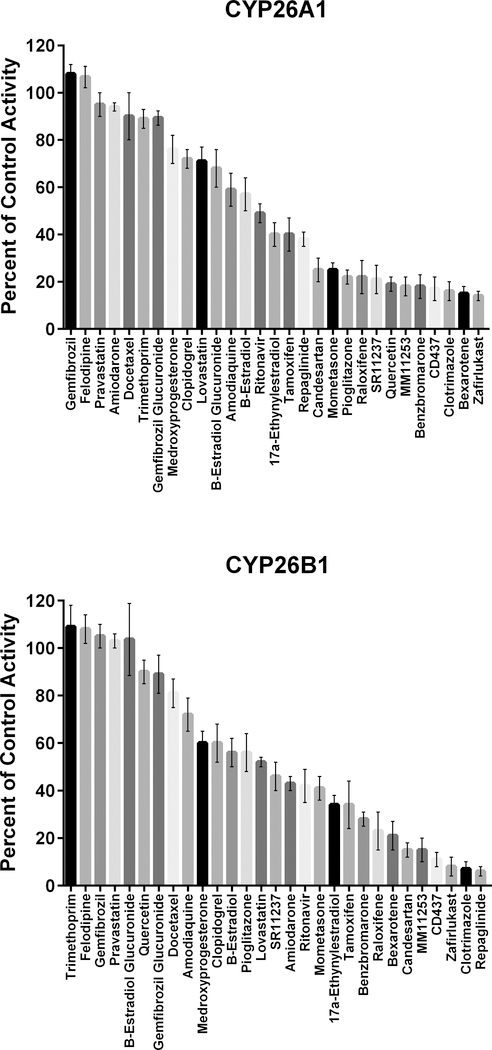
An initial set of 29 known CYP2C8 inhibitors was screened for inhibition of CYP26A1- or CYP26B1-catalyzed tazarotenic acid sulfoxide formation using a single inhibitor concentration (10 μM). Inhibition values ranged from no inhibition to greater than 90% inhibition (Figure 3). IC50 values were determined for 17 compounds which exhibited greater than 50% inhibition in the single concentration inhibition screen for either CYP26A1 or CYP26B1. Clotrimazole was the most potent inhibitor of CYP26 activity with IC50 values of 20 nM and 50 nM for CYP26A1 and CYP26B1, respectively (Table 2; Figure 4). The most potent inhibitors hypothesized to not inhibit through type II azole-heme interactions were zafirlukast for CYP26A1 (IC50 = 60 nM) and candesartan cilexetil for CYP26B1 (IC50 = 270 nM) (Figure 4). To determine whether the observed inhibition by candesartan cilexetil was due to the prodrug or degradation to candesartan, the stability of the prodrug in the three in vitro enzyme systems was assessed. Minimal degradation of candesartan cilexetil was observed in incubations with CYP26A1, CYP26B1 or CYP2C8 with only CYP2C8 showing any appreciable formation of the ester-hydrolyzed product (data not shown). While all of the inhibitors tested exhibited some degree of inhibition of both CYP26A1 and CYP26B1, benzbromarone (12.0-fold), fluconazole (28.3-fold), quercetin (39.9-fold) and zafirlukast (11.8-fold) were all identified as relatively selective inhibitors for CYP26A1 while repaglinide (12.6-fold) showed selectivity towards inhibition of CYP26B1.
Figure 3.
Correlation between previously reported CYP26 IC50 values using 9-cis-retinoic acid as a probe substrate and IC50 values generated using tazarotenic acid as a probe substrate for CYP26A1 (r2 = 0.78) or CYP26B1 (r2 = 0.62) in vitro activity in recombinant CYP enzymes. Lines represent unity, 3-fold and10-fold difference.
Table 2.
IC50 values for tazarotenic acid sulfoxide formation in recombinant CYP26 enzyme preparations by inhibitors of CYP2C8. Previously reported CYP2C8 IC50 data with the relevant probe substrate is also shown. IC50 values are shown ± standard error (N.R.: not reported).
| Inhibitor | CYP26A1 IC50 (μM) | CYP26B1 IC50 (μM) | CYP2C8 IC50 (μM) | Substrate | Reference |
|---|---|---|---|---|---|
| Benzbromarone | 0.63 ± 0.06 | 7.57 ± 4.93 | 0.38 (N.R.) | Montelukast (HLM) | (44) |
| Candesartan | 25.6 ± 25.8 | 58.3 ± 180 | 36.2 ± 1.7 | Amodiaquine | (51) |
| Candesartan Cilexetil | 0.41 ± 0.32 | 0.27 ± 0.06 | 0.496 ± 0.190 | Amodiaquine | (51) |
| Clotrimazole | 0.02 ± 0.01 | 0.05 ± 0.01 | 0.725 ± 0.116 | Amodiaquine | (51) |
| 17α-Ethynylestradiol | 2.24 ± 1.33 | 6.73 ± 3.77 | 6.54 ± 1.22 | Amodiaquine | (51) |
| Fluconazole | 0.70 ± 0.19 | 19.8 ± 3.21 | 48.9 (N.R.) | Amodiaquine | (50) |
| Itraconazole | 0.55 ± 0.09 | 0.16 ± 0.02 | 2.16 ± 0.41 | Paclitaxel | Unpublished Data |
| Mometasone | 0.90 ± 0.08 | 6.81 ± 1.19 | 0.813 ± 0.112 | Amodiaquine | (51) |
| Montelukast | 0.12 ± 0.02 | 0.61 ± 0.09 | 0.009 ± 0.001 | Amodiaquine | (51) |
| Pioglitazone | 0.93 ± 0.26 | 8.48 ± 1.19 | 11.7 ± 4.0 | Amodiaquine | (51) |
| Quercetin | 1.92 ± 0.39 | 76.2 ± 148 | 3.94 ± 0.64 | Amodiaquine | (51) |
| Raloxifene | 1.78 ± 0.77 | 3.28 ± 1.15 | 2.15 ± 0.90 | Amodiaquine | (51) |
| Repaglinide | 7.73 ± 2.69 | 0.61 ± 0.25 | 11.1 (N.R.) | Montelukast (HLM) | (44) |
| Ritonavir | 3.84 ± 2.82 | 2.56 ± 0.25 | 3.03 ± 1.14 | Amodiaquine | (51) |
| Rosiglitazone | 11.9 ± 1.02 | 8.47 ± 6.64 | 10.8 ± 3.1 | Amodiaquine | (51) |
| Tamoxifen | 21.4 ± 20.3 | 14.0 ± 1.11 | 3.34 ± 1.55 | Amodiaquine | (51) |
| Zafirlukast | 0.06 ± 0.02 | 0.71 ± 0.23 | 0.644 ± 0.273 | Amodiaquine | (51) |
Figure 4.
Single concentration (10 μM) inhibition screen using tazarotenic acid as a probe substrate of CYP26 activity. Inhibition values ranged from no inhibition to greater than 90% inhibition.
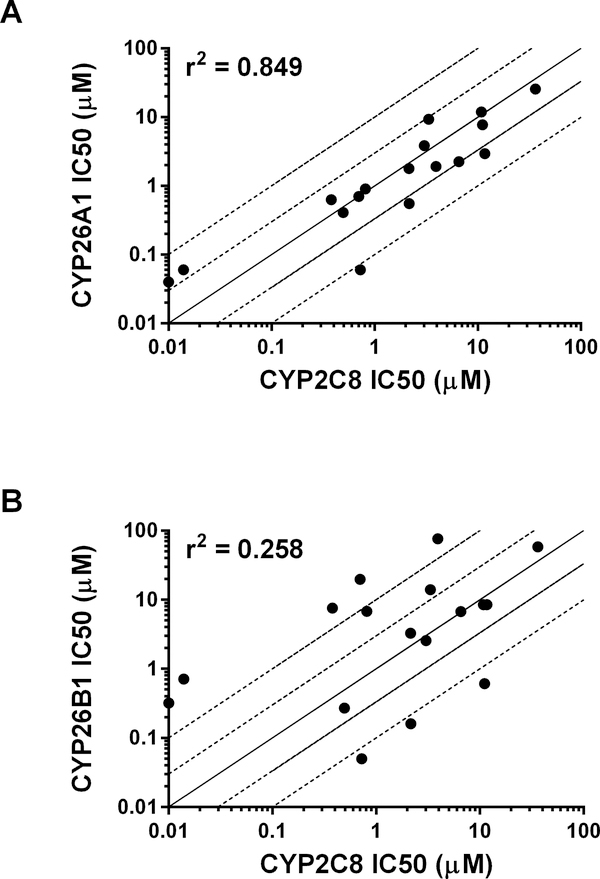
IC50 values obtained with the set of 17 compounds for CYP26A1 and CYP26B1 were then compared to previously reported literature CYP2C8 IC50 values (Table 2; references therein). A positive and statistically significant correlation was observed for CYP26A1 and CYP2C8 IC50 values (r2 = 0.849; Figure 5a). Only a weak correlation (r2 = 0.258) was observed between the IC50 values obtained for CYP26B1 and CYP2C8 (Figure 5b).
Figure 5.
In vitro IC50 curves for select CYP26A1 or CYP26B1 inhibitors using tazarotenic acid as a probe substrate. Data points represent the average of incubations conducted in triplicate and IC50 values were calculated using a three-parameter inhibition model with the Hill slope fixed to 1.
Computational Docking Simulations
Previous reports have implicated CYP2C8 in the metabolism of at-RA, the primary substrate of CYP26A1 and CYP26B1 (47, 48, 75). In order to compare the structural similarities between the active sites of CYP2C8 and either CYP26A1 or CYP26B1, homology models of the CYP26 isozymes were superimposed on the crystal structure of CYP2C8 (pdb 1PQ2). Comparison of the CYP26A1 homology model to CYP2C8 resulted in an RMSD value of 3.013 between the two protein structures. The RMSD value for the CYP26B1 protein structure and CYP2C8 was 4.624. Visual examination of the active sites of the three cytochrome P450 isozymes revealed carboxylic acid binding residues located in comparable regions of the active site of CYP2C8 (Gly98, Asn99, Ser100), CYP26A1 (Arg 86, Arg90) and CYP26B1 (Tyr372, Arg373) that have been suggested to interact with the carboxylic acid moiety of 9-cis-retinoic acid, at-RA, or tazarotenic acid (57, 60, 76, 77).
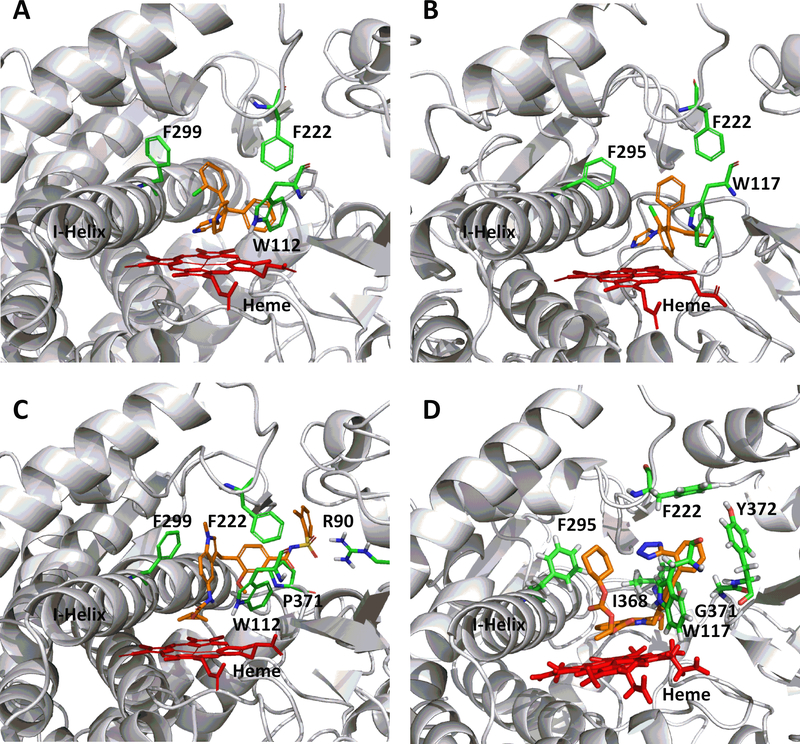
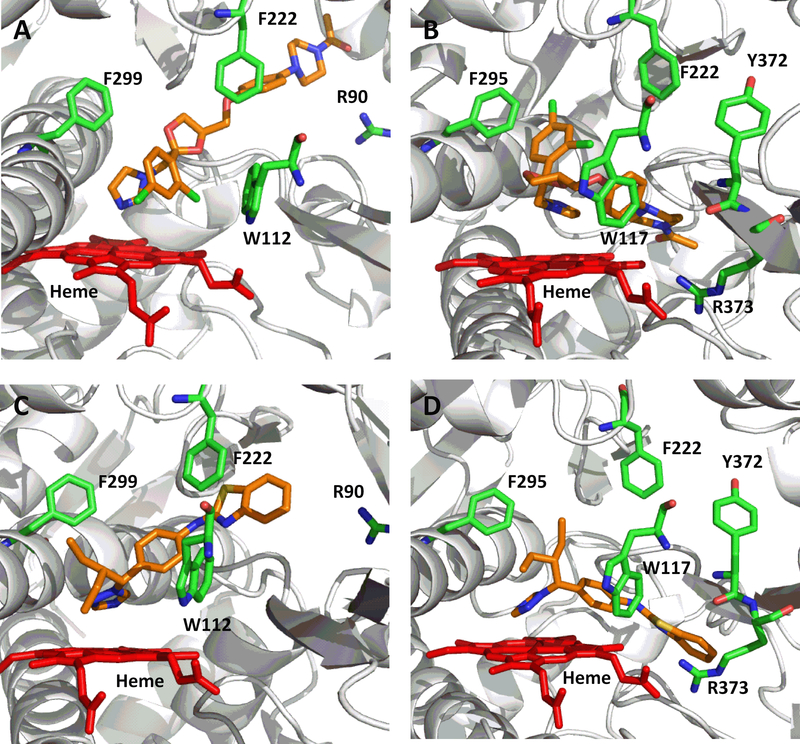
In order to rationalize the ligand binding of the known CYP2C8 inhibitors in the active sites of either CYP26A1 or CYP26B1, a number of computational docking experiments were performed. Docking simulations were carried out for the most potent azole-containing compound (clotrimazole for both CYP26A1 and CYP26B1) as well as zafirlukast for CYP26A1 and candesartan cilexetil for CYP26B1, two compounds hypothesized to not inhibit the enzymes through azole-heme interactions. As shown in Figures 6a and 6b, docking clotrimazole in the active sites of the CYP26A1 and CYP26B1 homology model predicted the sp2 nitrogen of the imidazole ring to be oriented toward the iron of the heme prosthetic group at a distance of 3.104 Å for CYP26A1 and 2.655 Å for CYP26B1. Docking scores were similar for both CYP26A1 (−8.602) and CYP26B1 (−7.148). When zafirlukast was docked in the active site of CYP26A1 (docking score = −11.688), the cyclopentyl moiety was predicted to be oriented towards the heme iron at an approximate distance of 3.316 Å (Figure 6c). Key active site interactions included π-stacking between the methylindole ring and F222 and F299, as well as between the tolyl ring and P371. Hydrogen bonding was predicted to occur between the sulfonyl oxygens of zafirlukast and R90. For CYP26B1 and candesartan cilexetil, a favorable docking score of −11.200 was achieved with the benzene ring of the benzimidazole moiety located approximately 3.424 Å from the heme iron (Figure 6d). The amino acid residues predicted to be involved in orienting candesartan cilexetil in the active site of CYP26B1 included Y372 (π-stacking interaction with the phenyl ring adjacent to the tetrazole moiety) as well as charged interactions between E116 and the tetrazole moiety. Computational docking studies designed to rationalize the binding interactions of other potential type II interactions (i.e., triazole containing compounds), suggested that ketoconazole and R115866 would also bind in a type II manner for both CYP26A1 and CYP26B1
Figure 6.
Correlation between previously reported CYP2C8 IC50 values and CYP26A1 (r2 = 0.849) or CYP26B1 (r2 = 0.258) IC50 values generated using tazarotenic acid as a probe substrate. Lines represent unity, 3-fold and10-fold difference.
Spectral Binding Studies
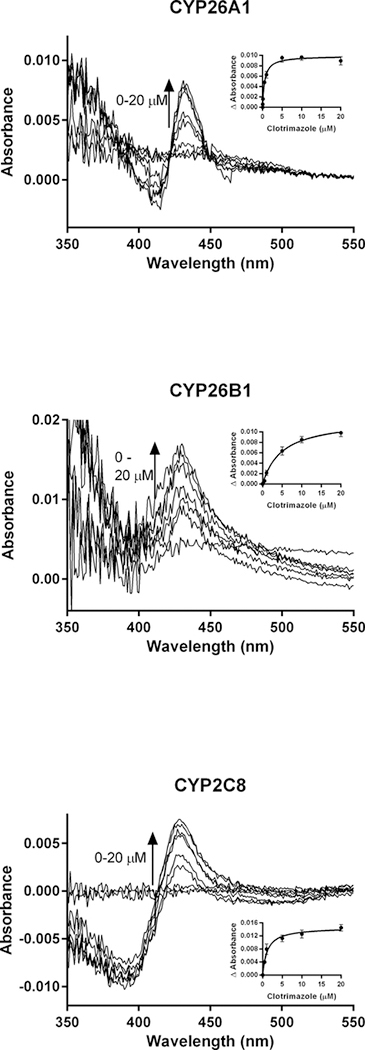
To further evaluate the results of the computational docking simulations with clotrimazole, zafirlukast and candesartan cilexetil, spectral binding studies were performed. Clotrimazole exhibited type II binding characteristics when incubated with CYP26A1, CYP26B1 and CYP2C8 as indicated by the observed maxima and minima of the UV-difference spectra (Figure 7). Spectral binding constants (Ks) were determined by nonlinear regression and were 533 nM, 4945 nM and 1574 nM for CYP26A1, CYP26B1 and CYP2C8, respectively (Table 3). No binding spectra could be obtained for zafirlukast or candesartan cilexetil in any of the systems tested (data not shown).
Figure 7.
Computational docking of clotrimazole (A, CYP26A1; B, CYP26B1), zafirlukast (C, CYP26A1) and candesartan cilexetil (D, CYP26B1) into the active sites of CYP26. The docking orientation of clotrimazole in the active sites of CYP26A1 and CYP26B1 suggests the potential for the imidazole moiety to inhibit the enzyme through type II binding interactions. Active site residues involved in the binding of zafirlukast in the active site of CYP26A1 (R90, W112, F222, and F299) and candesartan cilexetil in the active site of CYP26B1 (W117, F295, F299 and Y372) are similar to the active site residues known to be involved in retinoic acid binding for each isoform. Portions of the protein structure are not displayed for added clarity.
Table 3.
Spectral binding properties for clotrimazole in recombinant CYP26A1, CYP26B1 and CYP2C8. Standard error values are reported for IC50, fu and Ks data.
| CYP26A1 | CYP26B1 | CYP2C8 | |
|---|---|---|---|
| IC50 (nM) | 24.7 ± 8.1 | 50.1 ± 12.1 | 725 ± 116 |
| fu (IC50 assay) | 0.661 ± 0.042 | 0.430 ± 0.031 | 0.155 ± 0.024 |
| IC50,unb (nM) | 16.3 | 21.5 | 112 |
| Ks (nM) | 533 ± 71.8 | 4954 ± 640 | 1574 ± 569 |
| fu (spectral binding assay) | 0.025 ± 0.001 | 0.005 ± 0.0006 | 0.048 ± 0.002 |
| Ks,unb (nM) | 13.3 | 24.7 | 75.5 |
| Binding Mechanism | Type II | Type II | Type II |
As the clotrimazole spectral binding constants for CYP26A1 and CYP26B1 were approximately 22-fold and 99-fold higher than their respective IC50 values, the protein binding of clotrimazole under the relevant in vitro conditions was explored. Under the conditions used in the in vitro CYP26A1, CYP26B1 and CYP2C8 IC50 assays, clotrimazole had fu values of 0.661, 0.430 and 0.155, respectively. In the spectral binding assay, clotrimazole fu values were 0.025, 0.005 and 0.048 for CYP26A1, CYP26B1 and CYP2C8, respectively. When corrected for protein binding, the IC50,u and Ks,u values for clotrimazole were within two-fold of each other for each isozyme, suggesting that coordination of the imidazole nitrogen of clotrimazole to the heme iron is the most likely mechanism of clotrimazole inhibition for these three enzymes.
Calculation of Cmax,u / IC50
To characterize the potential clinical ramifications of the observed in vitro inhibition of CYP26A1 and CYP26B1 by known inhibitors of CYP2C8, reported clinical plasma Cmax values following typical oral or topical doses were obtained from the literature. Cmax values were corrected for plasma protein binding and compared to the in vitro IC50 values to obtain a Cmax,u / IC50 for CYP26A1 and CYP26B1 (Table 4). While total plasma concentrations following oral or topical administration for a number of the inhibitors exceeded their in vitro IC50 values, only clotrimazole and fluconazole exhibited maximum unbound concentrations which would suggest the potential for a meaningful interaction in vivo. Following topical administration, total skin concentrations of clotrimazole were reported to be 67.3 μM. Using plasma protein binding as a surrogate for the unbound fraction in the skin, Cmax,u / IC50 values for clotrimazole were 337 for CYP26A1 and 135 for CYP26B1. The predicted Cmax,u / IC50 values following 200 mg BID oral administration of fluconazole were 44.0 and 1.56 for CYP26A1 and CYP26B1, respectively.
Table 4.
Cmax,u / IC50 values for inhibitors of tazarotenic acid sulfoxidation. Cmax and fu data compiled from literature references as noted in the Materials and Methods.
|
Cmax,u/IC50 |
||||||
|---|---|---|---|---|---|---|
| Inhibitor | Oral Dose | Cmax (nM) | fu,plasma | Cmax, u (nM) | CYP 26A1 | CYP 26B1 |
| Benzbromarone | 100 mg | 9236 | 0.010 | 92.4 | 0.147 | 0.012 |
| Candesartan | 16 mg QD | 270 | 0.002 | 0.38 | < 0.001 | < 0.001 |
| Clotrimazole | 1% Topical | 67300* | 0.100 | 6730 | 337 | 135 |
| 17α-Ethynylestradiol | 30 μg | 0.5 | 0.002 | 0.00085 | < 0.001 | < 0.001 |
| Fluconazole | 200 mg BID | 34606 | 0.890 | 30794 | 44.0 | 1.56 |
| Itraconazole | 200 mg QD | 919 | 0.002 | 1.80 | 0.003 | 0.011 |
| Mometasone | 50 μg Inhaled | 0.05 | 0.010 | 0.0005 | < 0.001 | < 0.001 |
| Montelukast | 10 mg | 925 | 0.010 | 8.90 | 0.077 | 0.015 |
| Pioglitazone | 45 mg | 4489 | 0.010 | 38.0 | 0.048 | 0.005 |
| Quercetin | 500 mg TID | 50.9 | 0.009 | 0.45 | < 0.001 | < 0.001 |
| Raloxifene | 1 mg/kg | 1.05 | 0.050 | 0.50 | < 0.001 | < 0.001 |
| Repaglinide | 4 mg | 104 | 0.026 | 2.60 | < 0.001 | 0.004 |
| Ritonavir | 600 mg BID | 15258 | 0.010 | 150 | 0.040 | 0.060 |
| Rosiglitazone | 8 mg | 1673 | 0.002 | 3.40 | < 0.001 | < 0.001 |
| Tamoxifen | 10 mg BID | 323 | 0.020 | 6.40 | < 0.001 | < 0.001 |
| Zafirlukast | 20 mg BID | 1125 | 0.010 | 3.00 | 0.188 | 0.016 |
Represents reported clotrimazole concentration in skin (stratum corneum) following topical administration of a 1% clotrimazole cream formulation.
Discussion
Retinoic acid is a highly regulated signaling molecule that is involved in a host of dermatological, immunological and neurological functions through binding to the retinoic acid receptors and retinoid X receptors (12, 13, 78–82). As such, the metabolic pathways that are involved in the regulation of retinoic acid represent potential targets that can be exploited to alter concentrations of retinoic acid in vivo. Synthesis of retinoic acid begins with conversion of vitamin A (retinol) to retinal by alcohol dehydrogenases and short-chain dehydrogenases followed by the conversion of retinal to retinoic acid by retinaldehyde dehydrogenases (45, 83–85). Degradation of retinoic acid occurs through oxidation to 4-hydroxy-, 16-hydroxy-, and 18-hydroxyretinoic acid, which is catalyzed primarily by the CYP26-family (CYP26A1, CYP26B1 and CYP26C1) as well as by CYP2C8, CYP2C9 and CYP3A (14, 17, 18, 21, 25, 45, 47, 48, 75, 86–89). As such, the ability to modulate these pathways may prove to have a significant therapeutic benefit.
CYP2C8 was one of the first enzymes identified in the formation of 4-hydroxyretinoic acid and is the only drug metabolizing enzyme for which a crystal structure with a retinoic acid isomer bound in the active site exists (48, 57, 88). The enzyme is the major hepatic isoform involved in 13-cis-retinoic acid metabolism and can be inhibited by retinol and retinoic acid (45, 47, 90). Given the propensity for CYP26A1, CYP26B1 and CYP2C8 to both metabolize and to be inhibited by the same retinoids, the potential exists for the inhibitory pharmacophores of CYP2C8 and CYP26 to overlap. Indeed, when homology models of CYP26A1 and CYP26B1 built using CYP120 (pdb 2VE3) as a template were superimposed on the CYP2C8 crystal structure (pdb 1PQ2), RMSD values of 3.013 and 4.624 were calculated, respectively. Furthermore, closer inspection of the active sites of CYP26A1, CYP26B1 and CYP2C8 suggest the presence of carboxylic acid binding residues in similar spatial proximity to the heme prosthetic group. Previous work to solve the crystal structure of CYP2C8 with 9-cis-retinoic acid bound in the active site suggests Gly98, Asn99 and Ser100 are important residues in anchoring the carboxylate moiety of the retinoic acid molecule which undergoes catalysis (CYP2C8 simultaneously binds two molecules of 9-cis retinoic acid) while CYP26A1 or CYP26B1 homology models built off of various templates have indicated that the carboxylate of retinoic acid forms hydrogen bonds with Arg64 (91), Arg86 (76) or Arg90 (60, 77) for CYP26A1 and Arg95 and Ser369 (77, 92) or Tyr372 and Arg373 (60) for CYP26B1. The estimated active site volumes of CYP26A1 (918 Å3) and CYP26B1 (977 Å3) based on homology modeling are somewhat smaller than the volume of the active site measured from the crystal structure of CYP2C8 (1438 Å3), though it would appear they are large enough to accommodate larger xenobiotic compounds, similar to other CYP isoforms (56, 60).
In order to test the hypothesis of whether CYP26A1 and CYP26B1 were capable of binding xenobiotics with a similar pharmacophore profile as CYP2C8, a set of known CYP2C8 inhibitors was screened for inhibition activity against CYP26A1 and CYP26B1. Recently, tazarotenic acid has been identified as a substrate of CYP26A1 and CYP26B1 (60). To verify the use of tazarotenic acid sulfoxide formation as a probe for CYP26 activity, inhibition data was generated for known CYP26 inhibitors and compared to IC50 values previously obtained using 9-cis-retinoic acid. The observed r2 values suggest that tazarotenic acid is an appropriate probe substrate to assess the inhibition of CYP26A1 and CYP26B1 in vitro (Figure 2), though the possibility of substrate-dependent inhibition profiles cannot be ruled out. While the compounds rank-ordered in a similar fashion between the two assays, some notable differences were observed. Calculated IC50 values for CYP26A1 using tazarotenic acid as a probe substrate were lower than those using 9-cis-retinoic acid. Interestingly, the reverse was generally true for CYP26B1, with 9-cis-retionic acid IC50 values being lower than those generated using tazarotenic acid.
The inhibition profile of CYP2C8 has received a great deal of attention owing to its role in clinically relevant drug interactions. Inhibition of CYP2C8 is thought to be partially responsible for the observed drug interactions between fluvoxamine and rosiglitazone as well as between gemfibrozil and montelukast, rosiglitazone, pioglitazone, repaglinide, cerivastatin and loperamide (52–55, 93–96). To further characterize the drug interaction profile of CYP2C8, a significant amount of in vitro efforts have been reported, with compounds such as montelukast, candesartan cilexetil, zafirlukast and clotrimazole having sub-micromolar IC50s (44, 51). In the current study, multiple CYP2C8 inhibitors were identified as potent inhibitors of both CYP26 isoforms. Selective inhibitors of CYP26A1 (versus CYP26B1) included quercetin, fluconazole, benzbromarone, and zafirlukast while repaglinide was the only compound with a 10-fold or greater selectivity for CYP26B1 inhibition. The difference in inhibition profiles between the two enzymes suggests differences in the active site characteristics which lead to inhibitor binding as well as to the potential to identify novel chemical scaffolds with which to achieve selective inhibition of CYP26A1 or CYP26B1. When the IC50 values were compared to previously reported CYP2C8 IC50 values, a statistically significant correlation was observed for CYP26A1 (r2 = 0.849), suggesting that compounds which are inhibitors of CYP2C8 may also be inhibitors of CYP26A1. Perhaps further supporting the possibility for substrate-dependent inhibition profiles for CYP26 are the IC50 values observed for clotrimazole, fluconazole, quercetin and tamoxifen, four compounds previously reported to not be inhibitors of CYP26A1-catalyzed 4-hydroxyretinoic acid formation in vitro (97).
To further characterize the active site binding interactions that lead to inhibition of CYP26A1 and CYP26B1, the spectral binding characteristics of the most potent azole-containing compound for each enzyme, clotrimazole, were evaluated. In addition, the binding of the most potent inhibitor of CYP26A1 (zafirlukast) and CYP26B1 (candesartan cilexetil) which was hypothesized to not inhibit each enzyme through heme-azole interactions was characterized. Computational simulations with homology models of CYP26A1 or CYP26B1 predicted that clotrimazole would bind in the active site of each enzyme with the sp2 nitrogen of the imidazole ring oriented toward the heme, suggesting that the active site architecture is such that the imidazole is able to approach the heme iron (Figure 6a and 6b). Zafirlukast, which does not contain any structural moieties amenable to heme coordination, docked with its cyclopentyl moiety oriented towards the heme (Figure 6c). Interestingly, CYP26B1 docking of candesartan cilexetil, which contains a tetrazole moiety theoretically capable of coordinating to the heme iron, suggested that interactions with active site residues, rather than heme-azole coordination, were responsible for orienting candesartan cilexetil in the active site of CYP26B1 with the tetrazole moiety oriented in a distal fashion from the heme (Figure 6d). Indeed, when spectral studies were conducted, only clotrimazole exhibited a type II binding spectra for both CYP26A1 and CYP26B1, indicating that the imidazole nitrogen of clotrimazole was coordinated to the heme. Other known type II inhibitors of CYP26 activity in vitro include ketoconazole, R115866 and R116010 (39). Indeed, when docked in the active sites of either homology model, both ketoconazole and R115866 were oriented in such a manner as to support potential type II binding interactions.
The comparison of unbound inhibitor concentrations in vivo, [I], to in vitro IC50 or Ki values is a commonly used method to predict clinically relevant drug interactions. For reversible inhibitors, [I]/Ki values of between 0.1 and 1 suggest the possibility of a clinically relevant drug interaction while an [I]/Ki greater than 1 implies the interaction is likely (98–100). Using IC50 values as a surrogate for Ki, the ratio of unbound Cmax values for clotrimazole and fluconazole to their respective inhibition potencies suggest the potential for these two compounds to inhibit CYP26 activity either locally (clotrimazole) or systemically (fluconazole). The low bioavailability of clotrimazole implies that even with high skin concentrations of the drug, systemic effects are unlikely (101). The effect of antifungal drugs such as clotrimazole and fluconazole on retinoic acid concentrations has been previously reported, though the overall role of CYP26 in these interactions remains to be determined. For example, the Cmax and AUC of orally administered retinoic acid were shown to increase 6-fold and 4-fold, respectively, in a patient with acute promyelocytic leukemia upon co-administration of oral fluconazole (102). A second case study on a patient with the same form of leukemia receiving oral retinoic acid described the onset of pseudotumor cerebri, a CNS toxicity, upon administration of oral fluconazole, a condition which resolved after discontinuation of the fluconazole treatment (103). While inhibition of CYP2C8, CYP2C9 and CYP3A4 by fluconazole may also be involved in the reported drug interactions, the contribution of CYP26A1 or CYP26B1 cannot be ruled out, and may provide an plausible mechanism for the teratogenicity often associated with fluconazole in humans and animal models (104–107). In addition to clinical drug interactions, additional evidence exists in vitro and in pre-clinical species in regard to the effects of antifungal drugs on retinoic acid metabolism. For example, the combination of clotrimazole and at-RA has been shown to initiate cellular differentiation in retinoic acid-resistant cell lines and to inhibit retinoic acid metabolism in embryonic carcinoma cells, while a modest induction of murine CYP26 embryonic mRNA expression is observed after administration of teratogenic doses of fluconazole, perhaps in response to an increase in at-RA concentration (108–110). Similar to the clinical drug interactions observed between retinoic acid and fluconazole, while not definitive, the role of CYP26 in these in vitro interactions warrants further consideration. In terms of clinical drug interactions, however, additional experiments are necessary to determine whether the observed in vitro inhibition would translate to a clinical setting.
Conclusions
In conclusion, the results demonstrate that CYP26A1 and CYP26B1 are inhibited by many known inhibitors of CYP2C8. The overlap in inhibitory pharmacophores between CYP2C8 and CYP26A1 or CYP26B1 may be driven by similarities in the active site binding characteristics of each enzyme and may open the possibility to expand upon the known pharmacophores related to inhibition of retinoic acid metabolism. Further, the potential for inhibition of CYP26 to cause clinically relevant drug interactions suggests care should be taken when co-administering retinoic acid and potent inhibitors such as fluconazole or clotrimazole. Ultimately, the results expand upon the contributions of CYP26A1 and CYP26B1 to drug metabolism and drug interactions and should serve to increase the understanding of the enzymes as both a drug target and in regard to patient safety.
Figure 8.
Spectral binding results for clotrimazole with recombinantly expressed CYP26A1, CYP26B1 or CYP2C8, suggesting enzyme inhibition occurs through type II binding interactions with the heme. Ks,unb affinity constants for CYP26A1 (13.3 nM), CYP26B1 (24.7 nM) and CYP2C8 (75.5 nM) were determined through nonlinear regression analysis (inset figures) and corrected for nonspecific binding in the in vitro assays.
Figure 9.
Computational docking of ketoconazole (A, CYP26A1; B, CYP26B1) or R115866 (A, CYP26A1 or B, CYP26B1) supports the reported type II binding interactions observed for CYP26A1 and suggests a similar binding interaction will occur with CYP26B1. The sp2 hybridized nitrogen was located within 3 Å of the heme iron in all cases. Portions of the protein structure are not displayed for added clarity.
Acknowledgements
The authors thank Dr. Nina Isoherranen (University of Washington) for her insightful critique of our manuscript and Dr. Alex Zelter (University of Washington) for the expression and characterization of the recombinantly expressed CYP26A1 and CYP26B1 enzymes used in this research.
Funding Sources
This research in this manuscript was funded by Amgen Inc. (Thousand Oaks, CA), l’Institut National de la Santé et de la Recherche Médicale (INSERM), the National Institutes of Health National Institute of Aging [Grant R41AG046987] and by a RRIA award from the Michael J. Fox Foundation for Parkinson’s Research.
Abbreviations
- at-RA
all trans-retinoic acid
- Cmax,u
maximum unbound drug concentration
- Ks,u
unbound spectral binding constant
Footnotes
Declaration of Interest Statement
Philippe Diaz is the co-founder and chief scientific officer of Dermaxon. The authors declare no additional competing financial interests.
References
- 1.Clagett-Dame M, DeLuca HF. The role of vitamin A in mammalian reproduction and embryonic development. Annual review of nutrition. 2002;22:347–81. [DOI] [PubMed] [Google Scholar]
- 2.Lotan R Effects of vitamin A and its analogs (retinoids) on normal and neoplastic cells. Biochimica et biophysica acta. 1980;605(1):33–91. [DOI] [PubMed] [Google Scholar]
- 3.Maden M Retinoid signalling in the development of the central nervous system. Nature reviews Neuroscience. 2002;3(11):843–53. [DOI] [PubMed] [Google Scholar]
- 4.McCaffery P, Drager UC. Regulation of retinoic acid signaling in the embryonic nervous system: a master differentiation factor. Cytokine & growth factor reviews. 2000;11(3):233–49. [DOI] [PubMed] [Google Scholar]
- 5.Ross SA, McCaffery PJ, Drager UC, De Luca LM. Retinoids in embryonal development. Physiological reviews. 2000;80(3):1021–54. [DOI] [PubMed] [Google Scholar]
- 6.Sporn MB, Roberts AB. Role of retinoids in differentiation and carcinogenesis. Journal of the National Cancer Institute. 1984;73(6):1381–7. [PubMed] [Google Scholar]
- 7.di Masi A, Leboffe L, De Marinis E, Pagano F, Cicconi L, Rochette-Egly C, et al. Retinoic acid receptors: from molecular mechanisms to cancer therapy. Molecular aspects of medicine. 2015;41:1–115. [DOI] [PubMed] [Google Scholar]
- 8.Levin AA, Sturzenbecker LJ, Kazmer S, Bosakowski T, Huselton C, Allenby G, et al. 9-cis retinoic acid stereoisomer binds and activates the nuclear receptor RXR alpha. Nature. 1992;355(6358):359–61. [DOI] [PubMed] [Google Scholar]
- 9.Mangelsdorf DJ, Borgmeyer U, Heyman RA, Zhou JY, Ong ES, Oro AE, et al. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes & development. 1992;6(3):329–44. [DOI] [PubMed] [Google Scholar]
- 10.Mark M, Ghyselinck NB, Chambon P. Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annual review of pharmacology and toxicology. 2006;46:451–80. [DOI] [PubMed] [Google Scholar]
- 11.Altucci L, Leibowitz MD, Ogilvie KM, de Lera AR, Gronemeyer H. RAR and RXR modulation in cancer and metabolic disease. Nature reviews Drug discovery. 2007;6(10):793–810. [DOI] [PubMed] [Google Scholar]
- 12.Duester G Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134(6):921–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niederreither K, Dolle P. Retinoic acid in development: towards an integrated view. Nature reviews Genetics. 2008;9(7):541–53. [DOI] [PubMed] [Google Scholar]
- 14.Lutz JD, Dixit V, Yeung CK, Dickmann LJ, Zelter A, Thatcher JE, et al. Expression and functional characterization of cytochrome P450 26A1, a retinoic acid hydroxylase. Biochemical pharmacology. 2009;77(2):258–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ray WJ, Bain G, Yao M, Gottlieb DI. CYP26, a novel mammalian cytochrome P450, is induced by retinoic acid and defines a new family. The Journal of biological chemistry. 1997;272(30):18702–8. [DOI] [PubMed] [Google Scholar]
- 16.Ross AC, Zolfaghari R. Cytochrome P450s in the regulation of cellular retinoic acid metabolism. Annual review of nutrition. 2011;31:65–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thatcher JE, Isoherranen N. The role of CYP26 enzymes in retinoic acid clearance. Expert opinion on drug metabolism & toxicology. 2009;5(8):875–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thatcher JE, Zelter A, Isoherranen N. The relative importance of CYP26A1 in hepatic clearance of all-trans retinoic acid. Biochemical pharmacology. 2010;80(6):903–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson CH, Buttrick BR, Isoherranen N. Therapeutic potential of the inhibition of the retinoic acid hydroxylases CYP26A1 and CYP26B1 by xenobiotics. Current topics in medicinal chemistry. 2013;13(12):1402–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tay S, Dickmann L, Dixit V, Isoherranen N. A comparison of the roles of peroxisome proliferator-activated receptor and retinoic acid receptor on CYP26 regulation. Mol Pharmacol. 2010;77(2):218–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topletz AR, Thatcher JE, Zelter A, Lutz JD, Tay S, Nelson WL, et al. Comparison of the function and expression of CYP26A1 and CYP26B1, the two retinoic acid hydroxylases. Biochemical pharmacology. 2012;83(1):149–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Zolfaghari R, Ross AC. Cloning of rat cytochrome P450RAI (CYP26) cDNA and regulation of its gene expression by all-trans-retinoic acid in vivo. Archives of biochemistry and biophysics. 2002;401(2):235–43. [DOI] [PubMed] [Google Scholar]
- 23.White JA, Ramshaw H, Taimi M, Stangle W, Zhang A, Everingham S, et al. Identification of the human cytochrome P450, P450RAI-2, which is predominantly expressed in the adult cerebellum and is responsible for all-trans-retinoic acid metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(12):6403–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xi J, Yang Z. Expression of RALDHs (ALDH1As) and CYP26s in human tissues and during the neural differentiation of P19 embryonal carcinoma stem cell. Gene expression patterns : GEP. 2008;8(6):438–42. [DOI] [PubMed] [Google Scholar]
- 25.Taimi M, Helvig C, Wisniewski J, Ramshaw H, White J, Amad M, et al. A novel human cytochrome P450, CYP26C1, involved in metabolism of 9-cis and all-trans isomers of retinoic acid. The Journal of biological chemistry. 2004;279(1):77–85. [DOI] [PubMed] [Google Scholar]
- 26.Ahmad N, Mukhtar H. Cytochrome P450: a target for drug development for skin diseases. The Journal of investigative dermatology. 2004;123(3):417–25. [DOI] [PubMed] [Google Scholar]
- 27.Kuenzli S, Saurat JH. Retinoids for the treatment of psoriasis: outlook for the future. Current opinion in investigational drugs. 2001;2(5):625–30. [PubMed] [Google Scholar]
- 28.Miller WH, Jr. The emerging role of retinoids and retinoic acid metabolism blocking agents in the treatment of cancer. Cancer. 1998;83(8):1471–82. [DOI] [PubMed] [Google Scholar]
- 29.Njar VC. Cytochrome p450 retinoic acid 4-hydroxylase inhibitors: potential agents for cancer therapy. Mini reviews in medicinal chemistry. 2002;2(3):261–9. [DOI] [PubMed] [Google Scholar]
- 30.Njar VC, Gediya L, Purushottamachar P, Chopra P, Vasaitis TS, Khandelwal A, et al. Retinoic acid metabolism blocking agents (RAMBAs) for treatment of cancer and dermatological diseases. Bioorganic & medicinal chemistry. 2006;14(13):4323–40. [DOI] [PubMed] [Google Scholar]
- 31.Verfaille CJ, Borgers M, van Steensel MA. Retinoic acid metabolism blocking agents (RAMBAs): a new paradigm in the treatment of hyperkeratotic disorders. Journal der Deutschen Dermatologischen Gesellschaft = Journal of the German Society of Dermatology : JDDG. 2008;6(5):355–64. [DOI] [PubMed] [Google Scholar]
- 32.Gomaa MS, Bridgens CE, Illingworth NA, Veal GJ, Redfern CP, Brancale A, et al. Novel retinoic acid 4-hydroxylase (CYP26) inhibitors based on a 3-(1H-imidazol- and triazol-1-yl)-2,2-dimethyl-3-(4-(phenylamino)phenyl)propyl scaffold. Bioorganic & medicinal chemistry. 2012;20(14):4201–7. [DOI] [PubMed] [Google Scholar]
- 33.Le Borgne M, Marchand P, Le Baut G, Ahmadi M, Smith HJ, Nicholls PJ. Retinoic acid metabolism inhibition by 3-azolylmethyl-1H-indoles and 2, 3 or 5-(alpha-azolylbenzyl)-1H-indoles. Journal of enzyme inhibition and medicinal chemistry. 2003;18(2):155–8. [DOI] [PubMed] [Google Scholar]
- 34.Sun B, Liu K, Han J, Zhao LY, Su X, Lin B, et al. Design, synthesis, and biological evaluation of amide imidazole derivatives as novel metabolic enzyme CYP26A1 inhibitors. Bioorganic & medicinal chemistry. 2015;23(20):6763–73. [DOI] [PubMed] [Google Scholar]
- 35.Purushottamachar P, Patel JB, Gediya LK, Clement OO, Njar VC. First chemical feature-based pharmacophore modeling of potent retinoidal retinoic acid metabolism blocking agents (RAMBAs): identification of novel RAMBA scaffolds. European journal of medicinal chemistry. 2012;47(1):412–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun B, Song S, Hao CZ, Huang WX, Liu CC, Xie HL, et al. Molecular recognition of CYP26A1 binding pockets and structure-activity relationship studies for design of potent and selective retinoic acid metabolism blocking agents. Journal of molecular graphics & modelling. 2015;56:10–9. [DOI] [PubMed] [Google Scholar]
- 37.Gomaa MS, Armstrong JL, Bobillon B, Veal GJ, Brancale A, Redfern CP, et al. Novel azolyl-(phenylmethyl)]aryl/heteroarylamines: potent CYP26 inhibitors and enhancers of all-trans retinoic acid activity in neuroblastoma cells. Bioorganic & medicinal chemistry. 2008;16(17):8301–13. [DOI] [PubMed] [Google Scholar]
- 38.Gomaa MS, Bridgens CE, Aboraia AS, Veal GJ, Redfern CP, Brancale A, et al. Small molecule inhibitors of retinoic acid 4-hydroxylase (CYP26): synthesis and biological evaluation of imidazole methyl 3-(4-(aryl-2-ylamino)phenyl)propanoates. Journal of medicinal chemistry. 2011;54(8):2778–91. [DOI] [PubMed] [Google Scholar]
- 39.Thatcher JE, Buttrick B, Shaffer SA, Shimshoni JA, Goodlett DR, Nelson WL, et al. Substrate specificity and ligand interactions of CYP26A1, the human liver retinoic acid hydroxylase. Mol Pharmacol. 2011;80(2):228–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Coster R, Wouters W, Van Ginckel R, End D, Krekels M, Coene MC, et al. Experimental studies with liarozole (R 75,251): an antitumoral agent which inhibits retinoic acid breakdown. The Journal of steroid biochemistry and molecular biology. 1992;43(1–3):197–201. [DOI] [PubMed] [Google Scholar]
- 41.Karonen T, Neuvonen PJ, Backman JT. CYP2C8 but not CYP3A4 is important in the pharmacokinetics of montelukast. British journal of clinical pharmacology. 2012;73(2):257–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai XS, Yang LP, Li XT, Liu JP, Zhou ZW, Zhou SF. Human CYP2C8: structure, substrate specificity, inhibitor selectivity, inducers and polymorphisms. Current drug metabolism. 2009;10(9):1009–47. [DOI] [PubMed] [Google Scholar]
- 43.Totah RA, Rettie AE. Cytochrome P450 2C8: substrates, inhibitors, pharmacogenetics, and clinical relevance. Clinical pharmacology and therapeutics. 2005;77(5):341–52. [DOI] [PubMed] [Google Scholar]
- 44.VandenBrink BM, Foti RS, Rock DA, Wienkers LC, Wahlstrom JL. Evaluation of CYP2C8 inhibition in vitro: utility of montelukast as a selective CYP2C8 probe substrate. Drug metabolism and disposition: the biological fate of chemicals. 2011;39(9):1546–54. [DOI] [PubMed] [Google Scholar]
- 45.Marill J, Capron CC, Idres N, Chabot GG. Human cytochrome P450s involved in the metabolism of 9-cis- and 13-cis-retinoic acids. Biochemical pharmacology. 2002;63(5):933–43. [DOI] [PubMed] [Google Scholar]
- 46.Marill J, Idres N, Capron CC, Nguyen E, Chabot GG. Retinoic acid metabolism and mechanism of action: a review. Current drug metabolism. 2003;4(1):1–10. [DOI] [PubMed] [Google Scholar]
- 47.McSorley LC, Daly AK. Identification of human cytochrome P450 isoforms that contribute to all-trans-retinoic acid 4-hydroxylation. Biochemical pharmacology. 2000;60(4):517–26. [DOI] [PubMed] [Google Scholar]
- 48.Nadin L, Murray M. Participation of CYP2C8 in retinoic acid 4-hydroxylation in human hepatic microsomes. Biochemical pharmacology. 1999;58(7):1201–8. [DOI] [PubMed] [Google Scholar]
- 49.Rowbotham SE, Boddy AV, Redfern CP, Veal GJ, Daly AK. Relevance of nonsynonymous CYP2C8 polymorphisms to 13-cis retinoic acid and paclitaxel hydroxylation. Drug metabolism and disposition: the biological fate of chemicals. 2010;38(8):1261–6. [DOI] [PubMed] [Google Scholar]
- 50.Nath A, Zientek MA, Burke BJ, Jiang Y, Atkins WM. Quantifying and predicting the promiscuity and isoform specificity of small-molecule cytochrome P450 inhibitors. Drug metabolism and disposition: the biological fate of chemicals. 2010;38(12):2195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walsky RL, Gaman EA, Obach RS. Examination of 209 drugs for inhibition of cytochrome P450 2C8. Journal of clinical pharmacology. 2005;45(1):68–78. [DOI] [PubMed] [Google Scholar]
- 52.Backman JT, Kyrklund C, Neuvonen M, Neuvonen PJ. Gemfibrozil greatly increases plasma concentrations of cerivastatin. Clinical pharmacology and therapeutics. 2002;72(6):685–91. [DOI] [PubMed] [Google Scholar]
- 53.Honkalammi J, Niemi M, Neuvonen PJ, Backman JT. Dose-dependent interaction between gemfibrozil and repaglinide in humans: strong inhibition of CYP2C8 with subtherapeutic gemfibrozil doses. Drug metabolism and disposition: the biological fate of chemicals. 2011;39(10):1977–86. [DOI] [PubMed] [Google Scholar]
- 54.Niemi M, Backman JT, Granfors M, Laitila J, Neuvonen M, Neuvonen PJ. Gemfibrozil considerably increases the plasma concentrations of rosiglitazone. Diabetologia. 2003;46(10):1319–23. [DOI] [PubMed] [Google Scholar]
- 55.Tornio A, Niemi M, Neuvonen M, Laitila J, Kalliokoski A, Neuvonen PJ, et al. The effect of gemfibrozil on repaglinide pharmacokinetics persists for at least 12 h after the dose: evidence for mechanism-based inhibition of CYP2C8 in vivo. Clinical pharmacology and therapeutics. 2008;84(3):403–11. [DOI] [PubMed] [Google Scholar]
- 56.Schoch GA, Yano JK, Wester MR, Griffin KJ, Stout CD, Johnson EF. Structure of human microsomal cytochrome P450 2C8. Evidence for a peripheral fatty acid binding site. The Journal of biological chemistry. 2004;279(10):9497–503. [DOI] [PubMed] [Google Scholar]
- 57.Schoch GA, Yano JK, Sansen S, Dansette PM, Stout CD, Johnson EF. Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid. The Journal of biological chemistry. 2008;283(25):17227–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kerdpin O, Elliot DJ, Boye SL, Birkett DJ, Yoovathaworn K, Miners JO. Differential contribution of active site residues in substrate recognition sites 1 and 5 to cytochrome P450 2C8 substrate selectivity and regioselectivity. Biochemistry. 2004;43(24):7834–42. [DOI] [PubMed] [Google Scholar]
- 59.Melet A, Marques-Soares C, Schoch GA, Macherey AC, Jaouen M, Dansette PM, et al. Analysis of human cytochrome P450 2C8 substrate specificity using a substrate pharmacophore and site-directed mutants. Biochemistry. 2004;43(49):15379–92. [DOI] [PubMed] [Google Scholar]
- 60.Foti RS, Isoherranen N, Zelter A, Dickmann LJ, Buttrick BR, Diaz P, et al. Identification of Tazarotenic Acid as the First Xenobiotic Substrate of Human Retinoic Acid Hydroxylase CYP26A1 and CYP26B1. J Pharm Exp Ther. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherman W, Beard HS, Farid R. Use of an induced fit receptor structure in virtual screening. Chemical biology & drug design. 2006;67(1):83–4. [DOI] [PubMed] [Google Scholar]
- 62.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. Journal of medicinal chemistry. 2006;49(2):534–53. [DOI] [PubMed] [Google Scholar]
- 63.Gleiter CH, Morike KE. Clinical pharmacokinetics of candesartan. Clinical pharmacokinetics. 2002;41(1):7–17. [DOI] [PubMed] [Google Scholar]
- 64.Daley-Yates PT, Kunka RL, Yin Y, Andrews SM, Callejas S, Ng C. Bioavailability of fluticasone propionate and mometasone furoate aqueous nasal sprays. European journal of clinical pharmacology. 2004;60(4):265–8. [DOI] [PubMed] [Google Scholar]
- 65.Goodman Gilman. Goodman and Gilman’s The Pharmacological Basis of Therapeutics. 11th ed. Brunton LL, Lazo JS, Parker KL, editors. USA: McGraw-Hill Companies, Inc; 2006. [Google Scholar]
- 66.Saperstein S, Edgren RA, Lee GJ, Jung D, Fratis A, Kushinsky S, et al. Bioequivalence of two oral contraceptive drugs containing norethindrone and ethinyl estradiol. Contraception. 1989;40(5):581–90. [DOI] [PubMed] [Google Scholar]
- 67.Uchida S, Shimada K, Misaka S, Imai H, Katoh Y, Inui N, et al. Benzbromarone pharmacokinetics and pharmacodynamics in different cytochrome P450 2C9 genotypes. Drug metabolism and pharmacokinetics. 2010;25(6):605–10. [DOI] [PubMed] [Google Scholar]
- 68.DrugBank: Open Data Drug and Drug Target Database [Internet]. [cited May 19, 2015]. Available from: http://www.drugbank.ca/.
- 69.Deshpande. Evaluation of Topical Bioavailability of Clotrimazole Using DermatoPharmacoKinetic Method. Int J Sci Inv Today. 2013;2(3):216–25. [Google Scholar]
- 70.Boulton DW, Walle UK, Walle T. Extensive binding of the bioflavonoid quercetin to human plasma proteins. The Journal of pharmacy and pharmacology. 1998;50(2):243–9. [DOI] [PubMed] [Google Scholar]
- 71.Karonen T, Neuvonen PJ, Backman JT. The CYP2C8 inhibitor gemfibrozil does not affect the pharmacokinetics of zafirlukast. European journal of clinical pharmacology. 2011;67(2):151–5. [DOI] [PubMed] [Google Scholar]
- 72.Moon YJ, Wang L, DiCenzo R, Morris ME. Quercetin pharmacokinetics in humans. Biopharmaceutics & drug disposition. 2008;29(4):205–17. [DOI] [PubMed] [Google Scholar]
- 73.Walter-Sack I, de Vries JX, Ittensohn A, Kohlmeier M, Weber E. Benzbromarone disposition and uricosuric action; evidence for hydroxilation instead of debromination to benzarone. Klinische Wochenschrift. 1988;66(4):160–6. [DOI] [PubMed] [Google Scholar]
- 74.Buttrick BR. Characterization of selective and potent inhibitors of the human retinoic acid hydroxylases CYP26A1 and CYP26B1. Seattle, WA: University of Washington; 2012. [Google Scholar]
- 75.Marill J, Cresteil T, Lanotte M, Chabot GG. Identification of human cytochrome P450s involved in the formation of all-trans-retinoic acid principal metabolites. Mol Pharmacol. 2000;58(6):1341–8. [DOI] [PubMed] [Google Scholar]
- 76.Gomaa MS, Yee SW, Milbourne CE, Barbera MC, Simons C, Brancale A. Homology model of human retinoic acid metabolising enzyme cytochrome P450 26A1 (CYP26A1): active site architecture and ligand binding. Journal of enzyme inhibition and medicinal chemistry. 2006;21(4):361–9. [DOI] [PubMed] [Google Scholar]
- 77.Karlsson M, Strid Å, Sirsjö A, Eriksson LA. Homology Models and Molecular Modeling of Human Retinoic Acid Metabolizing Enzymes Cytochrome P450 26A1 (CYP26A1) and P450 26B1 (CYP26B1). Journal of Chemical Theory and Computation. 2008;4(6):1021–7. [DOI] [PubMed] [Google Scholar]
- 78.Asselineau D, Bernard BA, Bailly C, Darmon M. Retinoic acid improves epidermal morphogenesis. Developmental biology. 1989;133(2):322–35. [DOI] [PubMed] [Google Scholar]
- 79.Cunningham TJ, Duester G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nature reviews Molecular cell biology. 2015;16(2):110–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ransom J, Morgan PJ, McCaffery PJ, Stoney PN. The rhythm of retinoids in the brain. Journal of neurochemistry. 2014;129(3):366–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Raverdeau M, Gely-Pernot A, Feret B, Dennefeld C, Benoit G, Davidson I, et al. Retinoic acid induces Sertoli cell paracrine signals for spermatogonia differentiation but cell autonomously drives spermatocyte meiosis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(41):16582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raverdeau M, Mills KH. Modulation of T cell and innate immune responses by retinoic Acid. Journal of immunology. 2014;192(7):2953–8. [DOI] [PubMed] [Google Scholar]
- 83.Chen H, Howald WN, Juchau MR. Biosynthesis of all-trans-retinoic acid from all-trans-retinol: catalysis of all-trans-retinol oxidation by human P-450 cytochromes. Drug metabolism and disposition: the biological fate of chemicals. 2000;28(3):315–22. [PubMed] [Google Scholar]
- 84.Roos TC, Jugert FK, Merk HF, Bickers DR. Retinoid metabolism in the skin. Pharmacological reviews. 1998;50(2):315–33. [PubMed] [Google Scholar]
- 85.Zhang QY, Dunbar D, Kaminsky L. Human cytochrome P-450 metabolism of retinals to retinoic acids. Drug metabolism and disposition: the biological fate of chemicals. 2000;28(3):292–7. [PubMed] [Google Scholar]
- 86.Chen H, Fantel AG, Juchau MR. Catalysis of the 4-hydroxylation of retinoic acids by cyp3a7 in human fetal hepatic tissues. Drug metabolism and disposition: the biological fate of chemicals. 2000;28(9):1051–7. [PubMed] [Google Scholar]
- 87.Helvig C, Taimi M, Cameron D, Jones G, Petkovich M. Functional properties and substrate characterization of human CYP26A1, CYP26B1, and CYP26C1 expressed by recombinant baculovirus in insect cells. Journal of pharmacological and toxicological methods. 2011;64(3):258–63. [DOI] [PubMed] [Google Scholar]
- 88.Leo MA, Lasker JM, Raucy JL, Kim CI, Black M, Lieber CS. Metabolism of retinol and retinoic acid by human liver cytochrome P450IIC8. Archives of biochemistry and biophysics. 1989;269(1):305–12. [DOI] [PubMed] [Google Scholar]
- 89.Sonneveld E, van den Brink CE, van der Leede BM, Schulkes RK, Petkovich M, van der Burg B, et al. Human retinoic acid (RA) 4-hydroxylase (CYP26) is highly specific for all-trans-RA and can be induced through RA receptors in human breast and colon carcinoma cells. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research. 1998;9(8):629–37. [PubMed] [Google Scholar]
- 90.Yamazaki H, Shimada T. Effects of arachidonic acid, prostaglandins, retinol, retinoic acid and cholecalciferol on xenobiotic oxidations catalysed by human cytochrome P450 enzymes. Xenobiotica; the fate of foreign compounds in biological systems. 1999;29(3):231–41. [DOI] [PubMed] [Google Scholar]
- 91.Shimshoni JA, Roberts AG, Scian M, Topletz AR, Blankert SA, Halpert JR, et al. Stereoselective formation and metabolism of 4-hydroxy-retinoic Acid enantiomers by cytochrome p450 enzymes. The Journal of biological chemistry. 2012;287(50):42223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saenz-Mendez P, Elmabsout AA, Savenstrand H, Awadalla MK, Strid A, Sirsjo A, et al. Homology models of human all-trans retinoic acid metabolizing enzymes CYP26B1 and CYP26B1 spliced variant. Journal of chemical information and modeling. 2012;52(10):2631–7. [DOI] [PubMed] [Google Scholar]
- 93.Deng LJ, Wang F, Li HD. Effect of gemfibrozil on the pharmacokinetics of pioglitazone. European journal of clinical pharmacology. 2005;61(11):831–6. [DOI] [PubMed] [Google Scholar]
- 94.Jaakkola T, Backman JT, Neuvonen M, Neuvonen PJ. Effects of gemfibrozil, itraconazole, and their combination on the pharmacokinetics of pioglitazone. Clinical pharmacology and therapeutics. 2005;77(5):404–14. [DOI] [PubMed] [Google Scholar]
- 95.Karonen T, Filppula A, Laitila J, Niemi M, Neuvonen PJ, Backman JT. Gemfibrozil markedly increases the plasma concentrations of montelukast: a previously unrecognized role for CYP2C8 in the metabolism of montelukast. Clinical pharmacology and therapeutics. 2010;88(2):223–30. [DOI] [PubMed] [Google Scholar]
- 96.Niemi M, Tornio A, Pasanen MK, Fredrikson H, Neuvonen PJ, Backman JT. Itraconazole, gemfibrozil and their combination markedly raise the plasma concentrations of loperamide. European journal of clinical pharmacology. 2006;62(6):463–72. [DOI] [PubMed] [Google Scholar]
- 97.Foti RS, Honaker M, Nath A, Pearson JT, Buttrick B, Isoherranen N, et al. Catalytic versus inhibitory promiscuity in cytochrome P450s: implications for evolution of new function. Biochemistry. 2011;50(13):2387–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bjornsson TD, Callaghan JT, Einolf HJ, Fischer V, Gan L, Grimm S, et al. The conduct of in vitro and in vivo drug-drug interaction studies: a Pharmaceutical Research and Manufacturers of America (PhRMA) perspective. Drug metabolism and disposition: the biological fate of chemicals. 2003;31(7):815–32. [DOI] [PubMed] [Google Scholar]
- 99.Bjornsson TD, Callaghan JT, Einolf HJ, Fischer V, Gan L, Grimm S, et al. The conduct of in vitro and in vivo drug-drug interaction studies: a PhRMA perspective. Journal of clinical pharmacology. 2003;43(5):443–69. [PubMed] [Google Scholar]
- 100.Kosugi Y, Hirabayashi H, Igari T, Fujioka Y, Hara Y, Okuda T, et al. Evaluation of cytochrome P450-mediated drug-drug interactions based on the strategies recommended by regulatory authorities. Xenobiotica; the fate of foreign compounds in biological systems. 2012;42(2):127–38. [DOI] [PubMed] [Google Scholar]
- 101.Sawyer PR, Brogden RN, Pinder RM, Speight TM, Avery. Clotrimazole: a review of its antifungal activity and therapeutic efficacy. Drugs. 1975;9(6):424–47. [DOI] [PubMed] [Google Scholar]
- 102.Schwartz EL, Hallam S, Gallagher RE, Wiernik PH. Inhibition of all-trans-retinoic acid metabolism by fluconazole in vitro and in patients with acute promyelocytic leukemia. Biochemical pharmacology. 1995;50(7):923–8. [DOI] [PubMed] [Google Scholar]
- 103.Vanier KL, Mattiussi AJ, Johnston DL. Interaction of all-trans-retinoic acid with fluconazole in acute promyelocytic leukemia. Journal of pediatric hematology/oncology. 2003;25(5):403–4. [DOI] [PubMed] [Google Scholar]
- 104.Lopez-Rangel E, Van Allen MI. Prenatal exposure to fluconazole: an identifiable dysmorphic phenotype. Birth defects research Part A, Clinical and molecular teratology. 2005;73(11):919–23. [DOI] [PubMed] [Google Scholar]
- 105.Menegola E, Broccia ML, Di Renzo F, Giavini E. Antifungal triazoles induce malformations in vitro. Reproductive toxicology. 2001;15(4):421–7. [DOI] [PubMed] [Google Scholar]
- 106.Tiboni GM. Second branchial arch anomalies induced by fluconazole, a bis-triazole antifungal agent, in cultured mouse embryos. Research communications in chemical pathology and pharmacology. 1993;79(3):381–4. [PubMed] [Google Scholar]
- 107.Tiboni GM, Giampietro F. Murine teratology of fluconazole: evaluation of developmental phase specificity and dose dependence. Pediatric research. 2005;58(1):94–9. [DOI] [PubMed] [Google Scholar]
- 108.Kizaki M, Ueno H, Yamazoe Y, Shimada M, Takayama N, Muto A, et al. Mechanisms of retinoid resistance in leukemic cells: possible role of cytochrome P450 and P-glycoprotein. Blood. 1996;87(2):725–33. [PubMed] [Google Scholar]
- 109.Williams JB, Napoli JL. Inhibition of retinoic acid metabolism by imidazole antimycotics in F9 embryonal carcinoma cells. Biochemical pharmacology. 1987;36(8):1386–8. [DOI] [PubMed] [Google Scholar]
- 110.Tiboni GM, Marotta F, Carletti E. Fluconazole alters CYP26 gene expression in mouse embryos. Reproductive toxicology. 2009;27(2):199–202. [DOI] [PubMed] [Google Scholar]