Abstract
Protein-based drug delivery carrier has been one of the most employed modalities in the biopharmaceuticals. In this study, we have compared avidin and its two analogues, neutravidin and streptavidin, as nanocarriers for the delivery of biotin-labeled siRNA with the help of biotinylated cholesterol (targeting ligand) and protamine (condensing agent). These proteins have similar binding affinity to biotin but substantially difference in their physical and chemical characteristics. Here, we have shown how these characteristics affect the size, cellular uptake and activity of the avidin-based siRNA nanocomplex. In contrast to avidin and streptavidin nanocomplexes, neutravidin-based nanocomplex shows very low endosome entrapment and high cytoplasmic localization at extended times. High amount of the siRNA released in the cytoplasm by neutravidin-based nanocomplex at extended times (24 h) results in extensive and sustained PCBP2 gene silencing activity in HSC-T6 rat hepatic stellate cells. Neutravidin-based nanocomplex shows significantly low exocytosis in comparison to the streptavidin-based nanocomplex. Avidin-neutravidin- and streptavidin-based nanocomplexes are similar in size and had no significant cytotoxicity in transfected HSC-T6 cells or inflammatory cytokine induction in a whole blood assay. Compared to free siRNA, the neutravidin-based siRNA nanocomplex exhibits higher accumulation at 2 h in the liver of the rats with CC14-induced liver fibrosis. Neutravidin has therefore shown to be the most promising avidin analogue for the delivery of siRNA.
Keywords: Neutravidin, Avidin, Streptavidin, siRNA, Liver fibrosis, Conjugation, Nanocomplex, Biotin, disulfide bond
INTRODUCTION
Since the inception of RNA interference (RNAi), siRNA has proven to be the most specific and efficient molecule to knockdown a target gene. A tremendous amount of work has been devoted to explore this phenomenon for therapeutic purposes. A safe and efficient delivery of siRNA, however, is still the major stumbling block for its clinical translation regardless of its therapeutic potential.1 Lipids2, peptides3–5 polymers6 proteins7, 8 oligonucleotide9, 10 and inorganic materials11, 12 have been developed for siRNA delivery. Among these carriers, protein-based delivery system is a type of promising carriers because of its ease of construction, good solubility and low toxicity compared to synthetic materials.13 In our previous work, we developed a streptavidin-based nanocomplex to deliver the poly(rC) binding protein 2 (PCBP2) gene to hepatic stellate cells. PCBP2 encodes α-complex protein-2, αCP(2), which binds to the 3’ end of the collagen mRNA and increases its half-life during liver fibrogenesis, leading to accumulation of collagen in the liver. The streptavidin-based nanocomplex demonstrated a rapid silencing activity of the PCBP2 gene, but the silencing activity diminished at extended time points. This could be due to inefficient endosomal release and extensive exocytosis.8, 14, 15 Also, due to its bacterial origin, streptavidin may be arguably immunogenic, and several efforts have been made to develop less immunogenic analogs of streptavidin.16
Avidin, streptavidin, and neutravidin are functional and structural analogues that bind to biotin with extremely high affinity. Avidin is derived from eggs of oviparous vertebrates17, while streptavidin is derived from Streptomyces avidinii. Neutravidin is a chemically modified avidin without glycosylation. Although the three analogues share a similar tetramer structure and binding affinity to biotin, their physical and chemical structures are different. Depending on their specific physical and chemical characteristics, these avidin analogues are widely utilized in the fields of nanotechnology and biotechnology.18
For example, streptavidin and neutravidin lack the four mannose and three N acetyl glucosamine residue in each unit, which are present in avidin. In the absence of the carbohydrate moieties, the pI of neutravidin and streptavidin is slightly acidic, which prevents the nonspecific binding property of avidin (pI ~10). The higher pI of avidin is responsible for the positive charge at physiological pH and non-specific binding to the negatively charged molecules and surfaces, such as silica and cell membrane19. As described by Zhao et al20 and Nguyen et al19, avidin, neutravidin and streptavidin show positive cooperativity (affinity toward the biotin) with an increasing order (avidin<streptavidin<neutravidin). Nearly neutral charge on neutravidin keeps it from nonspecific protein-protein interaction.
With the knowledge of what we have previously reported8, in this study we compared the avidin-, neutravidin- and streptavidin-based nanocomplexes in delivering the PCBP2 siRNA to hepatic stellate cells. Protein-based delivery system is very critical and face challenges such as interactions with intrinsic proteins present in systemic circulation, pro-inflammatory response from innate immune system and cytotoxic effect. We therefore compared the serum stability, cellular uptake, silencing activity, exocytosis, and pro-inflammatory cytokine induction of these nanocomplexes. Biodistribution of the neutravidin-based nanocomplex in rats with CC14-induced liver fibrosis was evaluate using a small animal imaging system.
MATERIALS AND METHODS
Avidin, neutravidin and streptavidin were obtained from Pierce (Rockford, IL). Dulbecco’s phosphate buffered saline (DPBS) and protamine sulphate (Salmon × grade) were purchased from Sigma-Aldrich (St. Louis, MO). Dulbecco’s Modified Eagle’s Medium (DMEM), Opti-MEM reduced serum medium, scrambled siRNA (sense sequence: 5’-ACUACCGUUGUUAUAGGUGtt-3’), and Alexa Fluor 647-labeled siRNA were purchased from Invitrogen (Carlsbad, CA). GelRed™ was obtained from Biotium (Hayward, CA). Non-enzymatic cell dissociation reagent was purchased from MP Biomedicals LLC (Solon, Ohio). Annexin V–Propidium iodide apoptosis assay kit was obtained from Thermo Fisher Scientific (Grand Island, NY). iTaq™ Universal SYBR® Green One-Step Kit was purchased from Bio-Rad (Hercules, California). The rat hepatic stellate cell line (HSC-T6) was kindly provided by Dr. Scott L. Friedman (Mount Sinai School of Medicine, New York University).
Preparation of siRNA Nanocomplex
The siRNA nanocomplexes were prepared as we previously described.8 Biotin conjugated cholesterol was synthesized as we described,8 and the biotin labeled PCBP2 siRNA (sense sequence 5’-GUCAGUGUGGCUCUCUUAUtt-3’) was purchased from Gene Pharma (Shanghai, China). Briefly, the nanocomplex was formed by mixing biotin-siRNA, biotin-cholesterol and avidin/neutravidin/streptavidin in a 2:2:1 molar ratio. The complex was incubated for 10 min at room temperature and then condensed with protamine (N/P ratio of 10:1) for 30 min. Particle size and zeta potential of nanocomplexes were measured in HEPES buffer (pH 7.4) using a Malven Zetasizer Nano ZS (Malvern Instruments Ltd, United Kingdom). Silencing activity of the siRNA nanocomplex was examined as we reported.8
Nanocomplex Enhances Serum Stability of siRNA
siRNA nanocomplexes were incubated with 50% rat serum at 37°C for indicated time intervals, followed by electrophoresis through a 20 % native PAGE gel and visualization with GelRed™ staining under UV light. To further confirm that the siRNAs encapsulated inside nanocomplex are intact, the siRNA nanocomplexes treated with rat serum were incubated with 40 μM heparin and 100 mM DTT for 10 min to release the free siRNA from nanocomplex. Dissociated siRNA samples were electrophoresed in 2% agarose gel and 20% native PAGE and visualized with GelRed™ under UV light.
Silencing Activity of siRNA Nanocomplex
Silencing activity of the siRNA nanocomplexes on PCBP2 gene were evaluated in HSC-T6 cells as we described before.8, 14 Briefly, HSC-T6 cells were seeded in 24 well plates at a density of 50,000 cells/well and transfected with the avidin, neutravidin, and streptavidin siRNA nanocomplexes (100 nM siRNA) in Opti-MEM. Six or twenty-four hours after the transfection, the cells were harvested for RNA isolation. PCBP2 gene silencing activity was quantitated using real-time RT-PCR as we reported.8, 14
To evaluate whether serum proteins affect the silencing activity of nanocomplexes, HSC-T6 cells were also transfected with the avidin, neutravidin, and streptavidin siRNA nanocomplexes in DMEM medium supplemented with 10% FBS for 6 h.
Cellular Uptake of siRNA Nanocomplex
Cellular uptake of the siRNA nanocomplexes was examined by confocal microscopy and flow cytometry as described.5, 21 HSC-T6 cells were incubated with nanocomplexes for various time intervals and washed with PBS containing 1 mM heparin to remove nonspecifically bound nanocomplex. The cells were stained with LysoTracker DND 99, fixed with 10% formalin, and examined under a confocal microscope (Leica TCS SP5). Similarly, HSC-T6 cells were treated with nanocomplexes, washed with PBS containing 1 mM heparin, detached using trypsin, and centrifuged to recover the cells. The resulting cell pellet was washed, suspended, and subjected to fluorescence analysis using a BD FACS II flow Cytometer (Bectone Dickinson Instruments, Franklin Lakes, NJ).
Apoptosis and Necrosis Study
Apoptosis and necrosis of the cells transfected with nanocomplexes were evaluated using the Dead Cell Apoptosis Kit with Annexin V Alexa Fluor® 488 & Propidium Iodide (Thermo Fisher Scientific, Grand Island, NY) as described.22 After transfection with the avidin, neutravidin, and streptavidin nanocomplexes (100 nM PCBP2 siRNA) for 24 h, HSC-T6 cells were harvested using non-enzymatic cell dissociation agent and re-suspended in annexin-binding buffer at a concentration of 1×106 cells/mL. Five microliters of Alexa Fluor® 488 annexin V and 1 μL of PI working solution (100 μg/mL) were added to 100 μL of the cell suspension and incubated in dark for 15 min. After addition of 400 uL annexin-binding buffer, the samples were analyzed using a BD FACSCanto™ II Flow Cytometry Analyzer System (BD bioscience, San Jose, California). The excitation wavelength is 488 nm, and the emission wavelengths are 530 and 575 nm for Alexa Fluor 488 and Propidium Iodide, respectively. Untreated HSC-T6 cells were used as the negative control, and HSC-T6 cells treated with 2% Triton X-100 were used as the positive control.23
Exocytosis Study of the siRNA Nanocomplexes
Previously we developed a method to examine the exocytosis of siRNA nanocomplex from hepatic stellate cells.14 Briefly, HSC-T6 cells were seeded in a 96 well plate at a density of 5000 cells/well. The cells were transfected with nanocomplex containing Alexa Fluor 647-labeled siRNA for 6 h, followed by washing with DPBS for three times and replacement with fresh Opti-MEM medium. The fresh medium was then collected at various time intervals (3, 6, 18, 21 and 24 h) and treated with 40 μM heparin for 30 min to release free Alexa Fluor 647-siRNA from nanocomplex. Fluorescence of the siRNA samples was measured using a VICTOR × Multilabel Plate Reader (Perkin Elmer, Waltham, MA).
Inflammatory Cytokine Induction Study
Pro-inflammatory cytokine induced by the siRNA nanocomplexes in rat whole blood was examined as reported.24–26 Briefly, the siRNA nanocomplexes were incubated in 2 mL rat whole blood at 37°C for 24 h or 48 h. Untreated blood was used as a negative control. The plasma was extracted from the blood by centrifugation at 1500 g for 10 min. The concentrations of proinflammatory cytokine including IL6, IFNγ and TNFα were determined using Picokine ELISA kits according to the manufacturer’s protocol.
in-vivo Biodistribution Study
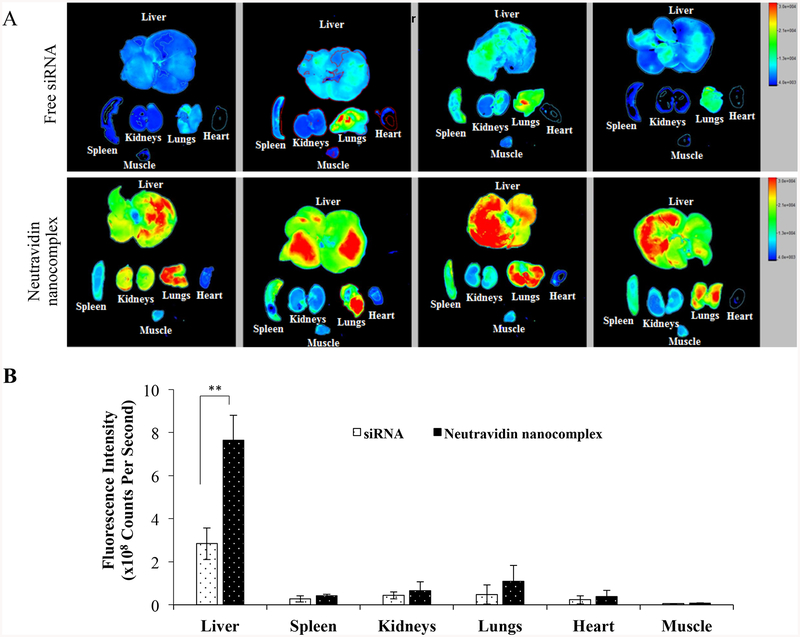
The animal protocol was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Missouri-Kansas City. Male Sprague Dawley rats were purchased from Charles River Laboratories, Inc. (Raleigh, NC) and housed in a temperature and humidity controlled room with a 12 h light-dark cycle. The 1:1 (v/v) mixture of CC14 and olive oil was intra-peritoneally (i.p) administered at a dose of 1 mL/kg twice a week for six consecutive weeks. The rats were then randomly divided into two groups. One group was intravenously (i.v.) injected with Cy5-labeled siRNA (0.1 mg/kg), and the second group was i.v. injected with neutravidin-based nanocomplexes encapsulating Cy5 labeled siRNA (0.1 mg/kg). After 2h, the rats were sacrificed and major organs including the liver, spleen, kidneys, lungs, heart, and muscle (thigh) were harvested and imaged using a Bruker MS FX PRO Imaging System (Billerica, MA).
Statistical Analysis
Statistical analysis was performed using a two-way analysis of variance (ANOVA) with Tuckey’s Post Hoc test. P<0.05 was considered statistically significant.
RESULTS
Characterization of the siRNA Nanocomplexes
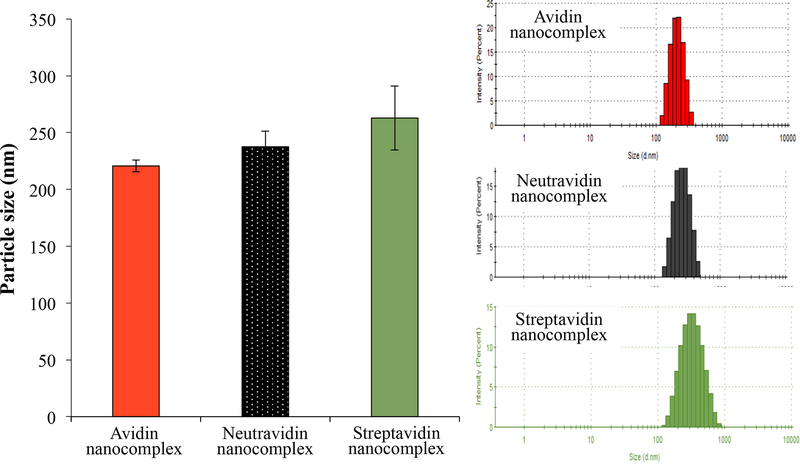
The hydrodynamic size and zeta potential of the siRNA nanocomplexes were measured using the Malven Zetasizer. The siRNA nanocomplexes were diluted in HEPES buffer (pH 7.4) during the characterization. As shown in Figure 1A, the particle sizes of the avidin, neutravidin, and streptavidin nanocomplexes are 225, 237, and 263 nm, respectively. The polydispersity index (PDI) values of the avidin, neutravidin, and streptavidin nanocomplexes are 0.103, 0.07, and 0.151, respectively. These results indicate that the avidin- and neutravidin-based nanocomplexes exhibit nearly monodisperse particles, while the streptavidin nanocomplex shows a relatively wide size distribution.
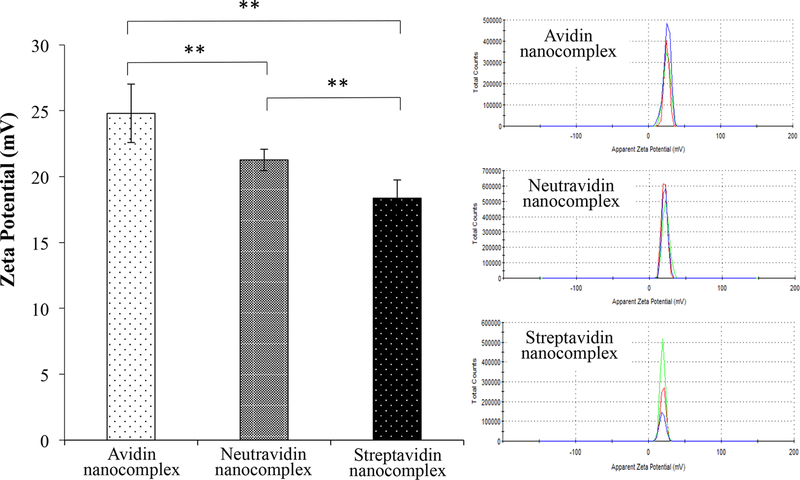
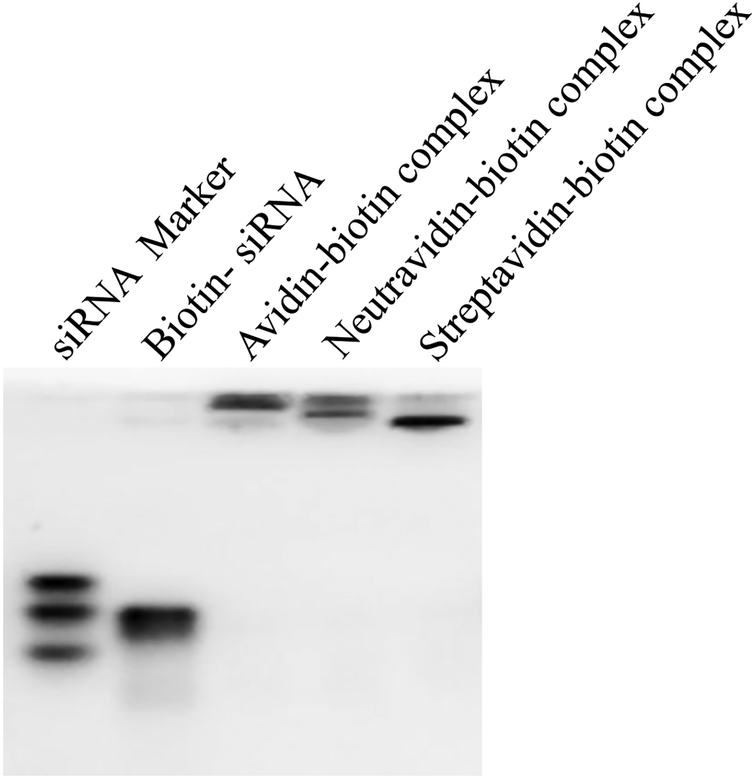
Figure 1.
Characterization of the avidin-, neutravidin-, and streptavidin-based siRNA nanocomplexes. (A) Particle size; (B) Zeta potential; (C) Complexation of biotin-siRNA, biotin-cholesterol, with avidin analogues.
Zeta potentials of the avidin, neutravidin, and streptavidin nanocomplexes are +25, +22, and +18 mV, respectively (Figure 1B). Obviously, cationic protamine contributes to the positive surface charge. The difference in zeta potential can be explained by the different isoelectric point (pI) of avidin, neutravidin and streptavidin. With a pI of ~5, streptavidin is slightly negatively charged under physiological condition, which neutralizes some of the protamine in nanocomplex. On the contrary, avidin has a basic pI of ~10 and therefore contributes more positive charges to nanocomplex, leading to a relatively high zeta potential. The zeta potential of neutravidin is also consistent with its slightly acidic pI 6.3.
Nanocomplex Enhances Serum Stability of siRNA
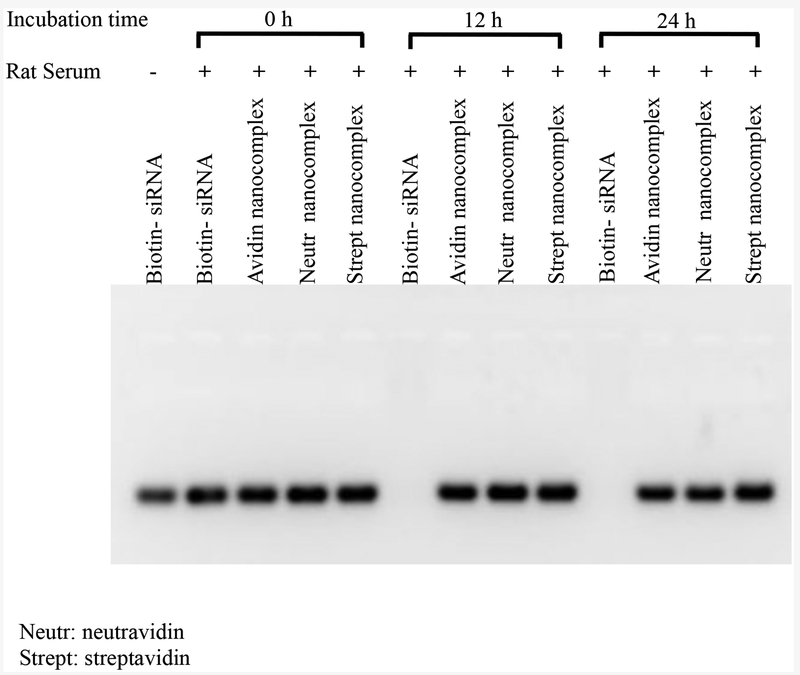
Because of the widely distributed nucleases in the body, stability is always a major concern in developing siRNA delivery systems. We therefore examined the serum stability of the avidin, neutravidin, and streptavidin nanocomplexes in 50% rat serum. After incubation in the serum, nanocomplexes were treated with heparin and DTT for 30 min to dissociate and release free siRNA from nanocomplexes. As Figure 2 illustrates, native siRNA is completely degraded in the serum after 12 h. On the contrary, the avidin, neutravidin, and streptavidin nanocomplexes protect the siRNA from degradation for up to 24 h.
Figure 2.
Nanocomplex enhances serum stability of siRNA. The avidin-, neutravidin-, and streptavidin-based nanocomplexes were incubated with 50% rat serum for 0, 12 and 24 h. The samples were incubated with 40 μM heparin and 100 mM DTT for 30 min to release free siRNA from the nanocomplex and then analyzed with 2% agarose gel.
Silencing Activity of Nanocomplexes
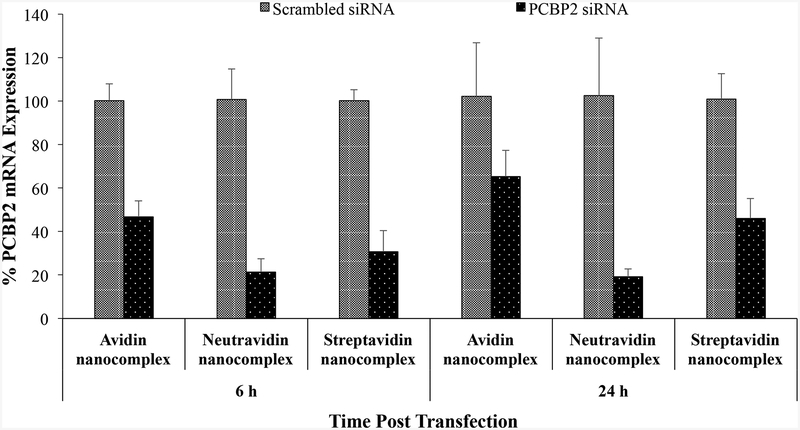
Having shown that the avidin, neutravidin, and streptavidin nanocomplexes can form nanoscale particles with siRNAs and efficiently protect them from serum degradation, we next compared their silencing activity in HSC-T6 cells at the mRNA level by real time RT-PCR. The cells were transfected with the avidin, neutravidin, and streptavidin nanocomplexes for 6 and 24 h. A scrambled siRNA was used as the negative control for each group. As show in Figure 3A, the neutravidin based nanocomplex exhibits the highest silencing activity at both time intervals (79% at 6 h and 81% at 24 h). The avidin based nanocomplex shows the lowest silencing activity among the three nanocomplexes. Moreover, both avidin and streptavidin based nanocomplexes show reduced silencing activity with time, which is consistent with our previous finding with the streptavidin nanocomplex.14 On the contrary, the neutravidin based nanocomplex exhibits sustained silencing activity over 24 h. The silencing activity of the neutravidin nanocomplex at 24 h post-transfection is similar to that at 6h post-transfection.
Figure 3.
(A) Silencing activity of avidin-, neutravidin- and streptavidin-based nanocomplexes (100 nM siRNA) at 6 and 24 h. (B). Silencing activity of avidin-, neutravidin- and streptavidin-based nanocomplexes in the presence of 10% FBS at 6 h. Nanocomplexes formulated with PCBP2 siRNA or scrambled siRNA were incubated with HSC-T6 cells in OptiMEM or DMEM supplemented with 10% FBS. Total RNA was isolated and silencing activity was evaluated by Real Time RT-PCR. Results are represented as the mean±SD (n=3).
Nonspecific protein binding is one of the barriers that may limit the cellular uptake and silencing activity of siRNA nanocomplex in the body. We therefore examined the silencing activity of nanocomplexes in DMEM supplemented with 10% FBS. As Figure 3B illustrates, addition of FBS in the medium does not compromise the silencing activity of neutravidin and streptavidin based nanocomplexes. Their silencing activities are similar to that in the Opti-MEM reduced serum medium (Figure 3A). However, the silencing activity of the avidin nanocomplex is dramatically reduced in the presence of FBS (Figure 3B), indicating its high protein binding with serum protein. This is consistent with the findings that avidin is more prone to nonspecific interaction because of its basic pI and glycosylation.19, 27 Whereas, serum proteins have negligible effect on the silencing activity of the neutravidin and streptavidin nanocomplexes because of their nearly neutral pI.
Cellular Uptake of Nanocomplexes
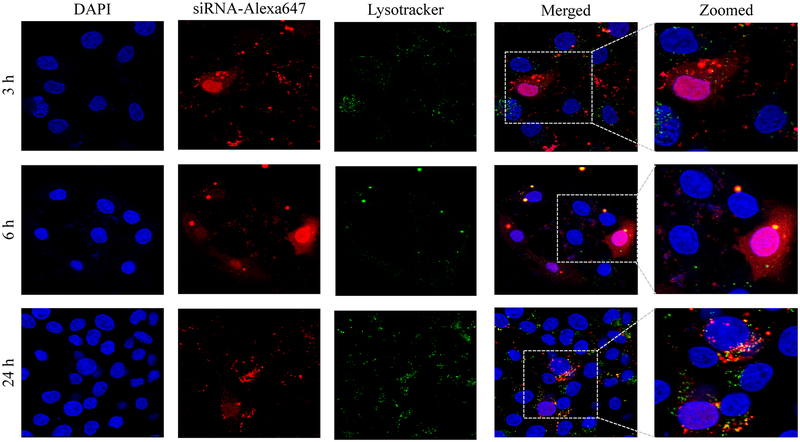
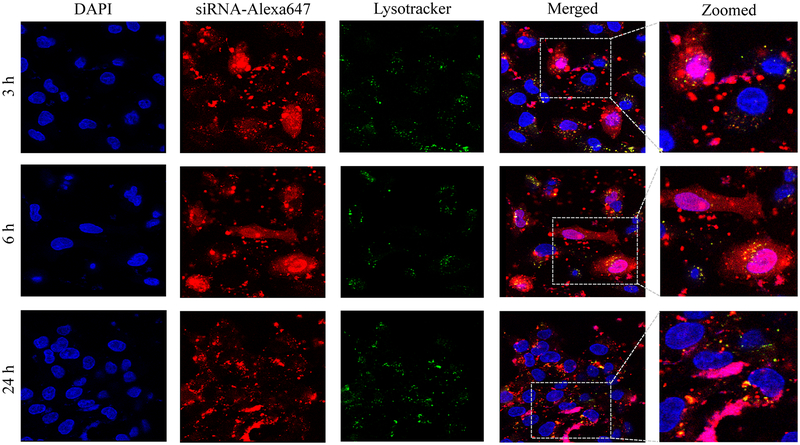
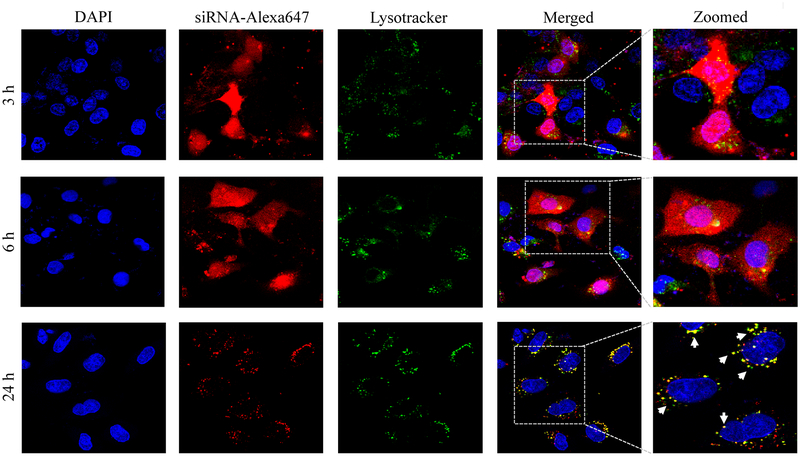
Confocal microscopy was used to compare cellular uptake of the avidin, neutravidin, and streptavidin nanocomplexes in HSC-T6 cells at 3, 6, and 24 h post-transfection. The avidin nanocomplex (Figure 4A) shows consistently lower uptake in comparison to neutravidin (Figure 4B) and streptavidin (Figure 4C) based nanocomplexes. The neutravidin nanocomplex shows the highest uptake over a 24-hour time period. This finding is consistent with the silencing activity results in Figure 3, where the avidin nanocomplex exhibits the lowest silencing activity, and the neutravidin nanocomplex exhibits the highest activity. Time courses of the cellular uptake for the three nanocomplexes are also different. Both the avidin (Figure 4A) and streptavidin (Figure 4C) nanocomplexes exhibit higher cellular uptake at early time points but much lower uptake at 24 h post-transfection. On the contrary, the neutravidin nanocomplex (Figure 4B) exhibits consistently high cellular uptake over 24 h. At extended time point (24 h), lysosomes (green) and siRNA (red) showed significant co-localization (indicated by white arrows) in the cells treated with streptavidin-based nanocomplex (Figure 4C). By contrast, neutravidin-based nanocomplex treated cells showed negligible co-localization of lysosomes and siRNA (Figure 4A).
Figure 4.
Cellular uptake of the avidin nanocomplex (A), neutravidin nanocomplex (B), and streptavidin nanocomplex (C) in HSC-T6 cells. Alexa Fluor-647 labeled siRNA was encapsulated in nanocomplexes and confocal microscopy was used to monitor the cellular uptake at different time intervals (3, 6, and 24 h).
Endosomal release of siRNA nanocomplex is always a major barrier for its silencing activity. We therefore examined co-localization of siRNA and lysosomes in the cells. As Figure 4 illustrates, the neutravidin nanocomplex shows the lowest endosomal entrapment inside the cells, while the streptavidin nanocomplex shows the highest endosomal entrapment.
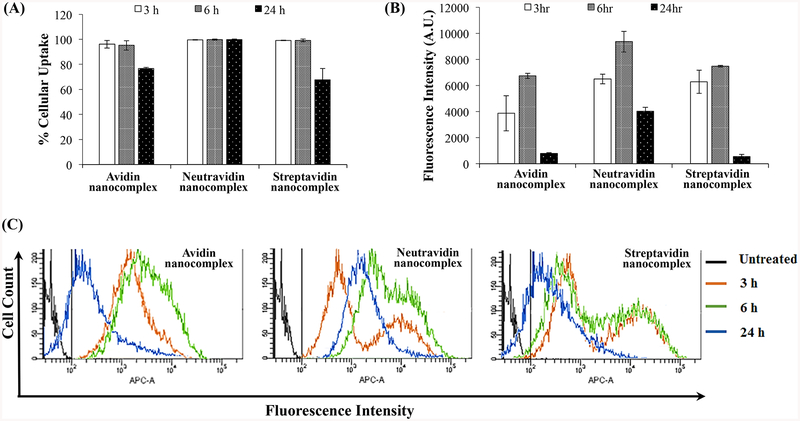
We further quantitated cellular uptake using flow cytometry analysis. As shown in Figure 5A, nearly 100% of the cells were transfected with the three nanocomplexes at 3 and 6 h post-incubation, indicating that nanocomplexes can rapidly deliver siRNA into the cells. However, the percent of transfected cells decreases at 24 h post-incubation in the cells treated with avidin and streptavidin nanocomplexes. Meanwhile, the neutravidin nanocomplex exhibits the same transfection efficiency throughout 24 h.
Figure 5.
Quantitative cellular uptake study of the avidin-, neutravidin- and streptavidin-based nanocomplexes. Cellular uptake of nanocomplexes encapsulating Alexa Fluor-647 labeled siRNA was quantitated using flow cytometry. (A) Percent of cells that take up the nanocomplex; (B) Fluorescence intensity of the cells at various time points; (C) Histogram Plot for the Alexa Fluor-647 siRNA mediated intensity peak shift by avidin-, neutravidin- and streptavidin-based nanocomplexes at various time points. The negative control gate was set at 102 APC-A (shown in black). APC-A is the equivalent fluorescent filter for the Alexa Fluor-647.
Neutravidin nanocomplexes showed significantly higher uptake in comparison to Avidin and Streptavidin based nanocomplexes at extended time point (24 h). Figure 5A shows that all nanocomplexes show rapid uptake, but differ quantitatively. As time progresses, Neutravidin nanocomplexes treated groups showed significantly higher Alexa Fluor 647-siRNA mediated fluorescence intensities (Figure 5B) in comparison to the other nanocomplexes (Avidin and Streptavidin nanocomplexes) at all the timepoints. At early timepoint (3h), Neutravidin and Streptavidin nanocomplexes showed similar fluorescence intensity but after 6h neutravidin nanocomplexes showed consistently higher uptake of Alexa Fluor 647-siRNA. Figure 5C clearly shows the peak shift of siRNA mediated fluorescence intensity with time for respective nanocomplex treated groups.
Exocytosis Study of Nanocomplexes
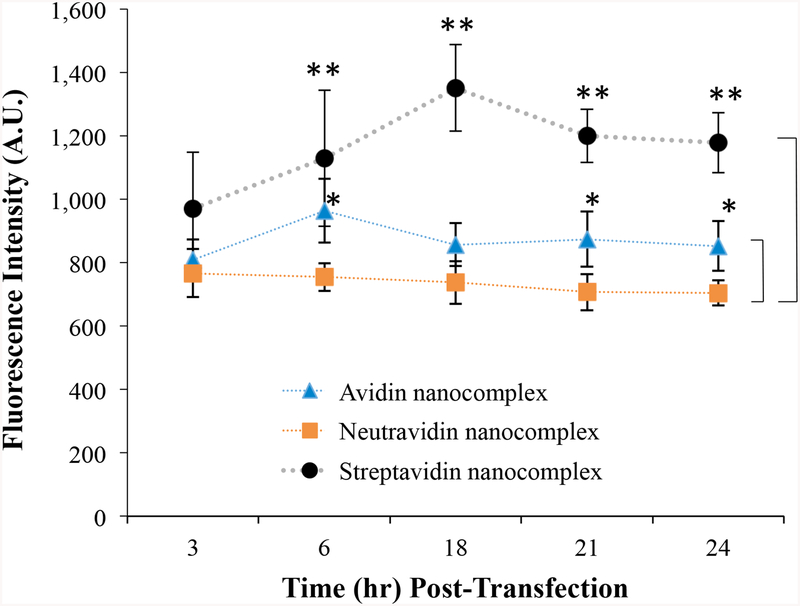
We have previously reported an intriguing phenomenon of exocytosis of streptavidin-based nanocomplex with the progression of time.14 However, cellular uptake studies such as confocal microscopy (Figure 4) and flow cytometry (Figure 5) indicated an extensive uptake of the neutravidin-based nanocomplex at extended time points. We therefore compared the exocytosis of the avidin-, neutravidin-, and streptavidin-based nanocomplexes in HSC-T6 cells. As shown in Figure 6, the neutravidin-based nanocomplexes exhibit the lowest exocytosis compared with the avidin- and streptavidin-based nanocomplexes, which explains the prolonged cellular uptake of siRNA at extended time points. The avidin-based nanocomplex showed lower exocytosis compared to the streptavidin-based nanocomplex (Figure 6).
Figure 6.
Exocytosis study of avidin-, neutravidin-, and streptavidin-based nanocomplexes. HSC-T6 cells were transfected with nanocomplex containing Alexa Fluor 647-siRNA for 6 h, and the medium was replaced with fresh OptiMEM medium. The fresh medium was collected at various time intervals and treated with heparin for 30 min and further analyzed for fluorescence intensity using a Victor × fluorescence plate reader. Results are represented as the mean ± SD (n = 3; * P ≤ 0.05; ** P ≤ 0.01).
Apoptosis and Necrosis Study
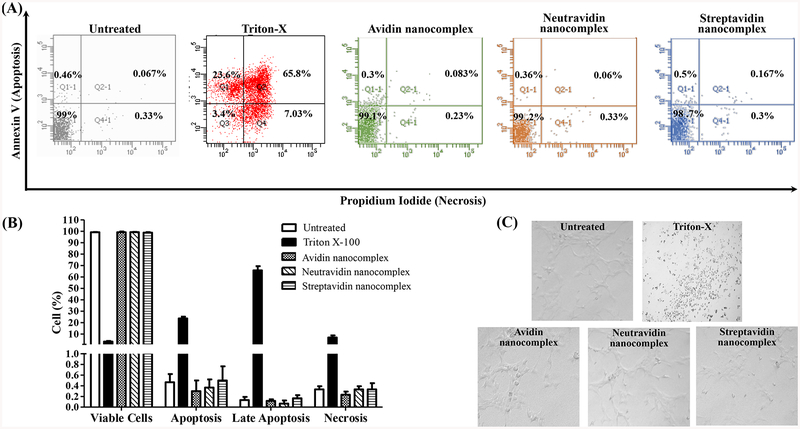
A safe siRNA delivery system should not induce nonspecific apoptosis and necrosis. We therefore evaluated the apoptotic effect of nanocomplexes in HSC-T6 cells. Untreated HSC-T6 cells were used as the negative control, and HSC-T6 cells treated with 2% Triton X-100 were used as the positive control. As illustrated in Figure 7A&B, all the three different nanocomplexes did not induce apoptosis, late apoptosis or necrosis in comparison to the untreated control group. By contrast, 2% Triton X-100 induced significant apoptosis and necrosis in HSC-T6 cells. Accordingly, morphology of the cells treated with nanocomplexes is similar to the untreated group and did not show any sign of wear or cell death that was observed in the cells treated with Triton (Figure 7C).
Figure 7.
Analysis of apoptosis and necrosis by propidium iodide staining and flow cytometry. HSC-T6 cells were treated with the avidin-, neutravidin- and streptavidin-based nanocomplexes (100 nM PCBP2 siRNA) for 24 h and then subjected to the analysis of apoptosis and necrosis. The acquisition data were divided into four quadrants according to the type of fluorescence emitted from the cells: Ql-1 calculates the percent of cells undergoing apoptosis (Annexin V), Q2–1 calculates the percent of cells undergoing late apoptosis or induced necrosis (Annexin V and Propidium Iodide), Q3–1 calculates the percent of cells with no fluorescence and Q4–1 calculates the percent of cells undergoing necrosis induced by the nanocomplex. Results are represented as the mean±SD (n=3).
Inflammatory Cytokine Induction Study
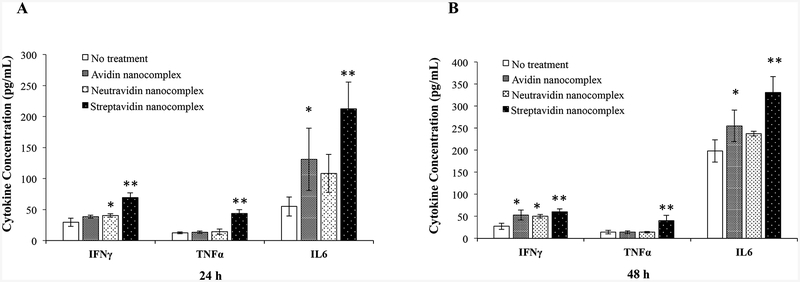
Nonspecific stimulation of immune system is always a concern for protein-based therapeutics and delivery systems. Cytokine and chemokine assays are commonly used to evaluate the immunotoxicity of nanoparticles or any sort of biopharmaceutical therapeutic agents.28 Recently, investigators have been using whole blood assay for the in-vitro analysis of immunogen-mediated cytokine responses.24, 26, 29, 30 Moreover, whole blood assay is preferred over the PBMC (Peripheral Blood Mononuclear Cell) assay because whole blood assay requires less blood volume for the analysis.31 Therefore, we incubated the whole blood of Sprague Dawley rats with nanocomplexes for 24 and 48 h. Plasma was then isolated from the whole blood, and the levels of primary proinflammatory cytokines were measured using a PicoKine™ ELISA assay kit (Bosterbio Pleasanton, CA).). IL6 and TNFa are considered very important cytokines among the whole pro-inflammatory cytokine family because they are involved in most inflammatory states and overexpressed even after a small infection or fever.32, 33 As shown in Figure 8, the streptavidin-based nanocomplex stimulate the highest expression of IFNγ, TNFα, and IL6 after 24 and 48 h incubation. On the contrary, the neutravidin-based showed the lowest stimulation of IFNγ, TNFα, and IL6 among the three nanocomplexes. Moreover, the cytokine expression levels of the cells treated with the neutravidin-based nanocomplex are similar to the negative control group, indicating low cytokine induction by the neutravidin-based nanocomplex.
Figure 8.
Inflammatory cytokine induction study of nanocomplexes in whole blood. Sprague-Dawley rat whole blood was incubated with the avidin-, neutravidin- and streptavidin-based nanocomplexes at 37°C for 24h (A) and 48h (B). The plasma was collected and quantified for the expression of IFNγ, TNF α and IL6 using ELSA kits. Results are represented as the mean ± SD (n=3; * P ≤ 0.05; ** P ≤ 0.01).
in-vivo Biodistribution Study
Neutravidin-based nanocomplex exhibited significantly high cellular uptake, better silencing efficacy, and insignificant inflammatory cytokine induction compared to avidin-and streptavidin-based nanocomplexes. These results suggest that neutravidin could be a potentially safe and efficient carrier for siRNA delivery. We thereafter studied the biodistribution of neutravidin-based nanocomplex encapsulating Cy5-labeled siRNA in rats with CC14-induced liver fibrosis (Figure 9). It has been previously reported that oligonucleotide shows the highest liver uptake at 30–90 min post-injection.34, 35 Moreover, the liver uptake of siRNA nanoparticles were compared with free siRNA at 60 and 120 min in rodents.36, 37 We therefore compared the distribution of free siRNA and neutravidin nanocomplex at 2 h post-injection. The free Cy5-labeled siRNA was rapidly eliminated from the body and showed very low liver accumulation at 2 h after tail vein injection. Compared to the free siRNA, the neutravidin-based nanocomplex increased the siRNA uptake in the liver by approximately 2.7 fold. On the other hand, the neutravidin-based nanocomplex and free siRNA did not show significant difference in the fluorescence in other major organs, such as the spleen, kidneys, lungs, heart and muscle. This result clearly demonstrated that the neutravidin-based nanocomplex does not have nonspecific binding after systemic administration and the biotin-cholesterol in the nanocomplex can efficiently guide the nanocomplex to fibrotic liver. This is in accordance with our previous finding, in which cholesterol was used to achieve targeted delivery of an oligonucleotide to fibrotic liver.38
Figure 9.
Biodistribution of the neutravidin-based siRNA nanocomplex in rats with CC14-induced liver fibrosis. Cy-5 labeled siRNA was used in this study. The rats were sacrificed two hours post tail vein injection, and major organs including the liver, spleen, kidneys, lungs, heart, and muscle were harvested for fluorescence imaging analysis using a Bruker MS FX PRO Imaging System. Fluorescence images of the major organs of four rats per group were presented in (A). (B). Region of interest (ROI) was determined by the Bruker molecular imaging software. Fluorescence intensities with respect to the area under the ROI were plotted for the liver, spleen, kidneys, lungs, heart and muscle. Results are represented as the mean ± SD (n=4; ** P≤0.01).
DISCUSSION
Protein-based nanocomplexes have a high potential for the targeted delivery of siRNA as a therapeutic agent. However, the protein carriers for the delivery of siRNA have been limited only to polycationic peptides, which have shown unfavorable biological and physiological properties when tested in vivo.39 In this work, we evaluated various avidin analogues and critically investigated their efficiency and safety for the delivery of siRNA to HSC. To perform this unbiased evaluation, all nanocomplexes were prepared and compared for physical properties under the same chemical and physiological conditions. The particle size analysis demonstrated that the avidin nanocomplexes were the smallest in size and had the highest positive zeta potential (Figure 1). Avidin has a pI value of 10, which suggests that at a physiological pH, it exhibits a net positive charge. Therefore, protamine and avidin synergistically contribute to the net positive charge of this nanocomplex, resulting in the smallest hydrodynamic size compared to the neutravidin- and streptavidin-based nanocomplexes. However, all nanocomplexes have sizes that were in the optimum range of 200–275 nm.
Avidin and the other tetravalent derivatives effectively formed stable nanocomplexes with biotinylated siRNA and biotinylated cholesterol (ligand). The serum stability studies demonstrated that the siRNA remained stable and condensed in nanocomplexes after prolonged incubation with serum. After 24 h of serum incubation, the amount of siRNA released from nanocomplexes by DTT and heparin was found to be stable (Figure 2), which demonstrated that all of the avidin analogues produced equally stable nanocomplexes. However, in contrast to the avidin- and streptavidin-based nanocomplexes, only the neutravidin-based nanocomplex demonstrated a sustained silencing activity through 24h (Figure 3A). This sustained silencing could be attributed to the efficient internalization and higher endosomal release.
Despite having similar serum stability and a smaller size, the avidin-based nanocomplex exhibited poor activity, which could be the result of their positive charge at physiological pH. The higher positive charge of avidin makes it more prone to nonspecific interactions with negatively charged molecules and proteins. As described by Trang T. Nguyen and co-workers, avidin, neutravidin and streptavidin show a cooperativity coefficient (η) greater than unity. This contributes to the tight interaction of avidin with biotin, but the relatively higher positive charge makes avidin more prone to non-specific interactions.40 We therefore tested for the non-specific binding and impact of serum protein interaction on the efficiency of the nanocomplex. Interestingly, the efficiency of the avidin nanocomplex was significantly compromised when they were incubated with HSC-T6 cells in the presence of 10% FBS for 6 h at 37° C. On the other hand, the neutravidin and streptavidin nanocomplexes showed consistent silencing activity after 6 h in the presence of serum, which suggested that neutravidin and streptavidin did not interact with the serum proteins. However, unlike the neutravidin nanocomplexes, the activity of streptavidin nanocomplex could not be sustained at later time points because of its high exocytosis, which could be due to its bacterial origin
In addition to its intrinsic protein-binding ability, the higher net positive charge of the avidin nanocomplexes contributed to the aggregation of these nanocomplex in the extracellular medium. As shown in Figure 4, the avidin nanocomplex (Figure 4A) has the lowest uptake compared to neutravidin (Figure 4B) and streptavidin (Figure 4C) nanocomplexes at all time points. In contrast, the neutravidin nanocomplexes showed a consistently higher cellular uptake, which was consistent with the sustained silencing activity at the extended time intervals. Interestingly, the groups treated with the neutravidin nanocomplex showed extremely low co-localization of the siRNA with the lysosomes, whereas the streptavidin-treated groups demonstrated higher siRNA co-localization with the lysosomes at the extended time points (24 h). Higher lysosomal co-localization of the streptavidin nanocomplexes in the cells could be due to the bacterial origin of streptavidin on the basis that the cells undergo auto-phagocytosis to degrade intrinsic or foreign proteins derived from bacteria.41 The cellular uptake and efficiency of various nanocomplexes were found to be greatly affected by their physical and chemical properties. The confocal microscopy results were confirmed by flow cytometry (Figure 5). This process revealed that the Alexa Fluor 647-based fluorescence remained significantly higher in case of the neutravidin nanocomplex-treated group at the later time points than that in the avidin and streptavidin nanocomplex-treated groups.
The activity of siRNA in any type of delivery system is compromised due to cellular recycling and exocytosis. We have previously demonstrated that exocytosis and cellular recycling negatively impacts the silencing activity of streptavidin based nanocomplex14. The exocytosis analysis revealed that the neutravidin nanocomplexes showed very low Alexa Fluor 647 associated fluorescence in the extracellular medium. In other words, the neutravidin nanocomplex showed significantly lower exocytosis than the streptavidin-based nanocomplex.
In addition to their therapeutic activities, biologies are required to be safe, and their administration must not cause any toxicity. Especially for protein therapeutics, toxicological studies are imperative and several efficient tools to evaluate the toxicity have been developed in the past decade.42 We therefore evaluated the cytotoxic effect of the avidin, neutravidin and streptavidin nanocomplexes in HSC-T6 cells in vitro. None of nanocomplexes showed significant apoptotic or necrotic effect on the cells compared to the untreated control group (Figure 7). However, the safety of nanoparticles cannot be determined using only apoptosis/necrosis (cytotoxicity) studies. The level of pro-inflammatory cytokines act as a marker of the nanoparticle-induced immunotoxicity and thus indicate the safety of a drug molecule or a delivery system.43 A potential immunotoxic response caused by a nanocarrier indicates that it is unsafe and therefore not a viable candidate for translational research. Among all of the cytokines, IL6, TNF α and IFN γ play vital roles in the primary inflammatory response of a body against any foreign agent.43, 44 Therefore, we tested all of nanocomplexes for the ability to increase the levels of these selected proinflammatory cytokines and found that the levels of the cytokines following administration of the neutravidin nanocomplexes were the same as the negative control group. However, the streptavidin nanocomplexes produced a statistically significant immunogenic response in comparison to the negative control group (Figure 8).
Regardless of promising in-vitro efficacy, many nanocarriers fail to succeed in-vivo because of their poor pharmacokinetics and targeting efficacy. Moreover, free siRNAs after administration have shown unfavorable PK profiles and extremely poor systemic stability.45 Groups treated with Free Cy5-siRNA was rapidly eliminated from the body and showed very low liver accumulation at 2 h after tail vein injection, In contrast, the neutravidin-based nanocomplex exhibited significantly higher siRNA uptake in the fibrotic liver, suggesting that this system can dramatically improve the serum stability and pharmacokinetic profile of siRNA in-vivo.
CONCLUSION
The siRNA nanocomplexes that were formulated using various avidin analogues showed similar morphological characteristics. The neutravidin-based nanocomplex demonstrated the most efficient and sustained silencing activity in-vitro. Moreover, this formulation showed the highest uptake at extended times compared to avidin- and streptavidin-based nanocomplexes. In addition, the neutravidin-based nanocomplex exhibited excellent liver retention in comparison to free siRNA. Hence, neutravidin is the best avidin analogue that can be potentially used as a nanocarrier for the delivery of siRNA to hepatic stellate cells. Future efforts will be made to increase siRNA loading and optimize particle size and zeta potential to further improve the pharmacokinetic profile of neutravidin-based siRNA nanocomplex.
ACKNOWLEDGMENTS
This work was supported by an award (1R01AA021510) from the National Institute of Health
Footnotes
Publisher's Disclaimer: This document is confidential and is proprietary to the American Chemical Society and its authors. Do not copy or disclose without written permission. If you have received this item in error, notify the sender and delete all copies.
REFERENCES
- 1.Cheng K; Mahato RI Biological and therapeutic applications of small RNAs. Pharmaceutical research 2011,28, (12), 2961–5. [DOI] [PubMed] [Google Scholar]
- 2.Akinc A; Goldberg M; Qin J; Dorkin JR; Gamba-Vitalo C; Maier M; Jayaprakash KN; Jayaraman M; Rajeev KG; Manoharan M; Koteliansky V; Rohl I; Leshchiner ES; Langer R; Anderson DG Development of Lipidoid-siRNA Formulations for Systemic Delivery to the Liver. Mol Ther 2009,17, (5), 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eguchi A; Meade BR; Chang Y-C; Fredrickson CT; Willert K; Puri N; Dowdy SF Efficient siRNA delivery into primary cells by a peptide transduction domain-dsRNA binding domain fusion protein. Nat Biotech 2009,27, (6), 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou KK; Pan H; Ratner L; Schlesinger PH; Wickline SA Mechanisms of Nanoparticle-Mediated siRNA Transfection by Melittin-Derived Peptides. ACS Nano 2013, 7, (10), 8605–8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qin B; Chen Z; Jin W; Cheng K Development of cholesteryl peptide micelles for siRNA delivery. Journal of controlled release : official journal of the Controlled Release Society 2013,172, (1), 159–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen M; Gao S; Dong M; Song J; Yang C; Howard KA; Kjems J; Besenbacher F Chitosan/siRNA Nanoparticles Encapsulated in PLGA Nanofibers for siRNA Delivery. ACS Nano 2012, 6, (6), 4835–4844. [DOI] [PubMed] [Google Scholar]
- 7.Choi K.-m.; Choi S-H; Jeon H; Kim I-S; Ahn HJ Chimeric Capsid Protein as a Nanocarrier for siRNA Delivery: Stability and Cellular Uptake of Encapsulated siRNA. ACS Nano 2011,5, (11), 8690–8699. [DOI] [PubMed] [Google Scholar]
- 8.Shukla RS; Tai W; Mahato R; Jin W; Cheng K Development of streptavidin-based nanocomplex for siRNA delivery. Molecular pharmaceutics 2013,10, (12), 4534–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu HY; Yu X; Liu H; Wu D; She JX Co-targeting EGFR and survivin with a bivalent aptamer-dual siRNA chimera effectively suppresses prostate cancer. Scientific reports 2016, 6, 30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binzel DW; Shu Y; Li H; Sun M; Zhang Q; Shu D; Guo B; Guo P Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Mol Ther 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khatri N; Baradia D; Vhora I; Rathi M; Misra A cRGD grafted liposomes containing inorganic nano-precipitate complexed siRNA for intracellular delivery in cancer cells. Journal of controlled release : official journal of the Controlled Release Society 2014,182,45–57. [DOI] [PubMed] [Google Scholar]
- 12.Park DH; Cho J; Kwon OJ; Yun CO; Choy JH Biodegradable Inorganic Nanovector: Passive versus Active Tumor Targeting in siRNA Transportation. Angewandte Chemie (International ed. in English) 2016,55, (14), 4582–6. [DOI] [PubMed] [Google Scholar]
- 13.Piradashvili K; Fichter M; Mohr K; Gehring S; Wurm FR; Landfester K Biodegradable Protein Nanocontainers. Biomacromolecules 2015. [DOI] [PubMed] [Google Scholar]
- 14.Shukla RS; Jain A; Zhao Z; Cheng K Intracellular trafficking and exocytosis of a multi-component siRNA nanocomplex. Nanomedicine : nanotechnology, biology, and medicine 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shukla RS; Qin B; Wan YJ; Cheng K PCBP2 siRNA reverses the alcohol-induced pro-fibrogenic effects in hepatic stellate cells. Pharmaceutical research 2011, 28, (12), 3058–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yumura K; Ui M; Doi H; Hamakubo T; Kodama T; Tsumoto K; Sugiyama A Mutations for decreasing the immunogenicity and maintaining the function of core streptavidin. Protein science : a publication of the Protein Society 2013,22, (2), 213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lesch HP; Kaikkonen MU; Pikkarainen JT; Yla-Herttuala S Avidin-biotin technology in targeted therapy. Expert opinion on drug delivery 2010, 7, (5), 551–64. [DOI] [PubMed] [Google Scholar]
- 18.Jain A; Cheng K The principles and applications of avidin-based nanoparticles in drug delivery and diagnosis. Journal of Controlled Release. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen TT; Sly KL; Conboy JC Comparison of the energetics of avidin, streptavidin, neutrAvidin, and anti-biotin antibody binding to biotinylated lipid bilayer examined by second-harmonic generation. Analytical chemistry 2012, 84, (1), 201–8. [DOI] [PubMed] [Google Scholar]
- 20.Zhao S; Walker DS; Reichert WM Cooperativity in the binding of avidin to biotin-lipid-doped Langmuir-Blodgett films. Langmuir 1993,9, (11), 3166–3173. [Google Scholar]
- 21.Chen Z; Jin W; Liu H; Zhao Z; Cheng K Discovery of Peptide ligands for hepatic stellate cells using phage display. Molecular pharmaceutics 2015,12, (6), 2180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H; Quan YY; Wang XP; Chen TS Gold Nanoparticles of Diameter 13 nm Induce Apoptosis in Rabbit Articular Chondrocytes. Nanoscale research letters 2016,11, (1), 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JS; Song KS; Lee JH; Yu IJ Evaluation of biocompatible dispersants for carbon nanotube toxicity tests. Archives of toxicology 2011,85, (12), 1499–508. [DOI] [PubMed] [Google Scholar]
- 24.Wolf B; Morgan H; Krieg J; Gani Z; Milicov A; Warncke M; Brennan F; Jones S; Sims J; Kiessling A A whole blood in vitro cytokine release assay with aqueous monoclonal antibody presentation for the prediction of therapeutic protein induced cytokine release syndrome in humans. Cytokine 2012, 60, (3), 828–37. [DOI] [PubMed] [Google Scholar]
- 25.Coch C; Lück C; Schwickart A; Putschli B; Renn M; Höller T; Barchet W; Hartmann G; Schlee M A Human In Vitro Whole Blood Assay to Predict the Systemic Cytokine Response to Therapeutic Oligonucleotides Including siRNA. PLoS ONE 2013,8, (8), e71057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez V; Mitjans M; Vinardell MP TNF alpha measurement in rat and human whole blood as an in vitro method to assay pyrogens and its inhibition by dexamethasone and erythromycin. Journal of pharmaceutical sciences 2004,93, (11), 2718–23. [DOI] [PubMed] [Google Scholar]
- 27.Marttila AT; Laitinen OH; Airenne KJ; Kulik T; Bayer EA; Wilchek M; Kulomaa MS Recombinant NeutraLite Avidin: a non-glycosylated, acidic mutant of chicken avidin that exhibits high affinity for biotin and low non-specific binding properties. FEBS Letters 2000,467, (1), 31–36. [DOI] [PubMed] [Google Scholar]
- 28.Dobrovolskaia MA; Germolec DR; Weaver JL Evaluation of nanoparticle immunotoxicity. Nature nanotechnology 2009,4, (7), 411–4. [DOI] [PubMed] [Google Scholar]
- 29.Bienvenu J; Monneret G; Fabien N; Revillard JP The clinical usefulness of the measurement of cytokines. Clinical chemistry and laboratory medicine 2000,38, (4), 267–85. [DOI] [PubMed] [Google Scholar]
- 30.Liu C; Zhang W; Yang H; Sun W; Gong X; Zhao J; Sun Y; Diao G A water-soluble inclusion complex of pedunculoside with the polymer beta-cyclodextrin: a novel anti-inflammation agent with low toxicity. PLoS One 2014,9, (7), e101761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deenadayalan A; Maddineni P; Raja A Comparison of whole blood and PBMC assays for T-cell functional analysis. BMC Research Notes 2013, 6, (1), 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheller J; Chalaris A; Schmidt-Arras D; Rose-John S The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 2011,1813, (5), 878–888. [DOI] [PubMed] [Google Scholar]
- 33.Liu C; Zhao J; Liu Y; Huang Y; Shen Y; Wang J; Sun W; Sun Y A novel pentacyclic triterpenoid, llexgenin A, shows reduction of atherosclerosis in apolipoprotein E deficient mice. Int Immunopharmacol 2016,40,115–124. [DOI] [PubMed] [Google Scholar]
- 34.Cheng K; Ye Z; Guntaka RV; Mahato RI Biodistribution and hepatic uptake of triplex-forming oligonucleotides against type alpha1(l) collagen gene promoter in normal and fibrotic rats. Molecular pharmaceutics 2005,2, (3), 206–17. [DOI] [PubMed] [Google Scholar]
- 35.Zhu L; Ye Z; Cheng K; Miller DD; Mahato RI Site-specific delivery of oligonucleotides to hepatocytes after systemic administration. Bioconjugate chemistry 2008,19, (1), 290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Wolf HK; Snel CJ; Verbaan FJ; Schiffelers RM; Hennink WE; Storm G Effect of cationic carriers on the pharmacokinetics and tumor localization of nucleic acids after intravenous administration. International journal of pharmaceutics 2007,331, (2), 167–75. [DOI] [PubMed] [Google Scholar]
- 37.Zheng M; Librizzi D; Kilic A; Liu Y; Renz H; Merkel OM; Kissel T Enhancing in vivo circulation and siRNA delivery with biodegradable polyethylenimine-graft-polycaprolactone-block-poly(ethylene glycol) copolymers. Biomaterials 2012,33, (27), 6551–8. [DOI] [PubMed] [Google Scholar]
- 38.Cheng K; Ye Z; Guntaka RV; Mahato RI Enhanced hepatic uptake and bioactivity of type alpha1(l) collagen gene promoter-specific triplex-forming oligonucleotides after conjugation with cholesterol. The Journal of pharmacology and experimental therapeutics 2006,317, (2), 797–805. [DOI] [PubMed] [Google Scholar]
- 39.Liu DV; Yang NJ; Wittrup KD A Nonpolycationic Fully Proteinaceous Multiagent System for Potent Targeted Delivery of siRNA. Molecular Therapy. Nucleic Acids 2014, 3, (5), e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen TT; Sly KL; Conboy JC Comparison of the Energetics of Avidin, Streptavidin, NeutrAvidin, and Anti-Biotin Antibody Binding to Biotinylated Lipid Bilayer Examined by Second-Harmonic Generation. Analytical chemistry 2012, 84, (1), 201–208. [DOI] [PubMed] [Google Scholar]
- 41.Ciechanover A Intracellular protein degradation: from a vague idea, through the lysosome and the ubiquitin-proteasome system, and onto human diseases and drug targeting (Nobel lecture). Angewandte Chemie (International ed. in English) 2005,44,(37), 5944–67. [DOI] [PubMed] [Google Scholar]
- 42.Vugmeyster Y; Xu X; Theil F-P; Khawli LA; Leach MW Pharmacokinetics and toxicology of therapeutic proteins: Advances and challenges. World Journal of Biological Chemistry 2012, 3, (4), 73–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elsabahy M; Wooley KL Cytokines as biomarkers of nanoparticle immunotoxicity. Chemical Society reviews 2013,42, (12), 5552–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolfram J; Zhu M; Yang Y; Shen J; Gentile E; Paolino D; Fresta M; Nie G; Chen C; Shen H; Ferrari M; Zhao Y Safety of Nanoparticles in Medicine. Current drug targets 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park J; Park J; Pei Y; Xu J; Yeo Y Pharmacokinetics and biodistribution of recently-developed siRNA nanomedicines. Advanced drug delivery reviews 2016,104, 93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]