Significance
The major histocompatibility complex (MHC), in particular, the human leukocyte antigen (HLA) loci encode key components of the immune system and include some of the most diverse genes in the human genome. Evolution of the HLA loci is generally thought to be driven by selection for increased diversity, i.e., against common haplotypes (negative frequency-dependent selection). Here, we compare the haplotype frequency and homozygosity distributions from the extensive National Marrow Donor Program registry to the predictions of different models of evolution and show that the data are best compatible with multiplicative fitness and rapid fitness decay of haplotypes in some populations. Selection against common haplotypes is not supported. The data cannot be explained by mating preferences or population substructure either.
Keywords: haplotype evolution, haplotype discovery, fitness decay, frequency-dependent selection, multiplicative fitness
Abstract
The major histocompatibility complex (MHC) is a central component of the vertebrate immune system and hence evolves in the regime of a host–pathogen evolutionary race. The MHC is associated with quantitative traits which directly affect fitness and are subject to selection pressure. The evolution of haplotypes at the MHC HLA (HLA) locus is generally thought to be governed by selection for increased diversity that is manifested in overdominance and/or negative frequency-dependent selection (FDS). However, recently, a model combining purifying selection on haplotypes and balancing selection on alleles has been proposed. We compare the predictions of several population dynamics models of haplotype frequency evolution to the distributions derived from 6.59-million-donor HLA typings from the National Marrow Donor Program registry. We show that models that combine a multiplicative fitness function, extremely high haplotype discovery rates, and exponential fitness decay over time produce the best fit to the data for most of the analyzed populations. In contrast, overdominance is not supported, and population substructure does not explain the observed haplotype frequencies. Furthermore, there is no evidence of negative FDS. Thus, multiplicative fitness, rapid haplotype discovery, and rapid fitness decay appear to be the major factors shaping the HLA haplotype frequency distribution in the human population.
The major histocompatibility complex (MHC), and in particular, the HLA (HLA) loci, have been the primary subject of numerous studies aimed at understanding the microevolutionary processes that generate and maintain genetic diversity (1–4). The MHC has been extensively characterized in many species and remains a potent model for examining the balance between stochastic and deterministic evolutionary processes (5). HLA is a multigene family encoding glycoprotein receptors that bind short peptides and present them to T lymphocytes which can mount an immune response upon recognition (6). The architecture of the MHC complex varies substantially across species (7, 8). However, in every species, the MHC contains at least one extremely polymorphic locus. In particular, the HLA (HLA) gene complex contains the HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 genes, which together account for over 15,000 distinct allele sequences described to date (9, 10). Accordingly, combinations of different alleles across HLA loci yield a vast, effectively infinite number of haplotypes.
Many distinct, although not necessarily mutually exclusive, mechanisms have been proposed to explain the enormous variability at the HLA loci. Because MHC molecules are central to the immune system, pathogen-driven selection for diversity (balancing selection) seems to be an obvious key factor, although the details of the proposed mechanisms differ (1, 2, 11–14). Additionally, MHC-dependent mate choice (assortative mating) (15–18) and sexual selection (19, 20) have been argued to shape the MHC diversity based, in particular, on evidence of MHC dependence on mother–fetal interactions and the apparent olfactory recognition of MHC haplotypes (21, 22). Population structure, which either emerges as a result of fluctuating selection driven by host–pathogen coevolution or is imposed by the environment, can maintain and also enhance diversity generated by other mechanisms (5, 23, 24).
Two alternative mechanisms have been proposed to underlie balancing selection. The first, negative frequency-dependent selection, is thought to be driven by the ability of rare MHC alleles to provide better protection against pathogens that might have adapted to evade high frequency MHC alleles, leading to selection against these common alleles (14, 25–28). An alternative mechanism, pathogen-driven overdominance, stipulates that heterozygotes, in which two variant alleles are expressed, are resistant to a broader range of parasites and therefore typically have greater fitness than homozygotes (11, 12, 25, 29).
Despite numerous reports on strong balancing selection at the HLA loci, elucidating the underlying mechanisms has proved difficult. A fundamental problem is that even in the studies that muster sufficient statistical power, there seem to be alternative explanations for the observed frequency and homozygosity distributions. Furthermore, competing mechanisms often confound the efforts to disentangle their effects (27).
Whereas balancing selection has long been observed at individual HLA loci (13, 30–32), there has been little exploration of the elements shaping the selection of haplotypes, or combinations of HLA alleles across loci. Recently, however, we have presented evidence that HLA haplotype frequencies are affected by strong positive frequency-dependent selection (33). We have showed there that the haplotype frequency family size distribution is best explained by a model containing a positive frequency-dependent selection, using the deviation of homozygosity and linkage disequilibrium measures, as well as from a fit to the full family size distribution.
Here, we quantitatively assess the mechanisms of selection that determine HLA haplotype frequency distributions by computing the likelihood of observing the HLA haplotype distribution in populations in the US Bone Marrow Donor Registry given different stochastic population models. This approach allows us to determine which of the selection mechanisms is best compatible with the observations. Comparison of several fitness functions indicates that the multiplicative form, in which low fitness haplotypes have a drastic effect on the fitness of a diploid genotype, is better compatible with the data than other fitness functions. Among the various evolutionary mechanisms considered—including overdominance, population structure, frequency-dependent assortative mating, frequency-dependent selection, and decaying fitness (34)—we found the strongest support for fitness decay. Our results indicate that a multiplicative fitness function, combined with exponential fitness decay of haplotypes over time in some populations, fits the data substantially better than any of the other explored models for populations where we were able to rank the models. For other populations, we were unable to distinguish between more complex models of selection and were only able to conclude that the multiplicative fitness function fitted the data best.
Results and Discussion
The Haplotype Frequency Resource.
Because haplotypic diversity is far greater than allelic diversity, an extensive population genetic reference panel for HLA haplotypes is essential to enable assessment of haplotype-level selection. High-resolution, five-locus (A ∼ C ∼ B ∼ DRB1 ∼ DQB1) HLA haplotype frequencies were estimated for 21 self-reported racial/ethnic categories using an expectation-maximization (EM) algorithm for 6.59-million-donor HLA typings from the National Marrow Donor Program registry (35, 36) (see SI Appendix for the detailed description, and a table of population acronyms, as well as detailed information for each population, with the haplotype frequencies and their homozygote frequencies, at https://github.com/louzounlab/HLA_data.git). For every distinct haplotype i, the resulting dataset includes the total number of copies ci and the number of homozygous individuals zi carrying two copies of haplotype i. Each of the 21 population samples is a snapshot of the evolutionary processes that affect the frequencies of the HLA alleles and the homozygosity for each allele. Therefore, given a population model, in which individuals are identified by pairs of HLA haplotypes, we can compute the likelihood of observing the frequency and homozygosity data as a snapshot of the population model in steady state. Population models with different mechanisms of selection can then be compared quantitatively. We have recently demonstrated the validity of the frequency estimates of HLA haplotypes and their adequacy for population structure modeling (33).
Models of Haplotype Frequency Evolution.
The key ingredient in evolutionary models of the haplotype frequency distribution is the function that assigns a fitness value to a genotype based on its constituent haplotypes. Models with selection, population structure, and/or assortative mating are compared with the neutral model in which mating is random and all individuals have the same fitness regardless of their genotype. In models with selection, we assume that each haplotype can be completely specified by a single continuously varying value h. A genotype is therefore characterized by a pair (h1, h2) of values. The fitness function maps the pair (h1, h2) to a positive Malthusian fitness f. We compare the neutral model, in which all individuals have the same fitness, to five models with selection and different forms of fitness function:
-
1)
additive, in which the fitness is f = h1 + h2;
-
2)
additive with overdominance, in which the fitness of a homozygote (h1, h1) is f = h1 (instead of 2h1) whereas the fitness of the heterozygote is f = h1 + h2;
-
3)
multiplicative, in which f = h1h2;
-
4)
hybrid, in which f = hmax − (hmax − h1) (hmax − h2)/hmax = h1 + h2 − h1h2/hmax; and
-
5)
hybrid as in 4 but with overdominance, so that homozygotes have fitness h1.
The hybrid model introduced the upper bound hmax on the haplotype contributions. This upper bound must exist in biologically plausible models simply because the growth rate is bounded from above. Without loss of generality, we set hmax = 1. A rescaling of time can transform any upper bound on h to unity. In addition, negative haplotype contributions are meaningless because the growth rate is positive by definition. We therefore postulate that h ∈ (0, 1].
All evolutionary models considered here share several elements. The population size N is fixed. The population evolves via discrete, nonoverlapping generations that consist of separate selection, mating, and mutation rounds. Each individual is characterized by a pair of haplotypes. During the selection round, a Wright–Fisher sample of the population is constructed whereby N individuals are selected with replacement, with probabilities proportional to their respective fitness values. The mating round consists of selecting N mating pairs as described in Materials and Methods. Each mating pair produces a single offspring that receives one random haplotype from each parent. During the subsequent mutation round, each haplotype of each individual is replaced with probability μ (hereafter, “haplotype discovery rate”) by a new haplotype for which the fitness contribution is selected at random from the uniform distribution.
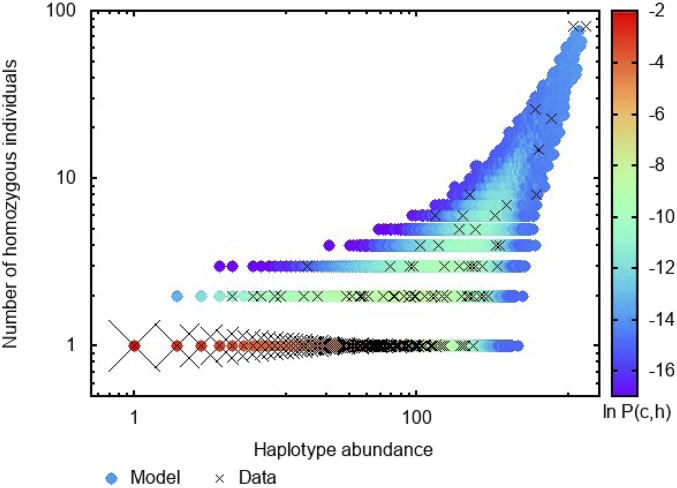
These “mutations” in the context of haplotypes are most likely recombination events creating new haplotypes rather than point mutations or deletions, or alternatively, the introduction of a haplotype from a foreign population. The stochastic population models are simulated in steady state and the joint probability distribution of the haplotype abundance and homozygosity is measured, allowing us to compute the likelihood of observing the data. Fig. 1 illustrates the joint probability distribution for haplotype abundance and homozygosity obtained under the model with multiplicative fitness (model 3 above) compared with data for one of the population groupings in the registry (see further details below). A detailed description of the optimization and examples for other populations including the data on the feasibility of testing each model with each population are presented in SI Appendix, section 4. The SI Appendix further describes all of the population groups from the registry that we used to fit our models (SI Appendix, section 5). Some of these include broad population categories made up of several subpopulations.
Fig. 1.
Model joint probability distribution P(c; h) measured via stochastic simulations for the model with the multiplicative fitness function and without additional selection mechanisms. Each circle corresponds to a pair (c,h) that occurred at least once among 105 uncorrelated snapshots of the population in steady state. The color represents ln P(c,h) (see color bar). Crosses are the (c,h) pairs observed in the HAWI (Hawaiian) sample of the MHC haplotype data. Cross size is proportional to the logarithm of the number of pairs present in the sample.
Multiplicative Fitness-Based Model Outperforms Other Models.
All populations were analyzed in equilibrium, after enough time steps not to be affected by the initial conditions (SI Appendix). Performing the likelihood maximization procedure (see Materials and Methods for details), we obtain the set of optimal model parameters and the estimate of the maximum log likelihood for each model–dataset pair. We compared the neutral model with the multiplicative fitness model for all populations in the dataset. The neutral model was found to be vastly inferior to the multiplicative model in all cases (SI Appendix, section 8). On average, introducing fitness variability increases the natural log likelihood by over 10,000. We therefore concentrate on the less dramatic differences between the models with alternative fitness functions that have the potential to illuminate specific evolutionary mechanisms. It was not feasible to run all combinations of models for all populations in the dataset, in particular, for the larger populations. SI Appendix, section 7 provides details of the populations for which different models were compared.
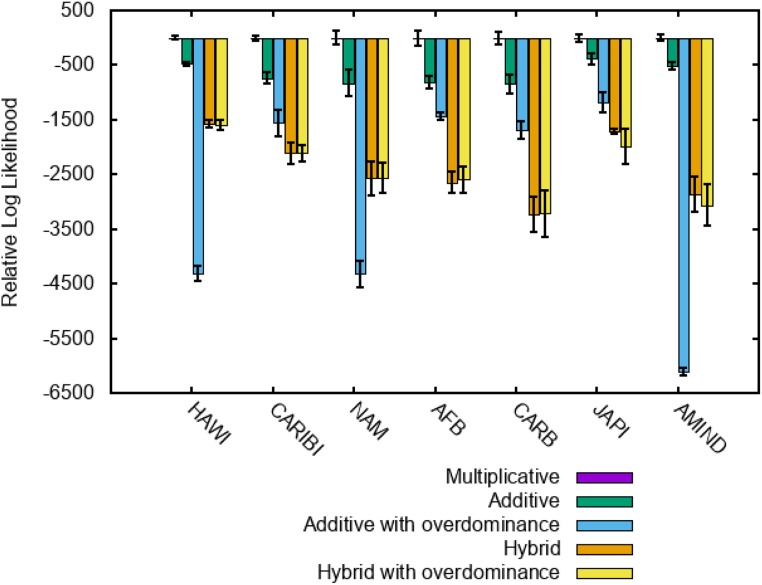
Fig. 2 shows the maximum log likelihood estimates for the models with the fitness functions described above relative to that of the model with the multiplicative fitness function (model 3). Clearly, the multiplicative fitness model, in which the fitness of a genotype is the product of its haplotype contributions, fits the haplotype frequency data far better than any of the other tested models in all included populations. In contrast, overdominance, which can be introduced in the additive and hybrid fitness models, clearly lacks support, and is the worst model for most populations.
Fig. 2.
Peak log likelihood for models with different fitness functions relative to that of the model with multiplicative fitness. For the model with multiplicative fitness, all relative log likelihood values are equal to zero, and accordingly, the respective bars are not visible. Not all populations were studied with all models. The population samples shown are only the ones simulated with the appropriate models.
Implications of the Model: Dominance of Deleterious Effects and High Haplotype Discovery Rate.
Under the premise that MHC molecules differ by their ability to serve in peptide presentation and that maximum diversity is beneficial, one would expect a model with the hybrid fitness function of the form h1 + h2 − h1h2 = 1 − (1 − h1) (1 − h2), under which it is sufficient to have one “good” allele, to be superior to other individuals. The finding that the data instead support the multiplicative fitness function implies that the reverse is true: one dominant low-fitness allele is sufficient to substantially suppress the fitness of an individual.
Another surprising finding of the model fits is the high value of the haplotype discovery rate μ which maximizes log likelihood. As shown in Fig. 3, in the model with multiplicative fitness, the estimated values of μ range from 0.1 to 0.25. The model with additive fitness yields lower estimates for μ in the 0.05–0.1 range, which could be considered more realistic values. However, the additive fitness model provides a significantly inferior fit to the data. This high discovery rate is a consistent feature of all models providing a good fit.
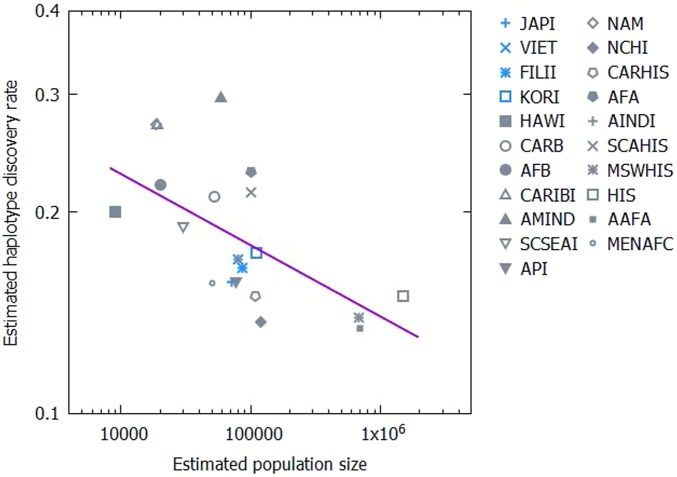
Fig. 3.
Negative correlation between the estimated population size N and haplotype discovery rate μ. Each point represents the fit of the model with multiplicative fitness to a sample labeled by a distinct ethnic identifier. The Spearman rank correlation coefficient is ρ = −0.704 with the P value of 0.0005. More genetically homogeneous island or peninsula populations are marked in gray.
Based on current estimates of recombination intensity in the HLA locus, and assuming that each recombination event yields a novel haplotype, roughly 3% of the gametes carry novel haplotypes (37). Thus, recombination can explain only about 20% of the inferred rate of haplotype discovery. Gene flow between populations potentially could be the dominant contribution to the high inferred μ values. If different populations are isolated from each other by distance or by a barrier, the rate of gene flow into a particular population is proportional to the length of the boundary that it shares with the surrounding semi-isolated populations. Under the assumption of an approximately constant population density, larger populations will have smaller relative gene influx because the length of the boundary scales sublinearly with the population size (in the simplest case, population size scales with the area occupied by the population). As shown in Fig. 3, the predicted negative correlation between the population size estimated by our model and the estimated haplotype discovery rate is indeed strong and significant (Spearman ρ = −0.704, P value = 0.0005). Although some populations are more deeply sampled than others, sample size in the registry scales approximately with the population sizes estimated by our model. Higher values of μ are observed in populations based on regional categories (for example, “Native North American,” “African Black”) that contain several geographically and linguistically isolated subgroups. These populations are likely to have a particularly long boundary across which the gene flow occurs, supporting our argument that μ approximates gene flow. Relatively homogeneous populations also seem to have relatively lower μ values (Fig. 3) although not all of the populations with a low μ value are homogeneous. However, the possibility also remains that the current estimates of recombination rates are far too low, and extremely high mutation and recombination rates could be the main mechanisms driving the evolution of the enormous amount of HLA polymorphism.
Selection in the Multiplicative Fitness Model.
We further compared four additional selection mechanisms that have been proposed to operate at the HLA locus. All three models extend the multiplicative fitness function that, as shown above, fits the data substantially better than the other tested functions. Obviously, not all combinations of fitness functions have been tested. The additional selection mechanism introduces a new parameter δ, which has different meanings in each of the three models.
In the first model with assortative mating, the procedure for selecting the mating pairs that give rise to the next generation is designed to suppress mating of individuals that carry the same haplotype. During the mating round, each member of the population receives a mate. The mate is selected at random. A mating probability factor p = e−δ1(f1 + f2) is computed, where f is the frequency of the haplotype if it is shared by the putative mating pair and 0 otherwise. The mating partner is kept with probability p. If the partner is not kept, another one is selected at random and the procedure is repeated until a mating partner is selected for every member of the population.
In the second model with population structure, the population has a one-dimensional ring topology. A mating partner is selected at a random distance, which is exponentially distributed with mean δ2. This length-scale δ2 sets the size of the interwoven local communities from which mating partners are likely to be selected.
In the third model with frequency-dependent selection, the fitness of an individual is the product of the intrinsic haplotype contributions h1h2 and the frequency-dependent factor e –δ3(f1 + f2), where f1 and f2 are the population frequencies of the two haplotypes. Positive δ enhances the fitness of common haplotypes more than that of rare haplotypes.
In the fourth model, the fitness decays as a function of time. The fitness of an organism is a product of its haplotype contributions and mating is random. The distribution of each haplotype to fitness decays exponentially with the number of generations since that haplotype was discovered. The initial contribution of a haplotype in its birth generation is drawn from a uniform distribution (0;1). In this model, δ4 is the decay rate of the haplotype fitness.
Positive Frequency-Dependent Selection and Fitness Decay.
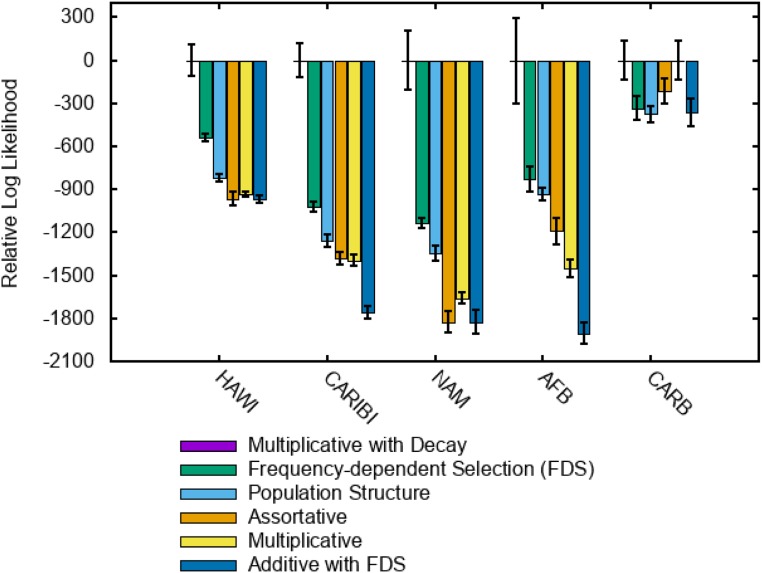
The results shown in Fig. 4 indicate that, when sufficient data are available, the assortative mating mechanism is not supported by the data, and the model with population structure is only weakly supported (for other populations, the parameter estimates were too noisy; see SI Appendix, section 9). In contrast, frequency-dependent selection is strongly supported, and the improvement over the models without frequency-dependent selection is highly significant. Surprisingly, however, the sign of the FDS on haplotypes is positive, in contrast to the observations for individual loci (33). As shown in Table 1, the magnitude of δ is moderate such that the fitness of a common haplotype with a frequency of 0.5 increases by about 10–15% as a result of FDS. However, for some populations, the best model by far is the one with fitness rapidly decaying over time. The optimal decay rate is of the order of 3–20 generations (Table 2) amounting to 50–500 y. In other words, a haplotype that was highly advantageous in the beginning of the previous century could have lost most of this advantage by now. For other populations, the optimal decay rate was 0. This can be either the solution of a convergence to a local minimum, or alternatively, might reflect a difference between populations with low variability/haplotype discovery rates (JAPI and AMIND) and populations with high variability.
Fig. 4.
Peak log likelihood for models with multiplicative fitness and additional mechanisms of selection. The values are compared with the multiplicative selection model with time decay (therefore, the relative likelihood values for this model are all equal to 0, and the corresponding bars are not visible). Accordingly, a value of 0 for any other model means a likelihood similar to that of the multiplicative selection with time decay model. Negative values represent a worse fit. Except for the additive model with FDS, all models are extensions of the multiplicative fitness model.
Table 1.
Estimated parameters for the model with multiplicative fitness and frequency-dependent selection
| Sample | N | μ | δ3 |
| HAWI | 6,000 ± 300 | 0.208 ± 0.002 | 0.27 ± 0.01 |
| CARIBI | 7,000 ± 500 | 0.168 ± 0.001 | 0.52 ± 0.02 |
| NAM | 13,700 ± 600 | 0.247 ± 0.001 | 0.20 ± 0.01 |
| AFB | 10,000 ± 600 | 0.282 ± 0.001 | 0.54 ± 0.01 |
| CARB | 17,000 ± 800 | 0.264 ± 0.001 | 0.49 ± 0.01 |
| SCSEAI | 17,400 ± 600 | 0.185 ± 0.001 | 0.16 ± 0.01 |
| JAPI | 29,000 ± 500 | 0.139 ± 0.001 | 0.072 ± 0.0004 |
| AMIND | 25,600 ± 700 | 0.127 ± 0.001 | 0.094 ± 0.0037 |
| FILLII | 56,800 ± 700 | 0.143 ± 0.001 | 0.058 ± 0.0017 |
| VIET | 34,100 ± 600 | 0.171 ± 0.001 | 0.112 ± 0.003 |
N, population size; μ, haplotype discovery rate; δ, strength of frequency-dependent selection.
Table 2.
Estimated parameters for the model with multiplicative fitness, rapid haplotype discovery and fitness decay
| Sample | N | μ | δ4 |
| HAWI | 8,100 ± 630 | 0.301 ± 0.02 | 2.06 ± 0.05 |
| CARIBI | 12,100 ± 1,800 | 0.487 ± 0.06 | 20.17 ± 0.03 |
| NAM | 24,600 ± 1,100 | 0.369 ± 0.01 | 17.37 ± 0.03 |
| AFB | 13,400 ± 1,500 | 0.308 ± 0.07 | 5.01 ± 0.03 |
| CARB | 35,660 ± 1,955 | 0.197 ± 0.01 | 0 |
| JAPI | 51,005 ± 2,019 | 0.13 ± 0.01 | 0 |
| AMIND | 31,001 ± 960 | 0.15 ± 0.1 | 0 |
| FILII | 71,420 ± 2,110 | 0.145 ± 0.01 | 0 |
The population samples were: HAWI, Hawaiian or Pacific Islander; CARIBI, Native Caribbean; NAM, Native American; AFB, African Black; CARB, Caribbean Black; SCSEAI, Southeast Asian; JAPI, Japanese; AMIND, Native North American; FILII, Filipino; VIET, Vietnamese.
So far, we combined more complex models with the multiplicative fitness function that our first round of simulations identified as providing the best match to the data. We additionally fitted a model combining an additive fitness function with FDS, but such a model was not supported by the data (Fig. 4).
Selection Against Low Fitness Haplotypes.
Our main findings are: 1) the multiplicative fitness model, in which the impact of deleterious haplotypes on the fitness of the diploid genotype, dominates the evolution of human MHC; 2) high rate of haplotype discovery; 3) fitness decay over time; and 4) if there is FDS, it is positive, i.e., rare haplotypes are selected against. We conjecture that these findings can be understood together if the fitness contribution of a haplotype is due, largely, to deleterious effects. Such effects can have multiple origins which might differ among populations, based on the history of their encounters with pathogens. Other factors that could affect fitness contributions of haplotypes might autoimmune effects, interactions between killer Ig-like receptors (KIR) on natural killer cells and their HLA ligands (38), or preferential gene flow with selection against specific genes (which might even be a bystander effect).
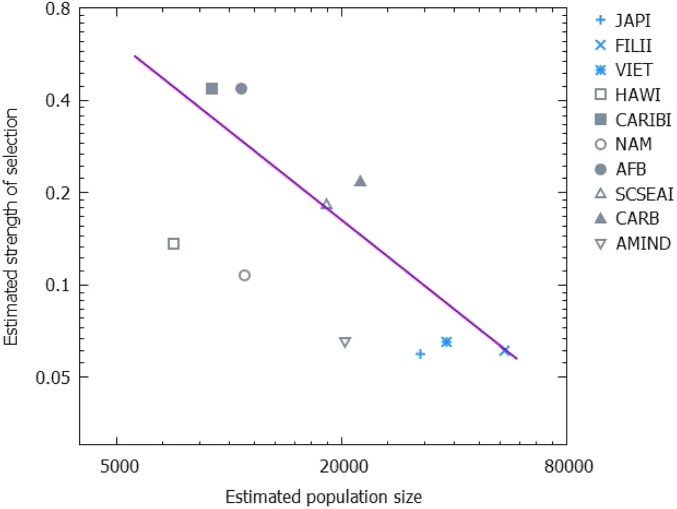
Among the putative mechanism(s) driving the observed selection on haplotypes, the KIR-HLA interactions seem to merit special attention. If the diversity of the molecules that trigger an inhibitory response, such as KIR, grows with the population size, adaptation in large populations would be hampered by this greater diversity, and accordingly, there will be a smaller fitness difference between common and rare haplotypes in larger populations. This conjecture predicts a negative correlation between the strength of the FDS and population size . As shown in Fig. 5, a strong and significant negative correlation is indeed observed in the data (Spearman ρ = −0.855, P value = 0.0016). If the KIR-HLA interactions affect fitness, a single haplotype that decreases the probability of a fetus formation can reduce the survival of any individual carrying this haplotype.
Fig. 5.
Dependency of the estimated selection strength on the population size. Estimated strength δ of positive frequency-dependent selection at the level of haplotypes is plotted against the estimated population size N. The Spearman rank correlation is ρ = −0.855 and the P value is 0.0016.
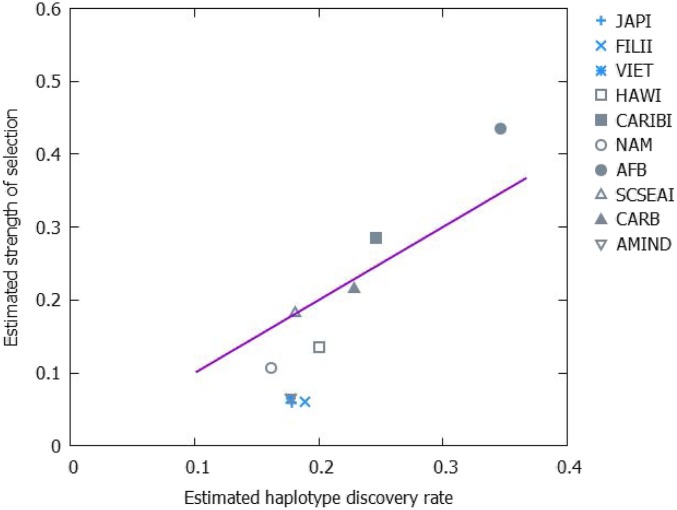
Moreover, if the fitness of HLA haplotypes is a function of their interactions with rapidly evolving KIR haplotypes, fitness could drop when the KIR haplotype distribution changes, which might result in a decline in the fitness of HLA haplotypes with time. High haplotype discovery rate entails stronger selection (more low-fitness haplotypes to eliminate; Fig. 6). The haplotype discovery rate is higher in small populations (Fig. 3), and therefore, in these populations, selection is also stronger (Figs. 5 and 6). It should be noted, however, that in some populations, in particular, AMIND, CARB, and JAPI, fitness decay over time is not supported by the data. In such populations, other factors could shape the haplotype frequency distribution, or else, the sample size could be too small to detect the effects of fitness decay.
Fig. 6.
Dependency of the estimated selection strength on the haplotype discovery rate. Estimated strength δ of positive frequency-dependent selection at the level of haplotypes is plotted against the estimated haplotype discovery rate. The populations are divided into heterogeneous (gray) and homogeneous (blue). The Spearman rank correlation is ρ = −0.703 and the P value is 0.022.
Allele-Based Models.
The model fits in this work so far assumed that each haplotype is associated with a fixed fitness contribution, regardless of what other haplotype it pairs with. Although this assumption seems plausible for complete haplotypes, it is less supportable at the level of individual alleles. The fitness value of particular alleles could depend on what other alleles comprise the haplotype. Nevertheless, it is instructive to perform the fitting of the allele level abundance and homozygosity data to the model with FDS, variability in the intrinsic fitness values of the allele variants and multiplicative fitness.
As in the case of haplotypes, the neutral model, in which all alleles have the same fitness and there is no FDS, can be confidently rejected. As shown in Table 3, the majority of sample/allele datasets do not seem to support FDS because the estimated strength is consistent with a zero value. The remainder of sample/allele combinations exhibits weak positive or negative FDS. The estimated strength of selection is an order of magnitude smaller than it is at the level of haplotypes. Therefore, it appears that the intrinsic variability of fitness contributions is mostly sufficient to explain the deviation from neutrality previously detected via the Ewens–Watterson homozygosity test (33).
Table 3.
The estimated strength of FDS at the level on individual alleles
| Allele | HAWI | CARIBI | NAM | AFB | CARB | AMIND |
| A | 0.0152 ± 0.0073 | −0.0121 ± 0.0053 | 0.0213 ± 0.0012 | −0.0059 ± 0.0028 | −0.0046 ± 0.0024 | 0.0023 ± 0.0006 |
| B | 0.0074 ± 0.0040 | −0.0156 ± 0.0075 | −0.0042 ± 0.0039 | −0.0048 ± 0.0032 | −0.0061 ± 0.0022 | −0.0036 ± 0.0021 |
| C | −0.0174 ± 0.0065 | −0.0140 ± 0.0044 | −0.0057 ± 0.0065 | 0.0038 ± 0.0029 | 0.0113 ± 0.0023 | 0.0050 ± 0.0026 |
| DRB1 | 0.0006 ± 0.0074 | −0.0147 ± 0.0055 | −0.0080 ± 0.0054 | −0.0067 ± 0.0034 | −0.0014 ± 0.0024 | −0.0037 ± 0.0012 |
| DQB1 | −0.0099 ± 0.0035 | −0.0057 ± 0.0057 | 0.0106 ± 0.0040 | 0.0088 ± 0.0036 | 0.0193 ± 0.0029 | 0.0024 ± 0.0004 |
Negative values of correspond to negative FDS.
Conclusions
To summarize, here we developed a quantitative framework for comparing an extensive data set of estimated HLA haplotype abundance and homozygosity values with predictions of evolutionary models. Under this framework, we compared models subject to distinct mechanisms of selection, alternative fitness functions and other factors, including FDS, population structure, and frequency-dependent assortative mating. We found that the data do not support fitness functions with overdominance. A multiplicative fitness function, whereby unfit haplotypes have a major effect on the genotype fitness, fits the data significantly better than all tested alternatives. Among additional mechanisms of selection, the data most strongly support a rapidly evolving fitness landscape for the populations where the fit was good enough to determine a mechanism. This finding implies that some haplotypes tend to have lower fitness and are selected against, even in the presence of other high fitness haplotypes. This fitness difference can be lost over time. As mentioned above, the simplest explanation for such fitness loss seems to involve interactions with other, rapidly evolving loci, such as KIR. In addition, recent theoretical analysis and empirical observations (31, 39) indicate that balancing selection in the HLA locus leads to accumulation of deleterious alleles in non-HLA genes associated with this locus (so-called “sheltered load” accumulation), thus incurring a fitness cost, which could translate into positive FDS.
Taken together, the results of this analysis imply that the observed frequency distributions of the HLA haplotypes are shaped by the trade-off between the high rate of new haplotype discovery, largely caused by extensive gene flow between populations and rapid haplotype fitness decay over time. However, these findings are not incompatible with pathogen-driven balancing selection at individual loci. The source of the deleterious effect of some haplotypes requires further exploration.
Materials and Methods
Haplotype Frequency Datasets.
Five-locus high resolution HLA A ∼ C ∼ B ∼ DRB1 ∼ DQB1 haplotype frequencies were estimated using the EM algorithm for 6.59-million-donor HLA typings from the National Marrow Donor Program registry (US) (36, 40). Given typing ambiguities, a large number of very low probability haplotypes emerge as artifacts from the EM. We removed low probability haplotypes by assigning each person in the sample a single pair of their most probable haplotypes with the remainder of their haplotype pair probability distribution discarded. The population haplotype frequencies were thus recalculated with a single haplotype pair assigned for each individual. Allele frequencies were derived as marginal sums of the haplotype frequencies.
Likelihood Computation and Comparison of Model Fits to the Data.
To compare the fits of the models to the data, we computed the likelihoods of observing different haplotype datasets, i.e., sets of (abundance, homozygosity) pairs, given a particular model. The likelihood is calculated using the joint probability P(c,z) that a random haplotype of a random individual in a sample occurs c times and there are z homozygous individuals in the sample which carry this haplotype. Given P(c,z), the log likelihood of observing a particular set of (abundance, homozygosity) pairs is approximated by
In general, the c and h values for different haplotypes are not independent. However, when c ≪ N, the coupling between different haplotypes is negligible, and therefore the product of individual probabilities P(c,z) can be used to calculate the likelihood.
P(c,z) is not analytically computable even under the simplest model. The approximate joint probability distribution is therefore computed via a stochastic simulation. An initial homogeneous population is evolved until steady state is reached according to the following criterion. The total number kt of distinct haplotypes in the population is measured every Nsamp = (log N)/μ generations. Two sets of measurements are compared: the last Nmeas measurements and the set of Nmeas measurements immediately preceding the last Nmeas. The steady state is considered to be reached when the hypothesis that the means of these two sets are the same cannot be rejected with significance level 10−4 using the two-sample t test; Nmeas = 105 is assumed throughout this study. Once the model is in steady state, we perform Nmeas measurements separated by Nsamp generations. In each measurement, the counts of (abundance, homozygosity) pairs are incremented for each haplotype of each individual in the population. These counts normalized by 2NNmeas approximate the joint probability distribution P(c,z).
To address the issue of numerical noise in the estimate of P(c,z), we use Gaussian kernel smoothing to compute the log likelihood ℒ Smoothing is performed in logarithmic space where z is incremented by unity to assure that it remains positive. The kernel has a single parameter λ which is the smoothing length scale in logarithmic space. We checked that the results are only weakly sensitive to λ which was accordingly fixed at λ = 0.02. Fig. 1 demonstrates the relative size of the smoothing scale λ and displays an example of the measured noisy joint probability distribution together with the sample data.
Gaussian kernel smoothing does not completely eliminate noise. The level of noise is higher when the (abundance, homozygosity) pairs that are present in the dataset are rarely observed in the simulations. When the haplotype discovery rate μ is high, haplotypes do not often reach high frequencies in the simulations and therefore the estimate of the joint probability distribution for high values of c is less reliable. However, the contributions of the high frequency haplotypes to the log likelihood are the most important. To deal with the higher noise in the more important region of parameter space, we estimated the model parameters that maximize the likelihood ℒ by fitting a general parabolic surface to the measured noisy log likelihood as a function of the model parameters. Before the parabolic fit, the model parameters were scaled so that their means are unity in the fitting region. The fit provides the estimates of the error in the maximum log likelihood as the mean deviation of the computed values from the fitted parabolic surface. The confidence intervals for the estimated model parameters are computed as the values at which the fitted log likelihood is reduced by three units. Not all populations and all models were studied because some of the populations were too large to simulate, and moreover, in some populations, the optimization did not converge (SI Appendix, section 7).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
Data deposition: Haplotype frequencies are available through http://frequency.nmdp.org. Detailed information on the frequencies and homozygosity is available on GitHub, https://github.com/louzounlab/HLA_data.git.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714436116/-/DCSupplemental.
References
- 1.Hughes A. L., Yeager M., Natural selection and the evolutionary history of major histocompatibility complex loci. Front. Biosci. 3, d509–d516 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Hughes A. L., Yeager M., Natural selection at major histocompatibility complex loci of vertebrates. Annu. Rev. Genet. 32, 415–435 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Piertney S. B., Oliver M. K., The evolutionary ecology of the major histocompatibility complex. Heredity 96, 7–21 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Spurgin L. G., Richardson D. S., How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. Biol. Sci. 277, 979–988 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buhler S., Sanchez-Mazas A., HLA DNA sequence variation among human populations: Molecular signatures of demographic and selective events. PLoS One 6, e14643 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein J., Natural History of the Major Histocompatability Complex (John Wiley & Sons, New York, 1986). [Google Scholar]
- 7.Kelley J., Walter L., Trowsdale J., Comparative genomics of major histocompatibility complexes. Immunogenetics 56, 683–695 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Shiina T., Blancher A., Inoko H., Kulski J. K., Comparative genomics of the human, macaque and mouse major histocompatibility complex. Immunology 150, 127–138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vandiedonck C., Knight J. C., The human Major Histocompatibility Complex as a paradigm in genomics research. Brief. Funct. Genomics Proteomics 8, 379–394 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gourraud P. A., et al. , HLA diversity in the 1000 genomes dataset. PLoS One 9, e97282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes A. L., Nei M., Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335, 167–170 (1988). [DOI] [PubMed] [Google Scholar]
- 12.Hughes A. L., Nei M., Maintenance of MHC polymorphism. Nature 355, 402–403 (1992). [DOI] [PubMed] [Google Scholar]
- 13.Takahata N., Satta Y., Klein J., Polymorphism and balancing selection at major histocompatibility complex loci. Genetics 130, 925–938 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slatkin M., Balancing selection at closely linked, overdominant loci in a finite population. Genetics 154, 1367–1378 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potts W. K., Manning C. J., Wakeland E. K., Mating patterns in seminatural populations of mice influenced by MHC genotype. Nature 352, 619–621 (1991). [DOI] [PubMed] [Google Scholar]
- 16.Chaix R., Cao C., Donnelly P., Is mate choice in humans MHC-dependent? PLoS Genet. 4, e1000184 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamiya T., O’Dwyer K., Westerdahl H., Senior A., Nakagawa S., A quantitative review of MHC-based mating preference: The role of diversity and dissimilarity. Mol. Ecol. 23, 5151–5163 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Winternitz J., Abbate J. L., Huchard E., Havlicek J., Garamszegi L. Z., Patterns of MHC-dependent mate selection in humans and nonhuman primates: A meta-analysis. Mol. Ecol. 26, 668–688 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Winternitz J. C., et al. , Sexual selection explains more functional variation in the mammalian major histocompatibility complex than parasitism. Proc. Biol. Sci. 280, 20131605 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jan Ejsmond M., Radwan J., Wilson A. B., Sexual selection and the evolutionary dynamics of the major histocompatibility complex. Proc. Biol. Sci. 281, 20141662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts T., Roiser J. P., In the nose of the beholder: Are olfactory influences on human mate choice driven by variation in immune system genes or sex hormone levels? Exp. Biol. Med. 235, 1277–1281 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Kromer J., et al. , Influence of HLA on human partnership and sexual satisfaction. Sci. Rep. 6, 32550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winternitz J. C., Wares J. P., Duplication and population dynamics shape historic patterns of selection and genetic variation at the major histocompatibility complex in rodents. Ecol. Evol. 3, 1552–1568 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minias P., Whittingham L. A., Dunn P. O., Coloniality and migration are related to selection on MHC genes in birds. Evolution 71, 432–441 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Takahata N., Nei M., Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics 124, 967–978 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slade R. W., McCallum H. I., Overdominant vs. frequency-dependent selection at MHC loci. Genetics 132, 861–864 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potts W. K., Slev P. R., Pathogen-based models favoring MHC genetic diversity. Immunol. Rev. 143, 181–197 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Apanius V., Penn D., Slev P. R., Ruff L. R., Potts W. K., The nature of selection on the major histocompatibility complex. Crit. Rev. Immunol. 17, 179–224 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Meyer D., Thomson G., How selection shapes variation of the human major histocompatibility complex: A review. Ann. Hum. Genet. 65, 1–26 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Key F. M., Teixeira J. C., de Filippo C., Andrés A. M., Advantageous diversity maintained by balancing selection in humans. Curr. Opin. Genet. Dev. 29, 45–51 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Lenz T. L., Spirin V., Jordan D. M., Sunyaev S. R., Excess of deleterious mutations around HLA genes reveals evolutionary cost of balancing selection. Mol. Biol. Evol. 33, 2555–2564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buhler S., Nunes J. M., Sanchez-Mazas A., HLA class I molecular variation and peptide-binding properties suggest a model of joint divergent asymmetric selection. Immunogenetics 68, 401–416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altera I., Gragert L., Fingerson S., Maiers M., Louzoun Y., HLA class I haplotype diversity is consistent with selection for frequent existing haplotypes. PLoS Comput. Biol. 13, e1005693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain K., Seetharaman S., Multiple adaptive substitutions during evolution in novel environments. Genetics 189, 1029–1043 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gragert L., Madbouly A., Freeman J., Maiers M., Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Hum. Immunol. 74, 1313–1320 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Slater N., et al. , Power laws for heavy-tailed distributions: Modeling allele and haplotype diversity for the national marrow donor program. PLoS Comput. Biol. 11, e1004204 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Bakker P. I., et al. , A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 38, 1166–1172 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiby S. E., et al. , Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J. Exp. Med. 200, 957–965 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Oosterhout C., A new theory of MHC evolution: Beyond selection on the immune genes. Proc. Biol. Sci. 276, 657–665 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bronson P. G., Mack S. J., Erlich H. A., Slatkin M., A sequence-based approach demonstrates that balancing selection in classical human leukocyte antigen (HLA) loci is asymmetric. Hum. Mol. Genet. 22, 252–261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.