Significance
R-SNARE (soluble N-ethylmaleimide–sensitive factor receptor) and the set of 3Q-SNAREs, anchored in separate membranes, can associate in trans in a coiled-coil tetramer. Proteoliposomes bearing only these SNAREs form abundant trans-complexes but fuse very poorly, while membrane association through even a synthetic tether allows rapid and functional SNARE complex assembly without catalysis by Munc18 or Munc13 family proteins. Tethering guides trans-SNARE associations to active conformations instead of the inactive trans-SNARE complexes which form in the absence of tethers.
Keywords: membrane fusion, yeast vacuoles, SNARE, HOPS
Abstract
R-SNAREs (soluble N-ethylmaleimide–sensitive factor receptor), Q-SNAREs, and Sec1/Munc18 (SM)-family proteins are essential for membrane fusion in exocytic and endocytic trafficking. The yeast vacuolar tethering/SM complex HOPS (homotypic fusion and vacuole protein sorting) increases the fusion of membranes bearing R-SNARE to those with 3Q-SNAREs far more than it enhances their trans-SNARE pairings. We now report that the fusion of these proteoliposomes is also supported by GST-PX or GST-FYVE, recombinant dimeric proteins which tether by binding the phosphoinositides in both membranes. GST-PX is purely a tether, as it supports fusion without SNARE recognition. GST-PX tethering supports the assembly of new, active SNARE complexes rather than enhancing the function of the fusion-inactive SNARE complexes which had spontaneously formed in the absence of a tether. When SNAREs are more disassembled, as by Sec17, Sec18, and ATP (adenosine triphosphate), HOPS is required, and GST-PX does not suffice. We propose a working model where tethering orients SNARE domains for parallel, active assembly.
Intracellular membrane fusion requires conserved proteins and lipids (1). Rab-family GTPases bind effector complexes which tether membranes to initiate docking (2). SNARE (soluble N-ethylmaleimide–sensitive factor receptor) proteins often consist of an N domain, a characteristic heptad-repeat SNARE domain, and a C-terminal membrane anchor (3). SNARE domains assemble into a 4-subunit complex of coiled-coils with each α-helical SNARE domain burying a largely apolar face into the interior of the complex. Arginyl and glutaminyl residues, termed the 0-layer, form a polar center of this otherwise apolar interior. The SNARE domains associate in parallel, with their N termini at one end of the coiled-coil and their C termini at the other (4–6), although an antiparallel association has been demonstrated as well (7, 8). SNAREs are in four conserved families, termed R, Qa, Qb, and Qc, and SNARE complexes have an RQaQbQc composition (9). SNARE complexes are either in trans, when anchored to two apposed membranes before fusion, or in cis on one membrane after fusion. SNAREs can assemble spontaneously, but their assembly can also be catalyzed by Sec1/Munc18 (SM) proteins, alone or as part of a multisubunit protein (10–12). Synaptic membrane fusion uses SNAREs and SM proteins, but also has additional proteins (e.g., synaptotagmin, complexin, and Munc13) which confer parallel assembly of the four SNARE domains (8), Ca2+ regulation, and speed. The interplay between fusion proteins has received extensive study, yet central aspects of how they cooperate for fusion are unclear.
We study membrane fusion with vacuoles (lysosomes) from Saccharomyces cerevisiae. Vacuoles undergo fission and homotypic fusion. At steady state, many yeast strains have one to three large vacuoles which are readily visualized in the light microscope. Mutants which block fusion but allow unimpeded fission allow cell growth on rich media with strikingly small vacuole morphology and were termed vam mutants (13). The VAM genes encode all six subunits of the HOPS (homotypic fusion and vacuole protein sorting) complex, which is required for homotypic fusion and vacuole protein sorting, as well as the vacuolar Rab-family GTPase Ypt7 and two of the SNAREs. Fusion also requires several proteins that are shared with trafficking in the exocytic pathway; being essential for growth, they were not identified by the VAM screen. Vacuole fusion has been studied in vivo, with the isolated organelle in vitro, and with purified proteins reconstituted into proteoliposomes of physiological lipid composition and fluidity. Fusion requires Ypt7 (14) and its large hetero-hexameric effector complex HOPS, comprised of Vps11, 16, 18, 33, 39, and 41p subunits (15–17). The Vps33 subunit is the SM family member of the vacuole and has been shown to have conserved grooves which bind the R- and Qa-SNARE domains, in parallel and in register (10). Vps39 and Vps41 subunits of HOPS have direct affinity for Ypt7 (18), explaining how HOPS and Ypt7 on each membrane can suffice for membrane tethering (19). Vacuoles have an R-SNARE (Nyv1), Qa-SNARE (Vam3), Qb-SNARE (Vti1), and Qc-SNARE (Vam7), referred to hereafter as simply R, Qa, Qb, and Qc. Each has a C-terminal apolar transmembrane anchor except for Qc, which has an N-terminal PX domain which contributes to membrane binding by its affinities for PI3P (20) and for HOPS (21). As purified, vacuoles have SNAREs as 4-SNARE complexes, subcomplexes, and as membrane-anchored individual SNAREs, although Qc is only present as part of SNARE complexes (22). Fusion begins with ATP (adenosine triphosphate)-dependent disassembly of the cis-SNARE complexes on each vacuole by Sec18/NSF and Sec17/αSNAP (23), freeing the SNAREs (including Qc; ref. 24) for later trans-SNARE complex assembly. The initial association of primed vacuoles, termed tethering, is mediated by HOPS and Ypt7 on each membrane (19, 21). SNAREs then assemble into trans-complexes linking R-SNAREs on one membrane to QaQbQc (3Q) complexes on its fusion partner (25). Fusion is enhanced by Sec17 and Sec18 binding to trans-SNARE complexes (26–28), which may provide a bulky wedge (29) and facilitate complete SNARE zippering (28).
Despite extensive study, it remains unclear when trans-SNARE complex assembly is spontaneous and when it is catalyzed by SNARE-binding sites on HOPS, including its SM subunit Vps33. It has been shown that coincubation of proteoliposomes bearing Ypt7 and the R-SNARE with other proteoliposomes bearing Ypt7 and the 3Q-SNAREs supports substantial trans-SNARE pairing but only very slow fusion (30). When HOPS was added after 30 min, there was a sudden burst of fusion at ∼100 times the prior rate, although HOPS only increased the level of trans-SNARE complex two- to fourfold (30). Proteoliposomes bearing concentrated SNAREs (1:2,000 molar ratio to lipid) spontaneously assembled more trans-SNARE complex than was seen after HOPS addition to protoliposomes with dilute SNAREs (1:16,000 molar ratio to lipid), yet the latter had a far greater rate of fusion. It was not clear in this study whether HOPS was using the accumulated trans-SNARE pairs for fusion or whether the new SNARE complexes which HOPS elicited were more active. If the latter, was the capacity of HOPS for SNARE recognition and binding essential for it to catalyze the assembly of new, active trans-SNARE complexes?
We now report that rapid fusion of membranes bearing R-SNARE to those with 3Q-SNAREs requires neither catalysis of SNARE complex assembly nor activation of the accumulated spontaneous trans-SNARE complexes by a membrane tether. R- and 3Q-SNARE proteoliposomes fuse almost as well with optimal levels of the synthetic tether GST-PX as with HOPS, even though GST-PX lacks binding sites for SNAREs. GST-PX is a dimeric protein (31) which relies on each PX domain in the dimer binding to PI3P in each proteoliposome to support fusion. Without GST-PX, there is only very slow fusion of coincubated R- and 3Q-SNARE proteoliposomes, but addition of GST-PX triggers rapid fusion, as had been seen for HOPS. Fusion remains fully sensitive to competition by added sR, a soluble form of the R-SNARE without its membrane anchor, and thus this fusion uses SNARE complex assembled after the addition of the GST-PX tether rather than stable preassembled trans-SNARE complex. GST-PX does not replace HOPS in the presence of Sec17 and Sec18, SNARE chaperones which may inhibit (32) or promote (27, 28) fusion. Thus, tethering is required for efficient assembly of R with 3Q-SNAREs into a fusogenic structure, and HOPS or GST-PX provides such tethering. The SNARE-recognition functions of HOPS are not required for productively assembling R- with the preassembled complex of 3Q-SNAREs if the membranes are first tethered. Based on pioneering studies with neuronal SNAREs (7), we propose a working model of antiparallel SNARE assembly without prior membrane tethering and functional, parallel assembly when membranes are first tethered.
Results
Separate proteoliposome stocks were prepared with either of two luminal fluorescent protein markers, Cy5-streptavidin or biotinylated-phycoerythrin. Fusion mixes the luminal compartments (33), and the ensuing binding of biotin to streptavidin brings the Cy5 and phycoerythrin fluorophores into intimate contact, assayed by FRET (fluorescence resonance energy transfer). No fusion is seen without a tethering agent when the proteoliposomes bear R- or 3Q-SNAREs at a molar ratio to lipids of 1:16,000 (Fig. 1A). To test whether tethering per se would suffice for fusion, we employed a recombinant fusion between GST, the inherently dimeric (31) glutathione transferase, and a PX domain which binds phosphatidylinositol 3-phosphate. The GST-PX dimer could bind PI3P on the R- and 3Q-SNARE proteoliposomes to tether them together. Fusion is supported by the addition of GST-PX (Fig. 1A). Fusion only relies on the capacity of PX to recognize PI3P and not some inherent fusogenic capacity of the PX domain, as fusion is also seen with GST-FYVE (Fig. 1B). GST-PX fusion is ablated by the Y42A mutation in the PX domain (Fig. 2A), removing its PI3P affinity (20). Fusion relies on the presence of PI3P on both the R- and 3Q- bearing proteoliposomes (Fig. 2B). Liberation of the PX domain from GST dimer by thrombin cleavage (SI Appendix, Fig. S2D) removes its capacity to support fusion (Fig. 2C).
Fig. 1.
GST-PX supports fusion through tethering. (A and B) Fusion reactions contained proteoliposomes bearing R or QaQbQc (3Q) at 1:16,000 SNARE:lipid molar ratios. Mixed proteoliposomes were incubated as described (34), but without albumin, glutathione, or DTT with 0–500 nM GST-PX (A) or GST-FYVE (B) for 40 min at 27 °C, and content mixing was assayed by FRET between the luminal markers (33). Kinetic curves in this figure are representative of n ≥ 3 experiments; means and SDs for each experiment are presented in SI Appendix, Fig. S1.
Fig. 2.
Membrane tethering is essential for fusion. Fusion of proteoliposomes bearing R or 3Q at 1:16,000 molar ratio to lipid. (A) Incubations were performed with 500 nM GST-PX (WT), GST-PX(Y42A), or their buffer for 40 min at 27 °C. (B) Reactions had 500 nM GST-PX as in A, but with R- and 3Q-proteoliposomes with or without PI(3)P as indicated. (C) GST-PX has a thrombin cleavage sequence between its GST and PX domains. Cleavage by thrombin was terminated by adding PMSF (phenylmethyl sulfonylfluoride) as indicated. Fusion incubations were supplemented with: buffer alone (black), GST-PX which had been incubated for 2 h at 4 °C with 1 unit/mL thrombin followed by addition of 200 μM PMSF to block further thrombin activity (orange), GST-PX incubated for 2 h at 4 °C with thrombin that had been premixed with PMSF (green), GST-PX (blue), or GST-PX incubated with PMSF without thrombin (red). Kinetic curves of content-mixing assays in this figure are representative of n ≥ 3 experiments; means and SDs for each experiment are presented in SI Appendix, Fig. S2.
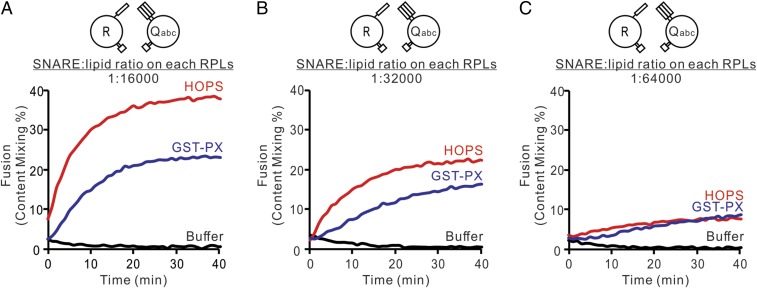
While GST-PX can tether membranes, it lacks the R- and Qa-SNARE domain recognition sites found in the Vps33 SM-family subunit of HOPS (10) as well as the HOPS affinity for Ypt7 (15); thus dimeric GST-PX has a purely tethering function. To compare the efficiency of fusion between R- and 3Q-proteoliposomes with optimal levels of HOPS or GST-PX, we prepared R- and 3Q-proteoliposomes with physiologically low 1:16,000, 1:32,000, and 1:64,000 molar ratio of SNAREs to lipid phosphorus and the HOPS receptor protein Ypt7 at 1:8,000 molar ratio to lipid (34). Optimal levels of HOPS and of GST-PX (SI Appendix, Fig. S4) gave comparable rates and extents of fusion (Fig. 3 and SI Appendix, Fig. S3), suggesting that HOPS largely functions here as a tether and that its SNARE recognition functions are not required in this intentionally simplified context, with the three preassembled Q-SNAREs and without Sec17 or Sec18.
Fig. 3.
GST-PX and HOPS are comparable tethers. (A–C) Fusion incubations had proteoliposomes bearing R or QaQbQc (3Q) at 1:16,000 (A), 1:32,000 (B), or 1:64,000 (C) SNARE:lipid molar ratios. Incubations were with 50 nM HOPS or 500 nM GST-PX for 40 min at 27 °C. All proteoliposomes had Ypt7-TM at a 1:8,000 protein:lipid molar ratio. Kinetic curves of content-mixing assays in this figure are representative of n ≥ 3 experiments; means and SDs for each experiment are presented in SI Appendix, Fig. S3.
trans-SNARE complex assembly can be competitively blocked by the addition of a soluble form of the R-SNARE, termed sR, which lacks its transmembrane anchor [Fig. 4, no sR (a, black) vs. with sR (b, gray); also ref. 35]. Ample trans-SNARE complexes have been shown to assemble during coincubation of R- and 3Q-SNARE proteoliposomes without tether (35); are these trans-SNARE complexes being used by the added GST-PX tether for fusion, or is fusion supported by the new trans-SNARE complexes assembled after GST-PX addition? R- and 3Q-SNARE proteoliposomes were preincubated together (Fig. 4, c, d; also see SI Appendix, Fig. S5) or separately (e, f) in the same buffer and at the same temperature for 30 min, then mixed with each other where separate and supplemented with buffer (c, e) or sR (d, f) immediately before GST-PX addition. Coincubation neither enhanced the kinetics of subsequent fusion after GST-PX addition (a vs. c) nor altered the sensitivity of the ensuing fusion to sR (b, d, and f), and thus fusion relied on the new trans-SNARE complexes rather than those which had been assembled before the addition of the tether.
Fig. 4.
Fusion induced by the GST-PX tether is from new trans-SNARE pairing. Fusion reactions had proteoliposomes bearing either the R or QaQbQc (3Q) at 1:16,000 SNARE:lipid molar ratio. Fusion incubations received 500 nM GST-PX at t = 0 min (a, b) or t = 30 min (c–f) with 1 μM soluble R-SNARE (sR) where indicated (b, d, and f). Content-mixing assays in this figure are representative of n ≥ 3 experiments; means and SDs for each experiment are presented in SI Appendix, Fig. S5.
Tethers Promote the Assembly of Functional trans-SNARE Pairs.
HOPS addition to Ypt7/R- and Ypt7/3Q proteoliposomes only increased trans-SNARE levels two- to fourfold (35), and the synthetic tether of dimeric GST-PX had a similar effect. With R- and 3Q-SNARE proteoliposomes at molar ratios of SNARE:lipid of 1:4,000, 1:8,000, and 1:16,000, we directly assayed the rates of fusion and the extent of association of R-SNARE with the Qa member of the 3Q complex in trans after incubation without any tether or with GST-PX (Fig. 5). There was a very slow linear rate of fusion in reactions with high SNARE concentrations (1:4,000 molar ratio to lipid) and without tether, but the fusion rate was higher with GST-PX (Fig. 5A). At each SNARE:lipid ratio, fusion was more rapid and more SNARE complex had assembled in the presence of GST-PX (Fig. 5 B and C, lanes 2, 4, and 6 vs. 1, 3, and 5). However, fusion was 23-fold more rapid for proteoliposomes with dilute SNAREs incubated with GST-PX than for proteoliposomes with concentrated SNAREs in the absence of GST-PX (Fig. 5B, lanes 6 vs. 1), while the latter actually had more trans-SNARE association than the former (Fig. 5C, lanes 1 vs. 6). The total SNARE complex measured here is the sum of the trans-SNARE complex, SNARE complex that had been trans but became cis as a result of fusion, and cis-SNARE complex which had never been trans but assembled after fusion from R- and Q-SNAREs which were brought into the same membrane by fusion. Thus, a greater portion of the total SNARE complex will be “trans” when there has been very little fusion (lane 1, no GST-PX) than when there has been fusion (lane 6, plus GST-PX), and the relative amount of truly trans-SNARE complex will be even less where more fusion had occurred (Fig. 5C, lane 6 relative to lane 1). The presence of even a synthetic tether therefore allows the assembly of trans-SNARE complexes which are far more functional.
Fig. 5.
Trans-SNARE complexes formed without a tether give far less fusion. (A) R- and 3Q-SNARE reconstituted proteoliposomes (RPLs) at 1:4,000, 1:8,000, or 1:16,000 SNARE:lipid molar ratio were incubated together without a tethering agent or with 500 nM GST-PX and assayed for fusion. (B) R- and 3Q-SNARE RPLs at SNARE:lipid molar ratios of 1:4,000, 1:8,000, or 1:16,000 were incubated together without tether (odd lanes) or with 500 nM GST-PX (even lanes) and assayed for the rate of fusion. (C) After a 10-min incubation, samples were solubilized in detergent and assayed for R bound to Qa as described (27). The signal intensity of condition 2 (1:4,000 proteoliposomes with 500 nM GST-PX) was normalized to 100%.
Is SNARE recognition by HOPS required in other contexts? GST-PX does not support the fusion of R- and 3Q-proteoliposomes in the presence of the physiological SNARE chaperones Sec17, Sec18, and ATP (Fig. 6 and SI Appendix, Fig. S6), which can disassemble trans-SNARE complexes (36) and block fusion without HOPS (37). Sec17, Sec18, and ATP are physiological factors which may largely keep SNAREs disassembled between cycles of docking and fusion, although the model reaction of fusion between liposomes bearing R-SNARE and 3Q-SNAREs has received extensive study. This model reaction, with preassembled 3Q-proteolipomes in the absence of Sec17 and Sec18, reveals that SNAREs which assemble spontaneously between untethered membranes are largely inactive for fusion, while it is tethering per se which permits the spontaneous assembly of fusion-competent trans-SNARE pairs.
Fig. 6.
HOPS is far better than GST-PX in fusion incubations with Sec17, Sec18, and ATP. Fusion reactions had proteoliposomes bearing Ypt7-TM at a molar ratio to lipid of 1:8,000 and either the R- or 3Q-SNAREs at 1:16,000 SNARE:lipid molar ratio. Fusion incubations received either buffer, 500 nM GST-PX, or 50 nM HOPS for 40 min at 27 °C (dotted lines) and Sec17 (300 nM), Sec18 (100 nM), and ATP (1 mM) where indicated (solid lines). Kinetic curves of content-mixing assays in this figure are representative of n ≥ 3 experiments; see SI Appendix, Fig. S6.
Discussion
SNAREs are essential for fusion. The fundamental discovery that R, Qa, Qb, and Qc SNAREs are complementary and can self-assemble in equimolar stoichiometry is a foundation for unraveling how Rabs, tethers, SM proteins, and SNARE chaperones cooperate with SNAREs and regulatory lipids for fusion. Do tethering factors merely spatially concentrate SNAREs by bringing membranes into close apposition to facilitate SNARE assembly in trans, and are SM proteins always required to catalyze rapid R and Qa SNARE assembly in trans? Our current findings show that tethering per se strikingly enhances the fusion potency of trans-SNARE complexes which form between R- and 3Q-SNARE complexes without further catalysis by SM protein functions. A working model of this is shown in Fig. 7. We suggest that the collision of membranes bearing R- and 3Q-SNAREs without prior tethering may commonly lead to antiparallel SNARE complexes (7) which do not support fusion, plus fewer parallel SNARE complexes which support the fusion seen at unphysiologically high SNARE levels (Fig. 7A). In contrast, the R- and 3Q-SNAREs on membranes with tethers (T in Fig. 7B) are oriented with all of their SNARE domain N termini distal to the bilayer and C termini proximal. Lateral movement of SNAREs along the surfaces of the bilayers of the two tethered membranes (Fig. 7B) will lead to SNARE collisions in which the SNARE domains are all parallel, giving parallel coiled-coil assemblies. It is possible that the initial, fusion-incompetent trans-SNARE complexes which form without the aid of a tether might then link the two membranes and thus fulfill the tethering function, allowing later trans-SNARE complexes to assemble in a fusion-competent conformation and thereby supporting the low rate of fusion seen with SNAREs alone. Direct assay of SNARE complex structure as formed in the presence or absence of tethers will be essential to test this model. This is technically challenging, as physiological levels of SNAREs are low, and trans-SNARE complexes are only a minor fraction of SNAREs (38) and are formed in the context of other proteins such as HOPS at the vertex ring domain where bilayers come into apposition (39).
Fig. 7.
Working model. (A) Membrane contact without prior tethering may give antiparallel trans-SNARE complexes which are inactive or even inhibitory. (B) Initial membrane contact by a tether (T) and lateral movement of oriented SNAREs in each membrane may favor parallel, active complex assembly. Some inactive antiparallel SNARE complexes, shown in A, may serve as a tether, allowing limited parallel SNARE complex assembly and slow fusion.
HOPS has direct affinity for vacuolar SNAREs (10, 21), but HOPS may bind SNAREs for purposes other than catalysis of R:Qa association. These might include 3Q assembly, SNARE complex protection from Sec17 inhibition (32), protection from disassembly by Sec17, Sec18, and ATP (36), or to provide protein bulk immediately adjacent to the trans-SNARE complexes at the fusion microdomain (29). Even at physiological 1:32,000 or 1:64,000 molar ratios of SNAREs to lipids (30), synthetic tethering by GST-PX gives comparable fusion to HOPS (Fig. 3), suggesting that there is little need for direct catalysis of functional R:3Q assembly once the 3Q-SNARE complex is assembled and tethering occurs.
Other pathways of SNARE assembly in which the Q-SNAREs are not preassembled may exploit the SM capacity to template R:Qa association. Munc18-1 has been shown by force microscopy (40) to catalyze the assembly of neuronal syntaxin (Qa) and synaptobrevin (R), forming an intermediate which can go on to full 4-SNARE domain complex upon association with SNAP-25 (Qb, Qc). In this same study, the recombinant Vps33 subunit of HOPS was shown to catalyze the assembly of vacuolar R- and Qa-SNAREs into a complex which can further assemble with Qb and Qc. In the context of membranes, SNARE complex formation does not always equate to fusion (Fig. 5; also ref. 30). Fusion can be reconstituted with proteoliposomes bearing R-SNARE and others bearing either one, two, or all 3Q-SNAREs plus hexameric HOPS and the remaining Q-SNAREs in soluble form without their membrane anchor (41). While HOPS supports each of these fusion reactions, fusion with Vps33 has not been reported. Further studies will be needed to establish conditions where HOPS templates the association of membrane-bound R- and Qa-SNAREs in trans as a favorable intermediate on the path to fusion.
Neuronal SNAREs can assemble in parallel or antiparallel, active or inactive, configurations (7, 8). Unc13 and Unc18 ensure the parallel assembly of the 3Q-SNARE domains of syntaxin and SNAP-25, and Unc13 further aligns the R-SNARE synaptobrevin (8). While there is no Unc13 for vacuoles, we now find that tethering aligns R- and 3Q-SNAREs for productive SNARE complex assembly, which we hypothesize to be a parallel conformation. Unc13 is also a tethering factor (42); perhaps this tethering activity per se favors parallel SNARE complex assembly in a similar manner to the GST-PX tether.
Experimental Procedures
Proteins and Reagents.
Most lipids were from Avanti Polar Lipids. Ergosterol was from Sigma, and PtdIns(3)P was from Echelon. Biotinylated R-phycoerythrin was purchased from Life Technologies, Cy5-streptavidin was from SeraCare, and underivatized streptavidin was from Thermo Fisher. Protein was assayed by the method of Bradford (43).
Protein expression and purification.
HOPS (19), vacuolar SNAREs (26, 33, 37, 44), GST-FYVE(Hrs) (45), and GST-PX(Vam7) (46) were isolated as described. Ypt7-TM, bearing the transmembrane anchor of Vam3, was derived from a construct kindly provided by Christian Ungermann, University of Osnabrück, Osnabrück, Germany.
Proteoliposome Preparation.
Proteoliposomes with mixed vacuolar lipids (VMLs) (37) were prepared as described (30) with or without Ypt7-TM at a 1:8,000 molar ratio to lipid and individual SNAREs at the indicated molar protein:lipid ratios. For VML (18:2) compositions, lipids in chloroform were mixed with β-OG in the following proportions: 47.6 mol % DLPC (1,2-Dioleoyl-sn-glycero-3-phosphocholine), 18 mol % DLPE (1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine), 18 mol % soy PI (l-α-phosphatidylinositol), 4.4 mol % DLPS (1,2-Dioleoyl-sn-glycero-3-phospho-l-serine), 2 mol % DLPA (1,2-Dioleoyl-sn-glycero-3-phosphate), 1 mol % 16:0 DAG (1,2-Dipalmitoyl-sn-glycerol), 8 mol % ergosterol, and 1 mol % PI(3)P [1,2-dipalmitoyl-sn-glycero-3-phospho-(1-myo-inositol-3-phosphate)].
Membrane Fusion Assay.
Fusion incubations (20 μL) bore Ypt7/R-SNARE proteoliposomes and Ypt7/Q-SNARE proteoliposomes (each 250 μM lipid) which had been separately preincubated for 10 min at 27 °C in RB150 [20 mM Hepes/NaOH, pH 7.4, 150 mM NaCl, 10% (vol/vol) glycerol], 5 μM streptavidin, 1 mM EDTA, and 10 μM GTP, then mixed with 1.25 mM MgCl2 to complete the exchange of Ypt7 to its GTP-bound form, required on the R-SNARE proteoliposomes for fusion (34). These preincubated proteoliposomes were mixed in equal parts, and 10-μL portions were placed in wells. A mixture of other proteins (10 μL; e.g., HOPS, GST-PX, GST-FYVE, Sec17, Sec18, and sSNAREs) were added at time 0. Content-mixing signals were measured in plates reader [PhycoE:Cy5 FRET: excitation (ex): 565 nm; emission (em): 670 nm; cutoff: 630 nm] every minute, using SpectraMax Gemini XPS (Molecular Devices) instrument. Maximal content mixing was measured by adding 0.2% (wt/vol) Thesit to parallel samples without streptavidin.
Coimmunoprecipitation.
R-SNARE, which coimmunoprecipitated with antibody to Qa, was measured as described (30). After assaying FRET for 30 min, samples were chilled, mixed with 10 volumes of 0 °C modified RIPA buffer [20 mM Hepes·NaOH, pH 7.4, 150 mM NaCl, 0.2% (wt/vol) BSA, 1% (vol/vol) Triton X-100, 1% (wt/vol) sodium cholate, 0.1% (wt/vol) SDS] bearing protein A magnetic beads (ThermoFisher) and 5 μM GST-R to prevent further association of wild-type R with Qa in the extract. After nutation (2 h, 23 °C), resin was thrice washed in 1 mL modified RIPA buffer. Proteins were recovered with 100 μL of SDS sample buffer (5 min, 95 °C). SDS/PAGE was followed by immunoblotting with antibody to R, Qa, or Qc. Bound protein was quantified by UN-SCAN-IT (Silk Scientific).
Supplementary Material
Acknowledgments
We thank Axel Brunger, Gustav Lienhard, and Charles Barlowe for stimulating discussions and Amy Orr for expert technical assistance. This work was supported by NIH grant R35GM118037.
Footnotes
Conflict of interest statement: W.W. and J.R. are coauthors on a 2017 Perspective article.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907640116/-/DCSupplemental.
References
- 1.Wickner W., Rizo J., A cascade of multiple proteins and lipids catalyzes membrane fusion. Mol. Biol. Cell 28, 707–711 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker R. W., Hughson F. M., Chaperoning SNARE assembly and disassembly. Nat. Rev. Mol. Cell Biol. 17, 465–479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jahn R., Scheller R. H., SNAREs—Engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7, 631–643 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Sutton R. B., Fasshauer D., Jahn R., Brunger A. T., Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Hanson P. I., Roth R., Morisaki H., Jahn R., Heuser J. E., Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell 90, 523–535 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Poirier M. A., et al. , The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat. Struct. Biol. 5, 765–769 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Weninger K., Bowen M. E., Chu S., Brunger A. T., Single-molecule studies of SNARE complex assembly reveal parallel and antiparallel configurations. Proc. Natl. Acad. Sci. U.S.A. 100, 14800–14805 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai Y., et al. , Molecular mechanisms of synaptic vesicle priming by Munc13 and Munc18. Neuron 95, 591–607.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fasshauer D., Sutton R. B., Brunger A. T., Jahn R., Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. U.S.A. 95, 15781–15786 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker R. W., et al. , A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349, 1111–1114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orr A., Song H., Rusin S. F., Kettenbach A. N., Wickner W., HOPS catalyzes the interdependent assembly of each vacuolar SNARE into a SNARE complex. Mol. Biol. Cell 28, 975–983 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parisotto D., et al. , An extended helical conformation in domain 3a of Munc18-1 provides a template for SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex assembly. J. Biol. Chem. 289, 9639–9650 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wada Y., Ohsumi Y., Anraku Y., Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. I. Isolation and characterization of two classes of vam mutants. J. Biol. Chem. 267, 18665–18670 (1992). [PubMed] [Google Scholar]
- 14.Haas A., Scheglmann D., Lazar T., Gallwitz D., Wickner W., The GTPase Ypt7p of Saccharomyces cerevisiae is required on both partner vacuoles for the homotypic fusion step of vacuole inheritance. EMBO J. 14, 5258–5270 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seals D. F., Eitzen G., Margolis N., Wickner W. T., Price A., A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. U.S.A. 97, 9402–9407 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wurmser A. E., Sato T. K., Emr S. D., New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 151, 551–562 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura N., Hirata A., Ohsumi Y., Wada Y., Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membranes and involved in the vacuolar assembly in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 272, 11344–11349 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Lürick A., et al. , Multivalent Rab interactions determine tether-mediated membrane fusion. Mol. Biol. Cell 28, 322–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hickey C. M., Wickner W., HOPS initiates vacuole docking by tethering membranes before trans-SNARE complex assembly. Mol. Biol. Cell 21, 2297–2305 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheever M. L., et al. , Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 3, 613–618 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Stroupe C., Collins K. M., Fratti R. A., Wickner W., Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 25, 1579–1589 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins K. M., Thorngren N. L., Fratti R. A., Wickner W. T., Sec17p and HOPS, in distinct SNARE complexes, mediate SNARE complex disruption or assembly for fusion. EMBO J. 24, 1775–1786 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayer A., Wickner W., Haas A., Sec18p (NSF)-driven release of Sec17p (α-SNAP) can precede docking and fusion of yeast vacuoles. Cell 85, 83–94 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Boeddinghaus C., Merz A. J., Laage R., Ungermann C., A cycle of Vam7p release from and PtdIns 3-P-dependent rebinding to the yeast vacuole is required for homotypic vacuole fusion. J. Cell Biol. 157, 79–89 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukuda R., et al. , Functional architecture of an intracellular membrane t-SNARE. Nature 407, 198–202 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Schwartz M. L., Merz A. J., Capture and release of partially zipped trans-SNARE complexes on intact organelles. J. Cell Biol. 185, 535–549 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zick M., Orr A., Schwartz M. L., Merz A. J., Wickner W. T., Sec17 can trigger fusion of trans-SNARE paired membranes without Sec18. Proc. Natl. Acad. Sci. U.S.A. 112, E2290–E2297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song H., Orr A., Duan M., Merz A. J., Wickner W., Sec17/Sec18 act twice, enhancing membrane fusion and then disassembling cis-SNARE complexes. eLife 6, 1–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Agostino M., Risselada H. J., Lürick A., Ungermann C., Mayer A., A tethering complex drives the terminal stage of SNARE-dependent membrane fusion. Nature 551, 634–638 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Zick M., Wickner W. T., A distinct tethering step is vital for vacuole membrane fusion. eLife 3, e03251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fabrini R., et al. , Monomer-dimer equilibrium in glutathione transferases: A critical re-examination. Biochemistry 48, 10473–10482 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Wang L., Ungermann C., Wickner W., The docking of primed vacuoles can be reversibly arrested by excess Sec17p (α-SNAP). J. Biol. Chem. 275, 22862–22867 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Zucchi P. C., Zick M., Membrane fusion catalyzed by a Rab, SNAREs, and SNARE chaperones is accompanied by enhanced permeability to small molecules and by lysis. Mol. Biol. Cell 22, 4635–4646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zick M., Wickner W., Improved reconstitution of yeast vacuole fusion with physiological SNARE concentrations reveals an asymmetric Rab(GTP) requirement. Mol. Biol. Cell 27, 2590–2597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zick M., Stroupe C., Orr A., Douville D., Wickner W. T., Membranes linked by trans-SNARE complexes require lipids prone to non-bilayer structure for progression to fusion. eLife 3, e01879 (2014). Correction in: eLife4, 08843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu H., Jun Y., Thompson J., Yates J., Wickner W., HOPS prevents the disassembly of trans-SNARE complexes by Sec17p/Sec18p during membrane fusion. EMBO J. 29, 1948–1960 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mima J., Hickey C. M., Xu H., Jun Y., Wickner W., Reconstituted membrane fusion requires regulatory lipids, SNAREs and synergistic SNARE chaperones. EMBO J. 27, 2031–2042 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collins K. M., Wickner W. T., Trans-SNARE complex assembly and yeast vacuole membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 104, 8755–8760 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L., Seeley E. S., Wickner W., Merz A. J., Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell 108, 357–369 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Jiao J., et al. , Munc18-1 catalyzes neuronal SNARE assembly by templating SNARE association. eLife 7, e41771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song H., Wickner W., A short region upstream of the yeast vacuolar Qa-SNARE heptad-repeats promotes membrane fusion through enhanced SNARE complex assembly. Mol. Biol. Cell 28, 2282–2289 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quade B., et al. , Membrane bridging by Munc13-1 is crucial for neurotransmitter release. eLife 8, e42806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bradford M. M., A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- 44.Thorngren N., Collins K. M., Fratti R. A., Wickner W., Merz A. J., A soluble SNARE drives rapid docking, bypassing ATP and Sec17/18p for vacuole fusion. EMBO J. 23, 2765–2776 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fratti R. A., Jun Y., Merz A. J., Margolis N., Wickner W., Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J. Cell Biol. 167, 1087–1098 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fratti R. A., Wickner W., Distinct targeting and fusion functions of the PX and SNARE domains of yeast vacuolar Vam7p. J. Biol. Chem. 282, 13133–13138 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.