Significance
To develop therapeutic strategies that enhance repair processes during central nervous system (CNS) injury, the molecular and cellular changes occurring at the injury site must be determined. However, precisely isolating the injury site, such as lesions in animal models of demyelination, has been difficult. We developed a simple and powerful method to track and analyze changes that occur within the lesion microenvironment over time. We demonstrated that neutral red-labeled lesions could be selectively isolated and processed for detailed analyses of transcripts, proteins, cell populations, and metabolites. The results of our study can be used to identify molecular pathways that promote CNS repair and the development of therapies to modulate neuroinflammation or enhance regeneration in demyelinating disorders such as multiple sclerosis.
Keywords: remyelination, demyelination, inflammation, animal models, vital dye
Abstract
Animal models of central nervous system (CNS) demyelination, including toxin-induced focal demyelination and immune-mediated demyelination through experimental autoimmune encephalomyelitis (EAE), have provided valuable insights into the mechanisms of neuroinflammation and CNS remyelination. However, the ability to track changes in transcripts, proteins, and metabolites, as well as cellular populations during the evolution of a focal lesion, has remained challenging. Here, we developed a method to label CNS demyelinating lesions by the intraperitoneal injection of a vital dye, neutral red (NR), into mice before killing. We demonstrate that NR-labeled lesions can be easily identified on the intact spinal cord in both lysolecithin- and EAE-mediated demyelination models. Using fluorescence microscopy, we detected NR in activated macrophages/microglia and astrocytes, but not in oligodendrocytes present in lesions. Importantly, we successfully performed RT-qPCR, Western blot, flow cytometry, and mass spectrometry analysis of precisely dissected NR-labeled lesions at 5, 10, and 20 d postlesion (dpl) and found differential changes in transcripts, proteins, cell populations, and metabolites in lesions over the course of remyelination. Therefore, NR administration is a simple and powerful method to track and analyze the detailed molecular, cellular, and metabolic changes that occur within the lesion microenvironment over time following CNS injury. Furthermore, this method can be used to identify molecular and metabolic pathways that regulate neuroinflammation and remyelination and facilitate the development of therapies to promote repair in demyelinating disorders such as multiple sclerosis.
The mechanisms of central nervous system (CNS) injury and repair have been extensively studied to identify strategies that promote regenerative processes and prevent neurological decline. In multiple sclerosis (MS), CNS lesions of the white matter, caused by repeated immune-mediated attacks and destruction of myelin, result in neurodegeneration and progressive disability (1–4). In the early stage of MS, endogenous CNS repair through remyelination takes place following demyelination and involves the recruitment, proliferation, and differentiation of oligodendrocyte precursor cells (OPCs) into myelin-producing oligodendrocytes (5–8). However, at the later, progressive stage, this regenerative process fails, with most lesions remaining demyelinated, leading to chronic axonal dysfunction and clinical deterioration (9–13). The mechanisms that lead to remyelination failure in MS remain unknown.
To understand the biology of remyelination and develop strategies to enhance this process in MS, several models of experimental demyelination are employed (14–19). One of the widely used approaches is focal injection of lysolecithin into the CNS white matter of rodents (15). The advantages of this model include robust remyelination, tractable lesion location, and the established time course of lesion evolution (20–23). However, the identification of a lesion site is limited to traditional methods such as histological or immunostaining and in situ hybridization on fixed tissue sections and precludes selective dissection of the entire lesion for quantitative analysis of transcripts, proteins, metabolites, and cell populations. A reliable strategy to rapidly identify and selectively dissect lesions from an intact unfixed tissue would significantly contribute to our understanding of the biochemical, molecular, and cellular composition of lesions over time.
Here, we developed a technique using neutral red (NR), a vital dye, to label CNS lesions in vivo in mouse models of experimental demyelination. NR incorporates into live cells and has a preference for cellular compartments with low intracellular pH, such as lysosomes in macrophages (24–26). We reasoned that NR would label areas of CNS injury in the live animal due to the enrichment of macrophages/microglia at lesion sites (27–30). We show that a single intraperitoneal (i.p.) injection of NR into experimentally demyelinated mice 2 h before sacrifice resulted in specific intense red labeling of demyelinated lesions on the intact CNS tissue during remyelination. NR injection successfully labeled focal lesions in lysolecithin-demyelinated spinal cord and corpus callosum, as well as multiple diffuse lesions in mice with experimental autoimmune encephalomyelitis (EAE). Taking advantage of its natural fluorescent characteristic, we detected NR staining in immunolabeled activated macrophage/microglia cells and a subpopulation of reactive astrocytes in demyelinated lesions. Importantly, we show that NR-labeled lesions can be selectively dissected out from unfixed CNS tissues, without the need for tissue sectioning, and processed for flow cytometry, RT-qPCR, and Western blot analyses. Furthermore, mass spectrometry analysis of NR-labeled lesions revealed a significant increase in several amino acids (AAs) and metabolites following demyelination, and their subsequent reduction during remyelination. This newly developed strategy will allow comprehensive analysis of the biochemical, molecular, and cellular composition of lesions over time, which could uncover novel mechanisms in immunoregulation and remyelination following CNS injury.
Results
Detection of NR in CNS Lesion following Demyelinating Injury.
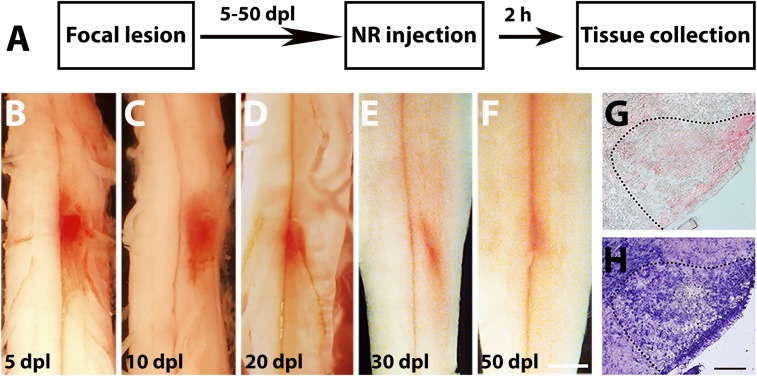
To detect and track a lesion site for quantitative analyses of microenvironmental changes occurring over the course of CNS injury, we developed a method using NR labeling. NR is a vital dye (31, 32), which has a preference for incorporating into cellular compartments with a low pH and has been used to determine cell viability and cytotoxicity (25, 26, 32). We hypothesized that administering NR into mice after spinal cord focal demyelination would result in the preferential staining of the demyelinated lesion in vivo, due to the enrichment of activated macrophages/microglia and astrocytes with high lysosomal content at the site (30, 33, 34). To determine whether NR selectively labels the lesion site, focal lysolecithin-induced demyelination was induced in the mouse ventral spinal cord white matter, and a single i.p. injection of the dye was performed 2–3 h before killing. No obvious adverse reactions were observed in mice after NR administration. The spinal cords were collected at 5, 10, 20, 30, and 50 d postlesion (dpl, Fig. 1A). These time points correspond to increased inflammation and OPC recruitment at 5 dpl, inflammation resolution and oligodendrocyte differentiation at 10 dpl, and remyelination completion after 20 dpl (22, 23, 35). We found that NR was highly visible on the intact ventral spinal cord in expected demyelinated lesion sites at 5, 10, and 20 dpl (Fig. 1 B–D). Moreover, no NR staining was detected outside of the lesion or in nonlesioned spinal cord tissue (Fig. 1 B–F). We observed a small area of weak NR staining at 30 and 50 dpl, suggesting complete inflammation resolution and lesion repair at these time points (Fig. 1 E and F). To confirm that NR labels demyelinated lesions, we generated cryosections of NR-labeled tissues. We detected NR labeling in the ventral white matter where the focal lesion was expected (Fig. 1G). The staining intensity was generally weaker in thin tissue sections (12 μm thick) compared with intact tissue. Compared with toluidine blue staining of adjacent tissue sections, we found that NR highlighted the same area of lesion as the toluidine blue stain, suggesting that NR labeled the demyelinated lesion boundary (Fig. 1 G and H). Therefore, NR injection shortly before tissue collection allows the rapid and precise detection of the lesion site.
Fig. 1.
Focal CNS lesion identified by NR labeling. (A) A schematic of experimental paradigm of a spinal cord lesion induced by lysolecithin injection and labeled with NR 2 h before killing at 5–50 dpl. (B–F) Bright-field representative images of NR-labeled lesion and unlabeled nonlesion tissue (n = 4–6 animals per time point examined) on the intact spinal cord at 5 (B), 10 (C), 20 (D), 30 (E), and 50 dpl (F). (G and H) Adjacent sections showing NR (G) and toluidine blue (H) staining of demyelinated area (dotted line). (Scale bars, B–F, 2 mm; G and H, 100 µm.)
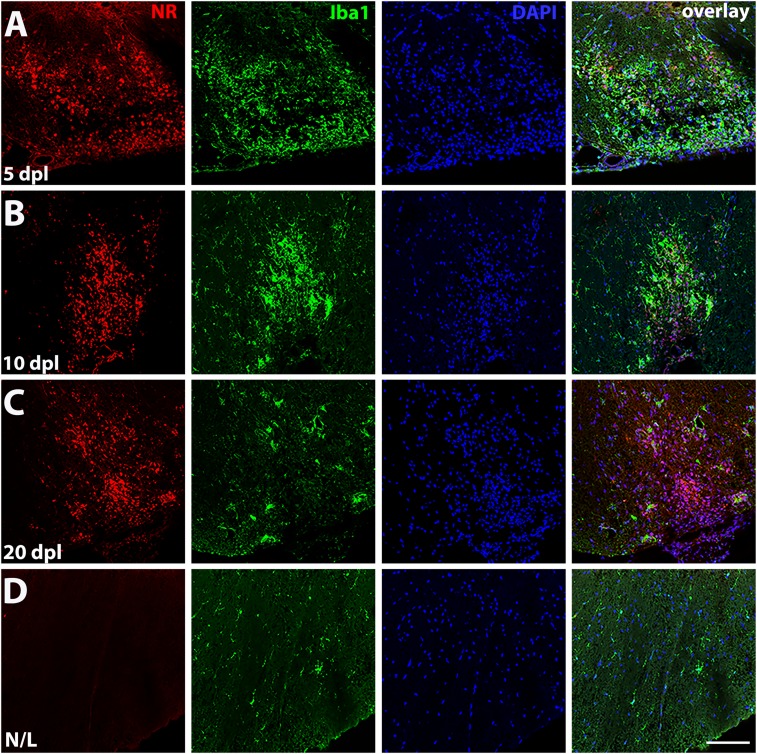
Previous reports have shown that NR can be used as a fluorescent probe with an excitation spectrum of 450–550 nm and a wide emission spectrum with the maximum range from 550 to 650 nm (31, 36). We utilized NR fluorescent properties combined with immunohistochemistry to identify labeled cell populations in lesions. We found that NR displayed intense fluorescence in the lesion at 5, 10, and 20 dpl, and mainly colocalized with activated macrophages/microglia labeled by the Iba1 marker (Figs. 2 A–C and 3C), whereas no NR labeling was detected in Iba1+ resting microglia with ramified appearance in the adjacent nonlesioned spinal cord (Figs. 2D and 3C). We further examined different types of macrophages/microglia labeled by NR in the lesion at 5 dpl, using iNOS for proinflammatory macrophages, CD68 for phagocytic macrophages, arginase 1 (Arg1) for antiinflammatory macrophages, and P2Y12 for resting microglia (SI Appendix, Fig. S1). We observed NR colocalization in iNOS+ (SI Appendix, Fig. S1A), CD68+ (SI Appendix, Fig. S1B), and Arg1+ (SI Appendix, Fig. S2A) macrophages/microglia, but not in P2Y12+ resting microglia (SI Appendix, Fig. S1C). We further analyzed Arg1 immunostaining at 10 and 20 dpl and found decreased numbers of Arg1+ cells at these time points, while NR staining was still observed in the lesion (SI Appendix, Fig. S2 B and C). To further determine that NR dye labels the original lesion area during remyelination, we performed myelin basic protein (MBP) immunostaining at 5, 10, and 20 dpl (SI Appendix, Fig. S3 A–C). We found that NR labeling remained detectable at the lesion, as confirmed by clustered DAPI labeling, and covered the remyelinating area with MBP staining at 20 dpl (SI Appendix, Fig. S3C).
Fig. 2.
Fluorescence detection of NR in demyelinating lesion. Representative confocal images showing NR fluorescence (red) colocalized with Iba1 (green), a marker of activated macrophages/microglia in a lesion, detected by enhanced DAPI staining (blue) at 5 (A), 10 (B), and 20 dpl (C) (n = 3 animals per time point examined). (D) An adjacent nonlesion side (N/L) shows no NR labeling in resting macrophages/microglia detected by Iba1 (green). (Scale bar, 100 μm.)
Fig. 3.
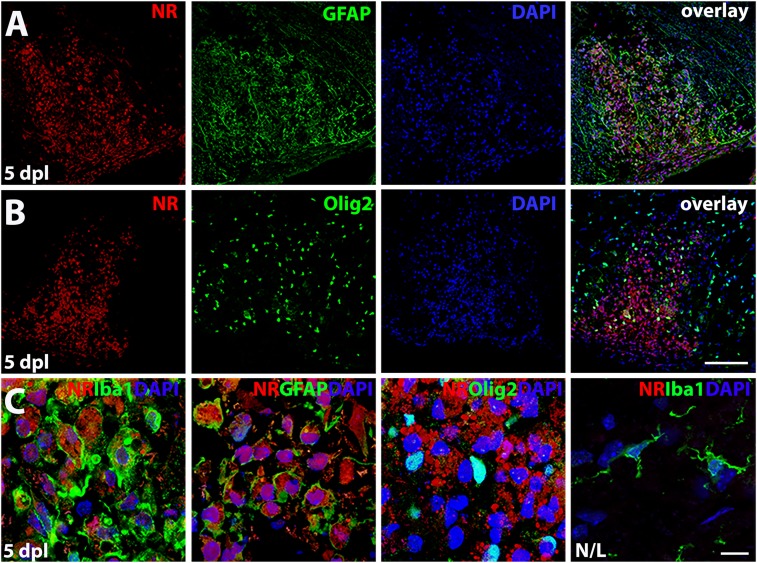
NR labels macrophages/microglia and reactive astrocytes but not oligodendrocytes. (A) NR fluorescence (red) and immunostaining (green) for reactive astrocyte marker GFAP or oligodendrocyte lineage cells, Olig2 (B) at 5 dpl. (C) High magnification images show intracellular NR staining in Iba1+ and GFAP+ cells. No NR labeling was detected in Olig2+ cells in lesion or Iba1+ resting microglia in nonlesion tissue at 5 dpl (n = 3 animals examined). (Scale bars, B, 100 µm; C, 10 µm.)
At 5 dpl, we also observed NR colabeling in a subpopulation of activated (or reactive) astrocytes marked by GFAP (Fig. 3A). The number of activated astrocytes was significantly decreased at 10 and 20 dpl, while NR staining remained visible in the lesion at these time points (SI Appendix, Fig. S4 B and C). At 5 dpl, NR-labeled astrocytes displayed a foamy cell morphology (Fig. 3C), compared with the fibrous-appearing astrocytes found in the lesions at later time points without the NR label. Moreover, we did not find any NR-labeled oligodendrocyte lineage cells marked by Olig2 in lesions, suggesting that NR does not incorporate into oligodendrocyte lineage cells (Fig. 3 B and C). The intense intracellular labeling in activated macrophages/microglia and reactive astrocytes likely corresponds to increased lysosomal activity in lesions. Moreover, the lack of NR labeling in oligodendrocyte lineage cells might relate to their higher intracellular pH (37) and lower lysosomal content (Fig. 3C). Hence, NR can be used for fluorescent detection of the lesion site and immunohistological analysis of the various cell populations present at different stages of lesion evolution.
Transcriptional and Protein Analyses of NR-Labeled Lesions.
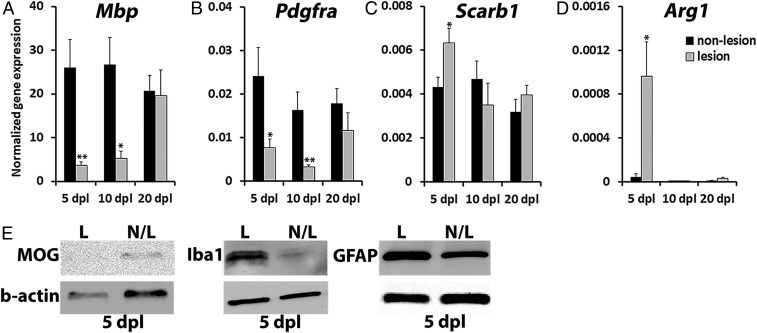
A major challenge in evaluating biochemical, molecular, and cellular changes during remyelination has been the difficulty to precisely isolate lesions from unfixed tissues. To characterize the molecular composition of lesions, NR-labeled lesions and adjacent unlabeled white matter tissue were selectively dissected from unfixed spinal cord and subjected to quantitative analyses of transcripts and proteins. RT-qPCR analysis of purified messenger RNA (mRNA) from isolated spinal cord tissues revealed a sevenfold reduction in gene expression for myelin basic protein (Mbp) at 5 dpl in NR-labeled lesions compared with adjacent nonlesioned tissues (n = 4–6 per group; P = 0.006, Fig. 4A), corresponding to the depletion of myelin in lesions after lysolecithin injection. However, Mbp expression in lesions gradually increased from 5 to 20 dpl, corresponding to increased remyelination in lesions over time (Fig. 4A). Interestingly, the expression of Pdgfrα, a marker of OPCs, was reduced by threefold compared with nonlesioned tissue at 5 dpl (n = 6 per group, P = 0.039, Fig. 4B), despite the expectation that OPC numbers would increase in lesions at this time point (23, 35). Pdgfrα expression was further decreased by 10 dpl, corresponding to the differentiation of recruited OPCs into oligodendrocytes as previously reported, and returned to control levels by 20 dpl (Fig. 4B). These results suggest that under the highly inflammatory demyelinating environment at 5 dpl, Pdgfra expression in OPCs may be lower than expected, but returns to homeostatic levels in the lesion upon myelin repair by 20 dpl.
Fig. 4.
Differential mRNA and protein expression in NR-labeled lesion. NR-labeled lesion and adjacent nonlesion tissue at 5, 10, and 20 dpl were dissected out and processed for RT-qPCR (A–D) or protein (E) analyses (n = 6–8 animals per time point examined). The expression of oligodendrocyte-specific genes, Mbp (A) and Pdgfrα (B), was normalized to B2m housekeeping gene; macrophage-specific Scarb1 (C) and Arg1 (D) mRNA levels were normalized to Ppia. (E) The amount of myelin oligodendrocyte glycoprotein (MOG), macrophage/microglia marker Iba1, and astrocyte marker GFAP in NR-labeled lesion (L) and unlabeled nonlesion (N/L) tissue at 5 dpl detected by Western blot. β-actin was used for loading control. Data presented as mean ± SEM. *P < 0.05, **P < 0.01.
We also examined the expressions of Scarb1 and Arg1, which are markers of activated macrophages/microglia, and found that both transcripts were significantly increased in NR-labeled lesions compared with nonlesioned white matter at 5 dpl (n = 6–8, P = 0.027, and P = 0.03) and returned to control levels by 10 dpl (Fig. 4 C and D). This result suggests that inflammation resolution has occurred by this time point and is consistent with published reports showing differential mRNA expression in lesions at different stages of remyelination (23, 35, 38–40).
To determine if changes in protein levels associated with remyelination and inflammation could be assessed in dissected NR-labeled lesions, we performed Western blot analysis on NR-labeled and adjacent nonlabeled tissue for the expression of myelin oligodendrocyte glycoprotein (MOG), Iba1, and glial fibrillary acidic protein (GFAP). We observed reduced MOG expression and increased Iba1 and GFAP expression in NR-labeled tissues compared with nonlabeled tissues at 5 dpl (Fig. 4E), consistent with the loss of oligodendrocytes and increase in activated macrophages/microglia and reactive astrocytes in the lesion at this time point (Fig. 4 A–D). Taken together, these data demonstrate that NR labeling allows for the quantitative assessment of lesion composition and can be used to characterize molecular signals over the course of CNS remyelination.
Flow Cytometry Analysis of Cell Populations in NR-Labeled Lesions.
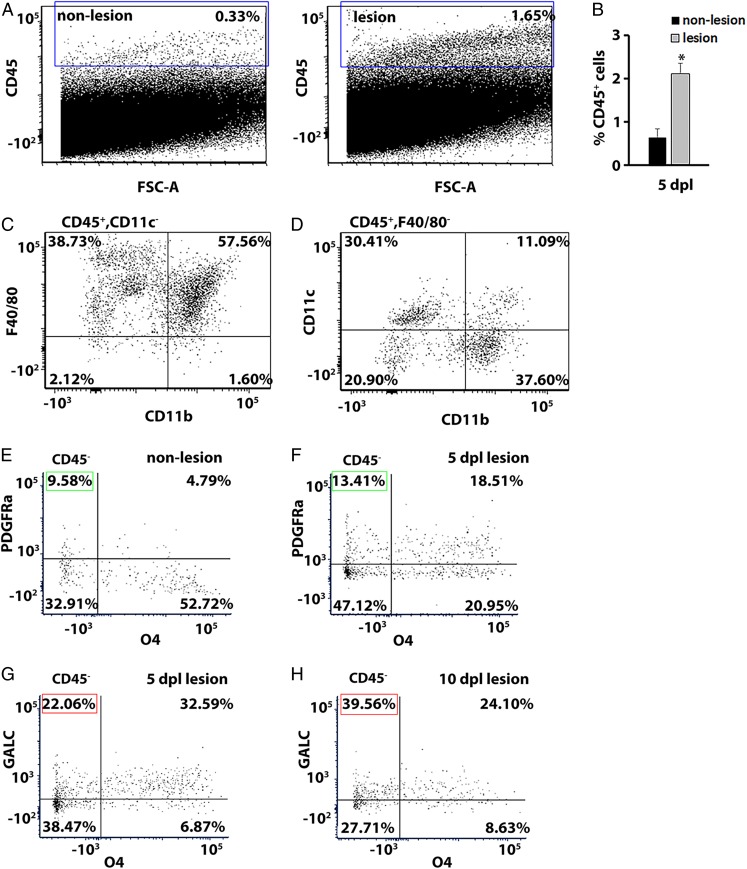
The evolution of a remyelinating lesion consists of dynamic shifts in cell populations from abundant proinflammatory macrophages/microglia and recruitment of OPCs during the initial stages of remyelination to inflammation resolution and increased oligodendrocyte differentiation at the later stages of myelin repair (2, 5, 7, 9, 12, 27, 39, 41). Since NR is delivered a few hours before killing, this method is not expected to induce cell toxicity and affect the inflammatory microenvironment or remyelination efficiency (Figs. 2–4). To identify cell populations present in the lesion, flow cytometry analysis was performed on dissected NR-labeled and adjacent nonlabeled tissue. The cells were dissociated and stained with anti-CD45, a marker of leukocytes, at 5 dpl. Flow cytometry analysis revealed a fivefold increase in the percentage of CD45+ cells in NR-labeled lesions compared with control (2.12 ± 0.23% NR labeled vs. 0.64 ± 0.2% nonlabeled; n = 3, paired t test P = 0.013; Fig. 5 A and B), consistent with increased inflammation at this time point. Among the CD45+CD11c− cell population present in the NR-labeled lesion at 5 dpl, the majority (57.56%) corresponded to F4/80+/CD11b+ macrophages (Fig. 5C). Moreover, among the CD45+ F4/80− cells, we detected mostly CD11b−CD11c+ lymphoid dendritic cells (30.41%) and CD11b+CD11c+ myeloid dendritic cells (37.60%) (Fig. 5D).
Fig. 5.
Flow cytometry analysis of immune cell populations in the CNS lesion. (A) Representative plots of flow cytometry analysis showing the percentage of CD45+ cells in NR-nonlabeled control (0.33%) and NR-labeled lesion (1.65%) tissue. (B) Quantification of CD45+ cells in the lesion compared with adjacent unlabeled nonlesion tissue at 5 dpl. Data are presented as mean ± SEM. *P < 0.05. (C) Representative plots showing a population of F4/80+ CD11b+ macrophage cells among the cells gated on CD45+ CD11c−, found in the lesion at 5 dpl. (D) Representative plots showing that among CD45+ F4/80− cells, CD11b− CD11c+ lymphoid dendritic cells and CD11b+ CD11c+ myeloid dendritic cells were detected in the lesion at 5 dpl. (E–H) Representative plots showing a population of CD45− oligodendrocytes labeled by Pdgfra (progenitor marker) and O4 (intermediate marker) in nonlesion (E) and at 5 dpl in lesion tissue (F). Green box indicates OPC population (Pdgfra+O4−). The percentage of mature oligodendrocytes was assessed by GALC at 5 dpl (G) and 10 dpl (H). Red box indicates mature oligodendrocytes (GALC+O4−).
To identify different populations of oligodendrocyte lineage cells present in the lesion, we further analyzed CD45− population using specific markers for different stages of oligodendrocyte maturation: Pdgfrα for OPCs, O4 for late progenitors/newly differentiated oligodendrocytes, and GALC for mature oligodendrocytes (42). Consistent with previously published studies, we observed an increase in OPCs marked by Pdgfrα (Pdgfrα+O4−) in the lesion at 5 dpl compared with nonlesion tissue (9.58% in nonlesion vs. 13.41% in lesion tissue, Fig. 5 E and F, respectively). Moreover, we found that the number of newly differentiated oligodendrocytes in the lesion (Pdgfrα−O4+) was reduced (52.72% in nonlesion tissue vs. 20.95% in the lesion, Fig. 5 E and F, respectively). Furthermore, we observed an increase in mature oligodendrocytes (GALC+O4−) in the lesion at 10 dpl, indicating oligodendrocyte differentiation and remyelination (22.06% at 5 dpl vs. 39.56% at 10 dpl, Fig. 5 G and H, respectively). These results demonstrate that we could successfully isolate OPCs and oligodendrocytes at various stages of differentiation in the NR-labeled lesion. Thus, the NR-labeling method can be used to quantify diverse cell populations present in the lesion and changes in cellular composition during remyelination.
Changes in Amino Acid and Metabolic Profiles during Remyelination.
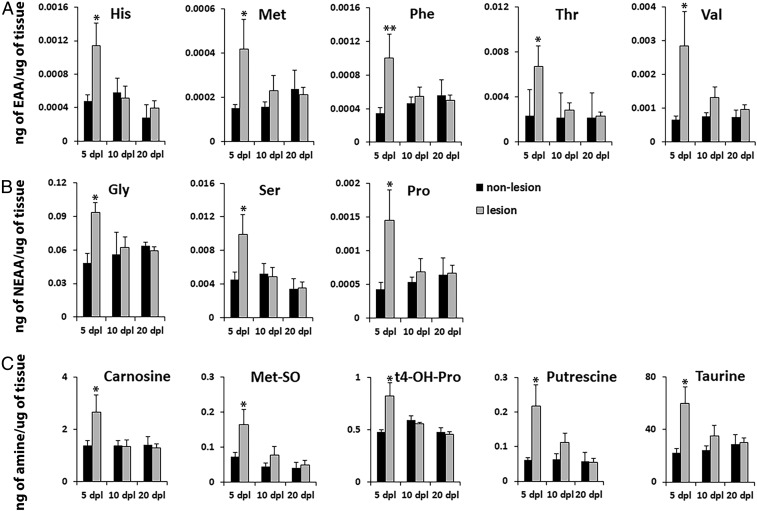
Metabolic profiling can be used to elucidate biochemical processes associated with cellular state and activity (43–45). To examine changes in AAs and other metabolites during remyelination, we performed mass spectrometry analysis of dissected NR-labeled lesions and adjacent nonlesioned tissue (Fig. 6). The NR dye has a distinct peak on a chromatogram of signal intensities among various metabolites, thus does not interfere with the identification of metabolites, making it suitable for mass spectrometry. We examined 21 AAs and found that 5 essential AAs, including histidine (His), methionine (Met), phenylalanine (Phe), threonine (Thr), and valine (Val) as well as 3 nonessential AAs, glycine (Gly), proline (Pro), and serine (Ser), were significantly increased in lesions at 5 dpl compared with adjacent nonlesioned tissue from the same spinal cord (n = 6–9 animals per group, P = 0.039 for His; P = 0.023 for Met; P = 0.0057 for Phe; P = 0.027 for Thr; P = 0.037 for Val; P = 0.048 for Gly; P = 0.026 for Pro; P = 0.047 for Ser; Fig. 6 A and B). Moreover, AA levels were reduced to baseline during remyelination at 10 and 20 dpl (Fig. 6 A and B). Corroborating our mRNA and protein profiles, which show up-regulation of inflammatory markers at 5 dpl (Fig. 4), these results suggest that increased AA levels in the lesion may fuel CNS inflammation during demyelination, while reduction in AAs might be necessary to promote inflammation resolution and remyelination.
Fig. 6.
Mass spectrometry generated metabolomics of the lesion environment during remyelination. NR-labeled lesion and adjacent unlabeled nonlesion tissue (n = 6–9 animals per group) were dissected out at 5, 10, and 20 dpl and processed for mass spectrometry of 21 AAs and 21 biogenic amines, using ultraperformance liquid chromatography electrospray ionization–tandem mass spectrometry analysis. Five essential AAs (A), 3 nonessential AAs (B), and 5 biogenic amines (C) were significantly increased in NR lesion compared with nonlesioned tissue at 5 dpl during active inflammation. All metabolite AAs and metabolite levels in NR lesion were reduced to control levels at 10 and 20 dpl during remyelination. Data are presented as mean ± SEM. **P < 0.01, *P < 0.05.
Following demyelinating injury, the lesion site undergoes metabolic changes that influence cell reprogramming and differentiation, leading to oligodendrocyte maturation and myelin production (45, 46). We performed mass spectrometry analysis measuring the levels of 21 biogenic amines in NR-labeled lesions vs. adjacent nonlesion tissue and identified several metabolites that were significantly increased in the lesion (Fig. 6C). Similar to the high level of AAs, such as Met and Pro, in lesions at 5 dpl (Fig. 6 A and B), we observed increased concentrations of methionine sulfoxide (Met-SO, P = 0.048, Fig. 6C), which is the oxidized form of Met and is considered a biomarker of oxidative stress, and hydroxyproline (t4-OH-Pro, P = 0.019, Fig. 6C), a derivative of Pro and a major component of collagen. The levels of putrescine, a product of AA breakdown, were also significantly up-regulated at 5 dpl (P = 0.021, Fig. 6C). We also detected an increase in the levels of taurine and carnosine in lesions (P = 0.012 and P = 0.046, respectively) at 5 dpl (Fig. 6C). Interestingly, taurine and carnosine have been previously shown to be synthesized and released by oligodendrocytes and suggested to regulate their differentiation (45, 47, 48). Together, these results show that NR-labeled lesions can be subjected to metabolomic studies, which can provide insights into biochemical changes in the lesion environment over the course of remyelination.
NR Labeling in Other Experimental Models of Demyelination.
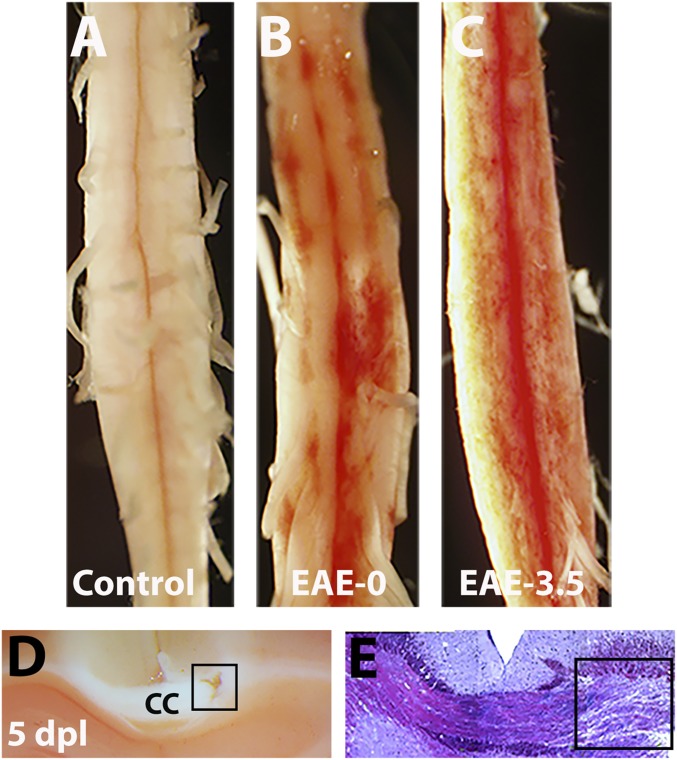
We next determined if NR labeling can be used in other models of experimental demyelination. EAE is an autoimmune-mediated inflammatory condition in the CNS that is characterized by demyelination and axonal injury and shares several key pathophysiological features of MS (16, 18). Mice that develop EAE display a typical profile of disease progression, in which a clinical score of 0.0 represents no obvious changes in motor function, while a score of 3.0–3.5 is given to animals that show complete hind limb paralysis, which usually appears by 16–18 d after EAE induction. To determine whether NR can also label lesions in the EAE model, we administered NR into mice with MOG-induced EAE at scores 0.0 or 3.5. As a control, NR was injected into wild-type mice without EAE induction. We found that in contrast to control mice that did not exhibit NR labeling (Fig. 7A), mice with EAE showed multiple sites of focal NR labeling throughout the ventral spinal cord at as early as score 0.0, demonstrating inflammatory cell activation days before the development of clinical disability (Fig. 7B). Additionally, we found the number of lesions and intensity of NR labeling increased in mice with a clinical score of 3.5 and appeared more widely distributed throughout the spinal cord (Fig. 7C). These results suggest that enhanced inflammation in the spinal cord contributed to exacerbated demyelinating injury and clinical severity. Moreover, the diffuse nature and variable intensity of NR labeling in lesions confirms the difficulty to reliably track remyelination in standard EAE models without a focal injury approach (49).
Fig. 7.
NR labels demyelinating lesion in the brain and EAE. (A–C) Representative images showing NR labeling of spinal cord from control mice without EAE (A), or with EAE at clinical score 0.0 (B) and 3.5 (C). Note that NR is not detectable in mice without EAE (control) but is detected in mice with EAE before clinical deficit. (D and E) NR labels lysolecithin-induced lesion (square) in the corpus callosum at 5 dpl. Bright field images showing NR (D) and toluidine blue staining (E) of demyelinated area.
To determine whether NR can label demyelinated lesions in the brain, stereotaxic injection of lysolecithin into the mouse corpus callosum was performed (50, 51), followed by NR injection before sacrifice at 5 dpl. In tissue sections, NR labeling could be detected in corpus callosum lesions (Fig. 7D), corresponding to lysolecithin-induced demyelination as confirmed by toluidine blue staining (Fig. 7E). These results demonstrate that NR labeling can be used to identify inflammatory lesions in mice following demyelinating injury and may be used to characterize the CNS inflammation and remyelination status in functional or therapeutic studies.
To determine if NR can label possible developmental damage of white matter tracts, we performed LPS injection to induce inflammatory responses in the young mice (postnatal day 14). This method led to the presence of NR-labeled macrophages/microglia marked by Iba1 immunoreactivity (SI Appendix, Fig. S5). However, we found that the number of these NR+ cells was low and sparsely located throughout the corpus callosum. Thus, using NR labeling in this model, or other models where the immune cell infiltration is diffuse and the lesion size is very small, may be challenging compared with the robust staining of lesions induced by focal toxin injection or EAE.
Discussion
The difficulty of rigorously tracking and quantifying the global metabolomic, molecular, and cellular changes that occur in lesions over the course of remyelination presents a major challenge in understanding the dynamic activities and shifts in glial and immune cell populations during lesion evolution. Previously, laser capture microdissection was one of the few methods available for selective lesion isolation from the unfixed tissue and has been used to track transcriptional and proteomic changes in tissue sections (35, 52). However, this technique is time consuming and costly, and may not yield adequate tissue quantity and quality for Western blots, flow cytometry, or metabolic analyses. Our newly developed method of labeling lesions with NR vital dye presents a major advantage in the field of experimental demyelination and CNS injury, as we now can precisely locate lesion site in fixed and unfixed tissue, which can be used for detailed biochemical, cellular, and molecular analyses.
In this study, we successfully performed immunohistochemistry, RT-qPCR, Western blot, and flow cytometry on NR-labeled lesions, confirming the corresponding profiles of remyelination stages in mice. We determined that NR can be used as a fluorescent probe to outline the lesion site and utilized its fluorescence characteristics to perform double labeling with several cellular markers previously found in the lesion. NR’s wide excitation and emission spectrums, ranging from 450 to 550 nm and from 500 to 700 nm, respectively, have to be considered for colocalization studies in fixed tissue (31, 36). However, a destaining procedure may be used to decrease background and reduce cross-talk between NR and other fluorescent probes (32). Our immunohistochemistry results showed that NR labels activated macrophages/microglia and reactive astrocytes, which are recruited to the lesion site for the clearance of myelin and cellular debris and are part of immunomodulatory mechanism after demyelination (27, 30, 41, 53). In contrast, NR is not detected in oligodendrocytes in lesions, or in cells in nonlesioned CNS tissues, thus showing selective labeling of proinflammatory cell types in demyelinated lesions.
The NR-labeling approach allows the selective isolation of lesions from freshly dissected, unfixed spinal cords for quantitative analyses of transcripts, proteins, metabolites, and cell populations. Consistent with previous reports (23, 35), we found that at 5 dpl the genes and proteins associated with inflammation were highly expressed, whereas the oligodendrocyte-specific markers were reduced at this stage. This pattern reverses when the lesion environment progresses to active remyelination stages at 10 and 20 dpl. Since NR does not interfere with the quality of mRNA isolation, this approach can be further used to generate comprehensive profiles of transcripts associated with inflammation and remyelination under normal and pathological conditions. Moreover, flow cytometry analysis of cell populations from NR-labeled lesion demonstrates enrichment in macrophages, lymphoid, and myeloid dendritic cells at 5 dpl. Importantly, we show that NR labeling is applicable to isolating and characterizing oligodendrocyte lineage cells at various stages of differentiation present in the lesion and enables tracking progression of remyelination associated with oligodendrocyte maturation in the lesion. These results provide a useful platform for further assessment of different cell populations in NR-labeled lesions.
Following NR administration in lysolecithin demyelination model, we observed an intense red staining at 5–20 dpl. We found that the lesion size remains detectable throughout the repair process (up to 20 dpl) because sufficient NR-labeled cells are detectable to outline the lesion boundary despite known reduction in inflammatory macrophages at 20 dpl. However, NR labeling was dramatically reduced and barely detectable by 30 or 50 dpl. These results suggest that NR will label the lesion site during the repair process but that the staining will diminish upon inflammation resolution and the completion of repair. Therefore, future studies would be necessary to confirm whether NR-labeling correlates with lesion size under various treatment strategies. In contrast to the easy detection and precise dissection of focal lesions labeled by NR, the isolation of individual NR-labeled lesions from intact spinal cords of mice with severe EAE (disability score 3.0–3.5) might be challenging as they display multiple sites of diffuse lesions throughout the entire spinal cord. It may be possible to isolate focal lesions from presymptomatic or less affected mice, since multiple sites of focal lesions could be detected throughout the ventral spinal cord at score 0 after EAE induction. However, examining individual lesions in this model may not provide meaningful insights due to the heterogeneity of lesion severity and the likely variability of remyelination status in each lesion. Importantly, NR labeling in EAE allows the gross assessment of the extent of inflammation in CNS lesions and may be applied to drug studies in multiple EAE models, including cortical or callosal inflammation in more aggressive EAE forms (54). Together, the NR-labeling approach would be highly valuable for future immunomodulatory drug studies in EAE and serve as an important complement to the analysis of protein/RNA/cellular composition in inflamed CNS tissues.
Using mass spectrometry analysis, we examined the levels of AAs and metabolites that might play a role in cell differentiation and reprogramming. We found significantly increased levels of several essential AAs at 5 dpl, suggesting that regulation of AA metabolism in demyelinated lesions may play a role in controlling the function and activity of proinflammatory cell types, such as T cell activation by phenylalanine (55). Another AA, methionine, and its metabolite, methionine sulfoxide, were also increased in the lesion at 5 dpl and have been previously shown to induce M1/classical macrophage activation, which is related to proinflammatory responses characterized by increased iNOS activity and TNF-α release (56). Interestingly, we also detected metabolites important for oligodendrocyte differentiation in NR-labeled lesion. For instance, it has been shown that carnosine is primarily synthesized by oligodendrocytes and the rate of its synthesis correlates with the differentiation of these cells in vitro (47, 48). Previous reports have also found that carnosine scavenges free radicals, inhibits inflammation in the CNS, and protects against ischemic and hypoxic brain damage through its antioxidative and antiapoptotic actions (57–59). Another metabolite, taurine, has also been shown to play a role in oligodendrocyte differentiation and suggested to increase the serine pool necessary for lipid biogenesis during remyelination (45, 60). Therefore, NR labeling could be used for the detailed biochemical, molecular, and cellular analyses of CNS lesions over time, revealing novel targets for developing therapeutic approaches to promote myelin repair. Currently, MS treatment aims to reduce inflammatory relapses but does not prevent irreversible neurological decline due to remyelination failure (61–63). This strategy could be combined with functional studies to uncover mechanisms of immunoregulation and remyelination and used to investigate drug targets for CNS repair.
Materials and Methods
Animals.
C57BL/6J mice of both sexes were purchased from The Jackson Laboratory and Charles River and were maintained on a 12-h/12-h light/dark cycle with food and water ad libitum. All experiments were performed in accordance with approved Institutional Animal Care and Use Committee protocols of Georgetown University.
Focal Spinal Cord Demyelination and NR Labeling.
Focal demyelination was induced by injecting 1% lysolecithin (Sigma-Aldrich) in PBS into the spinal cord ventral funiculus of 8- to 12-wk-old C57BL/6J mice of either sex as previously described (22, 64). At 5, 10, 20, 30, and 50 dpl, mice were i.p. injected with 500 µL of 1% NR (Sigma-Aldrich) in PBS for spinal cord lesion labeling and perfused intracardially with PBS or 4% paraformaldehyde (Sigma-Aldrich) (for immunohistochemistry) 2–3 h later. The NR-labeled lesion was detected on the ventral side of the freshly dissected spinal cord. Dissected labeled and nonlabeled tissue from the opposite site of a spinal cord were immediately processed or stored at −80 °C.
Immunohistochemistry.
Immunohistochemistry was performed as previously reported (23, 64) with modifications described in SI Appendix, Material and Methods. Before immunohistochemistry, sections were destained to reduce NR intensity with a solution of 50% ethanol/1% glacial acetic acid (Sigma-Aldrich) for 10 min as previously described (32). Sources and dilutions of primary antibodies were as follows: rabbit anti-Iba1 (1:400, Wako), rabbit anti-Olig2 (1:300, Millipore), mouse anti-GFAP (1:400, Sigma).
RNA Extraction, cDNA Synthesis, and RT-qPCR.
Total RNA was isolated using the TRIzol Reagent protocol (Life Technologies) and reverse transcribed into cDNA as described in SI Appendix, Material and Methods. Results were normalized against peptidylprolyl isomerase A (Ppia) and β-2-microglobulin (B2m) and were expressed as mean ± SEM. Ppia and B2m are recommended normalization factors for gene expression studies (65).
Flow Cytometry.
NR-labeled lesion and adjacent unlabeled nonlesion sites from the same spinal cord were dissected at 5 dpl and processed for flow cytometry as described in SI Appendix, Material and Methods. Cells were analyzed on the flow cytometer at the Georgetown Lombardi Comprehensive Cancer Center Flow Cytometry and Cell Sorting Shared Resource.
Western Blot.
NR-labeled lesion and unlabeled nonlesion sites were dissected from spinal cords at 5, 10, and 20 dpl as described above. The tissue was lysed in RIPA buffer (Millipore), separated by SDS/PAGE, and immunoblotted using the following antibodies: rabbit anti-Iba1 (1:500; Novus), rabbit anti-MOG (1:200; Santa Cruz Biotechnology), and rabbit β-actin (1:5,000 Abcam) for loading control. Proteins were detected using horseradish peroxidase-conjugated secondary antibodies and Pierce ECL Western blotting substrate.
Mass Spectrometry Analysis.
NR-labeled lesion and unlabeled nonlesion sites were dissected from spinal cords at 5, 10, and 20 dpl and processed for mass spectrometry analysis at the Proteomics & Metabolomics Facility at Georgetown University as described in SI Appendix, Material and Methods.
EAE.
EAE was induced using the EAE kit (Hooke Laboratories, catalog no.: EK-2110) according to the Hooke Laboratories protocol (http://hookelabs.com/protocols/eaeAI_C57BL6.html). The mice were scored according to the protocol from Hooke Laboratories and as described in SI Appendix, Material and Methods.
Corpus Callosum Lesion Using Lysolecithin.
Lysolecithin-induced demyelination of corpus collosum was performed as previously reported (50, 51) and described in SI Appendix, Material and Methods.
Statistical Analysis.
All statistics were performed using GraphPad Prism 6. Data are represented as mean ± SEM. Data significance was determined using 2-tailed Student’s t tests, or 1- and 2-way ANOVA with Tukey’s range test for post hoc analysis. EAE clinical score significance was determined using 1-way Wilcoxon Rank-Sum Test. Statistical significance is reported as *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Supplementary Material
Acknowledgments
This project was supported by a Harry Weaver Neuroscience Scholars Fellowship JF-1806-31381 from the National Multiple Sclerosis Society, NIH Grant 1R01NS107523-01, and Congressionally Directed Medical Research Programs/Department of Defense Grant W81XWH-17-1-0268 (to J.K.H.). We thank the Proteomics & Metabolomics Facility at Georgetown University, Dr. Amrita Cheema, and Khyati Mehta for their assistance with mass spectrometry analysis; Dr. Karen Creswell and Dan Xun for their assistance with flow cytometry; and all members of the J.K.H. laboratory for comments on this project.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819343116/-/DCSupplemental.
References
- 1.Compston A., Coles A., Multiple sclerosis. Lancet 359, 1221–1231 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Lassmann H., van Horssen J., Mahad D., Progressive multiple sclerosis: Pathology and pathogenesis. Nat. Rev. Neurol. 8, 647–656 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Dutta R., Trapp B. D., Relapsing and progressive forms of multiple sclerosis: Insights from pathology. Curr. Opin. Neurol. 27, 271–278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reich D. S., Lucchinetti C. F., Calabresi P. A., Multiple sclerosis. N. Engl. J. Med. 378, 169–180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franklin R. J., Ffrench-Constant C., Remyelination in the CNS: From biology to therapy. Nat. Rev. Neurosci. 9, 839–855 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Bradl M., Lassmann H., Oligodendrocytes: Biology and pathology. Acta Neuropathol. 119, 37–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J. K., et al. , Myelin regeneration in multiple sclerosis: Targeting endogenous stem cells. Neurotherapeutics 8, 650–658 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franklin R. J., ffrench-Constant C., Edgar J. M., Smith K. J., Neuroprotection and repair in multiple sclerosis. Nat. Rev. Neurol. 8, 624–634 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Franklin R. J., Why does remyelination fail in multiple sclerosis? Nat. Rev. Neurosci. 3, 705–714 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Irvine K. A., Blakemore W. F., Remyelination protects axons from demyelination-associated axon degeneration. Brain 131, 1464–1477 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Bramow S., et al. , Demyelination versus remyelination in progressive multiple sclerosis. Brain 133, 2983–2998 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Chamberlain K. A., Nanescu S. E., Psachoulia K., Huang J. K., Oligodendrocyte regeneration: Its significance in myelin replacement and neuroprotection in multiple sclerosis. Neuropharmacology 110, 633–643 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trapp B. D., Nave K. A., Multiple sclerosis: An immune or neurodegenerative disorder? Annu. Rev. Neurosci. 31, 247–269 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Kornek B., et al. , Multiple sclerosis and chronic autoimmune encephalomyelitis: A comparative quantitative study of axonal injury in active, inactive, and remyelinated lesions. Am. J. Pathol. 157, 267–276 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blakemore W. F., Franklin R. J., Remyelination in experimental models of toxin-induced demyelination. Curr. Top. Microbiol. Immunol. 318, 193–212 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Constantinescu C. S., Farooqi N., O’Brien K., Gran B., Experimental autoimmune encephalomyelitis (EAE) as a model for multiple sclerosis (MS). Br. J. Pharmacol. 164, 1079–1106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Procaccini C., De Rosa V., Pucino V., Formisano L., Matarese G., Animal models of multiple sclerosis. Eur. J. Pharmacol. 759, 182–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinman L., Zamvil S. S., Virtues and pitfalls of EAE for the development of therapies for multiple sclerosis. Trends Immunol. 26, 565–571 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Gallo V., Deneen B., Glial development: The crossroads of regeneration and repair in the CNS. Neuron 83, 283–308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeffery N. D., Blakemore W. F., Remyelination of mouse spinal cord axons demyelinated by local injection of lysolecithin. J. Neurocytol. 24, 775–781 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Huang J. K., Jarjour A. A., Ffrench-Constant C., Franklin R. J., Retinoid X receptors as a potential avenue for regenerative medicine in multiple sclerosis. Expert Rev. Neurother. 11, 467–468 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Keough M. B., Jensen S. K., Yong V. W., Experimental demyelination and remyelination of murine spinal cord by focal injection of lysolecithin. J. Vis. Exp., e52679 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Psachoulia K., et al. , IL4I1 augments CNS remyelination and axonal protection by modulating T cell driven inflammation. Brain 139, 3121–3136 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohkuma S., Poole B., Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc. Natl. Acad. Sci. U.S.A. 75, 3327–3331 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weeks B. A., Keisler A. S., Myrvik Q. N., Warinner J. E., Differential uptake of neutral red by macrophages from three species of estuarine fish. Dev. Comp. Immunol. 11, 117–124 (1987). [DOI] [PubMed] [Google Scholar]
- 26.Herbomel P., Thisse B., Thisse C., Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev. Biol. 238, 274–288 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Kotter M. R., Setzu A., Sim F. J., Van Rooijen N., Franklin R. J., Macrophage depletion impairs oligodendrocyte remyelination following lysolecithin-induced demyelination. Glia 35, 204–212 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Donnelly D. J., Gensel J. C., Ankeny D. P., van Rooijen N., Popovich P. G., An efficient and reproducible method for quantifying macrophages in different experimental models of central nervous system pathology. J. Neurosci. Methods 181, 36–44 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.David S., Kroner A., Repertoire of microglial and macrophage responses after spinal cord injury. Nat. Rev. Neurosci. 12, 388–399 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Cao L., He C., Polarization of macrophages and microglia in inflammatory demyelination. Neurosci. Bull. 29, 189–198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen G., Hanson C. L., Ebner T. J., Optical responses evoked by cerebellar surface stimulation in vivo using neutral red. Neuroscience 84, 645–668 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Repetto G., del Peso A., Zurita J. L., Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Protoc. 3, 1125–1131 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Mason J. M., et al. , IL-4-induced gene-1 is a leukocyte L-amino acid oxidase with an unusual acidic pH preference and lysosomal localization. J. Immunol. 173, 4561–4567 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Vogel D. Y., et al. , Macrophages in inflammatory multiple sclerosis lesions have an intermediate activation status. J. Neuroinflammation 10, 35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang J. K., et al. , Retinoid X receptor gamma signaling accelerates CNS remyelination. Nat. Neurosci. 14, 45–53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dubrovsky J. G., et al. , Neutral red as a probe for confocal laser scanning microscopy studies of plant roots. Ann. Bot. 97, 1127–1138 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boussouf A., Gaillard S., Intracellular pH changes during oligodendrocyte differentiation in primary culture. J. Neurosci. Res. 59, 731–739 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Mycko M. P., Papoian R., Boschert U., Raine C. S., Selmaj K. W., Microarray gene expression profiling of chronic active and inactive lesions in multiple sclerosis. Clin. Neurol. Neurosurg. 106, 223–229 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Kotter M. R., Li W. W., Zhao C., Franklin R. J., Myelin impairs CNS remyelination by inhibiting oligodendrocyte precursor cell differentiation. J. Neurosci. 26, 328–332 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mycko M. P., Brosnan C. F., Raine C. S., Fendler W., Selmaj K. W., Transcriptional profiling of microdissected areas of active multiple sclerosis lesions reveals activation of heat shock protein genes. J. Neurosci. Res. 90, 1941–1948 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Miron V. E., et al. , M2 microglia and macrophages drive oligodendrocyte differentiation during CNS remyelination. Nat. Neurosci. 16, 1211–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson A. P., Rodgers J. M., Goings G. E., Miller S. D., Characterization of oligodendroglial populations in mouse demyelinating disease using flow cytometry: Clues for MS pathogenesis. PLoS One 9, e107649 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaur P., et al. , Quantitative metabolomic and lipidomic profiling reveals aberrant amino acid metabolism in type 2 diabetes. Mol. Biosyst. 9, 307–317 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Altadill T., et al. , Enabling metabolomics based biomarker discovery studies using molecular phenotyping of exosome-like vesicles. PLoS One 11, e0151339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beyer B. A., et al. , Metabolomics-based discovery of a metabolite that enhances oligodendrocyte maturation. Nat. Chem. Biol. 14, 22–28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao C., Li W. W., Franklin R. J., Differences in the early inflammatory responses to toxin-induced demyelination are associated with the age-related decline in CNS remyelination. Neurobiol. Aging 27, 1298–1307 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Hoffmann A. M., Bakardjiev A., Bauer K., Carnosine-synthesis in cultures of rat glial cells is restricted to oligodendrocytes and carnosine uptake to astrocytes. Neurosci. Lett. 215, 29–32 (1996). [DOI] [PubMed] [Google Scholar]
- 48.Bakardjiev A., Carnosine and beta-alanine release is stimulated by glutamatergic receptors in cultured rat oligodendrocytes. Glia 24, 346–351 (1998). [PubMed] [Google Scholar]
- 49.Merkler D., Ernsting T., Kerschensteiner M., Brück W., Stadelmann C., A new focal EAE model of cortical demyelination: Multiple sclerosis-like lesions with rapid resolution of inflammation and extensive remyelination. Brain 129, 1972–1983 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Yamazaki R., Baba H., Yamaguchi Y., Unconventional myosin ID is involved in remyelination after cuprizone-induced demyelination. Neurochem. Res. 43, 195–204 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Sahel A., et al. , Alteration of synaptic connectivity of oligodendrocyte precursor cells following demyelination. Front. Cell. Neurosci. 9, 77 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han M. H., et al. , Proteomic analysis of active multiple sclerosis lesions reveals therapeutic targets. Nature 451, 1076–1081 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Miron V. E., Franklin R. J., Macrophages and CNS remyelination. J. Neurochem. 130, 165–171 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Mangiardi M., et al. , An animal model of cortical and callosal pathology in multiple sclerosis. Brain Pathol. 21, 263–278 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strasser B., Sperner-Unterweger B., Fuchs D., Gostner J. M., Mechanisms of inflammation-associated depression: Immune influences on tryptophan and phenylalanine metabolisms. Curr. Top. Behav. Neurosci. 31, 95–115 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Dos Santos L. M., et al. , Methionine and methionine sulfoxide treatment induces M1/classical macrophage polarization and modulates oxidative stress and purinergic signaling parameters. Mol. Cell. Biochem. 424, 69–78 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Berezhnoy D. S., et al. , Carnosine as an effective neuroprotector in brain pathology and potential neuromodulator in normal conditions. Amino Acids 51, 139–150 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Haus J. M., Thyfault J. P., Therapeutic potential of carbonyl-scavenging carnosine derivative in metabolic disorders. J. Clin. Invest. 128, 5198–5200 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mirzakhani N., et al. , Carnosine improves functional recovery and structural regeneration after sciatic nerve crush injury in rats. Life Sci. 215, 22–30 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Rosko L., Smith V. N., Yamazaki R., Huang J. K., Oligodendrocyte bioenergetics in health and disease. Neuroscientist 10.1177/1073858418793077 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang J. K., Franklin R. J., Regenerative medicine in multiple sclerosis: Identifying pharmacological targets of adult neural stem cell differentiation. Neurochem. Int. 59, 329–332 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Compston A., The basis for treatment in multiple sclerosis. Acta Neurol. Scand. Suppl. 183, 41–47 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Plemel J. R., Liu W. Q., Yong V. W., Remyelination therapies: A new direction and challenge in multiple sclerosis. Nat. Rev. Drug Discov. 16, 617–634 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Chamberlain K. A., Chapey K. S., Nanescu S. E., Huang J. K., Creatine enhances mitochondrial-mediated oligodendrocyte survival after demyelinating injury. J. Neurosci. 37, 1479–1492 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gong Z. K., et al. , Identification and validation of suitable reference genes for RT-qPCR analysis in mouse testis development. Mol. Genet. Genomics 289, 1157–1169 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.