Significance
Environmental water deficiency and osmotic stress have been proposed to trigger the opening of osmolality-sensitive OSCA channels, leading to downstream signaling cascades necessary for abiotic stress resistance. We report the structural and functional analysis of OSCA1.2 from the crop plant rice. By combining biochemical, biophysical, and computational studies, we derive a model of how OSCA1.2 could mediate transport pathway gating under osmotic stress. Structure and functional analyses provide a molecular framework for studying proposed mechanosensing mechanisms in plants.
Keywords: osmotic stress, channel, structure, cryo-EM, rice
Abstract
Sensing and responding to environmental water deficiency and osmotic stresses are essential for the growth, development, and survival of plants. Recently, an osmolality-sensing ion channel called OSCA1 was discovered that functions in sensing hyperosmolality in Arabidopsis. Here, we report the cryo-electron microscopy (cryo-EM) structure and function of an OSCA1 homolog from rice (Oryza sativa; OsOSCA1.2), leading to a model of how it could mediate hyperosmolality sensing and transport pathway gating. The structure reveals a dimer; the molecular architecture of each subunit consists of 11 transmembrane (TM) helices and a cytosolic soluble domain that has homology to RNA recognition proteins. The TM domain is structurally related to the TMEM16 family of calcium-dependent ion channels and lipid scramblases. The cytosolic soluble domain possesses a distinct structural feature in the form of extended intracellular helical arms that are parallel to the plasma membrane. These helical arms are well positioned to potentially sense lateral tension on the inner leaflet of the lipid bilayer caused by changes in turgor pressure. Computational dynamic analysis suggests how this domain couples to the TM portion of the molecule to open a transport pathway. Hydrogen/deuterium exchange mass spectrometry (HDXMS) experimentally confirms the conformational dynamics of these coupled domains. These studies provide a framework to understand the structural basis of proposed hyperosmolality sensing in a staple crop plant, extend our knowledge of the anoctamin superfamily important for plants and fungi, and provide a structural mechanism for potentially translating membrane stress to transport regulation.
Hyperosmolarity and osmotic stress are among the first physiological responses to changes in salinity and drought. Hyperosmolality triggers increases in cytosolic free Ca2+ concentration and thereby initiates an osmotic stress-induced signal transduction cascade in plants (1–4). Salinity and drought stress trigger diverse protective mechanisms in plants enabling enhanced drought tolerance and reduction of water loss in leaves.
Ion channels have long been hypothesized as sensors of osmotic stress. A candidate membrane protein named OSCA was isolated in a genetic screen for mutants that impair the rapid osmotic stress-induced Ca2+ elevation in plants (1). Arabidopsis thaliana OSCA1 (AtOSCA1) encodes a multispanning membrane protein that was reported to function in osmotic/mechanical stress-induced activation of ion currents. However, the underlying mechanisms and whether AtOSCA1 homologs include an ion-conducting Ca2+-permeable pore require further analysis. AtOSCA1 is a member of a larger gene family in Arabidopsis with 15 members (5), and with many homologs encoded in other plants and fungal genomes. Furthermore, evolutionary analyses have revealed that OSCA is distantly related to the anoctamin (ANO) superfamily, which includes the TMEM16 family of calcium-dependent ion channels and lipid scramblases (6).
Results
As we were interested in determining whether and how osmolality caused OSCA proteins to respond to osmotic stress in crop plants, we screened 5 such putative OSCA channels from rice, overexpressing them as tobacco etch virus (TEV) protease cleavable enhanced green fluorescent protein (GFP) fusions in Pichia pastoris. The Oryza sativa gene (annotated as OsOSCA1.2, GenBank KJ920372.1) was found to have both high levels of protein expression and desirable properties during purification, and was therefore chosen for further characterization. We purified the membrane protein to homogeneity (SI Appendix, Fig. S1A) and determined the oligomeric state of purified OSCA using size-exclusion chromatography coupled to multiangle laser light scattering (SEC-MALLS) analysis, revealing the detergent-solubilized protein to be a dimer (SI Appendix, Fig. S1B).
Functional Reconstitution of OsOSCA1.2.
Reconstitution of the OsOSCA1.2 purified proteins into droplet interface bilayers (DIBs) indicated the purified protein can mediate ion transport in this bilayer system (SI Appendix, Fig. S1 C–H). OsOSCA1.2 was active in symmetrical (cis/trans 150:150 mM KCl) ionic conditions in the absence of any other osmotically active solutes (SI Appendix, Fig. S1 C–E). In symmetrical ionic conditions, the current-to-voltage relation for OsOSCA1.2 was quite linear, yielding a unitary conductance of 284 ± 2 pS and showing no signs of current rectification. The unitary conductance is within the range of those reported recently for other reconstituted OSCA proteins (e.g., between 300 and 350 pS) in similar ionic conditions (5). Under nonsymmetrical ionic conditions (cis/trans 15:150 mM KCl), with an inwardly directed K+ gradient, the inward single-channel currents reversed (Erev) at about −26 mV, closer to the Nernst potential for K+ (EK+: −54 mV after correction for ionic activities), indicating a modest selectivity for K+ over Cl− as suggested by the calculated PK+/PCl− = 5 ± 1 (SI Appendix, Fig. S1I). The appearance of an infrequent 50% current amplitude subconductance state (SI Appendix, Fig. S1H) may be consistent with the proposed assembly of 2 cooperative subunits as confirmed by the dimeric nature of the OsOSCA1.2 protein inferred from SEC-MALLS (SI Appendix, Fig. S1B). The result is also consistent with other recent studies of OSCA proteins (7, 8). Overall, the above experiments suggest a transport function of the purified OsOSCA1.2 protein.
Structure of OsOSCA1.2.
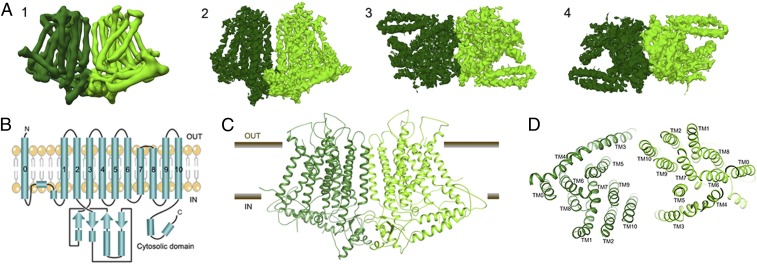
We determined a molecular structure of OsOSCA1.2 by single-particle cryo-electron microscopy (cryo-EM) to an overall resolution of 4.9 Å and local resolution in the membrane of 4.5 Å, revealing a dimer of C2 symmetry-related subunits (SI Appendix, Fig. S2). The overall dimensions of the protein are 140 Å × 55 Å × 85 Å. Each protomer is composed of 11 transmembrane (TM) spanning segments, associated extracellular and intracellular loops, and an intracellular soluble domain (Fig. 1A). All 11 TM helices and the soluble domain are well resolved in our cryo-EM maps, and large side chains provided suitable markers for ensuring proper sequence registration during atomic model building. (SI Appendix, Fig. S3). The final atomic model comprises 1,388 of the expected 1,424 residues with good geometry and an EMRinger (9) score of 0.89 (SI Appendix, Table S1).
Fig. 1.
Cryo-EM structure of the OsOSCA1.2 protein. (A, Left to Right) (1) Parallel to membrane plane view of unsharpened cryo-EM density map used for initial chain tracing and (2–4) sharpened 4.9-Å map used for model building and refinement [membrane plane view (2), extracellular view (3), and intracellular view (4)]. (B) Protein topology of OsOSCA1.2. The OsOSCA1.2 model is shown in the TM plane (C) and from the extracellular side (D).
According to the Transporter Classification (TC) Database (10), OsOSCA1.2 belongs to what is annotated as the calcium-permeable stress-gated cation channel family (TC accession no. 1.A.17.5) within the ANO superfamily (TC accession no. 1.A.17). This classification indicates that OsOSCA1.2 is distantly related to members of the ANO family (TC accession no. 1.A.17.1) for which high-resolution 3-dimensional structures are available (11, 12). Following a recently published bioinformatics approach (6), we had further predicted that OsOSCA1.2 had 11 TMs and the eighth hydrophobicity peak is composed of 2 TMs (Fig. 1B) based on hydropathy analysis and comparison of regions with the fungal homolog Nectria hematococca TMEM16 (NhTMEM16) (11). For convenience, we have kept the numbering convention of TMs consistent with NhTMEM16, and we thus refer to OsOSCA1.2’s additional N-terminal TM as TM0 (Fig. 1 C and D). Despite a relatively low degree of sequence similarity, we later confirmed that OsOSCA1.2 shares significant structural homology to the TMEM proteins with respect to 10 of the 11 TM regions, corresponding to TM1–TM10 in the mouse TMEM16A (mTMEM16A) structures (12).
TM0 threads from the extracellular N-terminal end of the protein through the membrane, linking to TM1 via an ∼50-residue strand that is likely conformationally flexible. This portion of the protein is the only region not fully resolved in our density maps (Fig. 1 B–D). A short helix on the cytoplasmic side then precedes TM1, and the C-terminal end of TM2 leads into the soluble cytosolic region of ∼170 residues. The remaining helices represent the ANO domain, encapsulating the predicted TM transport pathway. TM3 and TM4 are located on the outer edge of the TM region and are tilted with respect to the membrane. TM7 and TM8 are shorter in length and are the only TMs that do not span the entire length of the membrane, with the connecting loop (residues 578–583) being embedded in the membrane and consisting of hydrophobic residues.
The soluble domain is located on the intracellular side of the protein joining TM2 and TM3, and makes important structural contacts with the C terminus (Fig. 1 B–D). A core globular domain comprises a 4-stranded β-sheet buttressed by 2 short helices that interestingly forms a canonical RNA recognition motif (RRM) fold (13). Unlike true RNA binding RRM proteins, OsOSCA1.2 includes a fusion of a distinct 70-residue appendage between β-strands 2 and 3. These long extended helical arms protrude out from the RRM domain and are located proximal to and in the plane of what would be the inner-leaflet side of the plasma membrane.
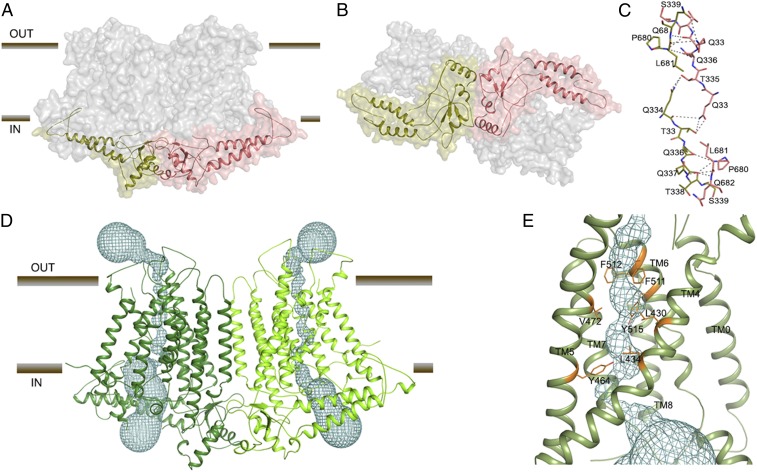
The dimer interface represents only a small percentage (∼2.5%) of the surface area of each protomer, burying a surface area of only ∼1,077 Å2, comprising interactions formed between the soluble domains (14) (Fig. 2 A and B). Interface residues Q334, T335, Q336, Q337, T338, S339, L681, Q682, and E683 from both subunits likely make several hydrogen bonds and hydrophobic interactions (Fig. 2C). Interestingly, the TMs from each subunit do not cause significant interactions contributing to the dimer interface. The orientation and offset of the 2 halves of the dimer create a large cavity between the 2 protomers, which, as predicted in recent AtOSCA structures (7, 8, 15), is likely filled with lipids when embedded in the cell membrane.
Fig. 2.
OsOSCA1.2 dimer interface and transport pathway. (A) OsOSCA1.2 surface representation. The TM domain is shown in gray, and the cytoplasmic domain is colored red and green. (B) View of OsOSCA1.2 from the cytoplasmic side. (C) Dimer interface residues. (D) Location of the predicted transport pathway in both subunits of OsOSCA1.2. The transport pathway is depicted as a cyan mesh. (E) Close-up view of the neck region, showing the residues “gating” the transport pathway.
The predicted transport pathway of OsOSCA1.2 is contained within each monomeric subunit (Fig. 2D), and formed between TM3 and TM7 as suggested by the topological similarities with mTMEM16A. Using the program HOLE to visualize a putative ion permeation pathway (16), the overall shape of the predicted transport pathway resembles an hourglass. The extracellular and intracellular vestibules are bridged by a narrow neck region that is about 25 Å long through the membrane. The putative pore has an opening more than 12 Å wide toward the extracellular side and narrows into the “neck” region ∼15 Å down the conduction pathway. The tightest juncture is ∼0.8 Å wide, suggesting that this transport pathway structure is in an inactive/closed conformation. The calculated transport pathway profile predicts that the residues F511, F512, and Y515 on TM6 and V472 and Y464 on TM5 form a gate that completely blocks the predicted transport pathway (Fig. 2E). All of the transport pathway-lining residues have good density except F511 (density to the β-carbon). The orientation of these residues in OsOSCA1.2 is similar compared with the corresponding pore-lining residues in AtOSCA1.1 and AtOSCA1.2, which further supports the transport pathway profile.
OsOSCA1.2 Computational Dynamics.
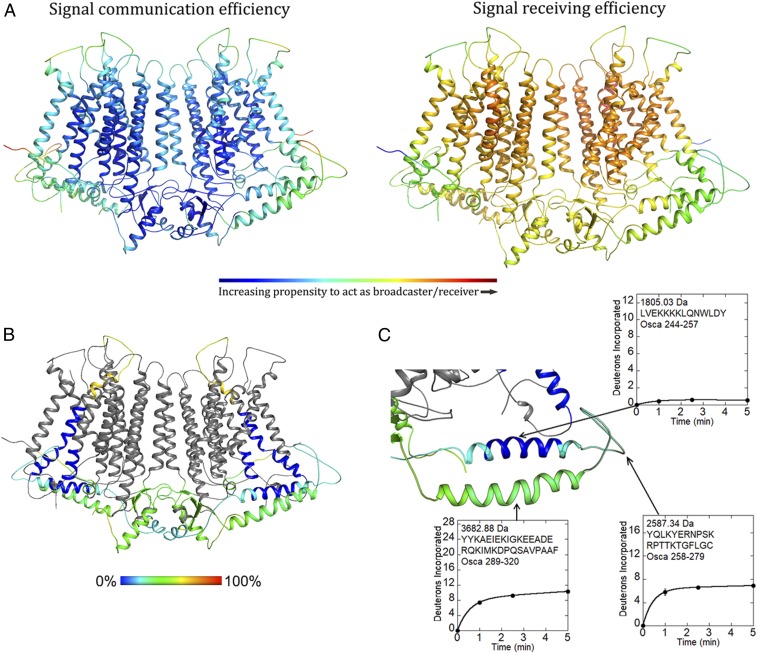
The DynOmics suite allows prediction and identification of candidate functional sites, signal transduction, and potentially allosteric communication mechanisms, leveraging rapidly growing structural proteomics data (17). The suite integrates 2 widely used elastic network models while taking account of the molecular environment like the lipid bilayer providing collective dynamics of structural resolved systems. We used DynOmics to do molecular dynamic simulations on our OsOSCA1.2 dimer model after embedding in the membrane, looking for regions that could potentially serve as functionally important sensors, broadcasters, and receivers (Fig. 3A). Our results revealed that the extended intracellular helical arms could communicate conformational perturbations, having the propensity to act as a broadcaster/receiver, extending to the central core sheet structure of the soluble domain and, more interestingly, TM6, which is proposed to be the transport pathway gating helix in related structures (7, 8, 11, 12, 15).
Fig. 3.
Computational and experimental dynamics of OsOSCA1.2. (A) Results of OsOSCA1.2 embedded in the membrane using the DynOmics suite. Panels show a color-coded map superimposed on the model illustrating signal communication (Left) and receiving (Right) efficiency. Regions that are colored red are more active, while those that are colored blue are inactive with regard to molecular dynamics prediction. (B) Relative uptake after 5 min of exchange. The structure is color-scaled and superimposed on the model. Regions colored gray yielded no detectable peptide fragments. (C) Close-up view of the extended and gating helix. Uptake plots for selected peptides are shown. Corresponding protein segments are outlined.
OsOSCA1.2 Experimental Dynamics.
To further understand and probe local conformational dynamics of OsOSCA1.2, we used hydrogen/deuterium exchange mass spectrometry (HDXMS). This approach utilizes the exchange that occurs between protons linked to amide bonds and protons or deuterium nuclei from solvent molecules to provide experimental information regarding regional solvent accessibility and dynamics. When a protein is added to a solution containing excess D2O, hydrogen/deuterium exchange occurs most rapidly for protons that are exposed to solvent and unconstrained by intermolecular hydrogen bonds. Protease digestion and chromatographic separation facilitate the quantification of deuterium nuclei incorporated throughout the protein as measured by mass spectrometry (18). For the hydrogen/deuterium exchange to occur in well-folded regions, a protein must sample exchange-competent conformations that expose amide protons. Thus, the uptake of deuterium over time reflects the local dynamics that individual regions of a protein undergo in solution (19).
HDXMS measurements using detergent-solubilized OsOSCA1.2 protein resulted in the identification of 32 peptides, which constitute 34.5% coverage of the molecule (Fig. 3B and SI Appendix, Fig. S4A), including the helical arms that were predicted to dynamically couple to the presumed gating helix TM6 and most likely to be responsible for sensing lateral tension in the membrane. The helix closest to the inner leaflet side of the membrane was covered by 2 peptides (corresponding to residues 244–257 and 245–257). Deuterium incorporation profiles revealed that this region was tightly protected from the exchange, indicative of rigid dynamics or association with a nearby surface (Fig. 3C). The following segment was also covered by 2 peptides (residues 258–279 and 258–286), which correspond to the C-terminal end of the protected helix and a nearby loop in our structure. This region was ∼25% saturated with deuterium nuclei at the earliest measured time point of 1 min, indicating rapid exchange associated with conformational flexibility. The remainder of this segment increased deuterium content by ∼5% over 5 min, suggesting conformational motions that gradually increased exposure to solvent. The helix farther from the membrane was covered by 3 peptides (residues 287–320, 289–320, and 305–320) and similarly displayed rapidly exchanging amides and ongoing deuterium exchange. Mass spectra from peptides corresponding to the unstructured loop and helix farther from the membrane all displayed bimodal deuterium uptake, which was more prominent among peptides corresponding to the loop (SI Appendix, Fig. S4B). The ongoing dynamics stand in sharp contrast to the rigidity of the helix (residues 241–266) closer to the membrane. Despite being spatially and sequentially near each other, these 2 intracellular helices have very different dynamic properties.
Topological Insertion of OSCA in Human Embryonic Kidney tsA201 Cells.
As the OSCA family includes an additional helix, TM0, we investigated the orientation of the protein in the cell membrane in mammalian human embryonic kidney (HEK) tsA201 cells. The topological prediction and structures suggested that the N and C termini of the molecule were on opposite sides of the membrane (Fig. 1B). We therefore made 2 C-terminal hemagglutinin (HA)-tagged (YPYDVPDYA) complementary DNA (cDNA) constructs of OsOSCA1.2 and AtOSCA1 in a pcDNA3.1 vector. HEK cell line tsA201 cells were transfected with both of these constructs (SI Appendix, Fig. S5 A and B); after 48 h, the cells were stained with anti-HA antibody conjugated to Alexa 488 without permeabilization (Materials and Methods). Our results suggest that the C terminus of the molecule was accessible only from outside the tsA201 cells.
Osmotic Stress Response of OsOSCA1.2.
It was suggested that the orthologs of OsOSCA1.2 in Arabidopsis, AtOSCA1 and AtOSCA1.2, are osmotic stress-responsive cation channels (1, 20). We therefore explored whether changes in the intracellular free Ca2+ concentration occur in response to osmolality changes in tsA201 cells using a ratiometric fluorescent indicator, Fura-2 AM (SI Appendix, Fig. S6). No obvious differences in intracellular calcium concentrations were observed between control and OsOSCA1.2-expressing tsA201 cells in response to hypoosmotic (168 mosmol⋅kg−1) and hyperosmotic (627 mosmol⋅kg−1) transitions (SI Appendix, Fig. S6 A and B; n > 23 OsOSCA1.2-expressing cells). In control experiments, rapidly induced fluorescence ratio changes were observed in response to exogenous adenosine 5′-triphosphate (ATP) in the same OsOSCA1.2-expressing tsA201 cells (SI Appendix, Fig. S6A), suggesting that Fura-2 effectively reported intracellular calcium changes in these experiments. The potential channel properties of OsOSCA1.2 were further investigated in Xenopus oocytes in response to hypoosmolality (∼112 mOsm/kg) and hyperosmolality (∼754 mOsm/kg). However, similar to water-injected oocytes, no large osmotic stress-induced currents were recorded in OsOSCA1.2-YFP– and OsOCSA1.2-StrepII–expressing oocytes (SI Appendix, Fig. S7 A–C). Experiments with OsOSCA1.2-YFP fusions showed plasma membrane localization of these proteins in Xenopus oocytes (SI Appendix, Fig. S7D). In positive controls, SLAC1-YC coexpressed with OST-YN showed large SLAC1-mediated anion currents (SI Appendix, Fig. S7 E and F).
Discussion
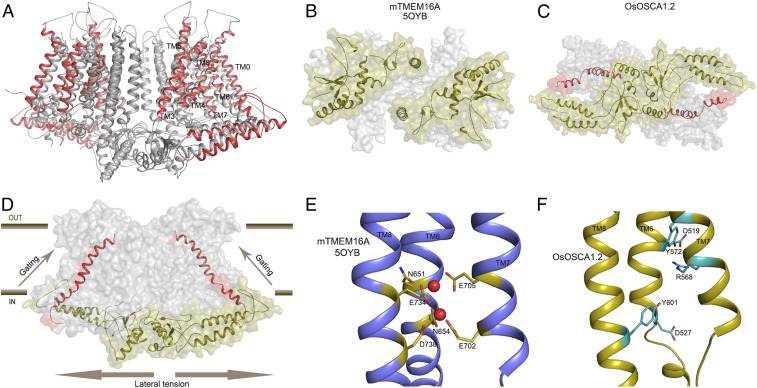
OsOSCA1.2 shares overall protein fold and topology with other recently determined homologous structures from A. thaliana (7, 8, 15). A superposition of these structures with OsOSCA1.2 showed a significant difference (root-mean-square deviation of ∼3–4 Å) for the transport pathway-lining helices (TM3–TM7), along with TM0 and TM8 (Fig. 4A). When comparing intracellular soluble domains, the extended helical arms of OsOSCA1.2 had noticeable differences compared with those of AtOSCA1 (Fig. 4A). These differences are likely due to a combination of conformational flexibility inherent in the detergent-solubilized protein and structural difference between species.
Fig. 4.
Structural comparisons of OsOSCA1.2 with other TMEM and OSCA structures. (A) Superposition of OsOSCA1.2 and AtOSCA1 (Protein Data Bank ID code 5YD1; ref. 8). TM1, TM2, TM9, and TM10 (shown in gray) close to the lipid-filled cleft are nearly superimposable and have little relative movement. Transport pathway-lining helices (TM3–TM7) showed significant movement, along with TM0 and TM8 (shown in red). (B) mTMEM16A soluble domains from the intercellular side are separated. (C) OsOSCA1.2 intracellular soluble domains are together and communicate with the gating helix TM6. (D) General mechanism of OsOSCA1.2 shown in the plane of the lipid membrane. Lateral tension on the inner leaflet side of the lipid bilayer causes a conformational change in the extended helices of the soluble domain, which is coupled to the gating helix TM6 opening of the transport pathway. (E) Calcium binding site residues of Ca2+-activated chloride channel mTMEM16A. Calcium ions are shown as red spheres. (F) Corresponding region of OsOSCA1.2 with charged and polar residues shown in cyan.
OsOSCA1.2 also shares structural homology to a Ca2+-activated chloride channel (mTMEM16A) and a phospholipid scramblase (NhTMEM16) of the TMEM family when comparing monomeric TM domain regions. However, they differ significantly in the regions of extracellular and intracellular loops and domains, as well as in the intermolecular packing arrangement of the respective dimers. The dimer interface of mTMEM16A buries less surface area (∼2%) compared with OsOSCA1.2, and most of the interactions are mediated through the TMs. The intracellular domains of mTMEM16A and NhTMEM16, which are formed by the N and C termini of the molecule, do not contribute to the formation of the dimer (Fig. 4B). In contrast, OsOSCA1.2 dimerizes mostly through interactions formed between the opposing intracellular soluble domains. This distinct dimeric packing resulted in a more pronounced offset between protomers that is ∼20 Å wider for OsOSCA1.2 compared with NhTMEM16 or mTMEM16A. In our OsOSCA1.2 structure, the extended helical arm in the intracellular soluble domain makes hydrophobic contacts with the loop connecting the gating helix TM6 (Fig. 4C), suggesting a possible role for the helical “arms” in sensing the membrane tension and, in turn, transmitting these conformational/mechanical changes to gate transport activity (Fig. 4D). This feature is missing in TMEM16 chloride channels, possibly as Ca2+ ions control gating.
Based on our present findings (SI Appendix, Figs. S6 and S7), a direct calcium-conducting function of OsOSCA1.2 and of AtOSCA1 remains an open question in need of further investigation. No clear osmotic stress-induced changes in intracellular calcium were found in OsOSCA1.2-expressing HEK cells, which may be due to improper expression (SI Appendix, Fig. S5). In Xenopus oocytes, although positive SLAC1 channel controls were confirmed and OsOSCA1.2 protein was expressed in the plasma membrane, no OsOSCA1.2-mediated currents were found in our Xenopus oocyte expression experiments (SI Appendix, Fig. S7 A–D). Purified OSCA1.2 reconstituted into liposomes showed discrete conducting states (SI Appendix, Fig. S1). Channel activity may be best investigated in the future in biological membranes in which regulatory proteins are included. Further research will be needed to investigate the transported substrates and selectivity of members of the plant OSCA family.
Our OsOSCA1.2 structure likely represents the nontransporting state conformation, as the narrowest domain of the predicted permeation pathway had a diameter of ∼0.8 Å and no tension, pressure, or osmolality mismatch was applied between the intracellular and extracellular sides of the OsOSCA1.2 protein during cryo-EM sample preparation (Fig. 2 D and E). Interestingly, when the OsOSCA1.2 structure was compared with the Ca2+-activated chloride channel mTMEM16A, we found a π-helical turn at TM6 near the “neck” region of the predicted transport pathway that might be associated with gating, in a similar fashion (Ca2+-activated α-to-π transition) to that observed in mTMEM16A (12, 21). The regulatory Ca2+ binding site composed of acidic and polar residues (E702 and E705 from TM7, E734 and D738 from TM8, and N651 and E654 from TM6) in the Ca2+-activated chloride channel mTMEM16A are well conserved in the TMEM16 family (Fig. 4E). However, compared with the corresponding region in OsOSCA1.2, the negatively charged residues D519 and E527 on TM6 and polar residue R568 on TM7 are located in spatially different positions (Fig. 4F). Therefore, OsOSCA1.2 will likely not specifically bind calcium ions in this region or provide specific selectivity for Ca2+.
Computational dynamic studies using our OsOSCA1.2 model and experiments using hydrogen/deuterium exchange with detergent-solubilized OsOSCA1.2 protein provide a potential molecular structural basis of how OsOSCA1.2 couples osmotic stress to induce transport gating in the membrane-spanning region. Taken together, both studies predict and support a model in which the extended helical arms (residues 241–266) have the mechanical rigidity and propensity to act as a broadcaster/receiver, transmitting conformational changes caused by lateral tension in the membrane to TM6 (Figs. 3 and 4F), which is linked to the predicted gating structure. In addition, information from HDXMS revealed the presence of bimodal deuterium exchange throughout the OsOSCA1.2 (Fig. 3 B and C and SI Appendix, Fig. S4B), most prominently within the helical arms (residues 258–320) and some extracellular loops (residues 489–511). The bimodal exchange is indicative of multiple correlated unfolding processes occurring in the observed regions (22). Interestingly, in each peptide where bimodal peaks were observed, the 2 peaks remained equal in intensity over the entire course of the experiment, suggesting at least 2 distinct conformational states occupied by the molecular ensemble at equilibrium in the resting state.
Several electrophysiological studies have used mammalian cells overexpressing OSCA channels to measure conductance gated by direct mechanotransduction or pressure (1, 5, 7). Interestingly, we found that the C terminus and presumably the entire cytosolic domain (residues 191–363) were on the outside when OSCA channels were overexpressed in mammalian cells (SI Appendix, Fig. S5). This finding suggests and could explain why channel opening due to changes in ion concentration may be impaired. As previous studies suggested Ca2+ conductance, we further probed ion flux using Fura-2, utilizing these cells as a proxy for calcium, but found no changes relative to controls for either hyperosmotic or hypoosmotic conditions (SI Appendix, Fig. S6). It remains unclear whether or not these ion channels can be gated by changes in ion concentration. Interestingly, analysis of the taxonomic distribution of different OSCA fragments suggests that TM0 is restricted to plants and that the cytosolic domain (residues 191–363; cytoL2) is probably distributed similar to the rest of the OSCA family (SI Appendix, Figs. S8–S10 and Table S1). In fungi and plants, the N terminus of TM0 is predicted to be on the outside, with the osmo-sensing cytosolic domain inside the cell (Fig. 1B). As such OSCA may be inserted in the membrane differently between mammalian and plant/fungi cells, which may be an important consideration in future functional studies.
Several important questions remain regarding these OSCA proteins and their role in plants. For example, the putative permeant substrates, including calcium and other possible substrates transported by OsOSCA1.2 or by other members within their greater family, are still unknown. It will certainly be a challenge to assign function to all of these proteins individually as these proteins are members of large gene families. Although we present a structure of OsOSCA1.2 along with computational and experimental dynamics, the detailed functional mechanism(s) coupling lateral tension in the membrane by OsOSCA1.2 to gating remains to be addressed in future studies. Furthermore, upstream regulatory proteins, the nature of the permeant substrate species, and how they are coupled to downstream signaling events remain to be determined.
Materials and Methods
Expression and Purification of OsOSCA1.2.
We cloned OsOSCA1.2 (GenBank KJ920372.1) and made TEV protease-cleavable GFP fusions into the pPICZc vector, and then tested expression in P. pastoris. Expression vectors were linearized using PmeI and electroporated into competent P. pastoris KM71H cells (Life Technology). The resulting transformants were cultured and induced in small scale to screen for target expression based on the intrinsic GFP fluorescence of cells and also from an anti-His Western blot of whole-cell lysate. OsOSCA1.2 was found to show both high levels of expression and desirable properties during purification (described below), and was therefore chosen for further characterization. Yeast clones selected for their high expression of OsOSCA1.2 were grown in minimal glycerol (4%) media, supplemented with 0.4% phosphoric acid and 0.024% trace metals at 28 °C in a New Brunswick BioFlo 415 system (Eppendorf). The pH of the media was titrated to pH 5 before inoculation and adjusted during the vegetative growth phase using 50% ammonium hydroxide. The dissolved oxygen (DO) was maintained at 10% minimally through cascaded agitation until a DO spike occurred. The fermentation culture was then induced at pH 5 by slow methanol addition for 16–18 h.
Cells were harvested and resuspended in cold lysis buffer [20 mM Tris⋅HCl (pH 8.0), 100 mM NaCl, 15% glycerol, 23.4 mM leupeptin, 7 mM E-64, 4 mM chymostatin, 14.5 mM pepstatin A, 1 mM phenylmethylsulfonyl fluoride, 25 mM benzamidine], and they were lysed by a single passage through a cell disruptor (TS-Series; Constant Systems, Inc.) at 40,000 psi. Cellular debris was removed by centrifugation (12,500 × g, 20 min, 4 °C), and the supernatant was continued onto a 38,400 × g spin for 4 h to fractionate the plasma membrane. The membrane fraction was resuspended in lysis buffer and frozen at −80 °C.
Membranes were solubilized with 1% n-dodecyl-β-d-maltopyranoside (DDM) and 0.1% sodium cholate for ∼90 min at 4 °C. Insoluble material was removed by centrifugation (38,400 × g, 60 min, 4 °C), and 15 mM imidazole was added to the supernatant before batch binding to Ni-NTA agarose resin (Qiagen). The bounded resin was sequentially applied to a gravity column housing and washed with buffer A [20 mM Hepes (pH 8.0), 150 mM NaCl, 0.03% DDM, 0.003% cholesteryl hemisuccinate], and an imidazole gradient was applied. Bound target protein was eluted with buffer A containing 300 mM imidazole, concentrated to ∼8 mL, desalted (HiPrep 26/10; GE Healthcare), and subjected to TEV protease digestion for 12 h at 4 °C. The TEV-digested sample was reapplied to Ni-agarose (Qiagen) to rebind the TEV protease and the C-terminal His-GFP tag. The collected OsOSCA1.2 was then concentrated to ∼1 mL and ultraspun at 95,000 rpm (TLA120.1 rotor) for 15 min at 4 °C. The sample was then applied to a Superdex 200 increase size-exclusion column (GE Healthcare) preequilibrated with 20 mM Hepes (pH 8.0), 150 mM NaCl, 0.06% n-undecyl-β-d-maltopyranoside, 0.2 mM Tris(2-carboxyethyl)phosphine, and 0.01% cholesteryl hemisuccinate, and run at 4 °C. Peak fractions off the SEC column were checked using sodium dodecyl sulfate/polyacrylamide gel electrophoresis and directly snap-frozen at a concentration of ∼3 mg/mL.
Determination of the Molecular Mass of OsOSCA1.2 Using SEC-MALLS.
SEC-MALLS was performed using a Superdex 200 Increase 10/300 GL size-exclusion column (GE Life Sciences) connected in series to a miniDAWN TREOS light-scattering detector and an Optilab T-rEX refractive index detector (Wyatt Technology). Purified OsOSCA1.2 was injected onto the column with 20 mM Hepes (pH 8.0), 150 mM NaCl, 0.03% n-dodecyl-β-d-maltopyranoside, 0.2 mM tris(2-carboxyethyl)phosphine, 0.001% sodium cholate, and 0.02% cholesteryl hemisuccinate, and run at 0.4 mL⋅min−1. The elution was monitored in-line with 3 detectors, and the molecular masses of the protein–micelle complex, the micelle, and the protein were calculated using ASTRA v.6 software (Wyatt Technology) in conjugate mode as previously described (23).
Functional Reconstitution of OsOSCA1.2 into DIBs.
Lipids of Escherichia coli extract polar (100600; Avanti) were dried under nitrogen and vacuum-desiccated for 1 h before resuspension in reconstitution buffer [10 mM Hepes (pH 7.4), 150 mM KCl] to a final concentration of 10 mg/mL. The resuspended lipids were incubated for a minimum of 20 min, followed by addition of 0.1% n-Decyl-β-d-maltopyranoside. The mixture was allowed to sit at room temperature for 30 min, followed by bath sonication for 5 cycles of 1 min of sonication and 2 min on ice. Purified OsOSCA1.2 was then mixed at a protein/lipid ratio of 1:500 (wt/wt) with the detergent-saturated liposomes. The protein was reconstituted by removal of detergents by the detergent-dilution method (24). OsOSCA1.2 containing proteoliposomes was resuspended to a final lipid concentration of ∼5 mg/mL, extruded through a 100-nm filter, and stored at −80 °C until use. Ion channel reconstitution into DIBs was done as detailed elsewhere (25–27). Briefly, a lipid asymmetrical droplet-droplet (∼200 nL) configuration (28) was obtained by placing OsOSCA1.2 containing proteoliposomes in E. coli extract polar lipids on the agar-coated head-stage Ag electrode and 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine/1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine/1,2-dioleoyl-sn-glycero-3-phosphate (1:1:0.5; 850457, 840034, and 840875, respectively; Avanti) liposomes on the reference electrode. The droplets were incubated in a hexadecane (H6703; Sigma) medium for 5–10 min to allow the formation of monolayers on each droplet. The 2 droplets were then brought into contact with each other, and the formation of bilayers was monitored using a triangular wave protocol. Incorporation of ion channels into the bilayer was detected as discrete fluctuations in current amplitude under voltage clamp conditions. Data were acquired using a Dagan 3900A amplifier and pCLAMP 10 software (Molecular Devices). Data were filtered at 1 kHz and sampled at 250 kHz using a Digidata 1440A system. Downward deflection traces represent inward currents (cis to trans), while upward deflections represent outward currents (trans to cis). Single-channel data analysis was performed using Clampfit 10 (Molecular Devices). The cation ionic concentrations of all solutions were verified experimentally via conductively coupled plasma emission spectrometry, and their ionic activities were calculated using GEOCHEM-EZ (29). All experiments were performed at room temperature.
EM Data Collection.
Quantifoil 1.2/1.3 Au (Quantifoil Micro Tools GmbH) or C-Flat 1.2/1.3 300 (Protochips) mesh grids were glow-discharged for 30 s at 30 mA (Emitech). Four microliters of OSCA1.2 at a concentration of 1.8 mg/mL was applied to the grids, blotted for 2.5 s at a relative humidity of 100%, and plunge-frozen in liquid ethane using an FEI Vitrobot Mark 2 system (FEI Company). 2 image sets were collected. The first dataset was collected using defocus phase contrast on an FEI Tecnai F30 microscope (FEI Company) operating at 300 kV with a K2 Summit camera (Gatan, Inc.) at a nominal magnification of 31,000× in superresolution mode with a pixel size of 0.636 Å using SerialEM software (Mastronarde Group). A total of 40 frames at 200 ms per frame were recorded for each image at a camera dose rate of 8 electrons per pixel per second. A total of 342,910 particles covering a defocus range from −0.8 to −2.8 μM were used to determine an initial 6.0-Å resolution map that allowed a polyalanine model to be built (Table 1). A higher resolution dataset was collected using an FEI Titan Krios equipped with a Volta phase plate (VPP), GatanEnergy filter, and K2 Summit camera (Gatan, Inc.). Data were collected at a nominal magnification of 105,000× in superresolution mode, and a total of 64,096 individual particle images at a fixed target defocus of −0.5 μm were used to determine the structure at a resolution of 4.9 Å (Table 1 and SI Appendix, Fig. S2). Sample micrographs from the VPP (Gctf fit of −0.5 μM) and defocus phase contrast (Gctf fit of −2.5 μM ) datasets are shown (SI Appendix, Fig. S11).
Table 1.
Cryo-EM data collection, 3-dimensional reconstruction, and model building
| Data collection and processing | Low-resolution data set | High-resolution data set |
| Microscope | FEI Tecnai F30 | FEI Titan Krios |
| Voltage, kV | 300 | 300 |
| Camera, mode | Gatan K2 Summit (40-frame superresolution movies) | Gatan K2 Summit (40-frame superresolution movies) |
| Target defocus, μm | −0.8 to −2.8 | −0.5 Volta phase plate |
| Pixel size, Å | 0.6355 | 0.6920 |
| Imposed symmetry | C2 | C2 |
| Electron dose, e-/Å2 | 45.5 | 40.8 |
| Initial particle images | 499,157 | 169,655 |
| Final particle images | 342,910 | 64,096 |
| Map resolution, Å | 6.0 | 4.9 |
| FSC 0.143 maximum local resolution, Å | 4.8 | 4.5 |
| Model building and refinement | ||
| Map sharpening B factor, Å2 | −800 | −400 |
| Protein residues, expected | 1,388 (1,424)* | 1,388 (1,424) |
| RMS Z score | ||
| Bond lengths, Z > 2 | 0.30 (2) | |
| Bond angles, Z > 2 | 0.44 (6) | |
| Validation | ||
| MolProbity score | 1.98 | |
| Clashscore | 7.99 | |
| EMRinger score | 0.82 | |
| Poor rotamers, % | 0.00 | |
| Ramachandran plot | ||
| Favored, % | 90.1 | |
| Outliers, % | 0.0 | |
FSC, Fourier shell correlation; RMS, root mean square.
Polyalanine model.
EM Data Processing.
The Tecnai F30 dataset, consisting of a total of 9,691 micrographs in 3 groups, was selected for initial processing after motion correction using MotionCor2 and contrast transfer function (CTF) estimation with Gctf. Non–dose-weighted micrographs were used for CTF estimation, and dose-weighted micrographs were used for all other processing. Approximately 1,000 manually picked particles from each group were used to generate 2× binned templates (2.542-Ǻ pixel size), which were used for autopicking in RELION. Autopicked particles were manually screened, and 499,167 particles were extracted for further processing in cryoSPARC. 2-dimensional classification and selection yielded 342,910 particles that were then used for initial model construction and auto-refinement. Auto-refinement and masking with C2 symmetry yielded a map with a resolution of 6.0 Ǻ by gold standard Fourier shell correlation (GSFSC) corrected for the effects of masking. Local resolution estimation in cryoSPARC indicated that the core regions have resolutions ranging from 4.5 to 6.0 Å. Sample defocus phase contrast 2-dimensional (2D) classes are shown (SI Appendix, Fig. S2B).
The Titan Krios dataset consisted of a total of 2,408 micrographs that were motion-corrected using MotionCor2, and CTFs were estimated using Gctf. Results were imported into RELION 2.1. A total of 1,126 corrected micrographs were selected for further processing after screening for excessive motion and poor or poorly estimated CTFs. A total of 1,134 particles were manually picked and classified in 2 dimensions, and the selected templates were used to auto-pick 372,278 particles. Further screening resulted in the selection of 650 micrographs containing 169,655 particles for additional processing. The 2× binned (2.76-Ǻ pixel size) particles were extracted and processed through 2 rounds of 2D classification and selection, resulting in 64,096 remaining particles. 3-dimensional auto-refinement with C2 symmetry using these particles and an initial model from the previous defocus contrast refinement yielded a model with a resolution of 7.4 Ǻ. Re-extraction with unbinned pixels and subsequent refinement led to no improvement in resolution at this stage. 3-dimensional classification into 10 classes and subset selection yielded 5 subsets (best single class, best 2 classes, best 5 classes, and all classes) that were used for another round of re-extraction and auto-refinement. The best resolution resulted from using all 64,096 particles and was unchanged at 7.4 Ǻ. Masking and postprocessing resulted in an estimated resolution of 6 Å. At this point, the 64,096 extracted particles were transferred to cryoSPARC, and subsequent processing was performed in cryoSPARC. 3-dimensional auto-refinement with C2 symmetry using all 64,096 particles and an initial model constructed using a subset of 16,438 selected particles resulted in a GSFSC estimated resolution of 4.9 Ǻ. The auto-refined, unsharpened map was further sharpened with a B-factors ranging from −350 to −600 out to a cutoff of 3.5 Ǻ for modeling, and a map with a B-factor of −530 was used for subsequent model building and refinement (SI Appendix, Fig. S2). Comparisons of VPP and defocus phase-contrast 2D classes are shown (SI Appendix, Fig. S2B).
Model Building and Refinement.
An initial polyalanine model was built using the 6.0-Å resolution map with multiple rounds of real-space refinement in Phenix/Coot (30, 31). To determine the absolute hand at this resolution, the initial and inverted models were utilized for molecular replacement using an X-ray diffraction dataset that extended to 9 Å in resolution. Only one model provided a solution to the molecular replacement search. Subsequent use of this initial model and the observation of the helical hand in the 4.9-Å resolution map further confirmed the correctness of the assigned hand. The full atomic model was built into the higher resolution map using multiple rounds of building and real-space refinement in Coot and Phenix. The density maps within the TM region were of sufficient quality to readily identify large aromatic side chains (SI Appendix, Fig. S3) and helped to confirm the correct sequence registration. Comparison with the recently determined structures of AtOSCA1.2 (7, 8, 15) further confirmed the correctness of our model despite the lower calculated overall resolution of our map.
Image Processing.
Motion-corrected projections with a pixel size of 1.271 Å (F30) and 1.384 Å (Titan Krios), with and without dose weighting, were constructed using MotionCor2 (32) with 2× binning and grouping. The CTF estimation was performed using Gctf (33), followed by manual selection to remove micrographs with poor or incorrectly fit CTF, poor astigmatism, and contamination. Manual and semiautomated particle picking was done using RELION 2.1 (34), followed by sorting and another round of manual overreading to remove low-quality micrographs. Subsequent refinements were carried out in RELION or cryoSPARC (35). Local resolution estimation was performed using cryoSPARC or ResMap (36).
HDXMS.
HDXMS measurements were made using a Synapt G2Si system (Waters Corporation). Deuterium exchange reactions were carried out by a Leap HDX PAL autosampler (Leap Technologies). The deuterated buffer was prepared by lyophilizing 10 mL of 20 mM Hepes (pH 8.0) and 150 mM NaCl. The lyophilized buffer was resuspended in 10 mL of 99.96% D2O immediately before use, to which was added powdered n-undecyl-β-d-maltopyranoside to a final concentration of 0.06% and cholesterol hemisuccinate to a final concentration of 0.01%. Each deuterium exchange time point (0 min, 1 min, 2.5 min, and 5 min) was measured in triplicate. For each measurement, 4 μL of protein at a concentration of 5 μM was mixed with 36 μL of D2O buffer at 25 °C. Deuterium exchange was quenched by combining 35 μL of the deuterated sample with 65 μL of 0.1% formic acid and 3 M guanidinium-HCl for 1 min at 1 °C. The quenched sample was then injected in a 50-μL sample loop and digested by an in-line pepsin column (Pierce, Inc.) at 15 °C. The resulting peptides were captured on a BEH C4 Vanguard precolumn at a flow rate of 400 μL⋅s−1, separated by analytical chromatography (Acquity UPLC BEH C4, 1.7 μM, 1.0 × 50 mm; Waters Corporation) using 7–85% acetonitrile in 0.1% formic acid over 7.5 min, and analyzed in a Waters Synapt G2Si quadrupole time-of-flight mass spectrometer following electrospray injection.
Data were collected in Mobility [Electrospray ionization positive ion detection (ESI+) mode, mass acquisition range of 200–2,000 mass-to-charge ratio (m/z), scan time of 0.4 s]. Continuous lock mass correction was performed using the infusion of leu-enkephalin (m/z = 556.277) every 30 s (mass accuracy of 1 ppm for calibration standard). For peptide identification, data were collected in MSE (mobility ESI+) mode. Peptide masses were identified following triplicate analysis of 10 μM OsOSCA1.2, and the data were analyzed using PLGS 2.5 (Waters Corporation). Peptide masses were identified using a minimal number of 250 ion counts for low-energy peptides and 50 ion counts for their fragment ions. The following parameters were used to filter peptide sequence matches: minimum products per amino acid of 0.2, a minimum score of 7, maximum monoisotopic mass (MH+) error of 5 ppm, and a retention time relative standard deviation of 5%, and the peptides had to be present in 2 of the 3 peptide identification runs collected. After identification in PLGS, peptides were analyzed in DynamX 3.0 (Waters Corporation). Deuterium uptake for each peptide was calculated by comparing the centroids of the mass envelopes of the deuterated samples with the undeuterated controls. To account for back-exchange and systematic autosampler sample handling differences, the uptake values measured at the 1-min time point were divided by 0.79. The longer 2.5-min and 5-min deuteration time point deuteration values were divided by 0.75. Data were plotted as a number of deuterons incorporated vs. time. The y-axis limit for each plot reflects the total number of amides within the peptide that can possibly exchange. Each plot includes the peptide MH+ value, sequence, and sequential residue numbering.
Production of OsOSCA1.2/AtOSCA1 Stable Expression Cell Line.
An epitope HA tag (YPYDVPDYA) was introduced onto the 5′ end (HA-OsOSCA1.2) or the 3′ end (OsOSCA1.2-HA, AtOSCA1-HA) of the full-length OsOSCA1.2 or AtOSCA1 cDNA by PCR. The cDNA was amplified using the PfuUltra II Fusion HS DNA Polymerase (Agilent Technologies, Inc.). PCR products were then subcloned into the pcDNA3.1 vector (Thermo Fisher Scientific). The cDNA inserts were verified by sequencing (GENEWIZ). After linearization of the vectors with PvuI enzyme, the vectors were transfected to HEK tsA201 cells (European Collection of Authenticated Cell Cultures) using Lipofectamine LTX with Plus Reagent (Thermo Fisher Scientific). Stable cell lines were selected 48 h posttransfection with 1 mg/mL Geneticin (Thermo Fisher Scientific).
Immunofluorescence Assay.
OsOSCA1.2/AtOSCA1-expressing cells were plated on poly-l-lysine–coated glass coverslips in 24-well plates. After 48 h, cells were fixed with 4% paraformaldehyde/phosphate-buffered saline (PBS). Cells were then blocked with 3% bovine serum albumin (BSA)/PBS for 30 min. Expressions of OsOSCA1.2/AtOSCA1 were detected with Alexa Fluor 488 anti-HA (16B12) antibody (BioLegend) in PBS or 1% BSA/PBS for 1 h. Samples were visualized on a fluorescence microscope (EVOS Cell Imaging Systems; Thermo Fisher Scientific).
Intracellular Ca2+ Imaging in HEK tsA201 Cells.
Intracellular Ca2+ concentration changes in tsA201 cells were observed using the ratiometric Ca2+ indicator dye Fura-2 (37). The mammalian cells were transfected with the pCDNA3.1 (control) or the pCDNA3.1-OsOSCA1.2-HA vector and were cultured on poly-l-lysine–coated, glass-bottomed, 35-mm dishes for 16–24 h. Cells were loaded with 5 μM Fura-2 AM (F1221; Invitrogen) in loading buffer (∼286 mOsm⋅kg−1; 130 mM NaCl, 3 mM KCl, 0.6 mM MgCl2, 0.1 mM CaCl2, 10 mM glucose, 10 mM Hepes, and adjusted to pH 7.4 with NaOH) and kept in the dark for 45 min, washed twice by assay buffer (∼286 mOsm⋅kg−1; 130 mM NaCl, 3 mM KCl, 0.6 mM MgCl2, 2 mM CaCl2, 10 mM glucose, 10 mM Hepes, and adjusted pH to 7.4 with NaOH) for 5 min each time, and then incubated with 1 mL of assay buffer for Ca2+ imaging. For hypoosmotic treatments, 1 mL of hypotonic buffer (3 mM KCl, 0.6 mM MgCl2, 2 mM CaCl2, 10 mM glucose, 10 mM Hepes, and adjusted to pH 7.4 using NaOH) was added into the original 1-mL assay buffer, resulting in a final osmolality of ∼168 mOsm⋅kg−1 of the incubation buffer. For ATP treatments, 20 μL of 1 mM ATP was added into the 2 mL of ∼168 mOsm⋅kg−1 incubation buffer, resulting in a final ATP concentration of 10 μM. For hyperosmotic treatments, 1 mL of hypertonic buffer (650 mM sorbitol, 130 mM NaCl, 3 mM KCl, 0.6 mM MgCl2, 2 mM CaCl2, 10 mM glucose, 10 mM Hepes, and adjusted to pH 7.4 with NaOH) was added, resulting in a final osmolarity of ∼627 mOsm⋅kg−1 in the incubation buffer. The osmotic concentrations of the buffers were determined by using a Wescor 5500 Vapor Pressure Osmometer. Time-resolved Fura-2 imaging was performed using an Eclipse TE300 inverted microscope equipped with a Plan Fluor 40×/1.30 oil objective (DIC H ∞/0.17 WD 0.2; Nikon), an ET-Fura2 filter set 79001 (EX340x, EX380x, ET510/80m; Chroma), a Mac 2002 System automatic controller, and a Cool SNAP HQ camera (Photometrics), and was guided by MetaFluor software version 7.0r3 (Molecular Devices). Fluorescence images excited at 340 nm or 380 nm were collected at a 200-ms exposure every 5 s. Emission ratios for 340/380-nm excitations in cells were processed and analyzed using Fiji (38).
2-Electrode Voltage-Clamp in Xenopus Oocytes.
The OsOSCA1.2 coding region was amplified using 5′-GGCTTAAUATGGCCACTGTTTCTGACATC-3′ and 5′-GGTTTAAUTCGTTCGTGTACAGTTTCAGGA-3′ primers and cloned into a pNB1 oocyte expression vector with C-terminal fusion of YFP or StrepII tag using the USER cloning method (39). OsOSCA1.2-YFP, OsOSCA1.2-StrepII, SLAC1-YC, and OST1-YN capped RNA (cRNA) were generated utilizing the mMESSAGE mMACHINE T7 Kit (Thermo Fisher Scientific) using linearized pNB1-OsOSCA1.2-YFP, pNB1-OsOSCA1.2-StrepII, pNB1-SLAC1-YC, or pNB1-OST1-YN plasmids.
Electrophysiological analyses in Xenopus oocytes were performed as reported in a previous report (40), with some modifications. Surgically extracted ovaries from Xenopus laevis were purchased from Nasco (LM00935) and Ecocyte Bio Science US treated with 25 mg/mL Collagenase D (11088882001; Roche Diagnostics) to release oocytes. The isolated oocytes were incubated in ND96 buffer (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM Hepes, adjusted pH to 7.4 with NaOH) overnight at 16 °C for recovery. Then, 25 ng cRNA of OsOSCA1.2-StrepII or 12.5 ng of cRNA of SLAC1-YC and OST1-YN was injected into oocytes and incubated in ND96 buffer at 16 °C for 2–3 d. 2-electrode voltage clamp recordings were performed using a Cornerstone (Dagan) TEV-200 amplifier and a Digidata 1440A low-noise data acquisition system controlled by Clampex 10.2 software (Molecular Devices). A 3 M KCl bath electrode was used to stabilize the bath electrode potential. Currents were recorded at 3-s voltage pulses ranging from +40 mV to −140 mV for OsOSCA1.2 or −160 mV for SLAC1 in −20-mV decrements followed by a +40-mV “tail” voltage. To investigate osmolality responses, oocytes were recorded in bath solutions with ∼217 mOsm⋅kg−1 (96 mM mannitol, 48 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM Hepes, adjusted pH to 7.4 with NaOH), ∼112 mOsm⋅kg−1 (48 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM Hepes, adjusted pH to 7.4 with NaOH), and ∼754 mOsm⋅kg−1 (596 mM mannitol, 48 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM Hepes, adjusted pH to 7.4 with NaOH). Osmolality buffers were exchanged by a peristaltic pump. For SLAC1-mediated current recordings, oocytes were incubated in bath solution with 100 mM NaNO3, 1 mM Ca (gluconate)2, 1 mM Mg (gluconate)2, 1 mM LaCl3, and 10 mM 2-(N-Morpholino)ethanesulfonic acid/Tris at pH 5.6.
Data Availability.
A cryo-EM map of OsOSCA1.2 has been deposited to the Electron Microscopy Data Bank under accession code EMD-20017. Atomic coordinates of OsOSCA1.2 have been deposited in the Protein Data Bank under ID code 6OCE. OSCA 1.2 has been incorporated to TCDB with accession 1.A.17.5.20.
Supplementary Material
Acknowledgments
Cryo-EM sample preparation and F30 data collection were performed at the University of Colorado Boulder EM Services Core Facility in the Department of Molecular, Cellular and Developmental Biology with the technical assistance of facility staff. We thank Valorie D. Bowman for assistance during data collection at the Purdue cryo-EM facility supported by National Institute of General Medical Sciences Grant U24 GM116789. This work was funded by National Science Foundation Plant Genome Research Program IOS-1444435 and, in part, supported by the NIH (Grant GM060396).
Footnotes
The authors declare no conflict of interest.
Data deposition: The cryo-electron microscopy map of Oryza sativa OSCA1.2 (OsOSCA1.2) has been deposited in the Electron Microscopy Data Bank under accession code EMD-20017. Atomic coordinates of OsOSCA1.2 have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 6OCE). OSCA 1.2 has been incorporated to Transporter Classification Database with accession 1.A.17.5.20 http://www.tcdb.org/search/result.php?tc=1.A.17.5.20.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900774116/-/DCSupplemental.
References
- 1.Yuan F., et al. , OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514, 367–371 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Knight H., Trewavas A. J., Knight M. R., Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 12, 1067–1078 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Stephan A. B., Kunz H. H., Yang E., Schroeder J. I., Rapid hyperosmotic-induced Ca2+ responses in Arabidopsis thaliana exhibit sensory potentiation and involvement of plastidial KEA transporters. Proc. Natl. Acad. Sci. U.S.A. 113, E5242–E5249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi W. G., Toyota M., Kim S. H., Hilleary R., Gilroy S., Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. U.S.A. 111, 6497–6502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murthy S. E., et al. , OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels. eLife 7, e41844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medrano-Soto A., et al. , Bioinformatic characterization of the Anoctamin Superfamily of Ca2+-activated ion channels and lipid scramblases. PLoS One 13, e0192851 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jojoa-Cruz S., et al. , Cryo-EM structure of the mechanically activated ion channel OSCA1.2. eLife 7, e41845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang M., et al. , Structure of the mechanosensitive OSCA channels. Nat. Struct. Mol. Biol. 25, 850–858 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Barad B. A., et al. , EMRinger: Side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saier M. H. Jr, et al. , The Transporter Classification Database (TCDB): Recent advances. Nucleic Acids Res. 44, D372–D379 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunner J. D., Lim N. K., Schenck S., Duerst A., Dutzler R., X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature 516, 207–212 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Dang S., et al. , Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature 552, 426–429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rai A., Suprasanna P., D’Souza S. F., Kumar V., Membrane topology and predicted RNA-binding function of the ‘early responsive to dehydration (ERD4)’ plant protein. PLoS One 7, e32658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Liu X., Wang J., Sun L., Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2. Nat. Commun. 9, 5060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smart O. S., Breed J., Smith G. R., Sansom M. S., A novel method for structure-based prediction of ion channel conductance properties. Biophys. J. 72, 1109–1126 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Chang Y. Y., Lee J. Y., Bahar I., Yang L. W., DynOmics: Dynamics of structural proteome and beyond. Nucleic Acids Res. 45, W374–W380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wales T. E., Engen J. R., Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 25, 158–170 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Truhlar S. M. E., Croy C. H., Torpey J. W., Koeppe J. R., Komives E. A., Solvent accessibility of protein surfaces by amide H/2H exchange MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 17, 1490–1497 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Hou C., et al. , DUF221 proteins are a family of osmosensitive calcium-permeable cation channels conserved across eukaryotes. Cell Res. 24, 632–635 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulino C., Kalienkova V., Lam A. K. M., Neldner Y., Dutzler R., Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM. Nature 552, 421–425 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Li M. J., Guttman M., Atkins W. M., Conformational dynamics of P-glycoprotein in lipid nanodiscs and detergent micelles reveal complex motions on a wide time scale. J. Biol. Chem. 293, 6297–6307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slotboom D. J., Duurkens R. H., Olieman K., Erkens G. B., Static light scattering to characterize membrane proteins in detergent solution. Methods 46, 73–82 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Mulligan C., et al. , The substrate-binding protein imposes directionality on an electrochemical sodium gradient-driven TRAP transporter. Proc. Natl. Acad. Sci. U.S.A. 106, 1778–1783 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang W. L., Chen M., Cronin B., Holden M. A., Bayley H., Asymmetric droplet interface bilayers. J. Am. Chem. Soc. 130, 5878–5879 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Leptihn S., et al. , Constructing droplet interface bilayers from the contact of aqueous droplets in oil. Nat. Protoc. 8, 1048–1057 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Syeda R., Holden M. A., Hwang W. L., Bayley H., Screening blockers against a potassium channel with a droplet interface bilayer array. J. Am. Chem. Soc. 130, 15543–15548 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Coste B., et al. , Piezo proteins are pore-forming subunits of mechanically activated channels. Nature 483, 176–181 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaff J., Schultz B., Craft E. J., Clark R. T., Kochian L. V., GEOCHEM-EZ: A chemical speciation program with greater power and flexibility. Plant Soil 330, 207–214 (2010). [Google Scholar]
- 30.Adams P. D., et al. , PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Q., et al. , Structure of mammalian endolysosomal TRPML1 channel in nanodiscs. Nature 550, 415–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheres S. H., A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grynkiewicz G., Poenie M., Tsien R. Y., A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 260, 3440–3450 (1985). [PubMed] [Google Scholar]
- 38.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nour-Eldin H. H., Hansen B. G., Nørholm M. H., Jensen J. K., Halkier B. A., Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 34, e122 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang C., Zhang J., Schroeder J. I., Two-electrode voltage-clamp recordings in Xenopus laevis oocytes: Reconstitution of abscisic acid activation of SLAC1 anion channel via PYL9 ABA receptor. Bio Protoc. 7, 2114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A cryo-EM map of OsOSCA1.2 has been deposited to the Electron Microscopy Data Bank under accession code EMD-20017. Atomic coordinates of OsOSCA1.2 have been deposited in the Protein Data Bank under ID code 6OCE. OSCA 1.2 has been incorporated to TCDB with accession 1.A.17.5.20.