Significance
This study uncovers a long noncoding RNA (lncRNA)-mediated mechanism underlying lung adenocarcinoma (LAD) metastasis. We here report that lncRNA LINC00673-v4 expression is up-regulated in LAD and is associated with disease progression. At the molecular level, LINC00673-v4 acts as a scaffold molecule that promotes the interaction between DDX3 and CK1ε and thus the phosphorylation of dishevelled, which subsequently activates WNT/β-catenin signaling and consequently causes aggressiveness of LAD. Treatment with antisense oligonucleotides against LINC00673-v4 strongly suppresses LAD metastasis in vivo.
Keywords: LINC00673-v4, metastasis, WNT/β-catenin, DDX3, CK1ε
Abstract
It is well recognized that metastasis can occur early in the course of lung adenocarcinoma (LAD) development, and yet the molecular mechanisms driving this capability of rapid metastasis remain incompletely understood. Here we reported that a long noncoding RNA, LINC00673, was up-regulated in LAD cells. Of note, we first found that LINC00673-v4 was the most abundant transcript of LINC00673 in LAD cells and its expression was associated with adverse clinical outcome of LAD. In vitro and in vivo experiments demonstrated that LINC00673-v4 enhanced invasiveness, migration, and metastasis of LAD cells. Mechanistically, LINC00673-v4 augmented the interaction between DDX3 and CK1ε and thus the phosphorylation of dishevelled, which subsequently activated WNT/β-catenin signaling and consequently caused aggressiveness of LAD. Antagonizing LINC00673-v4 suppressed LAD metastasis in vivo. Together, our data suggest that LINC00673-v4 is a driver molecule for metastasis via constitutively activating WNT/β-catenin signaling in LAD and may represent a potential therapeutic target against the metastasis of LAD.
Lung cancer is the most common type of cancer and a leading cause of deaths from cancer worldwide (1). Nonsmall cell lung cancer (NSCLC), which can be further subtyped into lung adenocarcinoma (LAD), lung squamous cell carcinoma (LSCC), large cell carcinoma (LCC), and other relatively less frequently diagnosed histological types, accounts for ∼85% of all lung cancer cases (2). Of note, LAD constitutes ∼50% of NSCLC cases and has become the most predominant histological subtype of lung cancer (3). Although currently available therapeutic strategies, including surgery, chemotherapy, radiotherapy, and targeting therapies, have achieved remarkable improvements in the past decades, the overall prognosis for NSCLC remains poor, with the 5-y survival rate being lower than 21% with all clinical stages combined, partly due to its usually aggressive clinical course presented (4, 5). Due to early local invasiveness and metastasis, LAD can spread to lymph nodes (LNs), contralateral lung, and distant organs such as bones and brain (6). Obviously, unidentified molecular events, and unknown molecules that mediate these events, might play pivotal roles in the development and progression of LAD aggressiveness. Efforts in addressing these issues would facilitate establishment of new effective therapeutic targets or prognostic biomarkers.
The WNT/β-catenin signaling is key to cancer development and progression and represents one of the best recognized cascades modulating tumor invasion and metastasis (7, 8). Accumulating evidence has implicated that constitutive activation of WNT/β-catenin signaling plays a pivotal role in LAD development and progression (9, 10). Moreover, clinically WNT/β-catenin pathway activation predicts increased risk of tumor recurrence in NSCLC patients (11). Of note, previous study by Nguyen et al. demonstrated that activation of WNT/β-catenin signaling correlated with a lower rate of metastasis-free survival of LAD patients, and that WNT/T cell factor (TCF) signaling mediated the metastatic capacity of LAD (12), further supporting a possible biological as well as clinical significance of WNT/β-catenin signaling in LAD.
It has been widely acknowledged that activation of WNT/β-catenin signaling is under sophisticated regulation (13). In the absence of WNT ligand stimulation, β-catenin is constitutively phosphorylated via interaction with glycogen synthase kinase (GSK-3) in a destruction complex composed of scaffold proteins Axin and adenomatous polyposis coli (APC), leading to ubiquitin-mediated degradation of β-catenin and an inactivated state of the signaling (14, 15). When a Wnt ligand binds the frizzled (FZD) receptor and coreceptors low-density lipoprotein receptor-related protein 5 or 6 (LPR5 or LPR6), the canonical WNT/β-catenin signaling is initiated, leading to phosphorylation of LPR5/6, which then recruits Axin and dishevelled (Dvl) to the plasma membrane, subsequently disrupting β-catenin destruction complex. Destabilization of the destruction complex prevents phosphorylation-dependent polyubiquitination and degradation of the β-catenin (16, 17). As a result, β-catenin translocates into the nucleus and induces transcription of downstream genes via interacting with members of the lymphoid enhancer-binding factor (LEF)/TCF family of transcription factors (18). In a subset of cancers, including colorectal cancer, activation of WNT/β-catenin signaling by stabilization of β-catenin is commonly caused by activating mutation in β-catenin or inactivating mutations of the APC (19). However, LAD rarely harbors somatic mutations that constitutively activate WNT/β-catenin signaling, suggesting that other mechanisms must be involved in promoting the hyperactivation of WNT/β-catenin signaling in this cancer type (20, 21).
Long noncoding RNAs (lncRNAs), a class of endogenous cellular RNAs longer than 200 nucleotides in size, lacking coding potential, play essential roles in many fundamental biological processes and human diseases (22–24). LncRNAs have emerged as new key players in cancer development and progression (23, 25), and studies have revealed that lncRNAs can regulate a variety of cancer-related signaling networks through their interaction proteins (23, 25). For example, lncRNA HULC directly binds the Y-box protein-1 (YB-1) and accelerates its phosphorylation via extracellular signal-regulated kinase (ERK), promoting the translation of silenced oncogenic mRNAs in hepatocellular cancer (26). Here we report that a lncRNA in LAD, LINC00673-v4, is highly prognostic for LAD and critical for WNT/β-catenin signaling activation. We show that LINC00673-v4 specifically binds DEAD box RNA helicase DDX3 and casein kinase 1ε (CK1ε), resulting in phosphorylation of Dvl and enhanced activation of WNT/β-catenin signaling. As a consequence, LINC00673-v4 promotes the aggressiveness of LAD.
Results
LINC00673-v4 Expression Is Up-Regulated in LAD and Correlates with Lymph Node Metastasis and Poor Prognosis.
Given the important roles of WNT/β-catenin signaling in LAD, we began our study by examining the possibility that lncRNA might be involved in the activation of WNT/β-catenin signaling in LAD. Using the Top/Fop flash luciferase assay, we assessed the effects of 10 of the most highly expressed lncRNA transcripts in LAD, as determined by integrated analysis of transcriptome sequencing data deposited at The Cancer Genome Atlas (TCGA) by Cabanski et al. (27), on WNT/β-catenin signaling. As shown in SI Appendix, Fig. S1, silencing ENST00000457958.6 led to significant suppression of the TCF/LEF activities in LAD cells while another 9 assessed lncRNAs showed no significant, or only weak effects, suggesting that ENST00000457958.6 might represent a main lncRNA player in the activation of WNT/β-catenin signaling in LAD.
Against the backdrop that our aforementioned data suggested a role of ENST00000457958.6 in WNT/β-catenin signaling, we performed the rapid amplification of cDNA ends (RACE) experiment and identified the 2,160 nt full-length ENST00000457958.6, as shown in SI Appendix, Fig. S2 B–D, further confirmed by Northern blotting analysis (SI Appendix, Fig. S2E). Of note, 100% identical sequence was released in the National Center for Biotechnology Information (NCBI) recently (LINC00673 transcript variant 4, NR_152515.1, https://www.ncbi.nlm.nih.gov/nuccore/NR_152515.1). Thus, we designated the transcript as LINC00673-v4. Human LINC00673 is located on chromosome 17 and includes five transcripts as shown in the NCBI (https://www.ncbi.nlm.nih.gov/gene/100499467). LINC00673-v4, one of the isoforms of LINC00673, contains three exons and is highly conservative across mammals (SI Appendix, Fig. S2A). Moreover, like most intergenic lncRNAs, LINC00673-v4 possesses a polyA tail and 5′ cap structure (SI Appendix, Fig. S2 D and F). Using RNA fluorescence in situ hybridization and cellular fractionation assays, LINC00673-v4 is found in the cytoplasm of LAD cells (Fig. 1A and SI Appendix, Fig. S2G). Analysis on the coding potential strongly indicated that LINC00673-v4 might lack protein-coding capacity (Fig. 1B and SI Appendix, Fig. S2H).
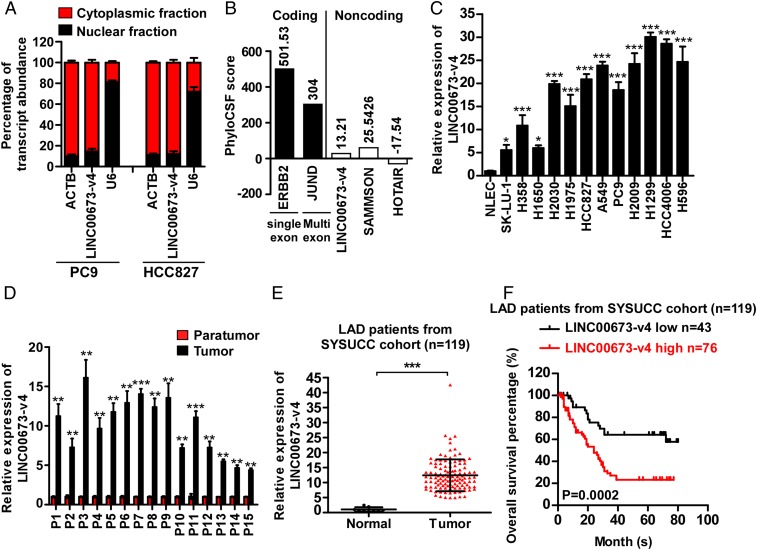
Fig. 1.
LINC00673-v4 is highly expressed in LAD with prognostic value. (A) The levels of U6 (nuclear control), ACTB (cytoplasmic control), as well as LINC00673-v4 were analyzed by qRT-PCR in nuclear and cytoplasmic fractions (each bar represents the mean ± SD derived from three independent experiments). (B) PhyloCSF analysis was performed to assess the maximum cerebrospinal fluid (CSF) scores of LINC00673-v4 and other known coding and noncoding RNAs. (C) The expression of LINC00673-v4 is elevated in LAD cell lines compared with primary normal lung epithelial cells (each bar represents the mean ± SD derived from three experiments, one-way ANOVA followed by Dunnett’s multiple comparison test. *P < 0.05, ***P < 0.001). (D) Analyses of LINC00673-v4 levels in LAD and adjacent nontumorous lung tissues by qRT-PCR (each bar represents the mean ± SD derived from three independent experiments, two-tailed Student’s t test. **P < 0.01, ***P < 0.001). (E) Analyses of LINC00673-v4 expression in Sun Yat-sen University Cancer Center (SYSUCC) LAD cohorts and normal lung tissues using qRT-PCR (two-tailed Student’s t test. ***P < 0.001). (F) Kaplan–Meier survival analysis of overall survival in SYSUCC LAD cohorts based on LINC00673-v4 expression.
We next employed The Atlas of Noncoding RNAs in Cancer (TANRIC) to investigate the expression of LINC00673 in LAD patients. As shown in SI Appendix, Fig. S3A, the level of LINC00673 in LAD was significantly elevated compared with that in the adjacent noncancerous tissues. The up-regulation of LINC00673 was also confirmed in our 119 cases of archived LAD tissue specimens using in situ hybridization (ISH) assay (SI Appendix, Fig. S3B). As data in the TANRIC dataset and ISH assay do not distinguish LINC00673 isoforms, we designed PCR primers (SI Appendix, Fig. S4A) to specifically detect only LINC00673-v4 in LAD cell lines, 15 paired tumor tissues and the adjacent benign lung tissues, and 119 cases of archived LAD tissues. We found that the expression of LINC00673-v4 was markedly up-regulated in LAD cell lines and tumor tissues (Fig. 1 C–E). Next, we investigated the abundance of transcript variants of LINC00673 in PC9, HCC827, and H2030 cell lines through qPCR using variant-specific primers against each transcript (SI Appendix, Fig. S4A). Our results showed that LINC00673-v4 was the most abundant transcript (SI Appendix, Fig. S4B) in the tested cells. The abundance of LINC00673-v4 was also validated in five cases of clinical LAD specimens (SI Appendix, Fig. S4C). These data together demonstrated a significant up-regulation of LINC00673-v4 in LAD, warranting further investigation on whether the up-regulated LINC00673-v4 plays a role in the development and progression of LAD.
Intriguingly, LINC00673-v4 is located within a chromosomal region that has been found to be commonly amplified in NSCLC, as shown by a previous study examining somatic copy number alterations (SCNAs) with high resolution using the Affymetrix 250K Sty I array (28) (SI Appendix, Table S1). In this current study, we verified the amplification of the LINC00673 gene in the TCGA dataset, and we found that the RNA expression level of LINC00673 positively correlated with its amplification status (SI Appendix, Fig. S3C). When further analyzing LINC00673 copy number variations in our collected patient samples (n = 119), increased copy numbers of LINC00673 were detected in 24 specimens (20.1%). In addition, the association of LINC00673-v4 expression level with its copy number was also confirmed in both LAD cell lines and LAD clinical tissues (SI Appendix, Fig. S3 D and E), together supporting the notion that gene amplification contributes to up-regulation of LINC00673-v4, at least in a certain fraction of cases.
To further understand the clinical significance of increased expression and copy number of the LINC00673 gene in LAD, correlation study was performed, and we found that LINC00673-v4 overexpression, and LINC00673 gene amplification as well, correlated with LN metastasis and patient prognosis (Fig. 1F and SI Appendix, Fig. S3F and Tables S2–S4; the cutoff value for distinguishing high versus low expression was selected using receiver operating characteristic [ROC] curve analysis). Consistently, the same correlations were found in the analysis of the TANRIC data (SI Appendix, Fig. S3G and Tables S5 and S6; determination of the cutoff value for predicting survival was performed using X-tile bioinformatics software).
LINC00673-v4 Promotes LAD Cell Invasion, Migration, and Metastasis.
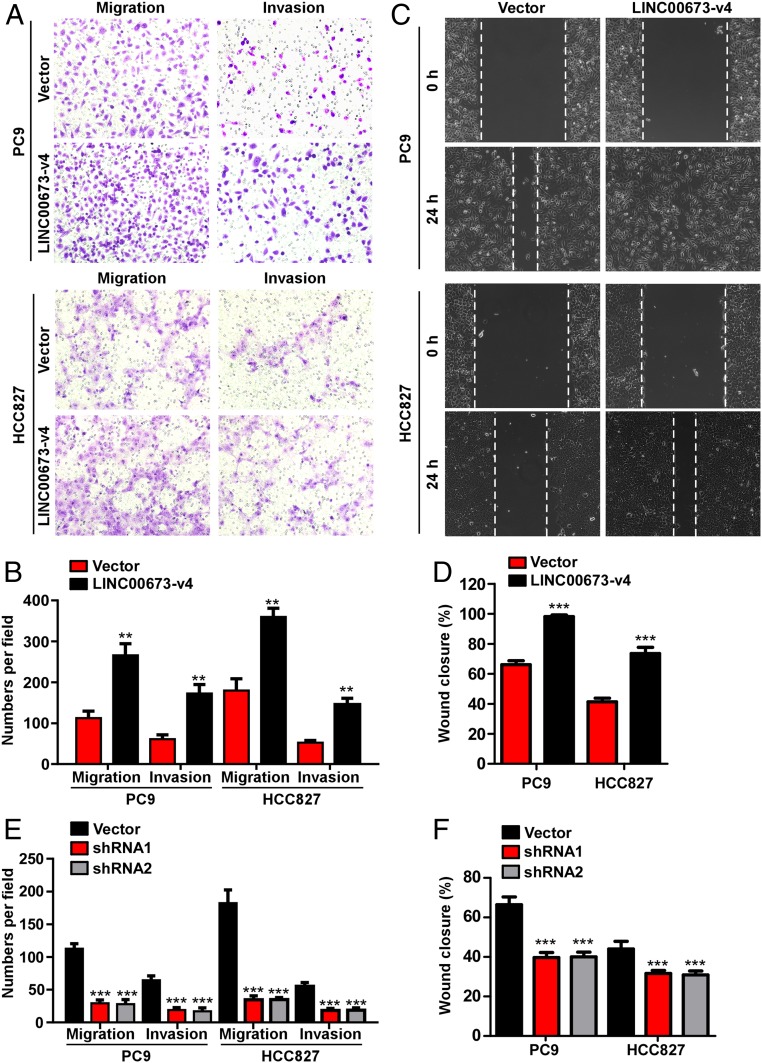
The strong association of LINC00673-v4 expression with LN metastasis and poor prognosis in LAD prompted us to investigate whether LINC00673-v4 promotes LAD cell invasion and migration. To this end, LAD cell lines (PC9, HCC827, and NCI-H2030 cell lines expressing intermediate LINC00673-v4 levels) with LINC00673-v4 stably overexpressed were constructed (SI Appendix, Fig. S5A). When tested for their invasiveness and migration, our results showed that ectopic overexpression of LINC00673-v4 in PC9, HCC827, and NCI-H2030 cells significantly increased the invasive and migratory capabilities of LAD cells as exhibited by Matrigel-coated or uncoated Transwell assays (Fig. 2 A and B and SI Appendix, Fig. S5B). Wound healing experiments showed that migration of LINC00673-v4–transduced cells was markedly enhanced compared with that of the vector-control cells (Fig. 2 C and D and SI Appendix, Fig. S5 C and D). In contrast, depletion of LINC00673-v4 (SI Appendix, Fig. S5A) inhibited cellular invasiveness and migration (Fig. 2E and SI Appendix, Fig. S5B). Moreover, knocking down LINC00673-v4 suppressed the wound healing capability of LAD cells (Fig. 2F and SI Appendix, Fig. S5 C and D).
Fig. 2.
LINC00673-v4 regulates LAD cell invasion and migration in vitro. (A) Representative micrographs of indicated invading or migrating cells analyzed by Matrigel-coated or noncoated Transwell assays, respectively. (B) Quantification of indicated invading or migrating cells analyzed by Matrigel-coated or noncoated Transwell assays, respectively (each bar represents the mean ± SD derived from three independent experiments, two-tailed Student’s t test. **P < 0.01). (C) Representative micrographs of wound healing assay of indicated cells. Wound closures were photographed at indicated time after wounding. (D) Quantification of wound closures of the indicated cells (each bar represents the mean ± SD derived from three independent experiments, two-tailed Student’s t test. ***P < 0.001). (E) Depletion of LINC00673-v4 inhibited invasion and migration of indicated cells (each bar represents the mean ± SD derived from three experiments, one-way ANOVA followed by Dunnett’s multiple comparison test. ***P < 0.001). (F) Knocking down LINC00673-v4 suppressed wound healing compared with the vector-control cells (each bar represents the mean ± SD derived from three experiments, one-way ANOVA followed by Dunnett’s multiple comparison test. ***P < 0.001).
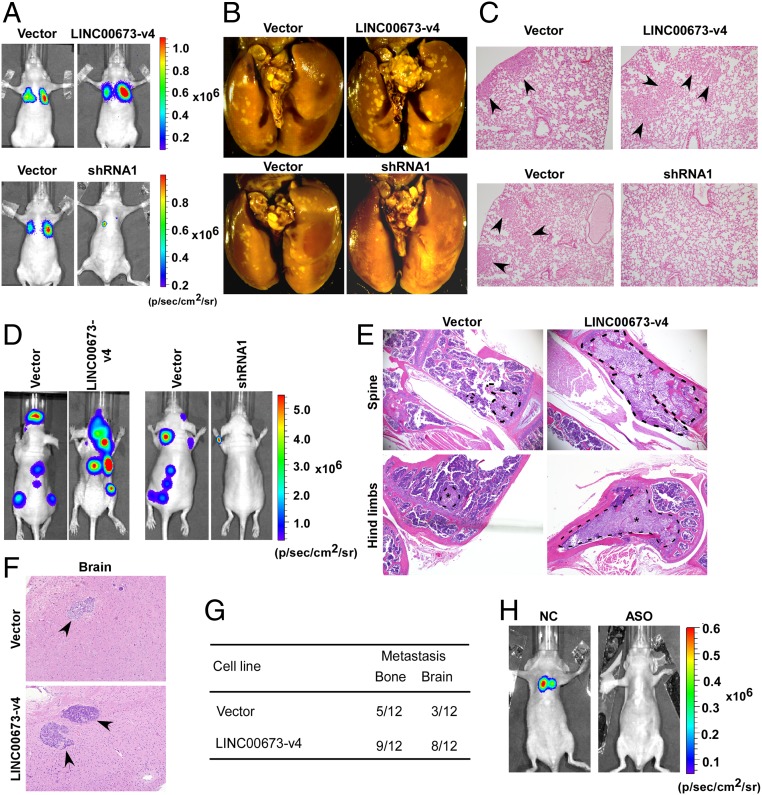
Further, the function of LINC00673-v4 was examined in vivo using experimental models of lung cancer metastasis in nude mice, with which PC9-pSin-Vector, PC9-LINC00673-v4, PC9-pSuper-Vector and PC9-LINC00673-v4–sh1 cells were injected into the lateral tail vein. As shown in Fig. 3 A and B and SI Appendix, Fig. S6A, LINC00673-v4–transduced PC9 cells showed significantly more pulmonary colonization than the vector-control cells, which was further confirmed by H&E staining (Fig. 3C). In contrast, the lung seeding activity of PC9 cells was significantly suppressed when LINC00673-v4 was depleted (Fig. 3 A–C and SI Appendix, Fig. S6A). Moreover, to further confirm the function of LINC00673-v4 in LAD metastasis, the intracardial injection metastatic model was employed. Specifically, genetically engineered cells were injected intracardially, and metastasis was determined by bioluminescence. As shown in Fig. 3D, the level of tumor burden was significantly increased in mice inoculated with LINC00673-v4–transduced PC9 cells, but was decreased for LINC00673-v4–silenced PC9 cells. Bone and brain metastases were revealed by histological analyses (Fig. 3 E–G and SI Appendix, Fig. S6 B and C). To test the efficacy of employing LINC00673-v4 antagonizing therapy in LAD, we also utilized an antisense LNA GampeRs for LINC00673-v4 to treat LAD metastatic models by directly i.v. injecting it into the lateral tail vein of the nude mice. The LINC00673-v4 antisense LNA GapmeRs-treated groups showed a significant reduction of metastatic burden compared with the antisense LNA GapmeRs negative control-treated group (Fig. 3H and SI Appendix, Fig. S6D). This result indicates that LINC00673-v4 GapmeRs represents a potential therapeutic approach for LAD metastasis. Taken together, our data indicate that LINC00673-v4 is a strong promoter molecule for LAD cell invasion, migration, and metastasis.
Fig. 3.
LINC00673-v4 promotes LAD cell metastasis in vivo. (A) Indicated cells were injected by tail vein into BALB/c nude mice for observation of lung metastases. Representative bioluminescence images are shown. (B) At the end of the experiments, mice were anesthetized and lung tissue specimens were collected, and the representative bright-field imaging of the lungs are shown. (C) Metastatic lesions in mice were confirmed by H&E staining. Arrows indicate metastatic nodules. (D) Genetically engineered cells were injected intracardially, representative bioluminescence images are shown. (E and F) Metastatic bone (E) and brain (F) lesions in mice were confirmed by H&E staining. (G) The number of mice in which the metastatic lesions were detected is summarized. (H) Antagonizing LINC00673-v4 suppressed LAD metastasis in vivo. Representative bioluminescence images are shown.
LINC00673-v4 Activates WNT/β-Catenin Signaling.
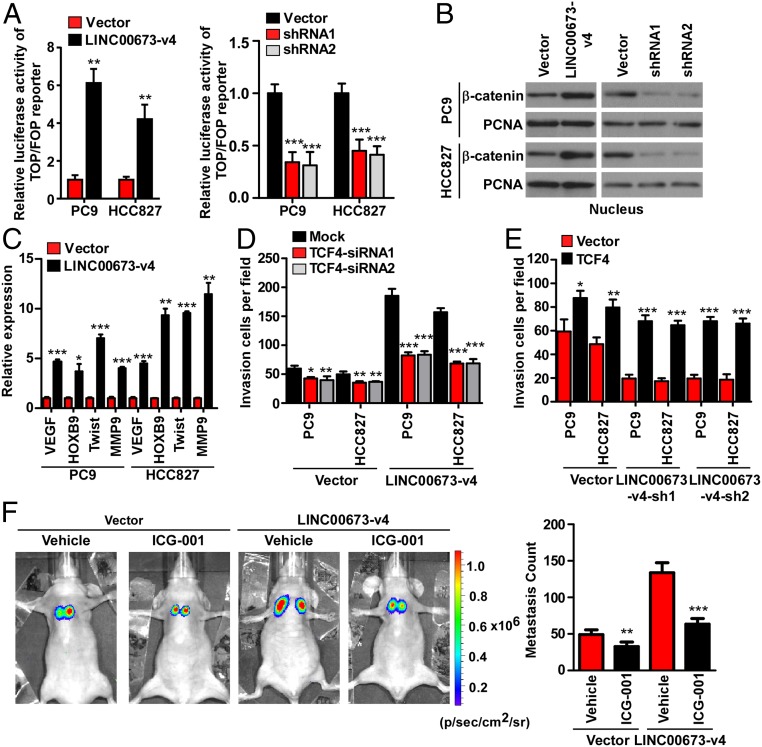
To further elucidate the role of LINC00673-v4 in WNT/β-catenin signaling, Top/Fop flash luciferase assays were performed in LINC00673-v4–overexpressing and –silenced LAD cells, respectively. As shown in Fig. 4A and SI Appendix, Fig. S7A, the TCF/LEF activity was significantly increased in cells overexpressing LINC00673-v4 and significantly decreased in cells with LINC00673-v4 silenced. Moreover, subcellular fractionation assays and immunofluorescent staining demonstrated that overexpression of LINC00673-v4 promoted the nuclear accumulation of β-catenin, whereas knockdown of LINC00673-v4 inhibited β-catenin nuclear accumulation (Fig. 4B and SI Appendix, Figs. S7B and S8). Next, we examined the effects of LINC00673-v4 on the expression of invasion-related genes downstream of the WNT/β-catenin signaling. As shown in Fig. 4C and SI Appendix, Fig. S7C, the expression levels of VEGF, HOXB9, Twist, and MMP9, were significantly up-regulated in cells overexpressing LINC00673-v4 but decreased in cells with LINC00673-v4 depleted.
Fig. 4.
LINC00673-v4 activates Wnt/β-catenin signaling. (A) Indicated cells were transfected with Top-flash or Fop-flash and Renilla pRL-TK plasmids and dual-luciferase assays were performed 48 h after transfection. The reporter activity was normalized by Renilla luciferase activity (Left: each bar represents the mean ± SD derived from three independent experiments, two-tailed Student’s t test; Right: each bar represents the mean ± SD derived from three experiments, one-way ANOVA followed by Dunnett’s multiple comparison test. **P < 0.01, ***P < 0.001). (B) Altered nuclear translocation of β-catenin in response to LINC00673-v4 overexpression in indicated cells was analyzed by Western blot (WB) analysis. Proliferating cell nuclear antigen was used as a loading control. (C) The expression levels of VEGF, HOXB9, Twist, and MMP9 were increased by ectopic expression of LINC00673-v4 (each bar represents the mean ± SD derived from three independent experiments, two-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001). (D) Quantification of invading cells using Transwell invasion assay in LINC00673-v4–overexpressing and vector-control cells with silencing of TCF4 (each bar represents the mean ± SD derived from three experiments, one-way ANOVA followed by Dunnett’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001). (E) Quantification of invading cells by overexpression of TCF4 in LINC00673-v4–silencing and vector-control cells (each bar represents the mean ± SD derived from three independent experiments, two-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001). (F) The effects of ICG-001, an inhibitor of Wnt signaling, on metastasis of PC9-LINC00673-v4 and PC9-Vector (two-tailed Student’s t test. **P < 0.01, ***P < 0.001).
We next investigated whether WNT/β-catenin activation plays a role in mediating LINC00673-v4–induced cellular invasion. First, we examined the effect of inhibiting WNT/β-catenin signaling, via depleting TCF4 or LEF1, on the invasion of LINC00673-v4–transduced PC9, HCC827, and H2030 cell lines. As indicated in Fig. 4D and SI Appendix, Fig. S7 D–F, LINC00673-v4–induced cell invasion was abrogated when TCF4 or LEF1 was silenced. We also assessed the effect of knocking down TCF4 or LEF1 on control vector-transduced cells. TCF4 or LEF1 knockdown resulted in a reduction in cell invasion, but not as significantly as in the LINC00673-v4–overexpressing LAD cells (Fig. 4D and SI Appendix, Fig. S7 D–F). Second, ectopic overexpression of TCF4 or LEF1 in LINC00673-v4–silenced PC9, HCC827, and H2030 cells rescued the ability of LAD cells to invade (Fig. 4E and SI Appendix, Fig. S7 D, G, and H). We also found that overexpression of TCF4 or LEF1 in empty vector-transfected LAD cells increased the invasiveness of the LAD cells, and these effects could be further enhanced in LINC00673-v4–silenced LAD cells (Fig. 4E and SI Appendix, Fig. S7 D, G, and H). Third, when we sought to further determine whether activated WNT/β-catenin signaling plays a role in LINC00673-v4–mediated prometastatic effects in a mouse model by employing a WNT/β-catenin signaling inhibitor, our in vivo experiments showed that the lung seeding activity of PC9-vector cells was suppressed by ICG-001 treatment, a Wnt signaling inhibitor, but the suppressive effect of ICG-001 on the lung seeding activity of PC9-vector cells is less potent than that on LINC00673-v4–overexpressing PC9 cells (Fig. 4F). Taken together, our results suggest that the activity of WNT/β-catenin signaling can be promoted by LINC00673-v4, which consequently augments invasion and metastasis of LAD cells.
LINC00673-v4 Interacts with DDX3 and CK1ε.
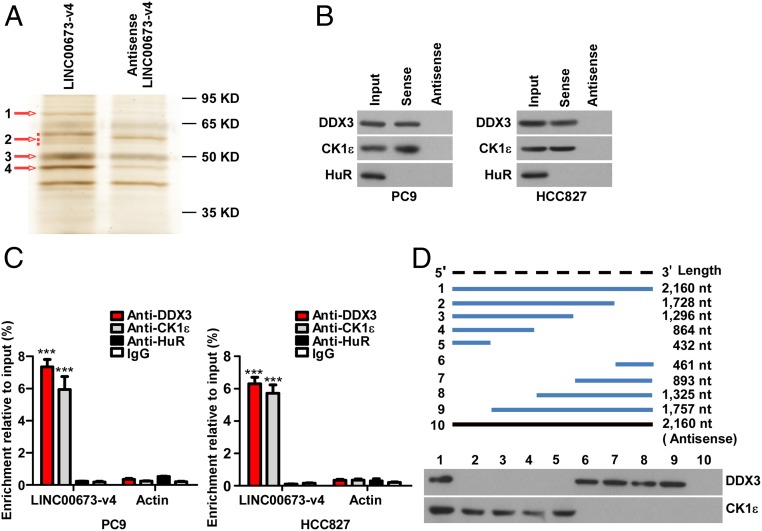
Previous studies have extensively shown that many lncRNAs exert their biological functions through physically interacting with protein molecules. To investigate the molecular mechanism underlying the role of LINC00673-v4 in modulating WNT/β-catenin signaling, we began our study by identifying intracellular binding proteins of LINC00673-v4. For this purpose, biotinylated LINC00673-v4, or its antisense RNA, was incubated with PC9 cell lysates, followed by a pull-down experiment. Several differentially displayed protein bands (bands 1–4 numerically labeled on the gel, arrowed in Fig. 5A) on the silver-stained polyacrylamide gel were identified. These differential bands in the gel were cut in slices and subjected to mass spectrometry (MS) analysis. After MS analysis of each band, a number of putative interacting proteins (unique peptides ≥2) in each band were identified (SI Appendix, Tables S7–S10). We noted that CK1ε, the top-ranked LINC00673-v4–interacting protein in band 4, has been suggested to play an important role in WNT/β-catenin signaling (SI Appendix, Fig. S9 and Table S10). To validate that LINC00673-v4 interacts with CK1ε, we performed RNA pull-down and immunoblotting using a CK1ε antibody, and the results showed that CK1ε was detected in the LINC00673-v4 protein complex but not in the control sample (Fig. 5B and SI Appendix, Fig. S10A). Moreover, the direct interaction was further verified by RNA immunoprecipitation (RIP) assay after UV cross-link (Fig. 5C and SI Appendix, Fig. S10B). Several other putative proteins with high peptide counts were also found in band 4, including amylase alpha 1A (AMY1A) that is involved in the glycosaminoglycan metabolism pathway, and ribosomal protein L4 (RPL4), a component of the 60S ribosome. It is of note that DDX3 interacts with CK1ε and stimulates its kinase activity, which promotes the phosphorylation of the Dvl and activation of WNT/β-catenin signaling (29, 30). Intriguingly, we found that DDX3 was among the putative LINC00673-v4–interacting proteins of band 1 (SI Appendix, Fig. S9 and Table S7). RNA pull-down followed by immunoblotting and RIP assay after UV cross-link were performed, and the results showed that LINC00673-v4 also directly interacted with DDX3 (Fig. 5 B and C and SI Appendix, Fig. S10 A and B). Consistent with the above results derived from the LAD cell lines, LINC00673-v4 was shown to interact with both DDX3 and CK1ε and activate WNT/β-catenin signaling in patient-derived primary LAD cells (SI Appendix, Fig. S11 A–D). In addition, we found several energy metabolism-related proteins in bands 2 and 3 with high peptide counts, including methylcrotonoyl-CoA carboxylase 2 (MCCC2), pyruvate kinase M1/2 (PKM), ATP synthase F1 subunit beta (ATP5B), glutamate dehydrogenase 1 (GLUD1), and ATP synthase F1 subunit alpha (ATP5A1) (SI Appendix, Tables S8 and S9). The interaction between these factors and LINC00673-v4 needs to be further validated.
Fig. 5.
LINC00673-v4 directly interacts with both DDX3 and CK1ε. (A) Imaging of RNA pull-down experiment followed by silver staining. (B) WB validation of proteins pulled down with LINC00673-v4. (C) RIP assays after UV cross-link were performed to verify the interaction of LINC00673-v4 with DDX3 and CK1ε (each bar represents the mean ± SD derived from three experiments, one-way ANOVA followed by Dunnett’s multiple comparison test. ***P < 0.001). (D) WB analysis of DDX3 and CK1ε pulled down by full-length or truncated LINC00673-v4.
To investigate whether LINC00673-v4 is not nonspecifically binding to just any RNA binding protein (RBP), we next investigated the interaction between LINC00673-v4 and an RBP (HuR) by RNA pull-down followed by immunoblotting assay and RIP assay after UV cross-link. Our results showed that HuR did not specifically interact with LINC00673-v4 (Fig. 5 B and C and SI Appendix, Figs. S10 A and B and S11 C and D).
To identify the intramolecular regions of LINC00673-v4 interacting with the two proteins involved in the interaction, eight fragments of LINC00673-v4 (1–1,728 nt, 1–1,296 nt, 1–864 nt, 1–432 nt, 1,729–2,189 nt, 1,297–2,189 nt, 865–2,189 nt, and 433–2,189 nt) were in vitro transcribed and biotinylated and used in pull-down assays with total protein extracts from PC9 cells. As shown in Fig. 5D, we found that segment 1,729–2,160 nt of LINC00673-v4 interacted with DDX3, while segment 1–432 nt of LINC00673-v4 interacted with CK1ε. Moreover, RNA folding analysis predicted that these two binding fragments contained stable stem-loop structures (SI Appendix, Fig. S11E), further underscoring the aforementioned deduced LINC00673-v4 regions interacting with DDX3 and CK1ε. In addition, our protein domain mapping experiment showed that the helicase ATP binding domain of DDX3 and the protein kinase domain of CK1ε interacted with LINC00673-v4 (SI Appendix, Fig. S12 A and B). Taken together, these data suggest that LINC00673-v4 is a direct interactive binding partner of both DDX3 and CK1ε.
LINC00673-v4 Functions as a Modular Scaffold in the WNT/β-Catenin Signaling Pathway.
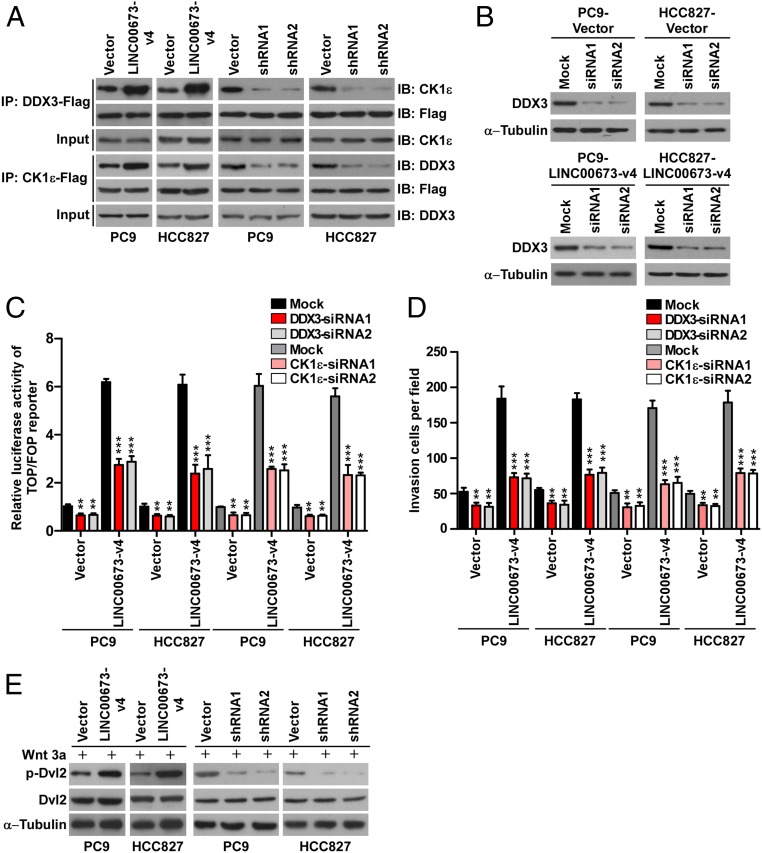
To further understand whether the binding of LINC00673-v4 with both DDX3 and CK1ε strengthens the interaction between DDX3 and CK1ε, we performed coimmunoprecipitation (co-IP) experiments and found that DDX3 and CK1ε could reciprocally interact with each other and that overexpression of LINC00673-v4 enhanced the interaction of DDX3 and CK1ε, whereas LINC00673-v4 knockdown attenuated this interaction in LAD cell lines and patient-derived LAD cells (Fig. 6A and SI Appendix, Figs. S10C and S11F), indicating that LINC00673-v4 level indeed augments the association between DDX3 and CK1ε. Moreover, immunofluorescence staining and in situ proximity ligation assay (PLA) also showed that interaction between DDX3 and CK1ε could be enhanced by LINC00673-v4 overexpression but was inhibited when LINC00673-v4 was depleted (SI Appendix, Fig. S13). These together suggest that LINC00673-v4 directly binds both DDX3 and CK1ε and strengthens their interaction.
Fig. 6.
LINC00673-v4 functions as a modular scaffold in the WNT/β-catenin signaling pathway. (A) Co-IP detection of the indicated proteins in PC9 and HCC827 cells with LINC00673-v4 overexpression or knockdown. (B) WB analysis of the transfection of siDDX3 in LINC00673-v4–overexpressing and vector-control PC9 and HCC827 cells. (C) Luciferase analysis of TCF/LEF transcriptional activity in indicated cells (each bar represents the mean ± SD derived from three experiments, one-way ANOVA followed by Dunnett’s multiple comparison test. **P < 0.01, ***P < 0.001). (D) Quantification of invading cells by Transwell invasion assay in LINC00673-v4–overexpressing and vector-control cells with depletion of DDX3 (each bar represents the mean ± SD derived from three experiments, one-way ANOVA followed by Dunnett’s multiple comparison test. **P < 0.01, ***P < 0.001). (E) The expression levels of p-Dvl2 and total Dvl2 were detected in indicated cells.
As it has been demonstrated that binding between DDX3 and CK1ε induces phosphorylation of Dvl and thereby enhances nuclear accumulation of β-catenin and activation of WNT/β-catenin signaling (29), we next sought to determine whether LINC00673-v4 interaction with both DDX3 and CK1ε is required for activation of WNT/β-catenin signaling and consequently, for the aggressiveness of LAD. To this end, we knocked down DDX3 or CK1ε in LINC00673-v4–overexpressing LAD cells and found that LINC00673-v4–induced WNT/β-catenin activation and increase of invasive potential in LAD cells could be inhibited by silencing DDX3 or CK1ε (Fig. 6 B–D and SI Appendix, Fig. S10 D–H). We also investigated the effect of knocking down DDX3 or CK1ε on control vector-transduced cells. DDX3 or CK1ε knockdown led to a reduction in WNT/β-catenin activation and cell invasion on control vector-transduced cells, but not as significantly as in the LINC00673-v4–overexpressing LAD cells (Fig. 6 B–D and SI Appendix, Fig. S10 D–H). We also observed that the phosphorylation of Dvl was increased in LINC00673-v4–overexpressing LAD cells, whereas silencing LINC00673-v4 expression decreased Dvl phosphorylation (Fig. 6E). Taken together, these data indicate that LINC00673-v4 may indeed act as a modular scaffold for the DDX3 and CK1ε complex to activate WNT/β-catenin signaling.
LINC00673-v4 Is Clinically Associated with WNT/β-Catenin Signaling in LAD.
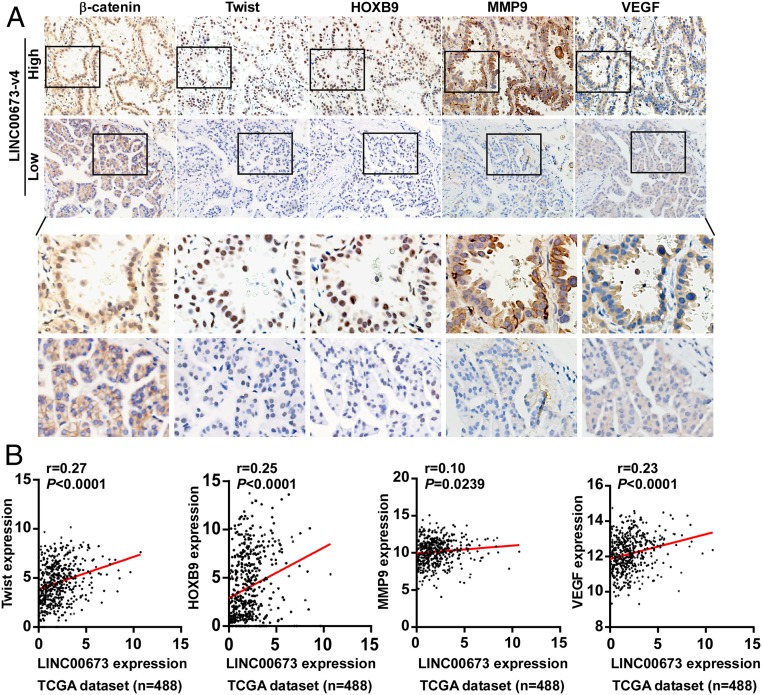
To further investigate whether the above findings were clinically relevant, we employed LAD clinical specimens to examine the expression of LINC00673-v4 and its correlation with hallmarks of WNT/β-catenin activation. As shown in Fig. 7A, positive correlations between LINC00673-v4 level and expression of nuclear β-catenin (r = 0.238, P = 0.009), Twist (r = 0.205, P = 0.025), HOXB9 (r = 0.197, P = 0.032), MMP9 (r = 0.191, P = 0.038), and VEGF (r = 0.202, P = 0.028) were found in LAD specimens (SI Appendix, Table S11). Additionally, we analyzed the data of the LAD patient cohort collected in the TANRIC database; as shown in Fig. 7B, high LINC00673 expression positively correlated with the levels of Twist, HOXB9, MMP9, and VEGF in LAD specimens, and vice versa, suggesting that in LAD patients, LINC00673-v4 clinically contributes to activation of the WNT/β-catenin signaling pathway, resulting in increased expression of nuclear β-catenin, VEGF, Twist, HOXB9, and MMP9.
Fig. 7.
LINC00673-v4 positively correlates with activation of Wnt/β-catenin signaling in clinical specimens. (A) Expression of LINC00673-v4 is associated with β-catenin localization and expression levels of MMP9, Twist, VEGF, and HOXB9. Two representative cases are shown (magnification 400×). (B) The association of LINC00673 and Twist, HOXB9, MMP9, as well as VEGF in 488 cases of LAD specimens was analyzed using TANRIC datasets (correlation was assessed using Spearman’s correlation coefficient).
Discussion
Our current study reports a prometastatic mechanism of LAD mediated by a lncRNA, LINC00673-v4, as evidenced by several lines of findings, including the identified up-regulation of LINC00673-v4 in LAD and its close correlation with the progression of LAD as well as clinical patient outcomes, in addition to the data from our mechanistic studies. At the molecular level, our study uncovers that LINC00673-v4 promotes cancer cell invasion, migration, and metastasis by serving as a scaffold molecule to overactivate WNT/β-catenin signaling.
Local invasion and distant metastasis are complex, multistep processes (31). Current understanding of how LAD cells invade neighboring tissue and disseminate to distant sites remains incomplete, and molecular targets for effective antimetastatic therapies are yet to be identified. Of particular note, activation of the WNT/β-catenin signaling pathway plays a vital role in the distant metastasis during LAD progression (12). Our present study demonstrates that LINC00673-v4 is an essential RNA regulator of WNT/β-catenin signaling. Our further in-depth analyses using mass spectrometry, RNA pull-down, and RIP assays have strongly suggested that LINC00673-v4 acts as a modular scaffold to directly interact with DDX3 and CK1ε, important modulators in WNT/β-catenin signaling activation. This notion is supported by several lines of experimental evidence. First, biochemical evidence revealed that LINC00673-v4 directly binds with CK1ε at its 5′ region and with DDX3 at its 3′ region. Second, the interaction between the DDX3 and CK1ε can indeed be promoted in LINC00673-v4–overexpressing cells and abrogated in cells with LINC00673-v4 depleted. Third, overexpression of LINC00673-v4 enhances the DDX3/CK1ε complex-induced Dvl phosphorylation and subsequent activation of WNT/β-catenin signaling, whereas silencing LINC00673-v4 inhibits these effects. Taken together, our data reported here reveal a lncRNA-based regulatory modality of the prometastatic WNT/β-catenin signaling.
It has been well documented that DDX3 interacts with CK1ε to enhance its kinase activity, leading to phosphorylation of Dvl and activation of WNT/β-catenin signaling in mammalian cells and malignant tumor cells (29, 32). During the activation of canonical Wnt signaling, secreted Wnt ligand-bound receptors recruit Dvl to the plasma membrane where they are activated. Phosphorylation of Dvl by CK1ε inactivates the destruction complex and results in stabilization and nucleus translocation of β-catenin (33). Notably, the oncogenic roles of DDX3 in various types of cancers, including NSCLC, have been demonstrated (34). Bol et al. found that DDX3 is overexpressed in NSCLC and associated with a poor clinical outcome (35). Moreover, DDX3 promotes cellular stemness and epithelial–mesenchymal transition and inhibits the sensitivity to EGFR-TKI in NSCLC cells (36). Our study extends the current understanding of the oncogenic roles of DDX3 and identifies a model that LINC00673-v4 scaffolds DDX3 and CK1ε to form a complex, which contributes to Dvl phosphorylation and subsequent activation of WNT/β-catenin signaling in LAD, highlighting an important lncRNA-protein kinase module that regulates β-catenin stabilization and aggressiveness of LAD.
It is particularly noteworthy that inhibition of DDX3 by RK-33, a small molecule inhibitor designed to bind the ATP binding site of DDX3 and thereby to abrogate its activity, suppresses Wnt signaling and the malignant phenotype in several types of cancers (37, 38). For instance, RK-33 in combination with radiation led to tumor regression in mouse models of lung cancer, indicating a potential therapeutic benefit of inhibiting DDX3 for cancers (35). However, whether such a treatment strategy can be challenged by drug resistance, which is commonly seen in most targeting therapies as well as chemotherapies, remains unknown. Interestingly, sensitivity of cancer cells to RK-33 inhibition of DDX3 appears to be associated with specific genetic alterations. In colorectal cancer cells, the highest RK-33 sensitivity was found in cancer cells with wild-type APC and a mutation in CTNNB1 (38). Given the pivotal role of LINC00673-v4 in DDX3/CK1ε complex and in the subsequent activation of WNT/β-catenin signaling, it would be of great interest to further investigate whether therapeutics targeting LINC00673-v4 may help overcome chemo resistance to DDX3 inhibitor(s) across various oncogenotypes.
It is noteworthy that LINC00673 has been implicated in the development and progression of NSCLC. Recently, LINC00673 has been shown to contribute to the bypass of Ras-induced NSCLC cell senescence by inhibiting p53 translation (39). Shi et al. demonstrated that the oncogenic role of LINC00673 in NSCLC was attributable to its suppression of NCALD via interacting with LSD1, a H3K4 histone demethylase (40). Furthermore, LINC00673 could function as a ceRNA to sponge miR-150–5p and thereby promote the proliferation and invasion of NSCLC cells (41). Moreover, LINC00673 promoted the aggressiveness of NSCLC cells via engaging in epigenetic gene silencing of HOX5 (42). Notably, these studies focused on LINC00673-v3, which is the second abundant transcript in LAD cells and specimens that we assessed. Here in our current study, we identified that LINC00673-v4 was the most abundant transcript in LAD and the importance of LINC00673-v4 in LAD cell invasion and metastasis. Despite the similarity among the sequences of LINC00673 transcripts, the difference between them may allow formation of sequence-specific structures and distinct structure-based interactions with proteins, DNAs, or RNAs. Thus, it can be rationally speculated that different LINC00673 transcripts might play distinct roles in the development and progression of NSCLC biologically via different modes of action.
Interestingly, the locus of LINC00673 largely overlaps with the locus of LINC00511. Based on the latest version of the Ensembl genome browser (Ensembl genome browser 96, http://asia.ensembl.org/index.html), 106 transcripts have been found to derive from LINC00511, including LINC00673-v1-5. Accumulating evidence has revealed the role of LINC00511 in the development and progression of cancers. For instance, LINC00511-213, one of the LINC00511 transcripts, was found to be up-regulated in NSCLC and to promote cell proliferation and invasion via binding histone methyltransferase enhancer of zeste homolog 2 (EZH2), acting as a modular scaffold of EZH2/PRC2 complexes to regulate the histone modification pattern on the p57 target gene (43). Due to the complexity of this gene locus, further studies are required to investigate the biological significance of this locus and its RNA products in cancers.
Of note, it is not impossible that LINC00673-v4 exerts its function via more than one mechanism or pathway. Also notably, we found that the majority of proteins identified in bands 2 and 3 with high peptide counts, including MCCC2, PKM, ATP5B, GLUD1, and ATP5A1, are involved in the processes of energy metabolism. It is of interest to note that reprogrammed energy metabolism plays important roles in cancer development and progression (44, 45). Thus, whether the oncogenic role of LINC00673-v4 also requires additional mechanisms via interacting these putative proteins remains to be clarified in future studies.
Accumulating evidence has indicated that specific lncRNA expression can correlate with clinical features in various types of cancers, supporting the utility of lncRNA in diagnosis and prognosis of the disease (46, 47). Here, we found that LINC00673-v4 was significantly up-regulated in LAD tissues. In our studies, it is noteworthy that the expression of LINC00673-v4 is closely associated with LN metastasis. It has been well recognized that progression of LAD can involve regional lymph nodes at early stages and that the degree of locoregional lymph node involvement is one of the crucial prognosis factors of the disease (48). Consistent with this notion, our results indicated that patients with high LINC00673-v4 had shorter survival time.
Given the clinical, functional, and mechanistic significance of LINC00673-v4 in LAD, we conclude that LINC00673-v4 and its regulatory role in WNT/β-catenin signaling is critical for the aggressive behavior of LAD, and LINC00673-v4 may potentially be an effective target for LAD therapy.
Materials and Methods
Cell Culture.
LAD cell lines, including HCC827, NCI-H1650, A549, NCI-H596, NCI-H1975, NCI-H1299, SK-LU-1, NCI-H358, NCI-H2009, HCC4006, and NCI-H2030 were purchased from the American Type Culture Collection (ATCC; Manassas, VA). PC9 were obtained from cell banks of the Shanghai Institutes of Biological Sciences (Shanghai, China). Cells were maintained in DMEM supplemented with 10% FBS (Corning, Corning, NY). All cell lines were authenticated by short tandem repeat (STR) DNA profiling and verified to be mycoplasma-free.
Bioinformatics Analysis.
RNAseq data for lncRNA expression of 58 pairs of LAD tissues versus paired adjacent noncancerous lung tissues and 488 cases of LAD tissues were mined from TCGA (https://cancergenome.nih.gov/) using TANRIC (https://ibl.mdanderson.org/tanric/_design/basic/index.html).
Statistical Analysis.
All statistical analyses were performed using the SPSS 20.0 statistical software package. Survival curves were analyzed by the Kaplan–Meier method, and a log-rank test was used to assess significance. The χ2 test was used to analyze the correlation between the expression levels of LINC00673-v4 and clinical parameters of patients. Comparisons between two groups were performed using Student’s t test. For pairwise multiple comparisons, one-way ANOVA followed by Dunnett’s multiple comparison test was used. Correlation between two groups was assessed by use of Spearman’s correlation coefficient. All error bars represent the mean ± SD derived from three independent experiments. P values <0.05 were considered statistically significant.
Study Approval.
Prior patient consent and approval from the Institutional Research Ethics Committee of Sun Yat-sen University were obtained for the use of these clinical materials for research purposes. All animal procedures were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University.
Data Availability.
All data generated and analyzed in this study are available with the paper and SI Appendix online.
Supplementary Material
Acknowledgments
This work was supported by the Natural Science Foundation of China (No. 81490752); the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (No. 81621004); Guangzhou Science and Technology Plan (No. 201803010039); “Guangdong Te Zhi Program” youth science and technology talent (Project No. 2015TQ01R281); and the Guangdong Natural Science Funds for Distinguished Young Scholars (No. 2014A030306023).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900997116/-/DCSupplemental.
References
- 1.Chen W., et al. , Cancer statistics in China, 2015. CA Cancer J. Clin. 66, 115–132 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Travis W. D., et al. ; WHO Panel , The 2015 World Health Organization classification of lung tumors: Impact of genetic, clinical and radiologic advances since the 2004 classification. J. Thorac. Oncol. 10, 1243–1260 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Imielinski M., et al. , Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller K. D., et al. , Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 66, 271–289 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Copur M. S., Crockett D., Gauchan D., Ramaekers R., Mleczko K., Molecular testing guideline for the selection of patients with lung cancer for targeted therapy. J. Clin. Oncol. 36, 2006 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Choi E. B., et al. , PARP1 enhances lung adenocarcinoma metastasis by novel mechanisms independent of DNA repair. Oncogene 35, 4569–4579 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Zhang X., et al. , Cytosolic TMEM88 promotes invasion and metastasis in lung cancer cells by binding DVLS. Cancer Res. 75, 4527–4537 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Cai J., et al. , MicroRNA-374a activates Wnt/β-catenin signaling to promote breast cancer metastasis. J. Clin. Invest. 123, 566–579 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Licchesi J. D., et al. , Epigenetic alteration of Wnt pathway antagonists in progressive glandular neoplasia of the lung. Carcinogenesis 29, 895–904 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ju Z., et al. , Global detection of molecular changes reveals concurrent alteration of several biological pathways in nonsmall cell lung cancer cells. Mol. Genet. Genomics 274, 141–154 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shapiro M., et al. , Wnt pathway activation predicts increased risk of tumor recurrence in patients with stage I nonsmall cell lung cancer. Ann. Surg. 257, 548–554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen D. X., et al. , WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell 138, 51–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polakis P., Drugging Wnt signalling in cancer. EMBO J. 31, 2737–2746 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubinfeld B., et al. , Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly. Science 272, 1023–1026 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Hart M. J., de los Santos R., Albert I. N., Rubinfeld B., Polakis P., Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta. Curr. Biol. 8, 573–581 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Ueno K., Hirata H., Hinoda Y., Dahiya R., Frizzled homolog proteins, microRNAs and Wnt signaling in cancer. Int. J. Cancer 132, 1731–1740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nam J. S., Turcotte T. J., Smith P. F., Choi S., Yoon J. K., Mouse cristin/R-spondin family proteins are novel ligands for the Frizzled 8 and LRP6 receptors and activate beta-catenin-dependent gene expression. J. Biol. Chem. 281, 13247–13257 (2006). [DOI] [PubMed] [Google Scholar]
- 18.McKendry R., Hsu S. C., Harland R. M., Grosschedl R., LEF-1/TCF proteins mediate wnt-inducible transcription from the Xenopus nodal-related 3 promoter. Dev. Biol. 192, 420–431 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Morin P. J., et al. , Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275, 1787–1790 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Ueda M., et al. , Mutations of the beta- and gamma-catenin genes are uncommon in human lung, breast, kidney, cervical and ovarian carcinomas. Br. J. Cancer 85, 64–68 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding L., et al. , Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455, 1069–1075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Y., Zhou T., Yu X., Xue Z., Shen N., The role of long non-coding RNAs in rheumatic diseases. Nat. Rev. Rheumatol. 13, 657–669 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Bhan A., Soleimani M., Mandal S. S., Long noncoding RNA and cancer: A new paradigm. Cancer Res. 77, 3965–3981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Botchkareva N. V. The molecular revolution in cutaneous biology: Noncoding RNAs: New molecular players in dermatology and cutaneous biology. J. Invest. Dermatol. 137, e105–e111 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Peng W. X., Koirala P., Mo Y. Y., LncRNA-mediated regulation of cell signaling in cancer. Oncogene 36, 5661–5667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li D., et al. , Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology 65, 1612–1627 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Cabanski C. R., et al. , Pan-cancer transcriptome analysis reveals long noncoding RNAs with conserved function. RNA Biol. 12, 628–642 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beroukhim R., et al. , The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cruciat C. M., et al. , RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-β-catenin signaling. Science 339, 1436–1441 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Dolde C., et al. , A CK1 FRET biosensor reveals that DDX3X is an essential activator of CK1ε. J. Cell Sci. 131, jcs207316 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steeg P. S., Metastasis suppressors alter the signal transduction of cancer cells. Nat. Rev. Cancer 3, 55–63 (2003). [DOI] [PubMed] [Google Scholar]
- 32.He T. Y., et al. , DDX3 promotes tumor invasion in colorectal cancer via the CK1ε/Dvl2 axis. Sci. Rep. 6, 21483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernatik O., et al. , Sequential activation and inactivation of Dishevelled in the Wnt/beta-catenin pathway by casein kinases. J. Biol. Chem. 286, 10396–10410 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bol G. M., Xie M., Raman V., DDX3, a potential target for cancer treatment. Mol. Cancer 14, 188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bol G. M., et al. , Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol. Med. 7, 648–669 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tyack P. L., Calambokidis J., Friedlaender A., Goldbogen J., Southall B., Formal comment on Schorr GS, Falcone EA, Moretti DJ, Andrews RD (2014) First long-term behavioral records from cuvier’s beaked whales (Ziphius cavirostris) reveal record-breaking dives. PLoS ONE 9(3): e92633. doi:10.1371/journal.pone.0092633. PLoS One 10, e0142287 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie M., et al. , RK-33 radiosensitizes prostate cancer cells by blocking the RNA helicase DDX3. Cancer Res. 76, 6340–6350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heerma van Voss M. R., et al. , Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer. Oncotarget 6, 28312–28326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth A., et al. , Targeting LINC00673 expression triggers cellular senescence in lung cancer. RNA Biol. 15, 1499–1511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi X., et al. , Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer. Oncotarget 7, 25558–25575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu W., et al. , Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol. Cancer 16, 118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma C., et al. , Long intergenic noncoding RNA 00673 promotes non-small-cell lung cancer metastasis by binding with EZH2 and causing epigenetic silencing of HOXA5. Oncotarget 8, 32696–32705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun C. C., et al. , Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol. Ther. Nucleic Acids 5, e385 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Fong M. Y., et al. , Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 17, 183–194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhai W., et al. , LncRNA-SARCC suppresses renal cell carcinoma (RCC) progression via altering the androgen receptor(AR)/miRNA-143-3p signals. Cell Death Differ. 24, 1502–1517 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao L., et al. , Long noncoding RNA LINC00092 acts in cancer-associated fibroblasts to drive glycolysis and progression of ovarian cancer. Cancer Res. 77, 1369–1382 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Schmid K., et al. , EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin. Cancer Res. 15, 4554–4560 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and analyzed in this study are available with the paper and SI Appendix online.