Significance
Although it is now possible to visualize single RNA molecules within cells by fluorescence in situ hybridization, the resulting signals are rather faint. Here we describe a method that produces amplified signals from single RNA molecules, while minimizing false-positive signals. Our probes enable detection of sparsely expressed mRNAs, and they are so specific that related mutant and wild-type mRNAs, differing from each other by as little as a single-nucleotide variation, yield distinct in situ signals in 2 distinguishable colors. This technology will aid studies of gene expression phenomena, such as allelic imbalance, and will be useful for the identification of mRNAs harboring somatic mutations in cancer cells, facilitating targeted therapy.
Keywords: cancer diagnostics, RNA imaging, mutation detection
Abstract
Amplification of signals by the hybridization chain reaction (HCR) is a powerful approach for increasing signal strength in single-molecule fluorescence in situ hybridization, but probes tagged with an HCR initiator sequence are prone to producing false signals. Here we describe a system of interacting hairpin binary probes in which the HCR initiator sequence is conditionally sequestered. The binding of these probes to a perfectly complementary target unmasks the initiator, enabling the generation of an amplified signal. This probe system can distinguish single-nucleotide variations within single mRNA molecules and produces amplified signals in situ for both mutant and wild-type variants, each in a distinguishable color. This technology will augment studies of imbalanced allelic expression and will be useful for the detection of somatic mutations in cancer biopsies. By tiling these probes along the length of an mRNA target, enhanced signals can be obtained, thereby enabling the scanning of tissue sections for gene expression utilizing lower magnification microscopy, overcoming tissue autofluorescence, and allowing the detection of low-abundance biomarkers in flow cytometry.
Advances in probe technologies now permit reliable detection of single RNA molecules by fluorescence in situ hybridization (sm-FISH) in cultured cells, tissue slices, and small intact model organisms (1, 2). Iterative rounds of hybridization and imaging allow the detection of dozens of different RNA species in the same cells (3, 4). Despite the use of multiple probes tiled over the length of mRNAs, signals in sm-FISH are rather faint, which necessitates imaging at high magnifications, thus rendering analyses of large tissue sections difficult. In flow cytometry–based FISH, low signal strengths limit the range of detectable biomarkers to abundantly expressed transcripts. In addition, RNA targets that differ from each other by only a single-nucleotide variation (SNV) are refractory to detection by sm-FISH, as only one of the many probes that are used binds to the mutated site. When single probes against SNVs are employed and detected with a microscopic setup that is sufficiently sensitive to detect the fluorescence of a single dye molecule, the majority of signals originate from the off-target binding of probes (5), although signals that are specific to an SNV can be recognized by their colocalization with signals from distinguishably colored sm-FISH probes that are hybridized to the same mRNA (5–7).
To increase signal strength in FISH, a number of signal amplification schemes that restrict the amplified signal to the site of the target have been developed. They include the hybridization chain reaction (HCR), branched DNA amplification, clamp-FISH, and rolling circle amplification (8–13). However, nonspecifically bound probes can also trigger amplification, giving rise to false signals that increase as the number of probes is increased (14).
Here we report a technique referred to as “high-fidelity amplified FISH” (amp-FISH), in which the target-bound probes generate amplified signals while the generation of signals from the nonspecifically bound probes is suppressed. This technique utilizes pairs of interacting binary probes that contain strategically placed hairpins. The hybridization of these probes to their target sequence, and their subsequent interaction with each other, drives changes in their conformations that lead to the release of a sequestered HCR initiator sequence, thereby enabling HCR to occur. We will first describe the basic features of these probes and demonstrate that they render signal generation target dependent. Then we will describe the use of a combination of 2 pairs of amp-FISH probes to generate amplified signals in distinguishable colors from single mRNA molecules that differ from each other by an SNV, using an experimental transcript produced from a transfected plasmid and an endogenous transcript that occurs naturally in cells. Finally, we will show how the tiling of multiple pairs of binary probe pairs leads to an enhancement in signal strength compared with conventional sm-FISH.
Results
Target-Mediated Unmasking of a Hidden HCR Initiator.
In the hybridization chain reaction, first described by Niles Pierce and his colleagues (8, 9), signals are generated by the polymerization of a pair of partially complementary, fluorescently labeled hairpins in situ at the target site on an mRNA molecule. Rather than using the mRNA target sequence to initiate HCR, a generic initiator sequence is appended to a unique target-specific probe that binds to the target (9). However, like any other hybridization probe, these probes sometimes associate nonspecifically with unrelated RNAs and with other components of the cellular matrix, generating false amplified signals. A split-probe strategy that reduces false signals has recently been described (14).
To ensure that only target-bound probes initiate HCR, we have designed an alternative scheme, high-fidelity amp-FISH, that combines 3 specificity-enhancing concepts in one design. First, we use target-specific binary probes that must hybridize next to each other on the target sequence to generate a signal (15, 16); second, one of these target-specific probes is a molecular beacon–like hairpin that undergoes a conformational reorganization only when bound to its intended target sequence (17); third, these 2 bound probes interact with each other, undergoing a target-mediated “toehold” strand-displacement reaction (18) that unmasks an otherwise sequestered HCR initiator sequence, which only occurs when the 2 probes are bound to the target at adjacent positions.
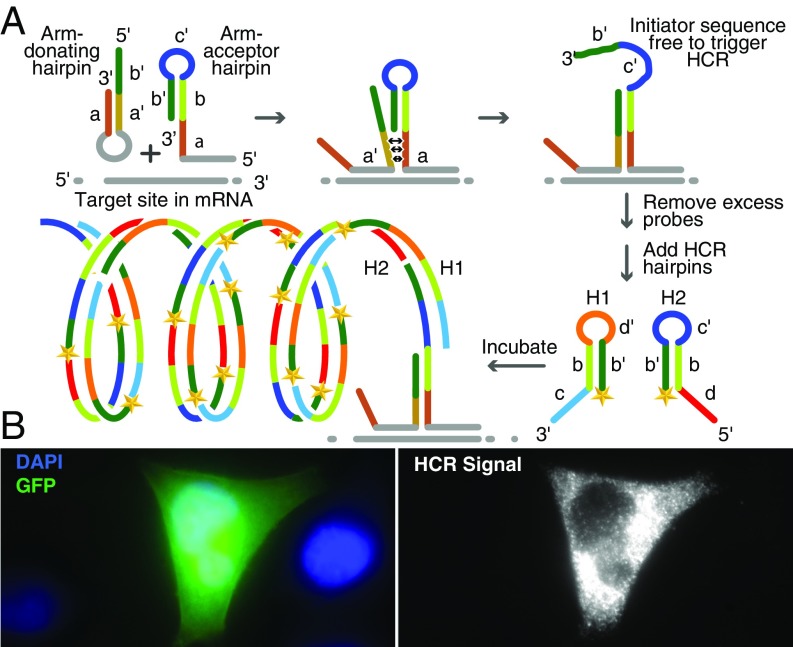
This basic system is shown in Fig. 1A. One of the probes is an “arm-donating hairpin,” which undergoes a molecular beacon–like conformational reorganization when it binds to its target sequence. In its unbound form, this probe’s mutually complementary “arm” sequences, a and aʹ, keep the molecule in a closed-hairpin conformation. However, when the probe sequence in the hairpin loop binds to its target sequence, the rigidity of the resulting probe-target hybrid forces these arm sequences to dissociate from each other. Only a perfectly complementary target sequence is able to drive this conformational change (19). The second probe, called an “arm-acceptor hairpin,” is designed to bind to an immediately adjacent target sequence to the right of the target sequence for the arm-donating hairpin. The arm-acceptor hairpin contains a copy of sequence a, which in the resulting tripartite nucleic acid complex is situated in close proximity to sequence a′ of the arm-donating hairpin; because these 2 sequences are complementary, they readily bind to each other. The binding of sequence a in one probe to sequence a′ in the other probe creates a toehold that is resolved by the displacement of sequence b′ in the arm-acceptor hairpin. This irreversible strand-displacement reaction causes sequence b′-c′ in the arm-acceptor hairpin to assume a single-strand conformation, which is then available to serve as the initiator sequence in HCR.
Fig. 1.
RNA detection in situ using a pair of high-fidelity amp-FISH probes. (A) Target-mediated release of a sequestered HCR initiator when a pair of amp-FISH probes hybridizes to its mRNA target sequence. The target mRNA and the target-specific regions of the probes are shown in gray. The intramolecular stem within the arm-donating hairpin dissociates upon the binding of this probe to its target sequence, and this probe’s arm sequence, a′, is positioned next to its complement in the adjacently hybridized arm-acceptor hairpin probe, which renders the HCR initiator sequence (b′-c′) single-stranded by strand displacement. After removal of excess probes, and after the addition of HCR hairpins labeled with a fluorophore (shown as a yellow star), a tandemly repeated, multiply labeled polymer of HCR hairpins is created at the site to which the amp-FISH probes are hybridized. (B) Creation of Cy5-labeled HCR signals initiated by a pair of amp-FISH probes designed to detect GFP mRNA synthesized in HeLa cells that were transfected with a plasmid encoding GFP. Left shows the cells as seen in the GFP and DAPI channels, and Right shows the same cells as seen in the Cy5 channel. The transfected cell in Left displays a green GFP fluorescence, whereas the neighboring cells, identified by their DAPI stained nuclei, are not transfected. When visualized with a Cy5 filter, the GFP positive cell in the middle displays strong HCR signals, but the two GFP-negative cells on either side display less than 1 spot per cell.
Significantly, after hybridization and removal of excess probes, any probe pairs that persist nonspecifically at unintended sites are unlikely to interact with each other because the strong intramolecular hybrids within the arm-donating hairpins prevent the probe pairs from interacting. Moreover, since the initiator sequence in nonspecifically bound arm-acceptor hairpins is sequestered within a strong hairpin, these probes are not able to initiate HCR by themselves.
To demonstrate the feasibility of this scheme, we designed a pair of probes for the detection of green fluorescent protein (GFP) mRNA, and we then probed HeLa cells transfected with a plasmid encoding GFP. This heterologous mRNA provided a convenient target for assessing the specificity of the probes. The cells were fixed, permeabilized, and then hybridized with a pair of amp-FISH probes specific for a particular sequence within the GFP mRNA. After the removal of excess probes, HCR hairpins labeled with Cy5 were added and allowed to interact with each other for 2 h to generate an amplified fluorescent signal, after which the unreacted HCR hairpins were removed by washing and the cells were imaged. These cells displayed large numbers of fluorescent spots produced by HCR, whereas the control cells that had not been transfected but had undergone the same procedure did not show any fluorescent spots (Fig. 1B).
To compare the performance of these masked binary hairpin probes with the performance of “passively tagged” probes, in which an HCR initiator sequence is appended to one end of a linear probe in the conventional manner (9), we hybridized the passively tagged probes to the same site in GFP mRNA as was used for the hybridization of the arm-acceptor hairpin. A comparison of the results obtained with the 2 kinds of probes showed that both systems yielded Cy5 fluorescent signals of similar magnitude (SI Appendix, Fig. S1A). However, an analysis of the nontransfected control cells revealed that the background signals were more intense when using the passively tagged probes than when using the masked binary hairpin probes, the latter being indistinguishable from the autofluorescence that occurs in cells not exposed to any fluorescent probes or to HCR hairpins (SI Appendix, Fig. S1B). Although the background signals were only modestly more intense in this experiment, where a single passively tagged probe was used, the intensity of the background signals increased dramatically when multiple passively tagged probes were used as described below and as noted by Choi et al. (14). Image analysis indicated that, on average, transfected cells expressed more than 100 fluorescent spots per cell irrespective of the probe system used, whereas nontransfected cells probed with passively tagged probes yielded 1.5 ± 0.6 spots per cell and binary hairpin probes yielded only 0.6 ± 0.3 spots per cell (P value = 0.008) (SI Appendix, Fig. S1 C and D).
Producing Amplified Signals in Distinguishable Colors for Single-Nucleotide Variations.
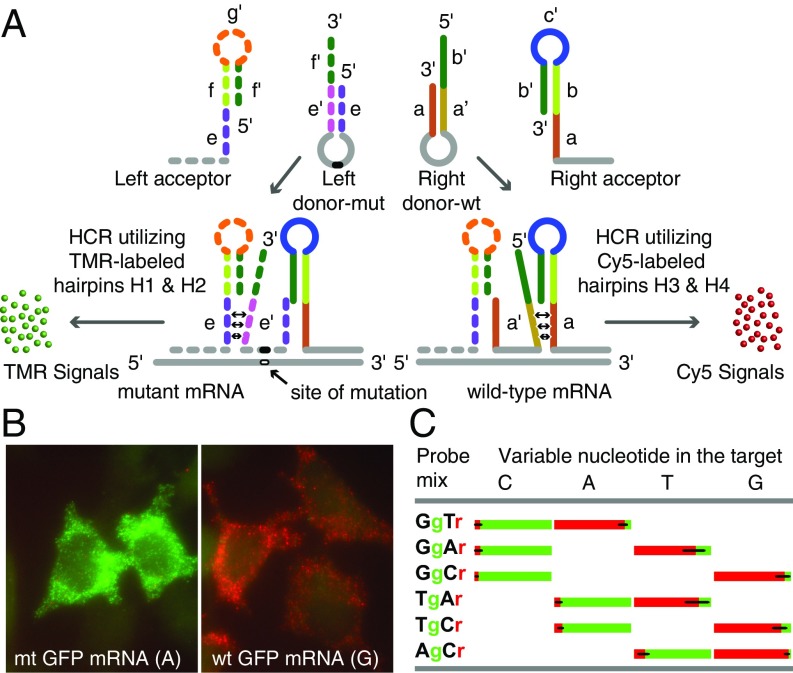
Exploiting the inherent high specificity of hairpin-shaped probes (19, 20), we designed a system that can distinguish an SNV, then produce an amplified signal in situ for a mutant target sequence in one fluorescent color, and simultaneously produce an amplified signal for a related wild-type target sequence in a different fluorescent color (Fig. 2A). This system utilizes 4 probes for the detection of SNVs: 2 arm-acceptor hairpins (a left acceptor and a right acceptor) that bind on either side of the sequence containing the SNV, and a pair of donor hairpins (a left donor and a right donor), only one of which binds to the target sequence containing the SNV that lies between the locations where the left acceptor and the right acceptor bind.
Fig. 2.
Generating amplified HCR signals from SNVs in single mRNA molecules using 4 interacting hairpin probes. (A) Depending on whether the target is mutant or wild-type, only one of the 2 donor hairpin probes binds to the target sequence in each mRNA, whereas both acceptor probes bind to it. The binding of a right donor-wt to a wild-type target sequence initiates a strand-displacement reaction in the right acceptor that leads to generation of a red HCR signal using Cy5-labeled HCR hairpins H3 and H4. The binding of a left donor-mut to a mutant target sequence initiates a strand-displacement reaction in the left acceptor that leads to generation of a green HCR signal using TMR-labeled HCR hairpins H1 and H2. (B) Examples of signals in either red or green channels after HCR. Cells were probed using a Tg-Cr probe mixture. This code identifies the discriminating nucleotide in the donor hairpin and lists the representative color of the HCR spots generated if that donor hairpin hybridizes to its target sequence. Cells expressing wild-type mRNAs yielded mostly red signals, and those expressing mutant mRNAs yielded mostly green signals. (C) Fractions of green and red spots from cells transfected with one of the 4 GFP variants that differed by the identity of the nucleotide (top) at one position. Each variant was probed using 3 pairwise combinations of the left and right donors (so that at least one donor was fully complementary to the target) together with common right and left acceptors. The bar graphs show the average percentage of spots seen in each color in single cells.
The right acceptor is similar in structure to the arm-acceptor hairpin in the basic binary probe system described above. Its hairpin is located at the 3′ end of its sequence, and it is designed to interact with the 5′ arm of the right donor probe. On the other hand, the left acceptor is different because it has a hairpin that is located at its 5′ end and is designed to interact with the 3′ arm of the left donor hairpin (Fig. 2A). Thus, the arm donor hairpins come in 2 flavors: a right donor hairpin, which only binds to a wild-type target sequence if it is present (right donor-wt) and whose hairpin arms dissociate upon hybridization to form a toehold with the right acceptor, leading to the freeing up of the right acceptor’s unique initiator sequence (b′-c′); and a left donor hairpin, which only binds to a mutant target sequence if it is present (“left donor-mut”) and whose hairpin arms dissociate upon hybridization to form a toehold with the left acceptor, leading to the freeing up of the left acceptor’s unique initiator sequence (f′-g′).
Because the 2 acceptor hairpins have unique HCR initiator sequences, they each initiate HCR with a different pair of distinguishably colored HCR hairpins. For example, as illustrated in Fig. 2A, wild-type target mRNAs generate a Cy5 signal using HCR hairpins H3 and H4, while mutant target mRNAs generate a tetramethylrhodamine (TMR) signal using HCR hairpins H1 and H2. Consequently, homozygous cells have spots in one fluorescent color and heterozygous cells have spots in 2 fluorescent colors, where the relative proportion of the differently colored spots reflects the relative levels of expression of the mRNAs generated from each allele.
We first optimized factors that are likely to influence the specificity of this probe system toward SNVs. The result of this optimization showed that either short probe (loop sequences in the right and left donors) lengths at a relatively low hybridization temperature (37 °C) in the absence of toehold sequences, or relatively longer probe lengths at a relatively higher hybridization temperature (50 °C) with a toehold sequence, enabled effective SNV discrimination (SI Appendix, Fig. S2). The first probe design was used for the detection of different GFP mRNA mutations, and the second probe design was used for the detection of a pair of SNVs in human epidermal growth factor receptor (EGFR) mRNAs.
For our GFP mRNA test system, we created a silent single-nucleotide mutation within the coding sequence of the GFP plasmid that we transfected into HeLa cells (substituting a guanosine residue, G, for an adenosine residue, A). Cells expressing either wild-type or mutant GFP mRNAs were hybridized with the full set of 4 probes (left acceptor, left donor-mut, right donor-wt, and right acceptor). After overnight hybridization followed by removal of excess probes by washing, HCR was carried out simultaneously using 4 HCR hairpins (one set, H1 and H2, for the generation of TMR signals on mutant targets; the other set, H3 and H4, for the generation of Cy5 signals on wild-type targets). Representative images confirmed that mutant mRNAs mainly generated TMR spots and wild-type mRNAs mainly generated Cy5 spots (Fig. 2B). The number of spots generated by the binding of the incorrect donor hairpin (i.e., green for wild-type targets and red for mutant targets) was so small as to be barely detectable in the resulting images.
To explore whether similar levels of discrimination can be achieved for the 2 other possible nucleotide substitutions, we created 2 additional GFP mutants at the same location (changing A in the wild-type mRNA target to either a uridine residue, U, or a cytidine residue, C), and we prepared a pair of donor hairpin probes for each mutation. We therefore had 4 different target plasmids differing from each other by the identity of the nucleotide at the same position, 4 left donor probes, and 4 right donor probes, each completely complementary to one of the 4 target sequences. Selecting one left donor probe and one right donor probe from each group, 6 different pairwise combinations of probes were created for the interrogation of each of the 4 targets. We repeated the experiment described above using 3 different pairwise combinations of probes for each of the 4 targets, such that each combination contained one probe that was fully complementary to the target and one that possessed a nucleotide that mismatched the corresponding nucleotide in the target. We counted all of the spots in each color (SI Appendix, Fig. S3). The relative proportion of the green and red spots observed in individual transfected cells is presented in Fig. 2C, which indicates that no matter which combination of probe pairs and targets was employed, the correct probe produced almost all of the spots. This represented an unprecedented level of SNV discrimination and at the same time produced amplified signals with low backgrounds.
Detecting the L858R Mutation in EGFR Transcripts.
To demonstrate that high-fidelity amp-FISH is able to detect SNVs in endogenous transcripts, which are expressed at much lower levels than transcripts that are produced by transfected plasmids, we decided to detect a somatic mutation, EGFR L858R, in which a G residue replaces a T residue in EGFR mRNA expressed in cancer cell lines. We utilized 3 human cell lines for this demonstration: the H1975 cell line, derived from a non-small-cell lung cancer and known to harbor this mutation in the EGFR gene (as a heterozygote); and HeLa and A431 cell lines that are wild-type with respect to this mutation (21). We confirmed the presence or absence of the EGFR L858R mutation in the mRNAs transcribed in these cell lines with the aid of real-time RT-PCR, utilizing SuperSelective primers (22) (SI Appendix, Fig. S4).
We designed a set of 4 allele-discriminating amp-FISH probes for the detection of the L858R mutation in EGFR mRNAs. In this set, a wild-type-specific donor probe was designed to yield Cy3 HCR signals and a mutant-specific donor probe was designed to yield Cy5 HCR signals. In addition to hybridizing the amp-FISH probes to the EGFR mRNAs at the site of the mutation, we simultaneously hybridized 48 Texas-Red (TR)–labeled sm-FISH probes to another region of EGFR mRNA so that their TR signals would identify the mRNA targets to which the amp-FISH probes would also bind.
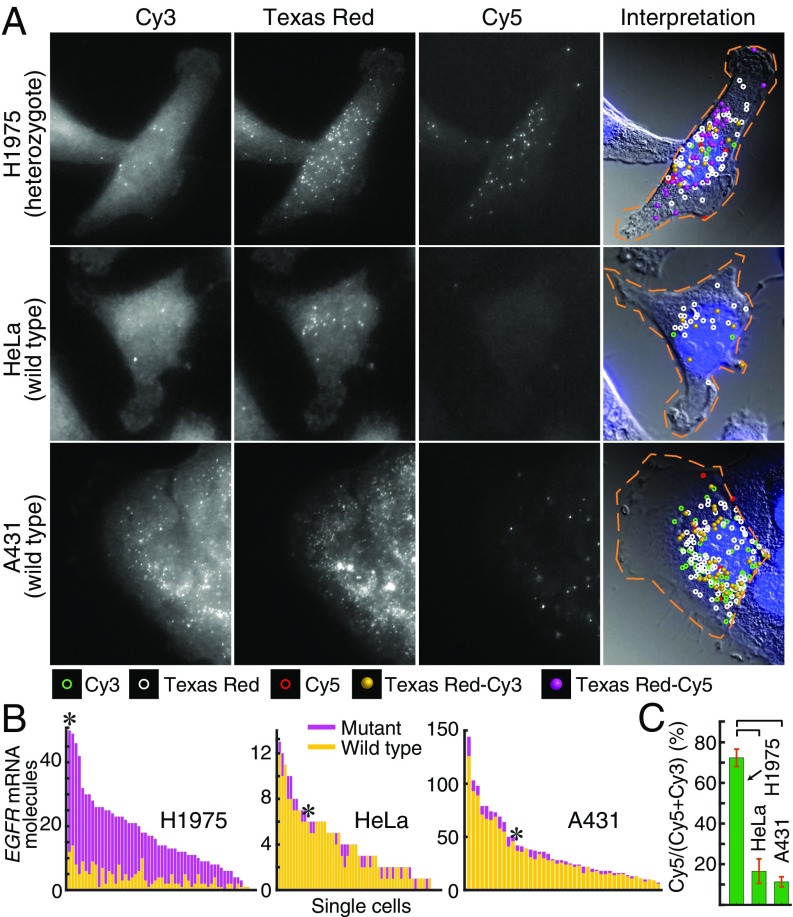
We acquired images of 3D optical sections and analyzed them using a custom image-processing algorithm to identify spots that were visible in more than one channel arising from the same mRNA molecules. This algorithm classified spots that were visible in 2 channels as being colocalized when they were so close to each other in 3 dimensions that their signals must have emanated from the same mRNA molecule (23, 24). A representative set of compressed z-stack images and the classification of the detected spots are presented in Fig. 3A. The average number of all spots from many cells is presented in SI Appendix, Table S1; the number of mutant and wild-type mRNAs detected in single cells is depicted in Fig. 3B; and their averages are presented in Fig. 3C.
Fig. 3.
Distinguishing the genotype of EGFR mRNA molecules with respect to the L858R mutation utilizing high-fidelity amp-FISH probes. (A) Representative images in 3 fluorescence channels of cells hybridized with 48 sm-FISH probes labeled with Texas Red and a set of 4 amp-FISH probes designed to identify wild-type mRNA molecules by their Cy3 HCR signals and mutant mRNA molecules by their Cy5 HCR signals. In the 3 panels on the left, merged z-stacks display the spot-like signals that each probe creates. In the panels on the right, a DIC image combined with DAPI stain is overlaid with the locations of spots detected in just one channel, or in various combinations of the 3 channels. Texas-Red–Cy3 spots identify wild-type mRNAs, and Texas-Red–Cy5 spots identify mutant mRNAs. (B) Histograms showing the number of Texas-Red–positive EGFR mRNA molecules identified as mutant (purple) or wild-type (yellow) in each analyzed cell (sorted in order of the sum of all spots in the cell). The histogram bars with an asterisk identify the cells shown. (C). Average number of Cy5 spots as a percentage of all HCR spots colocalized with Texas Red spots clearly distinguish the heterozygote cell line, H1975, from the homozygous wild-type cell lines, HeLa and A431. Horizontal brackets identify pairs of bars with statistically significant differences (P value < 1 × 10−5).
The percentage of Cy5 spots [Cy5/(Cy5 + Cy3)], considering only those spots colocalized with TR spots, revealed a clear distinction between cells expressing both mutant and wild-type mRNA (cell line H1975) and cells expressing only wild-type mRNA (cell lines HeLa and A531), and it pointed to the high specificity of the amp-FISH probe system (Fig. 3). Furthermore, the observed 72% mutant fraction in the H1975 cell line was consistent with a previous report that this cell line has undergone a 1.8-fold amplification of the mutant EGFR allele compared with a true diploid gene (implying a 3.6-fold amplification relative to the remaining wild-type EGFR allele) (25).
Although a fraction of Cy5 and Cy3 spots was not colocalized with the TR spots (potentially representing nonspecific signals), the percentage of Cy5 spots overall reflected the genotype of the cells (SI Appendix, Fig. S5). These observations indicated that amp-FISH probes were able to classify cells as being heterozygous mutant or homozygous wild-type solely by considering the spots generated by HCR without having to utilize sm-FISH signals as guides to identifying target-specific signals as in previous studies (5–7).
We also compared the performance of amp-FISH probes with an analogous pair of passively tagged probes and found that the latter produced a larger number of nonspecific spots and yielded poorer discrimination between the heterozygote mutant and wild-type cell lines, particularly when TR signals were not used as guides (SI Appendix, Fig. S5E). Consistent with this observation, an earlier study that utilized single-oligonucleotide probes without amplification showed that about 90–95% of those probes bound to off-target sites (5). The comparison of amp-FISH probe pairs with passively tagged probes also indicated that the efficiency with which the sequestered probes were converted into those that were able to initiate HCR upon binding to the target was very high (SI Appendix, Fig. S6).
Tiling Pairs of amp-FISH Probes over the Length of Target mRNAs to Enhance Signal Strength.
Another utility of amp-FISH probes is to enhance the signal strength in FISH assays by tiling multiple amp-FISH probe pairs over the length of the target mRNA without creating additional background. To explore this enhancement in signals, we prepared 24 pairs of right donor and right acceptor probes, each pair designed to hybridize to a different subsequence within EGFR mRNA. Each donor was synthesized separately, whereas the acceptors were synthesized by ligating a pool of the 24 target-specific oligonucleotides via click chemistry to a common oligonucleotide corresponding to the remainder of the acceptor sequence.
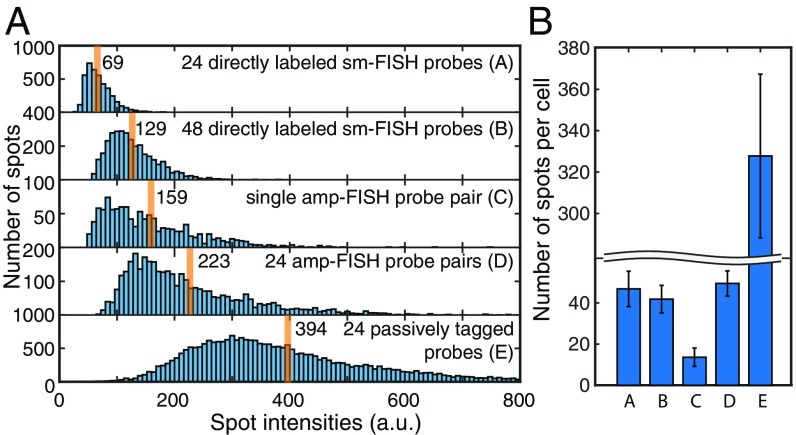
To quantify the enhancement in signal intensity due to the tiling of 24 pairs of amp-FISH probes over the EGFR mRNA, we probed EGFR mRNA in H1975 cells with 5 different probe sets, each labeled with Cy5: 24 directly labeled sm-FISH probes; 48 directly labeled sm-FISH probes (the former being a subset of the latter); a single pair of amp-FISH probes targeted to the wild-type version of the sequence at the L858R site; 24 amp-FISH probe pairs; and 24 passively tagged probes.
From z-stack images, we determined the intensities of the detected fluorescent spots (Fig. 4A and SI Appendix, Fig. S7). The 48 directly labeled probes yielded spots that were on average 1.9 times brighter than the spots produced by the 24 directly labeled probes, confirming that our computer image analysis algorithm was able to accurately measure spot intensities. The spots produced by the single amp-FISH probe pair were 2.3 times brighter than the spots produced by the 24 directly labeled sm-FISH probes and 1.2 times brighter than the spots produced by the 48 directly labeled sm-FISH probes.
Fig. 4.
Comparison of intensities and number of spots produced by 5 different probe sets for the imaging of EGFR mRNAs in H1975 cells. (A) Distribution of spot intensities of the brightest pixel within each spot (cumulative measurements: 55–70 cells) produced by each of the indicated probe sets. Orange lines identify the average spot intensity for each distribution. (B) Average number of spots per cell detected with each of the 5 probe sets. Probe sets A, B, D, and E were expected to detect all target mRNAs, whereas probe set C was expected to detect only the minor wild-type fraction of the target mRNAs.
Compared with the intensity of spots produced by the 24 directly labeled sm-FISH probes, the average intensity of the spots produced by the 24 amp-FISH probe pairs was 3.3 times brighter and the intensity of the spots produced by the 24 passively tagged probes was 5.7 times brighter. Why were the relative increases in signal intensity not proportional to the number of pairs of probes employed? The responsible factors likely include poor accessibility of some of the target sites in the mRNA to the probes; poor efficiency of interaction between the donor and acceptor pairs, resulting in inefficient unmasking of the HCR initiator sequence; and limited polymerization of the HCR oligonucleotides due to local crowding within the cross-linked cellular protein matrix.
We explored the variations in accessibility of the target sites using a set of 5 amp-FISH probe pairs either singly or in combinations. The results showed that individual probe pairs bound and signaled with relatively low and variable efficiencies, but exhibited some degree of cooperativity when used together (SI Appendix, Fig. S8 and Table S2). Due to the general plasticity of RNA secondary structures (26), the binding of a set of probes at one position on a target mRNA can potentially cause structural rearrangements that expose other binding sites in the mRNA, explaining the observed cooperativity. Consistent with previous observations (27), these data pointed to the relatively low accessibility of some target sites to individual probes as one of the reasons that signals did not increase in proportion to the number of probes. We also found that the presence of click links in the right acceptor probes did not significantly reduce signaling efficiency (SI Appendix, Fig. S8).
To explore whether a set of 24 amp-FISH probe pairs leads to the detection of all mRNA molecules present in a cell, we determined the average number of spots per cell produced by each of the 5 kinds of probes described above (Fig. 4B). This analysis revealed that the 24 amp-FISH probe pairs created about the same number of spots as produced by our gold-standard directly labeled sm-FISH probe set, whereas the passively tagged probe set produced far more spots than the number of EGFR mRNA molecules expected to be present in the H1975 cells (SI Appendix, Fig. S7). Assuming that the directly labeled probe sets yield accurate counts of mRNAs present in each cell (2), the greater number of spots seen with the passively tagged probe set suggests that a large fraction of those spots resulted from nonspecific sources.
Enhancing Signals in Flow Cytometry.
To explore the utility of the tiled amp-FISH approach in increasing signal intensity and signal-to-background ratios in flow-cytometric analyses, we selected as an experimental target interferon-γ (IFNγ) mRNA, which is expressed in peripheral blood mononuclear cells (PBMCs). These cells do not express IFNγ in their resting state, but when stimulated with phorbol 12-myristate 13-acetate (PMA) and ionomycin, ∼15% of the cells in the PBMC population respond by synthesizing IFNγ mRNA in 2 h (28). This system allows for an assessment of levels of both signal and background, utilizing the same cell population. For these experiments, we hybridized either resting or stimulated PBMCs with either 48 sm-FISH probes directly labeled with Cy5, 48 probes tagged with a passive HCR initiator, or 24 pairs of amp-FISH probes. Each probe set was specific for IFNγ mRNA. The cells hybridized with the 48 directly labeled sm-FISH probes were analyzed by flow cytometry immediately after removal of excess probes, whereas, the cells with the 48 passively tagged probes or with the 24 pairs of amp-FISH probes underwent HCR and removal of excess HCR hairpins before cytometry.
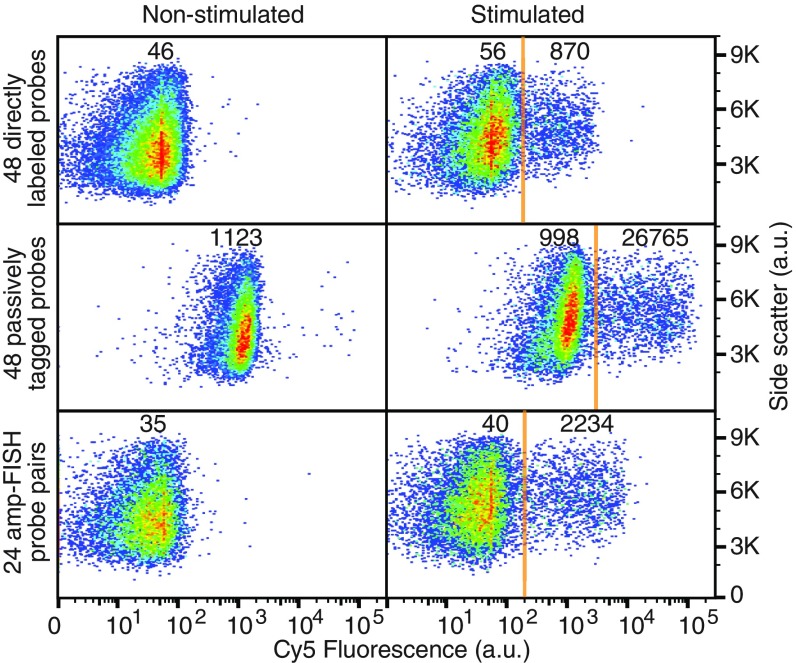
The results showed that the signal intensity in the cells that synthesized IFNγ increased about 30-fold as a consequence of HCR when using the 48 passively tagged probes compared with the signal intensity when using the 48 directly labeled sm-FISH probes. However, the intensity of the background signals in the cells that did not synthesize IFNγ also increased to a great extent (Fig. 5). The signal-to-background ratios for directly labeled probes, passively tagged probes, and amp-FISH probes were 17, 25, and 60, respectively. The 24 pairs of amp-FISH probes yielded 2.6-fold brighter signals than the 48 sm-FISH probes.
Fig. 5.
Comparison of signal and background levels in flow-cytometric assays for IFNγ mRNA utilizing 3 different probe sets. The fluorescence intensities of PMA/ionomycin-stimulated and nonstimulated PBMC populations are compared. The numbers shown represent mean Cy5 fluorescence intensities in the indicated populations.
In an alternative test system, in which GFP mRNA was used as the target in HeLa cells, an amp-FISH probe set containing 24 probe pairs (Dataset S1) produced 22-fold brighter signals compared with an sm-FISH probe set that contained 24 probes, and the same amp-FISH probe set produced 9-fold brighter signals compared with an sm-FISH probe set that contained 48 probes but yielded the same levels of background (SI Appendix, Fig. S9). Using the GFP mRNA target, we also found that, as the number of tiled probes increased, the signals generally increased, but a plateau was reached at 10 probe pairs. As in the EGFR and IFNγ systems described earlier, the background signals were larger for the passively tagged probes than they were for the amp-FISH probes in this system. Overall, the tiled amp-FISH probes yielded higher signal-to-background ratios than the tagged passively tailed probes (SI Appendix, Fig. S10).
Discussion
The amp-FISH probe system is an analytical tool that generates highly specific HCR signals in different colors for SNVs in situ. By counting the number of spots in each color in single cells, the relative expression levels of heterozygous alleles can be measured. This technology not only will be useful for studies of gene expression, such as allelic exclusion or imbalance; it also will be useful for the detection and identification of mRNAs transcribed from somatic mutations in cancer cells. In a number of situations, not only do cancers acquire mutations in key genes; these mutated genes are amplified, resulting in the synthesis of additional mutant mRNAs. Measurement of the ratio of mutant to wild-type mRNA (or naturally occurring SNVs) will enable analyses of gene amplification in cancer biopsies.
The results of the experiments described above indicate that a single amp-FISH probe pair yields spots that are about as bright as the spots produced by a traditional set of 48 sm-FISH probes, each labeled with a single dye moiety. Although we did not observe a proportional increase in spot intensity when tiling 24 pairs of amp-FISH probes across target mRNAs, we found that the spots were 3.3 times brighter than the spots produced by 24 singly labeled sm-FISH probes. Another measure of the gain in sensitivity was obtained in flow cytometry, where a 2.6-fold to 9-fold gain in sensitivity was observed with 24 amp-FISH pairs compared with the traditional 48 sm-FISH probes.
As a consequence of exploring different amp-FISH probe designs, we found that the use of toeholds decreased the specificity of SNV discrimination, but increased the efficiency of the unmasking of HCR initiator sequences. To obtain high specificity and efficiency for the detection of SNVs, we found that the hybridization temperature should be raised and that a 2-step hybridization should be performed when utilizing toeholds. Moreover, for general RNA detection, the use of multiple amp-FISH pairs with toeholds, as well as the tiling of amp-FISH pairs over the length of the target mRNA, ensures the detection of all mRNA molecules in a cell.
Because amp-FISH signals are created by the binding of 2 probes to unique sequences that must be contiguous, a number of applications are now feasible. Such applications include the detection of alternatively spliced transcripts, circular transcripts, and transcripts produced by fused genes. In addition, the inclusion of target-binding sequences containing modified nucleotides that have higher binding affinities in the amp-FISH probes will likely be advantageous for the detection of smaller targets, such as microRNAs in situ. Furthermore, by attaching one of the probes to an antibody that can recognize and bind to an RNA-bound protein, and then using a second probe that binds to the RNA in the immediate vicinity, it should be possible to detect RNA binding proteins at the sites where they are bound.
Materials and Methods
We performed either one-step hybridizations, in which both donor and acceptor probes were hybridized at the same time (GFP and IFNγ), or 2-step hybridizations, in which the acceptor probes were hybridized first and, after removal of the excess acceptor probes by washing, the donor probes were hybridized in a second step (EGFR).
The reported errors represented the 95% confidence intervals obtained from 35–90 cells. Hybridization reactions were carried out at 37 °C or 50 °C for 6–16 h. Synthesis, purification, and the self-annealing of probes is described in detail in SI Appendix, as are hybridization, washing of the hybrids, HCR, imaging, annotated sequences of all probes, image analysis, and statistical analysis.
Supplementary Material
Acknowledgments
We thank Fred Russell Kramer and Neeraja Syed for discussions and extensive help in preparation of the manuscript. This research was supported by NIH grants CA227291 and AI106036 and by a New Jersey Medical School Dean’s Biomedical Research Program Award.
Footnotes
Conflict of interest statement: Rutgers University receives royalties from the sale of prelabeled sm-FISH probes by LGC Biosearch Technologies, which markets them as Stellaris probes. A portion of these proceeds is distributed to S.T.’s laboratory for research support and to him personally. These proceeds do not influence the conclusions of this research. Part of the research described in this article is described in patent applications filed by Rutgers University.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814463116/-/DCSupplemental.
References
- 1.Femino A. M., Fay F. S., Fogarty K., Singer R. H., Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Raj A., van den Bogaard P., Rifkin S. A., van Oudenaarden A., Tyagi S., Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lubeck E., Coskun A. F., Zhiyentayev T., Ahmad M., Cai L., Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360–361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen K. H., Boettiger A. N., Moffitt J. R., Wang S., Zhuang X., RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levesque M. J., Ginart P., Wei Y., Raj A., Visualizing SNVs to quantify allele-specific expression in single cells. Nat. Methods 10, 865–867 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen C. H., van Oudenaarden A., Allele-specific detection of single mRNA molecules in situ. Nat. Methods 10, 869–871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ginart P., et al. , Visualizing allele-specific expression in single cells reveals epigenetic mosaicism in an H19 loss-of-imprinting mutant. Genes Dev. 30, 567–578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dirks R. M., Pierce N. A., Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. U.S.A. 101, 15275–15278 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi H. M., Beck V. A., Pierce N. A., Next-generation in situ hybridization chain reaction: Higher gain, lower cost, greater durability. ACS Nano 8, 4284–4294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F., et al. , RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Battich N., Stoeger T., Pelkmans L., Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat. Methods 10, 1127–1133 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Rouhanifard S. H., et al. , ClampFISH detects individual nucleic acid molecules using click chemistry-based amplification. Nat. Biotechnol. 37, 84–89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson C., Grundberg I., Söderberg O., Nilsson M., In situ detection and genotyping of individual mRNA molecules. Nat. Methods 7, 395–397 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Choi H. M. T., et al. , Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landegren U., Kaiser R., Sanders J., Hood L., A ligase-mediated gene detection technique. Science 241, 1077–1080 (1988). [DOI] [PubMed] [Google Scholar]
- 16.Tyagi S., Landegren U., Tazi M., Lizardi P. M., Kramer F. R., Extremely sensitive, background-free gene detection using binary probes and Qβ replicase. Proc. Natl. Acad. Sci. U.S.A. 93, 5395–5400 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyagi S., Kramer F. R., Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol. 14, 303–308 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Zhang D. Y., Chen S. X., Yin P., Optimizing the specificity of nucleic acid hybridization. Nat. Chem. 4, 208–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyagi S., Bratu D. P., Kramer F. R., Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16, 49–53 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Bonnet G., Tyagi S., Libchaber A., Kramer F. R., Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl. Acad. Sci. U.S.A. 96, 6171–6176 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawahara A., et al. , Molecular diagnosis of activating EGFR mutations in non-small cell lung cancer using mutation-specific antibodies for immunohistochemical analysis. Clin. Cancer Res. 16, 3163–3170 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Vargas D. Y., Marras S. A. E., Tyagi S., Kramer F. R., Suppression of wild-type amplification by selectivity enhancing agents in PCR assays that utilize superselective primers for the detection of rare somatic mutations. J. Mol. Diagn. 20, 415–427 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Batish M., van den Bogaard P., Kramer F. R., Tyagi S., Neuronal mRNAs travel singly into dendrites. Proc. Natl. Acad. Sci. U.S.A. 109, 4645–4650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vargas D. Y., et al. , Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell 147, 1054–1065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim Y., et al. , Spectrum of EGFR gene copy number changes and KRAS gene mutation status in Korean triple negative breast cancer patients. PLoS One 8, e79014 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kramer F. R., Mills D. R., Secondary structure formation during RNA synthesis. Nucleic Acids Res. 9, 5109–5124 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taneja K., Singer R. H., Use of oligodeoxynucleotide probes for quantitative in situ hybridization to actin mRNA. Anal. Biochem. 166, 389–398 (1987). [DOI] [PubMed] [Google Scholar]
- 28.Bushkin Y., et al. , Profiling T cell activation using single-molecule fluorescence in situ hybridization and flow cytometry. J. Immunol. 194, 836–841 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.