Significance
Identification of enzyme commission (EC) numbers is essential for accurately understanding enzyme functions. Although several EC number prediction tools are available, they have room for further improvement with respect to computation time, precision, coverage, and the total size of the files needed for EC number prediction. Here, we present DeepEC, a deep learning-based computational framework that predicts EC numbers with high precision in a high-throughput manner. DeepEC shows much improved prediction performance when compared with the 5 representative EC number prediction tools that are currently available. DeepEC will be useful in studying enzyme functions by implementing them independently or as part of a third-party software program.
Keywords: deep learning, DeepEC, enzyme commission number, EC number prediction, metabolism
Abstract
High-quality and high-throughput prediction of enzyme commission (EC) numbers is essential for accurate understanding of enzyme functions, which have many implications in pathologies and industrial biotechnology. Several EC number prediction tools are currently available, but their prediction performance needs to be further improved to precisely and efficiently process an ever-increasing volume of protein sequence data. Here, we report DeepEC, a deep learning-based computational framework that predicts EC numbers for protein sequences with high precision and in a high-throughput manner. DeepEC takes a protein sequence as input and predicts EC numbers as output. DeepEC uses 3 convolutional neural networks (CNNs) as a major engine for the prediction of EC numbers, and also implements homology analysis for EC numbers that cannot be classified by the CNNs. Comparative analyses against 5 representative EC number prediction tools show that DeepEC allows the most precise prediction of EC numbers, and is the fastest and the lightest in terms of the disk space required. Furthermore, DeepEC is the most sensitive in detecting the effects of mutated domains/binding site residues of protein sequences. DeepEC can be used as an independent tool, and also as a third-party software component in combination with other computational platforms that examine metabolic reactions.
High-quality and high-throughput prediction of enzyme commission (EC) numbers is essential for accurate understanding of enzyme functions and cellular metabolism overall. Such prediction is particularly important when, for example, annotating an increasing volume of (meta)genome sequence data (1), identifying catalytic functions of enzymes (2), establishing gene–protein–reaction associations in metabolism (3), designing a novel metabolic pathway (4), and building genome-scale metabolic networks in a high-throughput manner (5–7). In a genome annotation procedure, a protein sequence for a metabolic gene is assigned with EC numbers that correspond to a numerical classification scheme of enzymes based on relevant chemical reaction patterns (8, 9). Thus, EC number serves to associate a protein sequence with relevant chemical reactions. EC number consists of 4 level numbers, with each number separated by a period (i.e., a.b.c.d). The first to the fourth numbers correspond to class, subclass, sub-subclass, and serial number, respectively (Fig. 1). An EC number having all 4 level numbers for a protein sequence is the most specific annotation, which allows associating the protein sequence with specific chemical reactions; however, the first-level or the second-level EC numbers are usually considered to be insufficient annotation of a protein sequence.
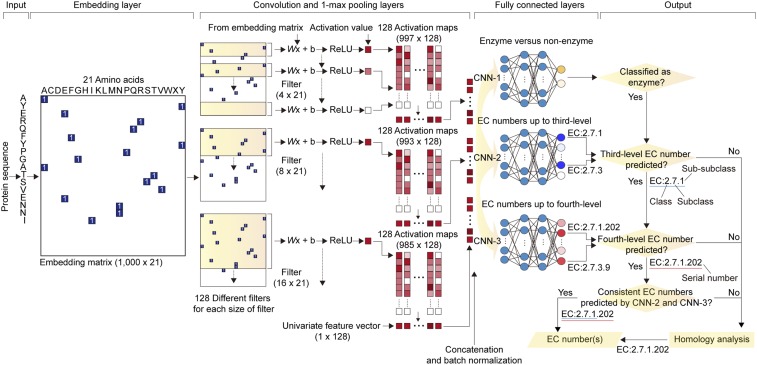
Fig. 1.
Overall scheme of DeepEC. DeepEC consists of 3 independent CNNs to classify whether an input protein sequence is an enzyme or not, using CNN-1, and to predict third- and fourth-level EC numbers using CNN-2 and CNN-3, respectively. Homology analysis is also implemented using DIAMOND (33) for EC numbers that cannot be classified by the CNNs. The 3 CNNs share the same embedding, convolution, and 1-max pooling layers, but have different fully connected layers to perform the 3 different tasks mentioned here. DeepEC generates final EC numbers as output only if the 3 CNNs generate consistent results: Binary classification of a protein sequence as an enzyme by CNN-1 and 2 EC numbers with consistent class (first number), subclass (second number), and sub-subclass (third number) generated from CNN-2 and CNN-3. Because the CNN-2 and CNN-3 are multilabel classification models, multiple EC numbers can be predicted for a given protein sequence. If a given protein sequence is classified as an enzyme by CNN-1, but is not assigned with specific EC numbers by CNN-2 and CNN-3, the homology analysis is subsequently conducted. See SI Appendix, Materials and Methods for details on the operation of CNNs, as well as implementation of homology analysis within DeepEC. ReLU stands for rectified linear unit.
As of September 2018, 6,238 fourth-level EC numbers have been defined in the ExPASy database (10). Because of the importance of EC number prediction in understanding enzyme functions, a number of relevant computational methods have been developed: PRIAM (11), EzyPred (12), CatFam (13), EnzML (14), EFICAz2.5 (15), EnzDP (16), SVM-prot (17), DEEPre (18), DETECT v2 (19), and ECPred (20). However, prediction performances of these tools have room for further improvement with respect to computation time, precision, and coverage for the prediction of EC numbers. Also, the EC number prediction tools should be locally installable to allow high-throughput prediction.
Here, we present DeepEC (https://bitbucket.org/kaistsystemsbiology/deepec), a deep learning-based computational framework that takes a protein sequence as an input and accurately predicts EC numbers as an output. DeepEC uses 3 convolutional neural networks (CNNs) as a major engine for the prediction of EC numbers, and also implements homology analysis for EC numbers that cannot be classified by the CNNs. DeepEC predicts EC numbers with high precision and can be implemented in a high-throughput manner through its local installation and faster computation. DeepEC can be used as an independent tool, as well as a third-party software component in combination with other computational platforms that examine metabolic reactions.
Results
Development of DeepEC.
For the development of DeepEC, a CNN was used among other deep learning methods because of its proven outstanding performance in detecting functional regions (e.g., motifs and/or domains) in a biological sequence (e.g., protein sequence) (21), which is highly relevant to the prediction of EC numbers. To develop DeepEC, a gold standard dataset covering 1,388,606 protein sequences and 4,669 EC numbers was prepared by processing protein sequences from both Swiss-Prot (22) (released February 2018) and TrEMBL (23) (released February 2018) datasets (SI Appendix, Materials and Methods and Figs. S1 A–C and S2).
DeepEC consists of 3 independent CNNs performing 3 different classification tasks for a single protein sequence given as an input. The first CNN, designated CNN-1, classifies whether a given protein sequence is an enzyme or a nonenzyme protein. The second and third CNNs, designated CNN-2 and CNN-3, predict the third- and fourth-level EC numbers, respectively. Here, CNN-2 and CNN-3 were designed to be multilabel classification models (i.e., activation of multiple output neurons at the same time) because one enzyme can have multiple EC numbers if it is a promiscuous enzyme. The 3 CNNs share the same embedding, convolution and 1-max pooling layers, but have different fully connected layers (Fig. 1). It is this fully connected layer in each CNN of DeepEC that performs one of the 3 different tasks mentioned here. EC numbers are generated as output from DeepEC for a given protein sequence only if the 3 CNNs generate consistent results: Binary classification of a protein sequence as an enzyme by the CNN-1 and generation of the third- and fourth-level EC numbers from the CNN-2 and CNN-3, respectively, with both having consistent class (first number), subclass (second number), and sub-subclass (third number; Fig. 1). Use of the multiple CNNs for DeepEC was considered to generate highly consistent and precise EC numbers as output: For 387,805 protein sequences in the testing dataset of the gold standard dataset (SI Appendix, Materials and Methods), there were 25,439 protein sequences, for which CNN-2 predicted correct third-level EC numbers, but CNN-3 failed to do so using the DeepEC with each CNN optimized.
CNN-1 was modeled (i.e., trained, validated, and tested) using 153,884 enzyme and nonenzyme protein sequences (SI Appendix, Materials and Methods and Fig. S3). CNN-2 and CNN-3 were modeled using a gold standard dataset (SI Appendix, Materials and Methods and Fig. S3). This modeling process was implemented by examining different values for 4 main hyperparameters of the 3 CNNs to maximize the classification performance of the DeepEC while avoiding overfitting of each CNN (SI Appendix, Materials and Methods and Dataset S1). The 4 hyperparameters include the window size of each filter, number of filters for each window size, number of hidden layers, and number of nodes in each hidden layer of the fully connected layer. A detailed description of the DeepEC development is given in SI Appendix, Materials and Methods.
For protein sequences that fail to be assigned with EC numbers by CNN-2 and CNN-3, although they are still classified as an enzyme by the CNN-1, they are given EC numbers from their homologous protein sequences having EC numbers through homology analysis (Fig. 1). It should be noted that the CNNs of DeepEC were modeled to predict EC numbers, for which 10 or more protein sequences are available in the gold standard dataset. In the gold standard dataset, 2,240 EC numbers are covered by fewer than 10 protein sequences each. Taken together, DeepEC can classify a total of 4,669 EC numbers for protein sequences, which is by far the greatest number of EC numbers covered by a single EC number prediction tool (Table 1).
Table 1.
Features of 6 different EC number prediction tools that are locally installable
| EC number prediction tool | Disk space required (GB) | Number of predictable EC numbers | Last update (year) |
| DeepEC | 0.045 | 4,669 | 2019 |
| CatFam | 2.072 | 1,653 | 2009 |
| DETECT v2 | 0.854 | 786 | 2018 |
| ECPred | 9.427 | 858 | 2018 |
| EFICAz2.5 | 24.682 | 2,757 | 2012 |
| PRIAM | 3.547 | 4,560 | 2018 |
Optimization of the Model Structure of DeepEC.
Upon construction, the EC number prediction performance of DeepEC was investigated as a function of the number of CNNs used, as well as a featurization method. First, prediction performance of DeepEC with the 3 CNNs was compared with that of DeepEC having a single CNN. Use of the 3 CNNs allows each CNN to perform a single classification task (i.e., binary classification of a protein sequence as an enzyme and generation of the third-level and fourth-level EC numbers), whereas use of a single CNN means it has to perform the 3 different tasks by itself. Such task allocation was expected to efficiently reduce the number of false-positive predictions (e.g., predicting EC numbers for a nonenzyme protein sequence). To validate this hypothesis, a negative testing was conducted by implementing DeepEC for the 22,168 nonenzyme protein sequences as inputs, for which EC numbers should not be predicted. Indeed, for the 22,168 nonenzyme protein sequences tested, only 150 fourth-level EC numbers were predicted by the DeepEC having 3 CNNs, whereas 4,852 fourth-level EC numbers were predicted by the DeepEC with one CNN (SI Appendix, Fig. S4). Also, a DeepEC having the fourth CNN that predicts second-level EC numbers was tested, but it did not give better prediction performance than the DeepEC having 3 CNNs. Finally, DeepEC having 5 CNNs, additionally predicting the first-level and the second-level EC numbers, also did not generate better prediction performance than the DeepEC with 3 CNNs (SI Appendix, Fig. S5). These results justify the use of 3 CNNs in DeepEC.
Next, the prediction performance when using one-hot encoding method was compared with another representative featurization method ProtVec (24). As DeepEC with the one-hot encoding method showed greater macro precision (0.953) and macro recall (0.735) values than those (0.891 and 0.656, respectively) obtained with the ProtVec method, the one-hot encoding method was used in all the implementations of DeepEC in this study (SI Appendix, Fig. S6).
Finally, for protein sequences annotated with multiple EC numbers, which are potential promiscuous enzymes, DeepEC showed reasonably high precision (0.940) and recall (0.905) values for those having 2 EC numbers (SI Appendix, Table S1). However, prediction performance of DeepEC decreased for the protein sequences having 3 (0.825 and 0.709 for precision and recall, respectively), 4 (0.775 and 0.720), and 5 (0.629 and 0.385) EC numbers. To further validate DeepEC, an in vitro enzyme assay was conducted for an enzyme from Escherichia coli, which was given different EC numbers from UniProt and DeepEC. For this, YgbJ was selected as a target enzyme, which is known as a putative l-threonate dehydrogenase (1.1.1.411) in UniProt, but was predicted to be 2-hydroxy-3-oxopropionate reductase or d-glycerate:NAD(P)+ oxidoreductase (1.1.1.60) by DeepEC. As a result of the enzyme assay (SI Appendix, Materials and Methods), YgbJ was shown to have activities for both d-glycerate and l-threonate (SI Appendix, Fig. S7). This enzyme assay suggests that YgbJ is likely a promiscuous enzyme, and therefore can be assigned with both 1.1.1.411 and 1.1.1.60. Although more enzyme assays need to be conducted to further rigorously validate DeepEC, the enzyme assay results presented here indicate that DeepEC seems reliable and can be used to complement other EC prediction tools to suggest alternative EC numbers.
Comparison of Prediction Performance of DeepEC with 5 Representative EC Number Prediction Tools.
Next, DeepEC was compared with the latest versions of 5 representative EC number prediction tools that are locally installable, including CatFam (13), DETECT v2 (19), ECPred (20), EFICAz2.5 (15), and PRIAM (11), with respect to their prediction performances. For a systematic comparison of prediction performances, 201 enzyme protein sequences were used as inputs, which were not used for the development of these 6 different tools; these enzyme protein sequences were obtained from the Swiss-Prot database released on August 2018. DeepEC showed both the greatest precision and recall values (0.920 and 0.455, respectively) compared with the other 5 EC number prediction tools: CatFam (0.880 and 0.327), DETECT v2 (0.804 and 0.203), ECPred (0.817 and 0.243), EFICAz2.5 (0.737 and 0.416), and PRIAM (0.809 and 0.356; Table 2). For another set of 2,310 enzyme protein sequences that were previously used for the development of these 6 different tools, obtained from the Swiss-Prot database released on March 2007, DeepEC still showed the greatest precision value (0.993; SI Appendix, Table S2). These results show that DeepEC predicts highly reliable (i.e., high-precision) EC numbers for a given protein sequence compared with the 5 available EC number prediction tools.
Table 2.
Prediction performances of 6 different, locally installable EC number prediction tools using 201 enzyme protein sequences as inputs, which were never considered for the development of all these tools (from the Swiss-Prot database released August 2018)
| EC number prediction tool | Precision | Recall | Run time (s) |
| DeepEC | 0.920 | 0.455 | 13 |
| CatFam | 0.880 | 0.327 | 47 |
| DETECT v2 | 0.804 | 0.203 | 5,480 |
| ECPred | 0.817 | 0.243 | 28,840 |
| EFICAz2.5 | 0.737 | 0.416 | 29,093 |
| PRIAM | 0.809 | 0.356 | 51 |
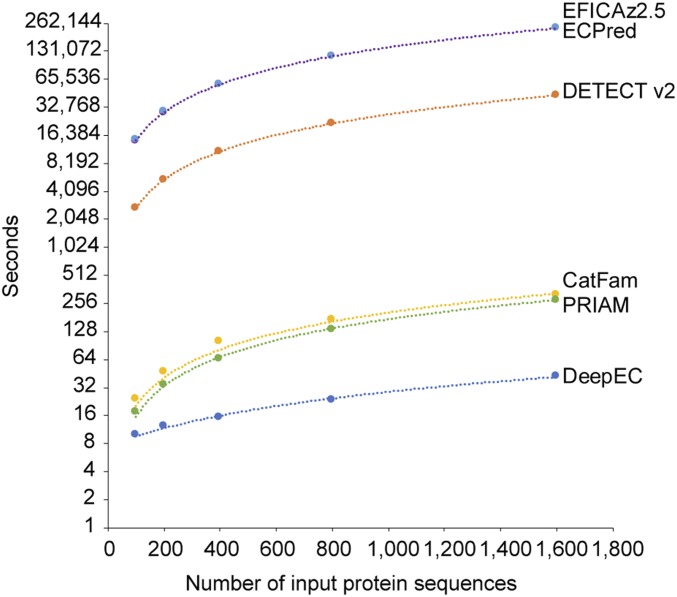
Importantly, DeepEC was the fastest among these tools for the prediction of EC numbers for the 3 different sets of protein sequences mentioned here: 201 enzyme protein sequences never used for the development of the 6 EC number prediction tools (Table 2), 2,310 enzyme protein sequences previously used for the development of the 6 tools (SI Appendix, Table S2), and 1,600 protein sequences randomly selected from the gold standard dataset (Fig. 2). Also, prediction of EC numbers for the entire set of 33,942,253 protein sequences available in 9,513 complete genomes (i.e., organisms) from the NCBI Genome database (https://www.ncbi.nlm.nih.gov/genome; as of May 2018) took 230 h (9.6 d) using DeepEC (see Dataset S2 for the list of organisms examined). A full list of EC numbers predicted for all these protein sequences is available at http://doi.org/10.5281/zenodo.2567339. In contrast, prediction of EC numbers for the same set of 33,942,253 protein sequences was estimated to take 1,874 h (78.1 d) using CatFam, 258,338 h (10,764.1 d) using DETECT v2, 1,359,575 h (56,649.0 d) using ECPred, 1,371,518 h (57,146.6 d) using EFICAz2.5, and 1,649 h (68.7 d) using PRIAM, based on their computation times measured (Fig. 2). Finally, DeepEC requires the smallest disk space, at 45 megabytes, which is substantially smaller than the other 5 tools, and which therefore makes DeepEC more suitable for its use as a light third-party software component. For a comparison, the required disk spaces are: CatFam, 2.1 gigabytes; DETECT v2, 854 megabytes; ECPred, 9.4 gigabytes; EFICAz2.5, 24.7 gigabytes; and PRIAM, 3.5 gigabytes (Table 1).
Fig. 2.
Computation time of DeepEC, CatFam, DETECT v2, ECPred, EFICAz2.5, and PRIAM. Six EC number prediction tools were used to predict EC numbers for 100, 200, 400, 800, and 1,600 protein sequences randomly selected from the gold standard dataset.
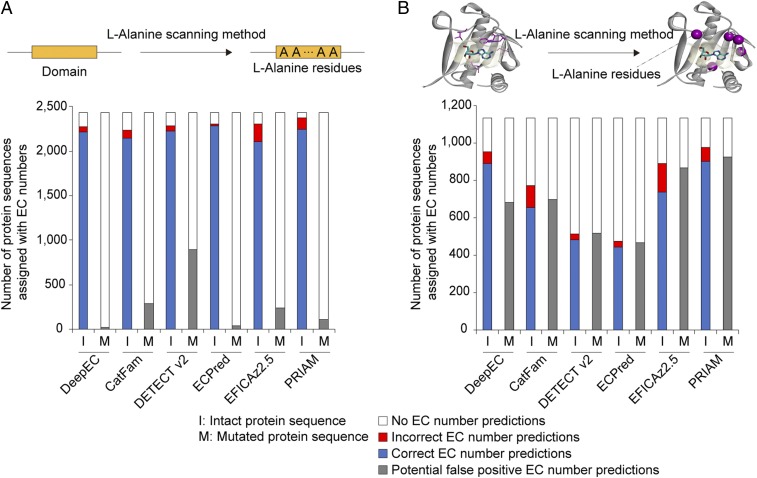
As another negative test, DeepEC was examined to determine whether it can still predict EC numbers for protein sequences mutated to have inactive domains. Being able to detect changes in enzymatic function as a result of mutations can be very useful when analyzing a set of homologous enzymes. For this test, DeepEC was again compared with CatFam, DETECT v2, ECPred, EFICAz2.5, and PRIAM. For a systematic comparison, 2,435 protein sequences from the gold standard dataset were used as inputs, which are associated with one of 487 EC numbers that can be commonly predicted by the 6 tools (see SI Appendix, Materials and Methods for details). Domains of the 2,435 protein sequences were detected by using PfamScan (25), and were subjected to the l-alanine scanning method (26) in which all the residues in a detected domain were substituted with l-alanine (Fig. 3A). Proteins having domains filled with l-alanine (hereafter known as mutated domains) are considered to have no functions. Finally, EC numbers of protein sequences with intact and mutated domains were predicted, using DeepEC. As a result, only 36 protein sequences with mutated domains (1.5% of the 2,435 protein sequences) were assigned EC numbers, using DeepEC. CatFam, DETECT v2, ECPred, EFICAz2.5, and PRIAM predicted EC numbers for a greater number of protein sequences with mutated domains: 294 (12.1%), 898 (36.9%), 39 (1.6%), 237 (9.7%), and 114 (4.7%) protein sequences with mutated domains, respectively (Fig. 3A). These results show that DeepEC, CatFam, DETECT v2, ECPred, EFICAz2.5, and PRIAM can all capture the effects of having mutated domains responsible for enzyme functions. However, DeepEC appeared to be the most sensitive in detecting the effects of mutated domains in protein sequences (i.e., the greatest difference between the blue and grey bars in Fig. 3A). Interestingly, DeepEC was also the most sensitive in capturing the effects of having binding site residues mutated using the l-alanine scanning method (SI Appendix, Materials and Methods and Fig. 3B). Proteins with binding site residues all replaced with l-alanine were also considered to have no functions. This classification task is considered to be more challenging than predicting EC numbers for protein sequences with mutated domains because a few binding site residues (11.0 amino acids on average for the 1,134 protein sequences examined; Fig. 3B) are physically separated from one another throughout the protein sequence, in contrast to domains having residues physically clustered in specific regions of the protein sequence.
Fig. 3.
Prediction of EC numbers for protein sequences having mutations using DeepEC, CatFam, DETECT v2, ECPred, EFICAz2.5, and PRIAM. (A) Prediction of EC numbers for 2,435 intact protein sequences (I on the x-axis) and 2,435 protein sequences having mutated domains where all of the domain residues were substituted with l-alanine residues through the l-alanine scanning method (26) (M on the x-axis). (B) Prediction of EC numbers for 1,134 intact protein sequences (I) and 1,134 protein sequences with their binding site residues all substituted with l-alanine (M). For A and B, see SI Appendix, Materials and Methods for preparation of the input protein sequences.
Use of DeepEC as a Third-Party Software Component for the Generation of Human Metabolic Reactions.
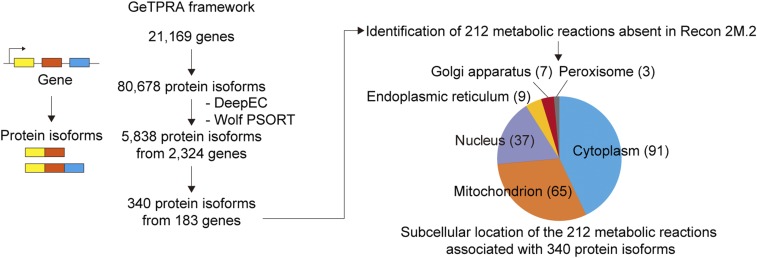
Finally, DeepEC was used as a part of a computational framework; namely, the gene–transcript–protein–reaction associations (GeTPRA) framework (27). The GeTPRA framework identifies metabolic reactions to be newly added to a human genome-scale metabolic model (GEM) if the reactions carry fluxes on their addition to the human GEM and have relevant experimental evidence. In this study, 80,678 human protein isoforms generated by 21,169 human genes obtained from Ensembl BioMart (28) were used as input for the GeTPRA framework. EFICAz2.5 was initially used for the GeTPRA framework to assign the protein sequences with EC numbers (27), but DeepEC was used in this study. As a result, a total of 5,838 protein isoforms generated from 2,324 human genes were predicted by DeepEC to have at least one EC number (Fig. 4 and Dataset S3). The newly predicted EC numbers for the protein sequences were used to retrieve metabolic reactions from the KEGG database (29), which were subsequently compartmentalized by predicting subcellular locations of the protein sequences, using Wolf PSORT (30). When these metabolic reactions were compared with those in Recon 2M.2 (27), a human GEM previously prepared using the GeTPRA framework with EFICAz2.5, 212 metabolic reactions mediated by 340 protein isoforms (encoded by 183 genes) appeared to be absent in the Recon 2M.2, although these reactions have experimental evidence available at UniProt (23), BRENDA (31), and the Human Protein Atlas (32) (Dataset S3). Thus, DeepEC can identify additional reactions through EC numbers that could not be predicted by other tools, and consequently allows more accurate reconstruction of GEMs. Also, this study demonstrates that DeepEC can be easily integrated with a third-party software program that requires the prediction of EC numbers.
Fig. 4.
Use of DeepEC as a third-party software component of the GeTPRA framework. In the GeTPRA framework (27), information on the predicted EC numbers and subcellular locations for 80,678 protein isoforms from 21,169 human genes was used to identify new flux-carrying metabolic reactions that can be considered for further studies on human metabolism and an update of existing human GEMs. Subcellular locations were predicted using Wolf PSORT (30). As another input of the GeTPRA framework, a human GEM Recon 2M.2 was used (27). The pie chart shows the subcellular location of 212 metabolic reactions that appeared to be absent in Recon 2M.2.
Discussion
In this study, we report the development of DeepEC that accurately predicts EC numbers for given protein sequences as input. DeepEC uses 3 different CNNs as a major engine, and also a homology analysis tool for the accurate prediction of EC numbers. DeepEC showed better prediction performance (i.e., precision value) than the 5 representative tools that are currently available, including CatFam, DETECT v2, ECPred, EFICAz2.5, and PRIAM. Also, DeepEC is faster and lighter than these 5 tools. DeepEC was found to be the most sensitive in capturing the effects of mutated domains and binding site residues among the tools compared as well. Taken together, DeepEC can serve as a powerful tool for the high-quality and high-throughput prediction of EC numbers, which should be useful for studying enzyme functions. DeepEC can be used as an independent tool, and also as a third-party software component in combination with other computational platforms that examine metabolic reactions.
Despite the improved prediction performance of DeepEC in comparison with the other 5 tools discussed in this study, there certainly exists room to improve. First, coverage of the gold standard dataset for each EC number needs to be much increased. As already seen in this study, the gold standard dataset covers each EC number with very different numbers of protein sequences; each of the 2,240 EC numbers in the gold standard dataset used in this study is covered by fewer than 10 protein sequences (SI Appendix, Materials and Methods). This was a reason that DeepEC could not predict the EC number 1.1.1.411 for YgbJ, which showed activities for both d-glycerate (1.1.1.60) and l-threonate (1.1.1.411) according to the in vitro enzyme assay; there were only 5 protein sequences for the EC number 1.1.1.411 in the gold standard dataset. Resolving such data imbalance could further improve the prediction performance of DeepEC in terms of the precision and coverage for the prediction of EC numbers. Also, availability of a negative control dataset (e.g., mutated enzyme protein sequences with their functions lost) would be extremely useful in improving the detection capacity of DeepEC for mutations in domains and binding site residues of protein sequences. Additional training of DeepEC with the negative control dataset would allow better detection of changes in the enzymatic function as a result of mutation. This feature can be especially useful when analyzing homologous enzymes, for example, from new (meta)genome data, which have mutations with previously unknown effects on enzyme functions. Above all, it will be important to use DeepEC in various settings, either independently or as part of a third-party software program, and to receive feedbacks from biochemists, enzymologists, and biotechnologists for more rigorous validation of DeepEC and future direction for its upgrades.
Materials and Methods
All the materials and methods conducted in this study are detailed in SI Appendix, Materials and Methods: Preparation of the gold standard dataset for DeepEC; optimizing the architecture of DeepEC; training CNNs of DeepEC; validating and testing CNNs of DeepEC; prediction of EC numbers for all the enzymes from 9,513 complete genomes, using DeepEC; preparation of enzyme and nonenzyme protein sequences for the modeling of CNN-1; preparation of protein sequences for the l-alanine scanning method that mutates domains and binding site residues; expression and purification of a putative l-threonate dehydrogenase; in vitro YgbJ assay; and development environment.
Data Availability.
Source code for DeepEC is available at https://bitbucket.org/kaistsystemsbiology/deepec. The list of EC numbers predicted for 33,942,253 protein sequences is available at http://doi.org/10.5281/zenodo.2567339.
Supplementary Material
Acknowledgments
We are grateful to Tong Un Chae, Jae Sung Cho, and Jiyong Kim for their contribution to enzyme assays. This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation of Korea. This work was also supported by the Bio & Medical Technology Development Program of the National Research Foundation of Korea funded by the Korean government, the Ministry of Science and ICT (NRF-2018M3A9H3020459).
Footnotes
Conflict of interest statement: S.Y.L. coauthored a community correspondence with C.D.M. and N.E.L., published in Molecular Systems Biology (2015) 11, 831 (DOI 10.15252/msb.20156157).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821905116/-/DCSupplemental.
References
- 1.Kodama Y., Shumway M., Leinonen R.; International Nucleotide Sequence Database Collaboration , The Sequence Read Archive: Explosive growth of sequencing data. Nucleic Acids Res. 40, D54–D56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedberg I., Automated protein function prediction–The genomic challenge. Brief. Bioinform. 7, 225–242 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Machado D., Herrgård M. J., Rocha I., Stoichiometric representation of gene-protein-reaction associations leverages constraint-based analysis from reaction to gene-level phenotype prediction. PLoS Comput. Biol. 12, e1005140 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finley S. D., Broadbelt L. J., Hatzimanikatis V., Computational framework for predictive biodegradation. Biotechnol. Bioeng. 104, 1086–1097 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim W. J., Kim H. U., Lee S. Y., Current state and applications of microbial genome-scale metabolic models. Curr. Opin. Syst. Biol. 2, 10–18 (2017). [Google Scholar]
- 6.Henry C. S., et al. , High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 28, 977–982 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Wang L., Dash S., Ng C. Y., Maranas C. D., A review of computational tools for design and reconstruction of metabolic pathways. Synth. Syst. Biotechnol. 2, 243–252 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webb E. C., Enzyme nomenclature 1992. Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes (Academic Press, San Diego, CA, 1992). [Google Scholar]
- 9.Hatzimanikatis V., Li C., Ionita J. A., Broadbelt L. J., Metabolic networks: Enzyme function and metabolite structure. Curr. Opin. Struct. Biol. 14, 300–306 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Gasteiger E., et al. , ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claudel-Renard C., Chevalet C., Faraut T., Kahn D., Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 31, 6633–6639 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen H. B., Chou K. C., EzyPred: A top-down approach for predicting enzyme functional classes and subclasses. Biochem. Biophys. Res. Commun. 364, 53–59 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Yu C., Zavaljevski N., Desai V., Reifman J., Genome-wide enzyme annotation with precision control: Catalytic families (CatFam) databases. Proteins 74, 449–460 (2009). [DOI] [PubMed] [Google Scholar]
- 14.De Ferrari L., Aitken S., van Hemert J., Goryanin I., EnzML: Multi-label prediction of enzyme classes using InterPro signatures. BMC Bioinformatics 13, 61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar N., Skolnick J., EFICAz2.5: Application of a high-precision enzyme function predictor to 396 proteomes. Bioinformatics 28, 2687–2688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen N. N., Srihari S., Leong H. W., Chong K. F., EnzDP: Improved enzyme annotation for metabolic network reconstruction based on domain composition profiles. J. Bioinform. Comput. Biol. 13, 1543003 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Li Y. H., et al. , SVM-prot 2016: A web-server for machine learning prediction of protein functional families from sequence irrespective of similarity. PLoS One 11, e0155290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., et al. , DEEPre: Sequence-based enzyme EC number prediction by deep learning. Bioinformatics 34, 760–769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nursimulu N., Xu L. L., Wasmuth J. D., Krukov I., Parkinson J., Improved enzyme annotation with EC-specific cutoffs using DETECT v2. Bioinformatics 34, 3393–3395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalkiran A., et al. , ECPred: A tool for the prediction of the enzymatic functions of protein sequences based on the EC nomenclature. BMC Bioinformatics 19, 334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alipanahi B., Delong A., Weirauch M. T., Frey B. J., Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 33, 831–838 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Bairoch A., Apweiler R., The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28, 45–48 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.UniProt Consortium , UniProt: A hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asgari E., Mofrad M. R., Continuous distributed representation of biological sequences for deep proteomics and genomics. PLoS One 10, e0141287 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finn R. D., et al. , Pfam: The protein families database. Nucleic Acids Res. 42, D222–D230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrison K. L., Weiss G. A., Combinatorial alanine-scanning. Curr. Opin. Chem. Biol. 5, 302–307 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Ryu J. Y., Kim H. U., Lee S. Y., Framework and resource for more than 11,000 gene-transcript-protein-reaction associations in human metabolism. Proc. Natl. Acad. Sci. U.S.A. 114, E9740–E9749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zerbino D. R., et al. , Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinsella R. J., et al. , Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database (Oxford) 2011, bar030 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horton P., et al. , WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 35, W585–W587 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schomburg I., et al. , BRENDA, the enzyme database: Updates and major new developments. Nucleic Acids Res. 32, D431–D433 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thul P. J., Lindskog C., The human protein atlas: A spatial map of the human proteome. Protein Sci. 27, 233–244 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buchfink B., Xie C., Huson D. H., Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source code for DeepEC is available at https://bitbucket.org/kaistsystemsbiology/deepec. The list of EC numbers predicted for 33,942,253 protein sequences is available at http://doi.org/10.5281/zenodo.2567339.