Significance
Cardiovascular disease represents a significant health challenge, with towering economic and social costs resulting from the high levels of associated morbidity and mortality. For conditions resulting from undesired blood clotting, treatment options remain limited and are accompanied by significant side effects. Using a rapid chemical ligation platform, 34 homogeneously modified variants of tick anticoagulant proteins were generated. Access to this synthetic protein library enabled key structure–activity relationships to be elucidated and revealed unexpected differences in the mechanism of thrombin inhibition by this group of otherwise closely related molecules. The synthetic platform reported here provides a unique means to expedite the generation and identification of polypeptide and protein therapeutic leads for clotting-associated diseases.
Keywords: ligation, protein, synthesis, thrombin, blood clotting
Abstract
Hematophagous organisms produce a suite of salivary proteins which interact with the host’s coagulation machinery to facilitate the acquisition and digestion of a bloodmeal. Many of these biomolecules inhibit the central blood-clotting serine proteinase thrombin that is also the target of several clinically approved anticoagulants. Here a bioinformatics approach is used to identify seven tick proteins with putative thrombin inhibitory activity that we predict to be posttranslationally sulfated at two conserved tyrosine residues. To corroborate the biological role of these molecules and investigate the effects of amino acid sequence and sulfation modifications on thrombin inhibition and anticoagulant activity, a library of 34 homogeneously sulfated protein variants were rapidly assembled using one-pot diselenide-selenoester ligation (DSL)-deselenization chemistry. Downstream functional characterization validated the thrombin-directed activity of all target molecules and revealed that posttranslational sulfation of specific tyrosine residues crucially modulates potency. Importantly, access to this homogeneously modified protein library not only enabled the determination of key structure–activity relationships and the identification of potent anticoagulant leads, but also revealed subtleties in the mechanism of thrombin inhibition, between and within the families, that would be impossible to predict from the amino acid sequence alone. The synthetic platform described here therefore serves as a highly valuable tool for the generation and thorough characterization of libraries of related peptide and/or protein molecules (with or without modifications) for the identification of lead candidates for medicinal chemistry programs.
Thrombosis is a common pathological process that underlies the development of ischemic stroke, myocardial infarction, pulmonary embolism, and venous thrombosis (1). Combined, these diseases are the leading cause of death and disability globally and are enormously costly to healthcare systems (2). Thrombosis arises from the formation of blood clots inside veins, arteries, or within the microvasculature, resulting in vessel occlusion and starvation of the tissues of oxygen, glucose, and other essential nutrients (3). The most common means of treating or preventing arterial thrombosis is through the use of antiplatelet agents (e.g., aspirin and clopidogrel) (4), whereas anticoagulants that target key blood coagulation proteases [including factor Xa (FXa) or thrombin] (5, 6) are primarily used to prevent or treat venous thrombosis. Antithrombotics are the most rapidly growing segment of the cardiovascular drug market, predicted to increase from $25 billion to >$40 billion by 2025. This growth is primarily driven by an aging population and rising rates of obesity leading to increased incidence of cardiovascular disease (7). However, currently approved antithrombotic drugs carry significant risk of bleeding and hemorrhage for a number of indications. As such, the development of new anticoagulants, particularly those with enhanced safety profiles, is urgently needed for improved prevention and treatment of thromboembolic disorders (8).
A number of potent and selective thrombin inhibitors are produced by hematophagous invertebrates as a means for the organisms to acquire a bloodmeal (9–12). While thrombin serves several roles in blood clotting, inhibition by these molecules prevents the proteolytic cleavage of soluble fibrinogen to insoluble fibrin, thus preventing the formation of clots (3). Given the potent thrombin inhibitory activity exhibited by these polypeptides, it is unsurprising that molecules derived from blood-feeding organisms have found utility in the clinic, with the archetypal leech-derived anticoagulant hirudin (and analogs thereof) used for a range of thromboembolic disorders, especially in patients with heparin-induced thrombocytopenia (13). Recently we have shown that the thrombin inhibitory activity of two tick-derived direct thrombin inhibitors (DTIs)––madanin-1 and chimadanin––is modulated by the posttranslational sulfation of conserved tyrosine residues (14). We have additionally shown that, unlike hirudin (15) and other tick (16–18) and mosquito (19, 20) DTIs that occupy the active site and exosite I (or fibrinogen binding site) of thrombin, these molecules possess an unusual binding mode whereby they straddle the active site and exosite II (or heparin binding site) (Fig. 1A). This work inspired the search for DTIs from other ticks, with a view to discovering modified polypeptide anticoagulants (21–23) that are described herein.
Fig. 1.
Exosite II-targeting tick-derived thrombin inhibitors. (A) Cartoon representation of the complex between thrombin (gray) and madanin-1 (yellow) (Protein Data Bank entry 5L6N; ref. 14). The side chains of residues from the exosite I (orange), exosite II (blue), and the catalytic triad (red) of thrombin and of the sulfotyrosine-containing motif of madanin-1 (yellow) are represented as sticks. (B) Multiple amino acid sequence alignment of tick salivary proteins. The amino acid sequence of the sulfotyrosine-containing natural anticoagulant madanin-1 (14) (Mad1 – Q86FP9) from Haemaphysalis longicornis is aligned with those of madanin-like 1 (MadL1 – Q4R1A5) and madanin-like 2 (MadL2 – Q4R1A2) from the same organism; hyalomin 1 (Hya1 – E2J6S1), hyalomin 2 (Hya2 – E2J6T8), and hyalomin 3 (Hya3 – E2J6R9) from H. marginatum rufipes (22); and And82 and And310 [reported by Francischetti et al. (23)] from D. andersoni. The acidic sulfation motif is highlighted in yellow, with potential sulfation sites shown in inverted type on a red background. Other strictly conserved residues are shown in inverted type on a magenta background and moderately conserved residues on an orange background. Proposed ligation junctions are shown in inverted type on a black background.
We initially employed a bioinformatic approach to identify seven proteins from other ticks (from the ixodid family) that possessed sequence similarity to madanin-1. This included madanin-like 1 and 2 (MadL1 and MadL2, 73 and 67% sequence identity to madanin-1, respectively) from Haemaphysalis longicornis, hyalomins 1–3 (Hya1, Hya2, and Hya3, 23, 28, and 21% sequence identity to madanin-1, respectively) from Hyalomma marginatum rufipes and andersonins 82 and 310 (And82 and And310, 24 and 29% sequence identity to madanin-1, respectively) from Dermacentor andersoni (Fig. 1). While unmodified variants of the hyalomin family have been shown to exhibit thrombin inhibitory activity (24, 25), based on our prior work on madanin-1 we predict that each is likely to be sulfated based on the presence of conserved tyrosine residues flanked by a highly acidic amino acid sequence––a quintessential motif for posttranslational sulfation (26). To date MadL1, MadL2 (14, 27), And82, and And310 (23) have only been characterized as putative thrombin inhibitors; however, based on the sequence alignment of these proteins (Fig. 1) we predicted that they are likely to be thrombin inhibitory proteins based on the highly conserved tyrosine sulfation motif that is also present in the hyalomins and madanin-1.
Because each of the seven tick-derived polypeptides bears two putative tyrosine sulfation sites, there are four possible sulfated variants (sulfoforms) that might exist for each. As such, in this work we sought to access a library of 28 tick-derived (sulfo)proteins via total chemical synthesis. Through the generation of this library of homogeneously modified proteins it was anticipated that the effect of amino acid sequence, and both the position and valency of sulfation, on the inhibition of thrombin and on anticoagulant activity could be determined. Significant interest is emerging in the generation of libraries of proteins for therapeutic discovery programs (28, 29). We envisaged that our approach to generate a focused synthetic library of modified proteins would provide the means to uncover key structure–activity relationships akin to small molecule-based medicinal chemistry.
Results
Synthesis of a Tick-Derived Sulfopeptide Library via Diselenide-Selenoester Ligation-Deselenization Chemistry.
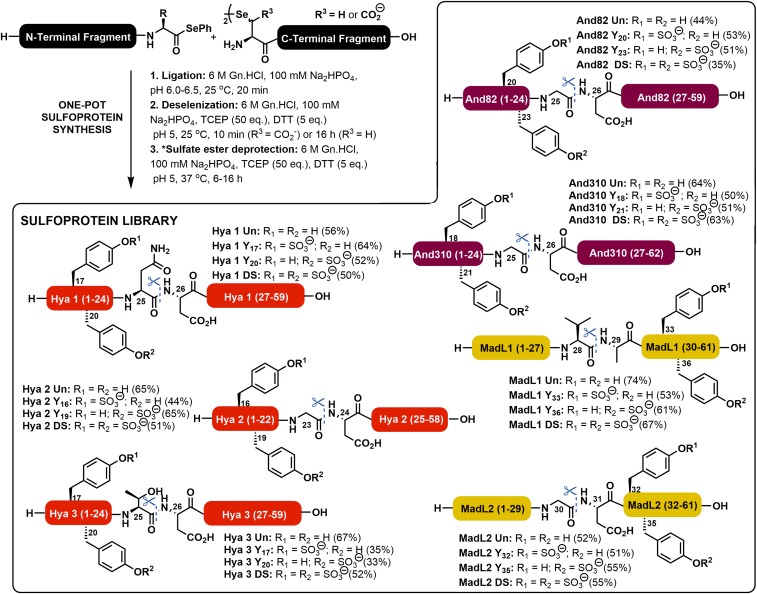
Given the number of (sulfo)proteins in the proposed library, a rapid and efficient means of accessing the targets was necessary. The length of the peptides (58–62 residues) meant that solid-phase peptide synthesis (SPPS) alone could not be used for efficient assembly. We therefore opted to disconnect each target into two fragments which could be fused through ligation chemistry. More specifically, we chose to employ the recently reported diselenide-selenoester ligation (DSL) methodology (30) for the assembly of the target sulfoprotein library. In its original form, the method enables the rapid ligation of a peptide bearing a C-terminal selenoester functionality with a peptide containing an N-terminal selenocystine moiety (the oxidized form of selenocysteine) to afford a native amide bond linking the two fragments. Importantly, the reactions are performed in aqueous media, are complete in minutes, and can be coupled with a one-pot chemoselective deselenization transformation to generate alanine in the place of selenocysteine at the ligation junction (30). With the exception of madanin-like 1 (MadL1), none of the target proteins possess appropriately positioned alanine residues for assembly through DSL at selenocystine. As such, for the majority of the targets we opted to employ DSL chemistry at β-selenoaspartate (24), a decision based on the number of aspartate residues located near the middle of each of the polypeptide sequences (Fig. 1B). Following the DSL reactions, in situ deselenization of the β-selenoaspartate could be used to generate the native (sulfo)protein targets.
Each of the requisite peptide selenoester and selenopeptide ligation partners were first prepared by modified Fmoc-strategy SPPS strategies (see SI Appendix for full synthetic details and SI Appendix, Figs. S1–S33). It should be noted that the sulfated tyrosine residues were incorporated into the N-terminal fragments [for sulfoforms of Hya1 (SI Appendix, Figs. S1–S5), Hya2 (SI Appendix, Figs. S6–S9), Hya 3 (SI Appendix, Figs. S10–S13), And82 (SI Appendix, Figs. S24–S28), and And310 (SI Appendix, Figs. S29–S33)] or C-terminal fragments [for MadL1 (SI Appendix, Figs. S14–S18) and MadL2 (SI Appendix, Figs. S19–S23)] as a preformed neopentyl (nP) sulfate ester protected building block (31, 32) during Fmoc-SPPS. This prevented the loss of the labile modification in the final acidic cleavage step (Fig. 2). Following the preparation of the suitably functionalized selenopeptide and peptide selenoester fragments, the library was next assembled via the proposed DSL-deselenization approach (SI Appendix, Figs. S34–S111). Briefly, diselenide-containing fragments were prepared as 10 mM solutions in ligation buffer (6 M Gn.HCl, 0.1 M Na2HPO4, pH 6.0–6.5) and were added to a solution of the selenoester-containing fragment in an equal volume of ligation buffer. For the synthesis of the hyalomins (SI Appendix, Figs. S34–S63) and the andersonins (SI Appendix, Figs. S88–S111), bearing two neopentyl-protected sulfated tyrosine residues in the selenoester-functionalized fragment, 10 vol.% N,N-dimethylformamide was added to the buffer to increase the solubility of the fragments. Upon mixing of the two fragments, the pH was adjusted to between 6.0 and 6.5 and the reaction was incubated at room temperature for 20 min, which led to completion of the ligation reactions in all cases (as judged by ultra performance liquid chromatography-mass spectrometry [UPLC-MS]). Upon completion, the ligation mixture was treated with an equal volume of a deselenization mixture containing 500 mM tris(2-carboxyethyl)phosphine and 50 mM DTT in ligation buffer. For the hyalomin (SI Appendix, Figs. S34–S63), andersonin (SI Appendix, Figs. S88–S111), and madanin-like 2 sulfoforms (SI Appendix, Figs. S76–S87), bearing a β-selenoaspartate residue at the ligation junction, deselenization was complete within 10 min and could be accomplished without hexane extraction of DPDS or sparging of the reaction mixture (normally required to prevent radical quenching or oxidation in the deselenization step, respectively). The rapid rate of the deselenization at β-selenoaspartate is a result of stabilization of the β-carbon–centered radical intermediate by the adjacent carboxylate moiety (24). Following the rapid DSL and deselenization steps, the reactions were further incubated for 8–16 h in the deselenization buffer to effect deprotection of the neopentyl sulfate esters. For madanin-like 1 (with selenocystine at the ligation junction; SI Appendix, Figs. S64–S75) both hexane extraction and thorough sparging was required to effect efficient deselenization, which required 16 h to reach completion. This prolonged deselenization step proceeded with concomitant deprotection of the neopentyl sulfate esters. Following the one-pot ligation–deselenization–deprotection sequence each of the (sulfo)proteins from the library were purified by reverse-phase HPLC (with a formic acid buffer to preserve the acid labile sulfate groups) to afford the target (sulfo)proteins in excellent yields (Fig. 2).
Fig. 2.
Chemical synthesis of a library of tick-derived (sulfo)proteins via one-pot DSL-deselenization chemistry. NB: Ligation junctions are shown as blue dotted lines with scissors. Isolated yields are over both the DSL and deselenization steps. R depicts the side chain of the C-terminal residue on the selenoester fragments, R1 and R2 depict sites of potential sulfation, and R3 depicts the side chain of the N-terminal diselenide residue. *This step is not necessary for madanin-like 1 due to the longer deselenization times at selenocystine (R3 = H) which lead to concomitant neopentyl sulfate ester deprotection.
In Vitro Thrombin Inhibitory and Anticoagulant Activity of Tick-Derived Sulfoproteins.
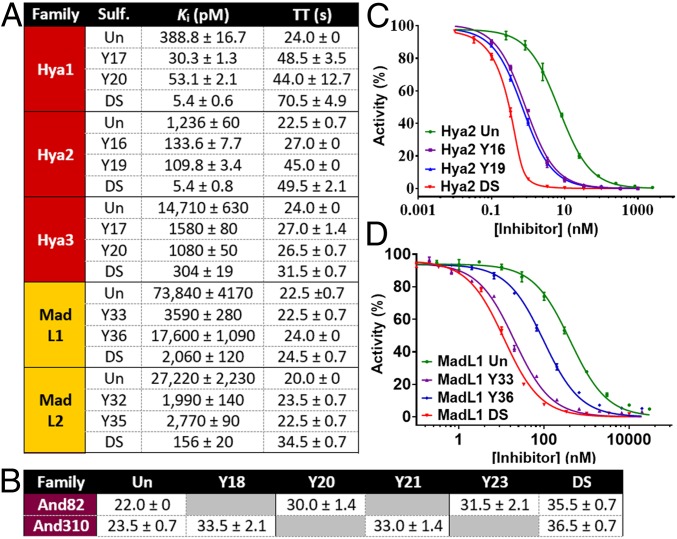
With the desired library of homogenously sulfated proteins in hand, we next investigated the inhibitory activity against α-thrombin using a well-established kinetic assay using the chromogenic substrate Tos-Gly-Pro-Arg-p-nitroanilide (SI Appendix, Fig. S112) (27). With the exception of And82 and And310, each of the synthetic proteins exhibited potent inhibition of α-thrombin, with the incorporation of the tyrosine sulfate posttranslational modifications leading to dramatic improvements in inhibitor potency (Fig. 3 and SI Appendix, Fig. S113 and Table S1). Monosulfation of the hyalomin family at Y16/17 or 19/20 led to an order of magnitude improvement in activity compared with the unsulfated proteins. Sulfation at Y17 and Y20 in Hya1 DS (Ki = 5.4 pM) provided nearly two orders of magnitude improvement in thrombin inhibition (Ki Hya1 Un = 389 pM), while double sulfation in Hya2 DS (Ki = 5.4 pM) led to a 2.5 orders of magnitude improvement in thrombin inhibitory activity over the nonmodified protein (Ki Hya2 Un = 1,236 pM). Interestingly, despite the high sequence similarity between the hyalomin family members, Hya3 Un (without sulfation) was significantly less potent (Ki Hya3 Un = 14.7 nM) compared with nonmodified Hya1 and Hya2. Nonetheless, sulfation at Y17 and Y20 provided almost two orders of magnitude improvement in thrombin inhibitory activity (Ki Hya3 DS = 304 pM). It should be noted that the doubly sulfated hyalomins exhibited similar inhibitory activity against mouse α-thrombin (SI Appendix, Fig. S114 and Table S2).
Fig. 3.
Thrombin inhibitory and anticoagulant activity of a library of synthetic tick-derived (sulfo)proteins. (A) Inhibition constants against human α-thrombin and TT for hyalomin and madanin-like (sulfo)proteins. (B) TT for andersonin (sulfo)proteins. Raw thrombin inhibitory data for (C) Hya2 and (D) MadL1 sulfoforms. TT measured at 10 nM inhibitor concentration; TT in the absence of an inhibitor = 19.5 s; all TT data are the mean of four replicates ± SEM. Un = unsulfated, DS = doubly sulfated.
Intrigued by the significantly lower thrombin inhibitory potency of Hya3 compared with the other hyalomins, we generated two chimeric proteins by crossing over the previously generated Hya1 and Hya3 fragments to determine which region of the protein is responsible for the drop in activity. Specifically, a Hya1/3 DS chimera was generated from the doubly sulfated Hya1 [1–25] selenoester and the Hya3 [26–59] diselenide dimer, while the Hya3/1 DS chimera was generated from the doubly sulfated Hya3 [1–25] selenoester and the Hya1 [26–59] diselenide dimer (SI Appendix, Figs. S115–S120). The thrombin inhibitory potency of each chimera was found to be intermediate to that of the two parent proteins (Ki Hya1/3 DS = 8.4 pM; Ki Hya3/1 DS = 42.6 pM), revealing a major contribution of the N-terminal half of Hya1 and Hya3 for their thrombin inhibitory activity (SI Appendix, Fig. S121).
Having determined the in vitro thrombin inhibitory activity of the proteins, we next measured the influence of the differentially sulfated hyalomins on coagulation using a thrombin time (TT) assay. The TT assay measures the time taken for human blood plasma to clot in the presence of an inhibitor after the addition of thrombin; the prolongation of TT therefore provides an indication of the anticoagulant potency. We observed a significant prolongation of TT to 32–71 s when 10 nM of the doubly sulfated hyalomins (Hya1-3 DS) were used (TT without inhibitor = 19.5 s). The monosulfated hyalomins prolonged TT less than the doubly sulfated homologs but more than the unmodified protein, consistent with the measured activity toward thrombin (Fig. 3 and SI Appendix, Table S1).
Despite the sequence homology with the hyalomin family, unmodified MadL1 Un (Ki = 73.8 nM) and MadL2 Un (Ki = 27.2 nM) were significantly less potent thrombin inhibitors than Hya1 Un and Hya2 Un (but similarly potent to Hya3 Un). Like the hyalomin family, monosulfation of MadL2 in MadL2 Y32 (Ki = 2 nM) or MadL2 Y35 (Ki = 2.8 nM) led to an order of magnitude improvement in thrombin inhibitory activity, while doubly sulfated MadL2 DS (Ki = 156 pM) showed two orders of magnitude improvement over the unmodified protein (Fig. 3 and SI Appendix, Table S1). In contrast to the other proteins studied in this series, the position of sulfation on MadL1 had a profound effect on potency. Specifically, sulfation at Y36 provided only a modest improvement in potency compared with the unmodified protein (Ki of MadL1 Y36 = 17.6 nM vs. 73.8 nM for MadL1 Un). However, sulfation at Y33 led to 1.5 orders of magnitude enhancement in activity (Ki of MadL1 Y33 = 3.6 nM). In this case the inclusion of two sulfate posttranslational modifications in MadL1 DS only led to a small additional improvement in potency (Ki = 2.1 nM). Interestingly, in this series of proteins only MadL2 DS caused a significant prolongation of TT (34.5 s at 10 nM), which reflects the significantly enhanced thrombin inhibitory activity of this sulfoprotein compared with the other members of the family (Fig. 3 and SI Appendix, Table S1).
Insights into the Binding Mode of the Tick-Derived Sulfoproteins.
To probe the thrombin binding mode of the sulfoproteins, the activity against human γ-thrombin of each of the doubly sulfated hyalomin and madanin-like variants was also assessed. γ-Thrombin results from the autolytic processing of the α-isoform that disrupts exosite I such that the affinity toward fibrinogen and exosite I-binding inhibitors is significantly reduced, while maintaining amidolytic activity toward small synthetic substrates. As such, reduced inhibitory activity of a molecule against γ-thrombin (compared with α-thrombin) is indicative of an involvement of exosite I in inhibitor binding. Both doubly sulfated madanin-like variants (MadL1 DS and MadL2 DS) retained activity against γ-thrombin, suggesting that these molecules, similar to madanin-1 (14), interact with the exosite II of thrombin (SI Appendix, Fig. S114 and Table S2). Despite the homology between the families of proteins, a 2–3 orders of magnitude loss in activity against γ-thrombin was observed with the doubly sulfated hyalomins (Hya1 DS, Hya2 DS, and Hya3 DS). This suggests a contribution of the interaction between the C-terminal region of the hyalomins and the exosite I of thrombin for the affinity and inhibitory activity of this group of anticoagulants (SI Appendix, Fig. S114 and Table S2). These studies further highlight the nuanced activity of this group of related thrombin-inhibiting proteins.
Anticoagulant Activity of the Hyalomin Sulfoproteins in an Activated Partial Thromboplastin Time Assay.
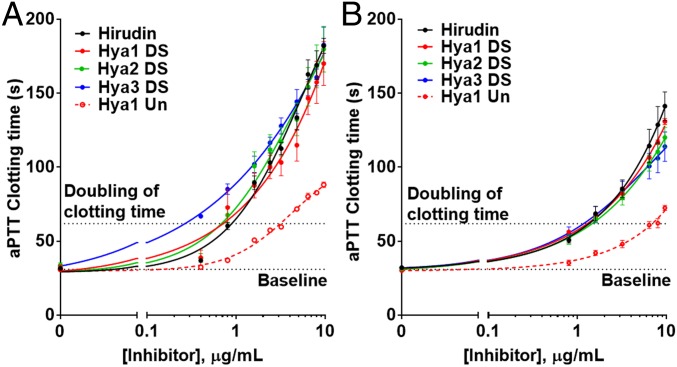
As a further measure of anticoagulant activity, we next assessed the synthetic hyalomin sulfoproteins in an activated partial thromboplastin time (aPTT) assay. The aPTT assay examines activation of the intrinsic and common pathways of the coagulation cascade through measurement of the time taken for fibrin clot formation upon initiation of the intrinsic pathway. We performed aPTT assays on both human and mouse plasma to test the potency of the sulfoproteins in the two organisms. The advantage of this assay over TT measurements is that anticoagulant activity of an inhibitor is assessed using thrombin generated via the intrinsic pathway rather than through exogenously added thrombin. A key parameter that is drawn from both the in vitro human and in vitro mouse aPTT assays is the dose of the anticoagulant that provides twofold prolongation of clotting time compared with the untreated control; this is deemed a therapeutic anticoagulant dose. Gratifyingly, a potent dose-dependent prolongation of the aPTT was observed for each of the sulfated tick-derived proteins with human plasma and each exhibited similar activity to the clinically approved hirudin, doubling aPTT at concentrations in the range 0.4–0.9 μg/mL (Fig. 4A). Consistent with the TT assay there was a significant improvement in anticoagulant activity with the sulfated versus unsulfated proteins, with the doubly sulfated Hya1 (Hya1 DS) variant showing twofold prolongation at ∼0.7 μg/mL vs. ∼3.3 μg/mL for Hya1 Un. The anticoagulant activities of the sulfated hyalomins were replicated in the aPTT assay on mouse plasma (Fig. 4B), consistent with the comparable inhibition exhibited against murine and human α-thrombin (vide supra).
Fig. 4.
Hyalomins prolong the aPTT. Dose–response curves for hirudin, Hya1 Un, Hya1 DS, Hya2 DS, and Hya3 DS in (A) human plasma (in vitro) and (B) mouse plasma (in vitro). Data points present mean ± SEM of 3–4 independent plasma samples for aPTT assay of each inhibitor. NB: Inhibitor concentrations are plotted on a log scale.
Unexpected Behavior of the Andersonins.
Despite the sequence similarity to the hyalomins and madanin-like proteins, the andersonins exhibited very different inhibitory profiles against thrombin. Specifically, while each of the And82 and And310 variants showed initial thrombin inhibition in the chromogenic assay, reactivation of the enzyme was observed after 25–40 min (SI Appendix, Fig. S122). This gave a kinetic profile that could not be fitted to a competitive inhibition model, preventing determination of a Ki value for these proteins. This behavior was thought to be due to cleavage of the andersonins by thrombin. Investigation of the putative thrombin digestion products of each of the andersonins by mass spectrometry led to the identification of thrombin cleavage sites: two in And310 (K40-L41 and R43-H44) and three in And82 (R10-S11, R40-L41, and R43-T44) (SI Appendix, Fig. S123). The presence of multiple cleavage sites provides an explanation for the considerable loss in activity exhibited by the andersonins upon incubation with thrombin as the resultant small fragments would have poor affinity for the enzyme. We subsequently explored the kinetics of cleavage on each of the And310 sulfoforms and showed that in each case, both proteolytic events were complete within 10 min (SI Appendix, Fig. S124). These data were consistent with the rapid deactivation of the inhibitors observed in the chromogenic assay. These findings raise the intriguing question of why thrombin inhibitors with such short half-lives are generated by the D. andersoni ticks.
Interestingly, all eight synthetic sulfoforms within the andersonin family were capable of prolonging coagulation in a TT assay, with the doubly sulfated homologs And82 DS and And310 DS having the greatest anticoagulant effect (TT = 35.5–36.5 s at 10 nM; Fig. 3A and SI Appendix, Table S3). This result may seem at odds with the cleavage and reactivation of thrombin observed in the chromogenic assay; however, the shorter time scales of the TT assays, together with the known rates of proteolytic cleavage of the inhibitors by thrombin, meant that these experiments were likely measuring the anticoagulant activity before completion of the proteolytic inactivation.
Engineering Andersonins Resistant to Thrombin Cleavage.
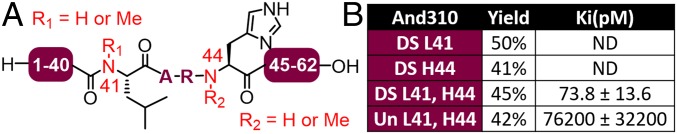
To prevent the proteolytic inactivation of the andersonins we investigated whether we could synthetically engineer backbone N-methylated variants (at the identified cleavage sites) for the generation of stable thrombin inhibitors (33). Toward this end, we proposed three differentially backbone methylated variants of And310: one bearing N-methylation at L41, a second bearing N-methylation at H44, and a third N-methylated at both sites. In this case we targeted the doubly sulfated variant of And310 DS which exhibited the most potent (albeit marginally) anticoagulant activity in the TT assay (Fig. 3A). The requisite backbone methylated diselenide-functionalized fragments could be accessed by Fmoc-SPPS using an on-resin methylation strategy (SI Appendix, Figs. S125–S127), while the same doubly sulfated And310 (1–25) selenoester fragment prepared for the parent proteins could be reused (SI Appendix, Fig. S32) (34). These fragments were subjected to the one-pot DSL–deselenization–deprotection sequence employed earlier and afforded the desired differentially backbone methylated variants And310 DS L41, And310 DS H44, and And310 DS L41 H44 in good yields (Fig. 5 A and B and SI Appendix, Figs. S128–S136). We also prepared a doubly backbone methylated but unsulfated variant (And310 Un L41 H44) as a control protein for the thrombin inhibition and anticoagulant studies (SI Appendix, Figs. S137–S139).
Fig. 5.
Noncleavable engineered variants of And310 display increased anticoagulant activity. (A) Simplified structure of methylated And310 variants; (B) thrombin inhibitory activity of backbone N-methylated And310 variants against human α-thrombin. ND: Inhibition constants could not be determined due to thrombin cleavage.
With the backbone methylated sulfoproteins in hand, we next interrogated the stability of each of the variants in the presence of thrombin by UPLC-MS. Cleavage was prevented at each of the individually methylated sites, with the doubly methylated variant completely resistant to proteolysis up to 8 h (SI Appendix, Fig. S140). We next assessed the thrombin inhibitory activity of the backbone methylated constructs. Only the doubly methylated variants of And310, And310 Un L41 H44 and And310 DS L41 H44, displayed seemingly interpretable kinetic profiles, although they could not be fitted to the Morrison model for tight binding inhibitors. In theory, the sigmoid-shaped onsets observed for these progress curves (SI Appendix, Fig. S141 A and B) are admissible under conditions of high enzyme concentration that do not apply here (35), or else for slow-onset inhibition due to enzyme isomerization (36). Since increasing concentrations of inhibitor exacerbate the sigmoid shape, a mechanism in which the inhibitor binds to the productive, nonisomerized enzyme was proposed (SI Appendix, Fig. S142 A and B) and numerically validated (SI Appendix, Fig. S141 A and B). The sulfated homolog And310 DS L41 H44 was significantly more potent (Ki = 73.8 ± 13.6 pM; SI Appendix, Fig. S142C) than And310 Un L41 H44 (Ki = 76,200 ± 32,200 pM; SI Appendix, Fig. S142C), further reflecting the importance of the tyrosine sulfate modifications for modulation of activity. Both And310 DS L41 H44 and And310 Un L41 H44 were also assessed in the aPTT assay with human plasma with twofold prolongation of clotting time observed at ∼7 μg/mL for doubly sulfated And310 DS L41 H44 vs. at ∼35 μg/mL for unsulfated And310 Un L41 H44 (SI Appendix, Fig. S143).
Conclusions
In summary, seven putative tick-derived thrombin inhibitors have been identified that possess two conserved sites of posttranslational tyrosine sulfation. To investigate the thrombin inhibitory and anticoagulant activity of this group of proteins, as well as the effect of tyrosine sulfation, a library of 28 (sulfo)proteins was proposed. Each member of the target library was efficiently accessed through chemical synthesis using one-pot DSL-deselenization technology (30) and enabled the determination of key structure–activity data. Several members of the synthetic library possessed potent thrombin inhibitory activity, the most active being doubly sulfated Hya1 and Hya2 (Ki = 5.4 pM) that were more than two orders of magnitude more potent than the unsulfated proteins. The anticoagulant activity was also modulated by the sulfation state (as judged by TT and aPTT assays) with the doubly sulfated variants of all library members showing the most potent activity.
Despite similarities in size and overall amino acid sequence, library members displayed wide-ranging thrombin inhibitory and anticoagulant potencies (across four orders of magnitude). Of particular note was the nearly two orders of magnitude difference between the closely related doubly sulfated hyalomins Hya1 DS and Hya3 DS. Through the synthetic generation of two hybrid sulfoproteins (Hya1/3 DS and Hya3/1 DS) we show that this difference in potency was owing to variation in the N-terminal sequence of the hyalomins.
Equally unexpected was the finding that And82 and And310 were rapidly degraded by thrombin due to multisite cleavage leading to reversal of an initially potent thrombin inhibitory profile. This intriguing instability of the andersonins is likely countered by the continuous infusion of active inhibitor by ticks during feeding. However, it cannot be ruled out that the cleavage fragments possess other still unidentified biological roles. The rapid deactivation of And310 was abrogated by the generation of a synthetic backbone dimethylated variant that was resistant to proteolysis and led to picomolar inhibition of thrombin, albeit through an isomerization-mediated mechanism.
Through evolutionary divergence, nature often produces a large range of closely related proteins with potent biological activities. This is clearly evident in thrombin-inhibiting anticoagulants produced in the saliva of blood-feeding organisms (21). We have revealed surprising variation in potency and the molecular mechanisms of action of three families of ixodid tick proteins and have identified potent anticoagulants for future therapeutic investigation. However, these represent only a small fraction of potential therapeutic leads from the more than 800 different tick species known. Given the efficiency of the DSL-deselenization platform developed in this work, one could imagine using the tick salivary proteome as the starting point for the development of modified synthetic protein leads in a manner analogous to that used in small molecule medicinal chemistry to rapidly develop privileged “hits” from nature.
Methods
Full details for peptide synthesis, ligation reactions, thrombin inhibition, TT, and aPTT assays, as well as kinetic and mass spectrometry analysis of thrombin cleavage, can be found in SI Appendix. Animal procedures were performed in accordance with the Australian code of practice for the care and use of animals for scientific purposes and were approved by the University of Sydney Animal Ethics Committee (Project 2017/1197).
Supplementary Material
Acknowledgments
This work was funded by a National Health and Medical Research Council Project Grant (1120941). This work was funded in part by national funds through Fundação para a Ciência e a Tecnologia (Portugal) in the form of Contract DL 57/2016/CP1355/CT0011 (to J.R.-R.) and by the European Regional Development Fund (Grant Norte-01-0145-FEDER-000012). The Proteomics Scientific Platform of i3S, supported by the Portuguese Mass Spectrometry Network, part of the National Research Infrastructures Roadmap (ROTEIRO/0028/2013; LISBOA-01-0145-FEDER-022125), is also acknowledged.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905177116/-/DCSupplemental.
References
- 1.Benjamin E. J., et al. , Heart disease and stroke statistics—2019 update: A report from the American Heart Association. Circulation 139, e56–e528 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Wendelboe A. M., Raskob G. E., Global burden of thrombosis: Epidemiologic aspects. Circ. Res. 118, 1340–1347 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Lippi G., Franchini M., Targher G., Arterial thrombus formation in cardiovascular disease. Nat. Rev. Cardiol. 8, 502–512 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Michelson A. D., Antiplatelet therapies for the treatment of cardiovascular disease. Nat. Rev. Drug Discov. 9, 154–169 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Gross P. L., Weitz J. I., New anticoagulants for treatment of venous thromboembolism. Arterioscler. Thromb. Vasc. Biol. 28, 380–386 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Ginsberg J. S., Management of venous thromboembolism. N. Engl. J. Med. 335, 1816–1828 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Fan P., et al. , Recent progress and market analysis of anticoagulant drugs. J. Thorac. Dis. 10, 2011–2025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fredenburgh J. C., Gross P. L., Weitz J. I., Emerging anticoagulant strategies. Blood 129, 147–154 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Corral-Rodríguez M. Á., Macedo-Ribeiro S., Pereira P. J. B., Fuentes-Prior P., Leech-derived thrombin inhibitors: From structures to mechanisms to clinical applications. J. Med. Chem. 53, 3847–3861 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Corral-Rodríguez M. Á., Macedo-Ribeiro S., Barbosa Pereira P. J., Fuentes-Prior P., Tick-derived Kunitz-type inhibitors as antihemostatic factors. Insect Biochem. Mol. Biol. 39, 579–595 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Koh C. Y., Modahl C. M., Kulkarni N., Kini R. M., Toxins are an excellent source of therapeutic agents against cardiovascular diseases. Semin. Thromb. Hemost. 44, 691–706 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Ribeiro J. M., Francischetti I. M., Role of arthropod saliva in blood feeding: Sialome and post-sialome perspectives. Annu. Rev. Entomol. 48, 73–88 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Lee C. J., Ansell J. E., Direct thrombin inhibitors. Br. J. Clin. Pharmacol. 72, 581–592 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson R. E., et al. , Tyrosine sulfation modulates activity of tick-derived thrombin inhibitors. Nat. Chem. 9, 909–917 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Rydel T. J., et al. , The structure of a complex of recombinant hirudin and human alpha-thrombin. Science 249, 277–280 (1990). [DOI] [PubMed] [Google Scholar]
- 16.Koh C. Y., et al. , Crystal structure of thrombin in complex with S-variegin: Insights of a novel mechanism of inhibition and design of tunable thrombin inhibitors. PLoS One 6, e26367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macedo-Ribeiro S., et al. , Isolation, cloning and structural characterisation of boophilin, a multifunctional Kunitz-type proteinase inhibitor from the cattle tick. PLoS One 3, e1624 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Locht A., et al. , The ornithodorin-thrombin crystal structure, a key to the TAP enigma? EMBO J. 15, 6011–6017 (1996). [PMC free article] [PubMed] [Google Scholar]
- 19.Figueiredo A. C., et al. , Unique thrombin inhibition mechanism by anophelin, an anticoagulant from the malaria vector. Proc. Natl. Acad. Sci. U.S.A. 109, E3649–E3658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pirone L., et al. , Functional analyses yield detailed insight into the mechanism of thrombin inhibition by the antihemostatic salivary protein cE5 from Anopheles gambiae. J. Biol. Chem. 292, 12632–12642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mans B. J., Louw A. I., Neitz A. W. H., Evolution of hematophagy in ticks: Common origins for blood coagulation and platelet aggregation inhibitors from soft ticks of the genus ornithodoros. Mol. Biol. Evol. 19, 1695–1705 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Francischetti I. M. B., Anderson J. M., Manoukis N., Pham V. M., Ribeiro J. M. C., An insight into the sialotranscriptome and proteome of the coarse bontlegged tick, Hyalomma marginatum rufipes. J. Proteomics 74, 2892–2908 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francischetti I. M. B., Sá-Nunes A., Mans B. J., Santos I. M., Ribeiro J. M. C., The role of saliva in tick feeding. Front. Biosci. 14, 2051–2088 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell N. J., et al. , Accelerated protein synthesis via one-pot ligation-deselenization chemistry. Chem 2, 703–715 (2017). [Google Scholar]
- 25.Jablonka W., et al. , Identification and mechanistic analysis of a novel tick-derived inhibitor of thrombin. PLoS One 10, e0133991 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monigatti F., Gasteiger E., Bairoch A., Jung E., The sulfinator: Predicting tyrosine sulfation sites in protein sequences. Bioinformatics 18, 769–770 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Figueiredo A. C., de Sanctis D., Pereira P. J. B., The tick-derived anticoagulant madanin is processed by thrombin and factor Xa. PLoS One 8, e71866 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gates Z. P., et al. , Xenoprotein engineering via synthetic libraries. Proc. Natl. Acad. Sci. U.S.A. 115, E5298–E5306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam K. S., et al. , A new type of synthetic peptide library for identifying ligand-binding activity. Nature 354, 82–84 (1991). [DOI] [PubMed] [Google Scholar]
- 30.Mitchell N. J., et al. , Rapid additive-free selenocystine-selenoester peptide ligation. J. Am. Chem. Soc. 137, 14011–14014 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Simpson L. S., Widlanski T. S., A comprehensive approach to the synthesis of sulfate esters. J. Am. Chem. Soc. 128, 1605–1610 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Stone M. J., Payne R. J., Homogeneous sulfopeptides and sulfoproteins: Synthetic approaches and applications to characterize the effects of tyrosine sulfation on biochemical function. Acc. Chem. Res. 48, 2251–2261 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Chatterjee J., Gilon C., Hoffman A., Kessler H., N-methylation of peptides: A new perspective in medicinal chemistry. Acc. Chem. Res. 41, 1331–1342 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Biron E., Chatterjee J., Kessler H., Optimized selective N-methylation of peptides on solid support. J. Pept. Sci. 12, 213–219 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Pinto M. F., et al. , Enzyme kinetics: The whole picture reveals hidden meanings. FEBS J. 282, 2309–2316 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Baici A., Kinetics of Enzyme-Modifier Interactions (Springer, Wien, 2015). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.