Significance
Vibrio species are causal agents of important infectious diseases in humans and animals. Here, we have studied Vibrio infections in an ecologically relevant model host, oyster, for which natural virulent Vibrio strains are available. We have shown that virulent Vibrio species use different mechanisms to evade host cellular defenses. These mechanisms relied on distinct molecular determinants shared between strains of the same Vibrio species, but not between different species. The different mechanisms nonetheless converged on the same crucial outcome, immune cell lysis. Such cytotoxicity enabled virulent strains to cause systemic infections. Species-specific mechanisms that actively target host immunity could be more common than thought among Vibrio species, including human pathogens.
Keywords: T6SS, toxin, dual RNA-seq, cytolysis, pathogenesis
Abstract
Vibrio species cause infectious diseases in humans and animals, but they can also live as commensals within their host tissues. How Vibrio subverts the host defenses to mount a successful infection remains poorly understood, and this knowledge is critical for predicting and managing disease. Here, we have investigated the cellular and molecular mechanisms underpinning infection and colonization of 2 virulent Vibrio species in an ecologically relevant host model, oyster, to study interactions with marine Vibrio species. All Vibrio strains were recognized by the immune system, but only nonvirulent strains were controlled. We showed that virulent strains were cytotoxic to hemocytes, oyster immune cells. By analyzing host and bacterial transcriptional responses to infection, together with Vibrio gene knock-outs, we discovered that Vibrio crassostreae and Vibrio tasmaniensis use distinct mechanisms to cause hemocyte lysis. Whereas V. crassostreae cytotoxicity is dependent on a direct contact with hemocytes and requires an ancestral gene encoding a protein of unknown function, r5.7, V. tasmaniensis cytotoxicity is dependent on phagocytosis and requires intracellular secretion of T6SS effectors. We conclude that proliferation of commensal vibrios is controlled by the host immune system, preventing systemic infections in oysters, whereas the successful infection of virulent strains relies on Vibrio species-specific molecular determinants that converge to compromise host immune cell function, allowing evasion of the host immune system.
Vibrionaceae (hereafter vibrios) are ɣ-proteobacteria ubiquitous in oceans and marine coastal environments. They are free-living or associated with organic particles (sea snows) and organisms (1), in the latter case establishing a wide range of mutualistic, commensal, and pathogenic interactions with their hosts. The partnership between the luminous bacterium Vibrio fischeri and the Hawaiian bobtail squid Euprymna scolopes has been used to study mutualistic symbiosis in animals (2, 3). In contrast, some Vibrio species, including Vibrio cholerae, cause disease in humans (4). Other Vibrio species cause disease in wild and farmed marine animals including corals, shrimp, mollusks, and fishes, having significant ecological and economic consequences (5–7). In this context, the oyster Crassostrea gigas provides a model for studying the interactions of Vibrio species with an ecologically relevant host, specifically to understand their disease-causing properties (8).
In oysters, maintenance of homeostatic interactions between the microbiota and host immunity is poorly understood (9), and can be disrupted by abiotic and biotic factors, including pathogenic agents. For example, in oysters with Pacific oyster mortality syndrome, a Herpes virus was shown to cause immune suppression, enabling microbiome dysbiosis and proliferation of bacteria such as Vibrio crassostreae (10). This species and other closely related species of the Splendidus clade (e.g., Vibrio splendidus, Vibrio tasmaniensis, and Vibrio cyclitrophicus) have been repeatedly isolated from diseased oysters, and their virulence confirmed in experimental oyster infections (11–14).
Most oyster–vibrio interactions have been studied by using a single strain, V. tasmaniensis LGP32, as a model. This strain is a facultative intracellular pathogen of hemocytes, oyster immune cells (15). Several LGP32 genes conferring resistance to reactive oxygen species (ROS) and heavy metals were shown to be required for protection against killing by hemocytes (16). Moreover, inside phagosomes, the LGP32 strain undergoes membrane remodeling that confers resistance to host antimicrobial peptides (17). This strain was shown to be cytotoxic to hemocytes (16), but the mechanisms causing cytotoxicity remain unknown, and it remains unclear whether other strains belonging to V. tasmaniensis or to the Splendidus clade as a whole are also intracellular pathogens and cytotoxic to hemocytes. Within the Splendidus clade, virulence is an ancestral trait that has been lost in several species (12). The ancestrally acquired r5.7 gene, which encodes for an exported protein of unknown function, is necessary for virulence across the entire Splendidus clade (12, 14). The r5.7 gene is absent from V. tasmaniensis, although this species includes virulent strains, suggesting that virulence relies, at least in part, on alternative mechanisms. Thus, to understand the basis of Vibrio-driven diseases, it is necessary to identify the specific determinants of virulence of the species belonging to the Splendidus clade and their potential host target(s).
An effective strategy to characterize different virulence pathways is to compare the genomes of closely related strains with potentially distinct virulence mechanisms. Complementary information can be obtained through global analyses of bacterial and host transcriptional responses, i.e., dual transcriptomics. Indeed, bacterial colonization has been shown to have substantial transcriptional effects on host and microorganisms. Thus, simultaneously measuring host and pathogen gene expression by using dual RNA-seq provides useful insights into the physiological changes that occur during the course of an infection, as well as the response of such changes to each other (18), which can be tested by using gene knock-out experiments.
In the present study, we have characterized the within-host cellular and molecular mechanisms leading to colonization success in oysters infected with 2 Vibrio species belonging to the Splendidus clade, V. tasmaniensis and V. crassostreae. By using an integrative approach, we provide a cellular and molecular description of the changes occurring in host and bacterial physiologies during the infection. Our results show that all virulent strains from both species actively repress host cellular defenses by killing hemocytes and colonize oyster tissues. We have found that the process of immune evasion and colonization is accompanied by the repression of a series of antibacterial defenses and manipulation of metal and ROS homeostasis. Interestingly, we have also identified distinct genetic determinants required for bacterial virulence and cytotoxicity in both vibrio species. Taken together, our results show that vibrios have acquired species-specific virulence mechanisms that converge in a common end, i.e., alteration in key cellular defenses in oysters.
Results and Discussion
Differential Cytotoxicity of Virulent Vibrio Strains.
Upon being phagocytized, the virulent V. tasmaniensis LGP32 strain causes high levels of hemocyte cytotoxicity (16). To determine if hemocyte cytotoxicity is a common feature of V. tasmaniensis and V. crassostreae, hemocytes were exposed to 4 virulent strains of each species, as well as a closely related but nonvirulent strain as a control (Fig. 1A). Only virulent strains induced hemocyte lysis (Fig. 1B). Nonetheless, the V. crassostreae J2-9 strain induced hemocyte lysis significantly more rapidly than the V. tasmaniensis LGP32 strain (Fig. 1C), Furthermore, treatment of hemocytes with cytochalasin D, which inhibits phagocytosis, strongly reduced the cytotoxicity of all V. tasmaniensis strains (RM-ANOVA P value < 0.001; Fig. 1B) but had no effect on V. crassostreae cytotoxicity. V. crassostreae J2-9 secretion products alone were unable to induce cytotoxicity (SI Appendix, Fig. S1), prompting us to ask whether cellular contact was required for J2-9 to kill hemocytes. Transwell experiments revealed that J2-9 cytotoxicity was significantly reduced in the absence of contact with hemocytes (RM-ANOVA P value < 0.01; Fig. 1D). Thus, although both species have major lytic effects on oyster hemocytes, their cytotoxicity is exerted through different mechanisms.
Fig. 1.
Species-specific mechanisms of cytotoxicity in V. crassostreae and V. tasmaniensis. (A) Multilocus sequence typing tree of V. crassostreae and V. tasmaniensis with closely related nonvirulent strains. The tree was reconstructed with maximum-likelihood method from a concatenated alignment of 6 core genes (gyrB, hsp60, mreB, recA, rpoD, and topA) representing 10,626 sites. Node support was assessed with 100 bootstrap replicates. The strain cytotoxicity, which correlates with strain virulence (B), is indicated by a red circle, whereas absence of these phenotypes is indicated by a green circle. A filled orange square indicates the presence of the r5.7 gene, which encodes for a known virulence factor. The effect of the presence of T6SS systems on the different replicons (1, on chromosome 1; 2, on chromosome 2; P, on a plasmid) is indicated by a filled blue square. (B–D) Cytotoxicity of vibrios on a monolayer of hemocytes was monitored by the SYTOX Green assay for 15 h. Cells were infected with bacteria at a multiplicity of infection (MOI) of 50:1. (B) Maximum cytotoxicity exerted by V. tasmaniensis or V. crassostreae strains in the presence (gray bars) or absence (back bars) of cytochalasin D, which was used as an inhibitor of phagocytosis. Error bars represent the SD of the mean (RM-ANOVA, P < 0.001). All virulent strains are cytotoxic. Cytotoxicity appears to be dependent on phagocytosis for V. tasmaniensis virulent strains only. The cytotoxicity exerted by V. crassostreae was found to be higher than that of V. tasmaniensis. (C) Time-course analysis of cytotoxicity of J2-9 (black diamond), LGP32 (black circle), J2-8 (gray diamond), and LMG20 012T (gray circle) strains. J2-9 cytotoxicity is faster than LGP32 cytotoxicity. J2-8 and LMG20 012T are noncytotoxic. The error bar represents the SD of the mean (RM-ANOVA, P < 0.05). (D) Maximum cytotoxicity for J2-9 strain was observed when in contact with hemocytes (black bars) vs. a Transwell plate without hemocyte contact (gray bars). The contact between hemocytes and vibrios is necessary for significant expression of J2-9 cytotoxicity. The error bar represents the SD of the mean (RM-ANOVA, P < 0.01; SI Appendix, Fig. S1).
Virulent Vibrios Escape Hemocyte Aggregation and Cause Systemic Infection.
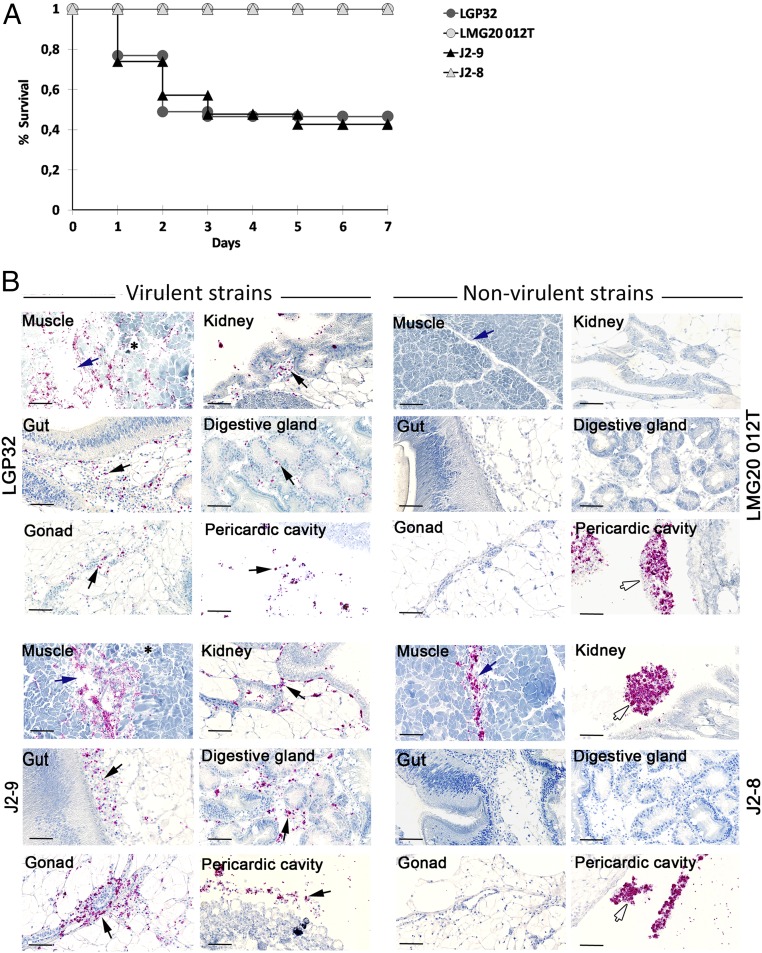
To explore the pathogenesis of the virulent strains, we compared their histopathological effects versus those of nonvirulent strains. Strains were first injected into oysters from the same biparental family to reduce host genetic variability. Virulent strains (V. tasmaniensis LGP32 and V. crassostreae J2-9) induced mortality within 1 d and reached a plateau after 2–3 d (Fig. 2A). No significant differences were observed in the kinetics and rate of mortality induced by either of the strains, with ∼40% of oysters surviving after 3 d (log-rank test, P = 0.802; Fig. 2A). No mortalities were recorded in oysters injected with nonvirulent strains (LMG20012T, J2-8; Fig. 2A). The similarity in virulence of the 2 strains was confirmed in 2 other families of oysters, suggesting that virulence phenotypes do not depend on the genetic background of the oysters (SI Appendix, Fig. S2).
Fig. 2.
Virulent strains LGP32 and J2-9 colonize oyster connective tissues and cause damage to the adductor muscle (SI Appendix, Fig. S2). (A) Kaplan–Meier survival curves were generated from a biparental family of juvenile oysters (Decipher family 14) injected with the virulent LGP32 or J2-9 or the nonvirulent LMG20 012T or J2-8 (7.5 × 107 CFU per oyster). Mortalities were counted daily over 7 d in 2 tanks containing 20 oysters each. Only J2-9 and LGP32 induced significant mortalities in oysters. (B) Tissue localization and histopathology of oysters infected with vibrios. Juvenile oysters were injected with 7.5 × 107 CFU of GFP-expressing LGP32, GFP-expressing J2-9, GFP-expressing LMG20012T, or GFP-expressing J2-8. After 8 h, oyster tissues were fixed and stained with hematoxylin. Vibrios were stained on histological sections with anti-GFP antibodies coupled to alkaline phosphatase (red staining). Photographs were taken with a NanoZoomer scanner. Virulent vibrios J2-9 and LGP32 were found in the pericardial cavity and in the connective tissue surrounding the kidney, the gut, the digestive gland, and the gonad (black arrow). Histological damage was observed in the adductor muscle near the site of injection, where virulent vibrios were abundant (blue arrow). No histological damage was recorded in oysters infected with nonvirulent strains. J2-8 was controlled by hemocyte aggregates in the muscle, the kidney, and the pericardial cavity, whereas LMG20012T was controlled by hemocyte aggregates in the pericardial cavity only (white arrow). (Scale bar: 20 µm.)
Bacterial load and tissue damages were analyzed 8 h after injection, before the first mortality occurred. Similar bacterial loads were observed for virulent and nonvirulent vibrios, with 1 copy of the vibrio genome per 25–50 copies of the oyster genome (SI Appendix, Table S1). However, localization of bacteria and damages to oyster tissues differed markedly between virulent and nonvirulent strains. Both virulent strains, LGP32 and J2-9, spread between muscle fibers and caused tissue disorder with loss of cellular integrity in the adductor muscle around the injection site (Fig. 2B). Moreover, a systemic and massive colonization of the conjunctive tissue was observed around the kidney, the digestive gland, the gonad, and under the basal lamina of the gut (Fig. 2B). In contrast, the nonvirulent strains LMG20012T and J2-8 did not cause any detectable tissue damage beyond the lesion caused by the needle injection in the muscle. These strains were not detected in histological sections or were restricted to the needle path within the adductor muscle (Fig. 2B). Instead, they were found in large hemocyte aggregates in the pericardial cavity, and, for J2-8, the kidney, aggregates that were not observed in oysters injected with virulent strains (Figs. 2B and 3A). These cell aggregates are reminiscent of extracellular DNA traps [referred to as Neutrophil extracellular traps (NETs) in mammals], which can bind and neutralize pathogenic bacteria and microorganisms, a defense mechanism conserved across species and recently observed in mollusks (19). In oysters, such DNA traps were shown to contain antimicrobial H1- and H5-like histones active against vibrios (20). Thus, our data suggest that virulent vibrios escape from hemocyte control and invade the oyster conjunctive tissue, whereas nonvirulent vibrios are contained within hemocyte aggregates, which we hypothesize prevent tissue colonization and associated tissue damage. In vivo, both virulent strains were highly cytotoxic to hemocytes. Indeed, V. crassostreae J2-9, but not the nonvirulent J2-8 strain, caused significant damage to circulating hemocytes within 8 h of oyster infection (Fig. 3B), consistent with previous observations for V. tasmaniensis LGP32 (16). In histological sections, V. crassostreae J2-9 was mainly observed outside hemocytes whereas V. tasmaniensis LGP32 and the nonvirulent strains J2-8 and LMG20012T were observed to have been heavily phagocytized (Fig. 3A). Therefore, our in vivo data are consistent with the cell contact-dependent and phagocytosis-dependent cytotoxicity observed in vitro for V. crassostreae and V. tasmaniensis, respectively (Fig. 1).
Fig. 3.
Virulent strains cause damage to hemocytes and escape hemocyte aggregates in vivo. (A) Juvenile oysters were injected with 7.5 × 107 CFU of GFP-expressing LGP32, GFP-expressing J2-9, GFP-expressing LMG20012T, or GFP-expressing J2-8. After 8 h, oyster tissues were fixed and stained with hematoxylin. Photographs of hemocytes were taken in histological sections by using differential interference contrast (DIC) microscopy. Hemocytes and vibrios appear in blue and red, respectively. J2-9 is present inside and outside the hemocytes, whereas LGP32 and the nonvirulent vibrios are massively phagocytized. Hemocyte aggregates are observed with nonvirulent vibrios only. (Scale bar: 10 µm.) (B) Confocal microscopy of hemolymph collected from animals infected with V. crassostreae J2-9 or the nonvirulent control J2-8. Juvenile oysters were injected with 1.5 × 108 CFU of GFP-expressing vibrios. For each experimental condition, hemolymph was withdrawn from 5 oysters 2 h, 4 h, and 8 h after injection. Hemocytes were observed by confocal microscopy on Cyto-Spin glass slides after the staining of nuclei and actin cytoskeleton with DAPI and phalloidin-TRITC, respectively. In oysters injected with J2-9, J2-9 cells were often found associated with hemocyte lysis (mostly observed at 4 h and 8 h; white arrow), which was absent in J2-8–injected oysters at any time point. In contrast to J2-9, the nonvirulent J2-8 was massively phagocytized at 2 h and 4 h, often inside hemocyte aggregates; it became almost undetectable in the oyster hemolymph at 8 h, whereas J2-9 was still present (SI Appendix, Fig. S3). (Scale bar: 10 µm.) Similar observations of in vivo hemocyte lysis have been recorded for LGP32 in a previous study (16).
Virulent Vibrios Bypass Host Defenses.
The outcome of an infection depends on a series of complex host and bacterial factors that coordinate and fine-tune host–pathogen interactions (21, 22). Herein, by implementing dual RNA-seq in an in vivo infection, we were able to analyze the mechanisms underpinning colonization success by vibrios in pathological and nonpathological contexts.
We focused on host responses by comparing the transcriptomes of oysters injected with vibrios (virulent strains LGP32 or J2-9 or nonvirulent strains LMG20012T or J2-8) versus that of mock-infected oysters (injected with sterile seawater). Oysters were killed 8 h after the infection and before the onset of mortality. From 3 independent experimental infections of the same family of oysters, we observed significant differential expression [false discovery rate (FDR) < 0.05] for 331 host genes in response to vibrios (Dataset S1). RNA-seq data were validated by qRT-PCR (correlation R2 = 0.92; linear regression test, P < 0.001; SI Appendix, Fig. S4).
All vibrios induced an immune response in oysters independently of their virulence. Among the 331 genes differentially expressed in response to vibrios, 129 responded similarly to all strains (Fig. 4A), with 25.6% of them being involved in immunity (Fig. 4B and Dataset S1). We observed an overrepresentation of transcripts from 3 major immune signaling pathways, the interleukin 17 (IL-17), tumor necrosis factor (TNF), and Toll-like receptor (TLR) pathways (Dataset S1). This includes transcripts homologous to TLR13 and TLR4, which have been shown to recognize bacterial 23S ribosomal RNA and LPS, respectively, in mammals (23, 24). IL-17 is also recognized as a key player in the immune response of oysters against bacterial infections (25). Consequently, all vibrio infections induced oyster antimicrobial defenses mainly based on antimicrobial peptides (big defensins) (26) (SI Appendix, Fig. S5), ROS (dual oxidase DUOX expression; Dataset S1), and heavy metals (metallothioneins and zinc transporters; Dataset S1). These antimicrobial functions are believed to control bacterial infections in oysters (10, 16) and more generally microbiome homeostasis across animal species (27, 28).
Fig. 4.
Vibrio virulence status dictates the oyster transcriptional responses to infection. Transcriptional responses of juvenile oysters (fulsib family Decipher 14) to virulent or nonvirulent strains of the Splendidus clade were compared. In 3 independent experiments, groups of 30 oysters were injected with 7.5 × 107 CFU of LGP32, LMG20012T, J2-9, J2-8, or sterile seawater (mock-infected oysters). RNA-seq was performed on pools of 30 oysters collected 8 h after injection for each experiment under each condition. Transcriptomes of vibrio-infected oysters were compared with mock-infected oysters. RNA-seq results showed a significant modulation of 331 genes (Benjamini–Hochberg procedure, which controls FDR, P < 0.05). (A) Venn diagram showing the number of modulated genes in oysters injected with each vibrio strain. Virulent vibrios LGP32 and J2-9 induce a greater transcriptomic response than nonvirulent vibrios. (B) Functional categories of differentially expressed genes. Histograms show the percentage of differentially expressed genes that respond similarly to all vibrio infections, to virulent vibrios only (J2-9 and LGP32), or specifically to strains J2-9 and LGP32. Induced genes are shown in pink, and repressed genes are shown in blue. (C) Heat map representing the mean of differential expression (log2 fold change) of genes significantly induced (red) or repressed (blue) in 3 independent experiments (DESeq2: FDR < 0.05). Data show that oyster transcriptomes cluster according to vibrio virulence status. SI Appendix, Fig. S5 provides antimicrobial peptide expression (Dataset S1).
However, a set of shared antibacterial responses was specifically repressed in oysters infected by the virulent V. tasmaniensis LGP32 and V. crassostreae J2-9 strains, suggesting a common strategy of immune evasion. Overall, a much more extensive response was induced against infection by virulent vibrios (132 differentially expressed host genes) compared with that by nonvirulent strains (20 differentially expressed host genes: Fig. 4A). This response clustered according to vibrio virulence rather than their phylogenetic relatedness (Fig. 4C). Host responses were similarly altered by both virulent strains (57 genes; Fig. 4A) encompassing 4 main functional categories: metabolism, cell proliferation and differentiation, immunity, and extracellular remodeling (Fig. 4B and Dataset S1). Immune genes involved in antibacterial defenses were repressed (Dataset S1). These included the NADPH-mediated production of ROS, C-type lectin-mediated antibacterial immunity, and Deleted in Malignant Brain Tumors 1 (DMBT-1)-mediated mucosal immunity. We recently demonstrated the role that repression of ROS in oyster hemocytes plays in survival of Vibrio tasmaniensis (16). C-type lectin domain family 4 proteins, which display aggregation and direct antibacterial properties in oysters (29), are involved in the maintenance of microbiome homeostasis (30, 31). The DMBT-1 mucosal fluid protein functions in mucosal immunity, involving bacterial recognition and countering invasion (32), and has been shown to suppress the virulence of P. aeruginosa (33). These antibacterial defenses may play a role in controlling commensal bacteria, and their repression by virulent vibrios likely represents an important strategy to bypass the host defense mechanisms and avoid clearance.
Oyster responses to vibrios revealed the existence of virulence mechanisms specific to each species. V. tasmaniensis LGP32 infection was characterized by multiple specific host responses (48 genes) falling into 5 main categories: metabolism, stress response, cell proliferation and differentiation, metal homeostasis (mainly induced genes), and extracellular matrix remodeling (both induced and repressed genes; Fig. 4C and Dataset S1). The LGP32-specific modulation of genes belonging to “cytoskeleton dynamics” is consistent with the phagocytosis-mediated cytotoxicity of LGP32 (16). Likewise, specific induction of zinc transporters and metallothioneins revealed the involvement of “metal homeostasis” in LGP32 pathogenesis (Fig. 4C and Dataset S1). On the contrary, specific responses to V. crassostreae J2-9 (27 genes) mostly fell into the “immunity” category, representing 33% of the specific responses to J2-9 (Fig. 4 B and C). Immune genes were generally repressed, particularly those encoding C1q domain-containing (C1qDC) proteins (Dataset S1), a family of complement-related proteins, which, in oysters, can serve as opsonins that enhance vibrio phagocytosis (34). Their repression is consistent with the low levels of phagocytosis of V. crassostreae J2-9 (Fig. 3A). Genes in the category “cell signaling and adhesion” tended to be repressed by intracellular LGP32, whereas they were induced by J2-9. Thus, even though virulence appeared as the major criterion discriminating oyster responses to vibrios, specific signatures were also observed in the host response to distinct Vibrio species.
Overall, our oyster transcriptomics analyses suggest that the ability of virulent vibrios to escape hemocyte control and colonize oysters does not rely on the escape from immune recognition but rather on an active repression of a set of antibacterial defenses. Moreover, we found that vibrio strategies to bypass host defenses rely, at least in part, on a species-specific alteration of host responses.
Vibrio Gene Expression Support Species-Specific Virulence Mechanisms.
Global changes in bacterial gene expression can be monitored to explore mechanisms driving colonization success during pathogenesis. We used bacterial RNA-seq to analyze the transcriptomes of virulent vibrios (LGP32 and J2-9) in infected oysters. V. tasmaniensis LGP32 and V. crassostreae J2-9 showed substantial transcriptomic reprogramming inside the host, with 46.7% (2,266 genes) and 50.1% (2,815 genes) differentially expressed genes, respectively, compared with time 0 (before injection; fold change ≥2, FDR < 0.05; Fig. 5A). Overall within-host transcriptomes were comparable between the 2 virulent strains in terms of functional category distribution. A common group of orthologous genes differentially expressed during oyster infection was identified, which encompassed 340 repressed and 501 induced genes (Fig. 5 B and C and Dataset S2). Most induced genes encoded proteins involved in “carbon compound and carbohydrate utilization” and “amino acid transport and metabolism” (Fig. 5C), reflecting a major metabolic shift in response to the host as a new environment (35). “Defense mechanisms” and “metal homeostasis” were also strongly induced in the Vibrio species, particularly genes encoding efflux machineries (multidrug, copper, and zinc transporters), siderophores, and iron uptake systems (Fig. 5C and Dataset S2). Therefore, gene expression of both virulent strains in oyster is consistent with a competitive behavior in a hostile environment that is poor in the essential iron but rich in zinc, copper, and other antimicrobials (Fig. 4C and SI Appendix, Fig. S4).
Fig. 5.
Responses to host-imposed stresses revealed by within-host bacterial RNA-seq. Within-host transcriptomes of V. tasmaniensis LGP32 and V. crassostreae J2-9 (8 h after infection) were compared with bacterial transcriptomes before the infection (controls) in 3 independent experiments. Only those genes whose expression varied significantly by more than 2-fold were considered. (A) Number of bacterial genes significantly induced and repressed during host colonization. The proportion of differentially expressed genes over the entire transcriptome of LGP32 and J2-9 is given as a percentage of the total number of genes for each genome. Approximately half of the total bacterial transcriptome is altered during host colonization. Repressed and induced genes are showed in blue and purple, respectively. (B) Venn diagram showing the core response of pathogens during colonization (340 repressed genes and 501 induced genes, not including genes with an unknown function). The diagram was built on common orthologous genes differentially expressed upon host colonization in J2-9 and LGP32. (C) Functional distribution of the bacterial genes during LGP32 and J2-9 colonization process (genes without annotated function were not included; SI Appendix, Figs. S6 and S7 and Dataset S2).
V. tasmaniensis LGP32 and V. crassostreae J2-9 also displayed species-specific responses to colonization, which were mostly clearly observed when considering the 200 most highly induced genes of both strains (SI Appendix, Fig. S6). The V. tasmaniensis LGP32 response was characterized by a strong induction of genes involved in cell wall biogenesis, transport, regulation, and redox homeostasis (SI Appendix, Fig. S6). This result is in agreement with the known protective remodeling of vibrio membranes, and strong induction of antioxidants inside the phagosomes of oyster hemocytes (16, 17). Several adhesins, including mshM (VS_0329) and the type IV pilin assembly protein pilB (VS_2548), were induced (Dataset S3), which may favor LGP32 attachment to hemocytes and promote phagocytosis, as previously shown for the major adhesin OmpU (15). Various expression patterns were observed for genes encoding putative RTX toxins. For example, the gene VS_II0512 was highly expressed inside and outside the host whereas VS_1394 was induced during colonization (Dataset S3). Oyster colonization had significant effects on the expression of genes encoding type 6 secretion systems (T6SS; reviewed in refs. 36–40) localized on the first (T6SS1) and second (T6SS2) chromosomes in V. tasmaniensis. T6SS is a multiprotein secretion system responsible for the transport and delivery of toxins (effectors) into recipient cells (39). It has been shown to contribute to pathogenicity toward plant and animal cells as well as to take part in interbacterial competition. T6SS2 genes were expressed only inside the host (Dataset S4), whereas T6SS1 genes were highly expressed outside the host and decreased to the levels of T6SS2 expression inside oysters (similar numbers of normalized counts; Dataset S4). The high preinfection expression levels of T6SS1 and the within-host expression of T6SS2 were confirmed over 24 h in oysters experimentally infected with V. tasmaniensis LGP32 (SI Appendix, Fig. S7).
The response of V. crassostreae J2-9 to host colonization was characterized by a strong induction of adhesion genes (SI Appendix, Fig. S7), including a type IV pilus cluster, further supporting the role of attachment of the bacteria to the host target cells during the infection process (Dataset S2). The r5.7 gene (VCR9J2v1_730268) encoding an exported protein of unknown function, which has been previously shown to be necessary for virulence in V. crassostreae (12, 14), was one of the most highly induced virulence genes in J2-9 inside the host (Dataset S3). A number of genes encoding putative RTX toxins were highly expressed outside and inside the host, whereas other toxins and toxin secretion systems were expressed only inside the host (Dataset S3). In particular, a NAD-arginine ADP ribosyltransferase (putative toxin, VCR9J2v1_730029) and 2 toxin secretion membrane fusion proteins (VCR9J2v1_740022, VCR9J2v1_740023) were expressed only inside the host (Dataset S3). Genes encoding a T6SS, carried by the virulence plasmid pGV1512 (11), were induced in oyster (Dataset S4). Genes related to the mobile genetic elements (Fig. 5C), particularly those involved in conjugative transfer, were very highly induced in J2-9 during oyster colonization (Dataset S2), suggesting that horizontal gene transfer between infecting V. crassostreae could occur during host colonization.
Taken together, cellular and molecular data highlight 2 strategies evolved by virulent vibrios in the Splendidus clade: V. tasmaniensis acts as an intracellular pathogen that subverts cellular functions and kills oyster phagocytes as part of its pathogenic process, whereas V. crassostreae acts as an extracellular pathogen that uses cell contact-dependent cytotoxicity to overcome the cellular defenses of its host. These results indicate that, within the Splendidus clade, distinct virulent species have acquired the capacity to compromise hemocyte functions to mount successful infections.
Species-Specific Virulence Converges to Impair Host Defenses.
To confirm our conclusions that V. tasmaniensis LGP32 and V. crassostreae J2-9 use species-specific virulence determinants, we sought to identify genes responsible for virulence and cytotoxicity by using a gene knock-out approach. Candidate genes were selected based on their high expression or induction levels upon oyster colonization (Datasets S3 and S4). None of the mutants showed impaired bacterial growth in culture media (SI Appendix, Figs. S8A and S9A).
We knocked out potential adhesins and toxins. In V. tasmaniensis LGP32, the inactivation of an adhesin (mshM, VS_0329), a type IV pilin assembly protein pilB (VS_2548), and putative RTX toxins (VS_II0512 and VS_1394) did not significantly attenuate virulence in the experimental infection of oysters (SI Appendix, Fig. S8B). Similarly, in V. crassostreae J2-9, inactivation of a NAD-arginine ADP ribosyltransferase (VCR9J2v1_730029) and a membrane fusion protein involved in toxin secretion (VCR9J2v1_740022) had no significant effect on virulence (SI Appendix, Fig. S9B).
We next examined the role of T6SS in both virulent strains, as this secretion system expressed by LGP32 and J2-9 is well known for the delivery of effectors (toxins) into recipient cells (39). T6SS is encoded by cluster(s) of 15–20 genes that includes components of a contractile nanomachine, as well as regulators and effectors (36–40). The constituents of the phage-like subassembly are required for T6SS function (SI Appendix, Fig. S10). The VipA/B proteins act as a phage contractile sheath. The hemolysin coregulated proteins (Hcp) form a tail-tube. The valine-glycine repeat protein G (VgrG), a fusion of gp5 and gp27 phage-like domains, forms a cell-puncturing tip. Proteins from the proline-alanine-alanine-arginine repeat (PAAR) superfamily interact with VgrG, forming the canonical tip of the puncturing device complex. Effectors can be delivered to the target cell as a complex with VgrG/PAAR spikes or through the Hcp channel. The C-terminal domains of several VgrG and PAAR proteins (referred to as “evolved” VgrG or PAAR proteins) are predicted to have various enzymatic activities toxic for prokaryotic and eukaryotic cells.
In V. tasmaniensis LGP32, the constitutively expressed T6SS1 was identified in 3 other virulent strains of the species, but was absent from the nonvirulent strain (Fig. 1A, SI Appendix, Fig. S10, and Dataset S4). In all virulent strains, vgrG and paar-encoded proteins do not contain C-terminal effector domains (SI Appendix, Figs. S10–S12). However, all of them contain an evpP ortholog (VS_2123 in LGP32), as revealed by searching for potential T6SS effectors in a public database (http://db-mml.sjtu.edu.cn/SecReT6/). In the intracellular pathogen Edwardsiella tarda, evpP encodes a non-VgrG non-PAAR T6SS effector that is involved in cell invasion (41) and inhibition of the host immune response (42). Together these data suggest a role for T6SS1 in V. tasmaniensis virulence.
As we predicted, inactivation of the T6SS1 vipA gene resulted in a substantial loss of virulence in oyster experimental infections and cytotoxicity toward oyster hemocytes (P < 0.01; Fig. 6). This is reminiscent of the Burkholderia sp. T6SS5/T6SS1, which is essential for virulence in the host (43). Thus, our data indicate that the phagocytosis-dependent cytotoxicity of V. tasmaniensis toward immune cells requires the intracellular secretion of T6SS1 cytotoxic effectors. Likewise, studies by Ma and Mekalanos reported a role for the V. cholerae T6SS in inducing macrophage death (44), whose activation required bacterial phagocytosis by the host cells (45). Together, these examples indicate that phagocytes are a natural target for T6SS in Vibrio species. They demonstrate that T6SS functions to repress immune defenses in different hosts during critical intracellular stages of infection. Like in other Gram-negative bacteria adopting intracellular stages, we show here that T6SS plays a critical role in the success of vibrio infections (42, 43).
Fig. 6.
Species-specific virulence factors are needed for immune cell lysis. (A) Virulence of V. crassostreae (Upper) and V. tasmaniensis (Lower) isogenic mutants was assessed by intramuscular injection of juvenile oysters. Mortalities were recorded at day 1. Data are expressed as percentage of mortality induced by the wild-type strains. (B) Cytotoxicity of vibrio isogenic mutants lacking virulence genes was determined on monolayers of hemocytes and monitored by the SYTOX Green assay. Cells were infected with bacteria at an MOI of 50:1. Maximum cytotoxicity displayed by vibrios is shown here. Error bars represent the SD of the mean (RM-ANOVA, P < 0.001; SI Appendix, Figs. S8 and S9).
T6SS2 in V. tasmaniensis LGP32 was induced upon oyster colonization and reached maximum expression in dead oysters (SI Appendix, Fig. S7). However, this T6SS2 is absent from 2 other V. tasmaniensis virulent strains (J5-9 and J0-13; Fig. 1A and Dataset S4). Inactivation of T6SS2 did not alter LGP32 virulence and cytotoxicity (Fig. 6), indicating that it does not have a direct effect on host–pathogen interactions. Closer examination of the T6SS2 gene cluster revealed the absence of an evolved vgrG gene and the lack of a paar gene in proximity to vgrG. However, 7 genes encoding PAAR proteins and putative AHH endonucleases were present in tandem in a proximal region of the T6SS2 (SI Appendix, Figs. S10B and S11). This suggests that T6SS2 may play a role in interbacterial competition during colonization of oyster tissues, as shown for V. fischeri in its mutualistic association with squid (46). It could also be needed to exploit host resources after host death.
In V. crassostreae J2-9, the single T6SS was induced upon host colonization. This T6SS, carried by the plasmid pGV1512, is present in all V. crassostreae virulent strains (Fig. 1A and Dataset S4). As the encoded PAAR and VgrG proteins lack C-terminal effector domains (SI Appendix, Figs. S10 and S11), and because we were unable to identify any putative T6SS effector, the function of T6SS in V. crassostreae cannot presently be predicted. Even though it has previously been demonstrated that pGV1512 is necessary for J2-9 virulence (11), we did not observe any alteration in the cytotoxicity of J2-9 upon the loss of pGV1512 (Fig. 6B). Therefore, we have no evidence that T6SS could be involved in V. crassostreae cytotoxicity toward oyster hemocytes. Instead, one of the most highly induced J2-9 virulence genes inside the host, r5.7 (Mage VCR9J2v1_730268; GenBank accession no. WP_048663858.1), was demonstrated to be necessary for cytotoxicity (P < 0.001; Fig. 6), and was also required for cytotoxicity in V. crassostreae strain J5-5 (SI Appendix, Fig. S13). Previous work has shown that R5.7 alone is not toxic toward oysters and suggested that the protein interacts with the external surface of Vibrio and/or its cellular targets in oysters (12). r5.7 is a virulence gene widely distributed across the Splendidus clade, and we show here that it is a key determinant of V. crassostreae cell contact-dependent cytotoxicity to hemocytes, yet has been lost in V. tasmaniensis species. Thus, our results show that, in virulent species of the Splendidus clade in which the ancestral virulence gene r5.7 has been lost, other mechanisms have been acquired (T6SS1 in V. tasmaniensis) that specifically target immune cells through cytotoxic mechanisms that enable successful host colonization.
Conclusion
Although cytotoxicity is widespread among Vibrio species pathogenic for humans and animals and is known to be important in disease progression, it has not been clearly associated with immune repression. Here, we demonstrate that the fate of vibrio infections in oysters, a natural host for marine Vibrio species, is determined by the specific mechanisms that have evolved in virulent strains to induce cytotoxicity. These mechanisms impair the host cellular defenses that normally prevent vibrios from causing systemic colonization of host tissues. Cytotoxicity toward immune cells was shown to be dependent on distinct molecular determinants present in V. crassostreae and V. tasmaniensis, and other virulent species of Vibrio have been shown to be equipped with different specific virulence mechanisms. For example, in the Splendidus clade, MARTX toxins are found exclusively in V. splendidus and are necessary for virulence toward oysters (12). In more distant species from the Anguillarum clade, Vibrio aestuarianus uses a metalloprotease to evade hemocyte control (47, 48) and cause septicemia in oysters (49). Thus, Vibrio species that are pathogenic for oysters have evolved a diverse array of molecular mechanisms that all converge on the attenuation of the cellular defenses of their host, enabling subsequent tissue colonization (Fig. 7). As these mechanisms have been acquired independently during evolution in diverse species pathogenic for oysters, they could be a good indicator of the adaptation of these pathogenic species to oysters as a niche (50). Beyond oyster pathogens, a specific mechanism of phagocyte targeting has also been reported in V. cholerae (45). We believe more attention should be paid to Vibrio cytotoxicity in repressing host immunity, as this may be an insufficiently explored determinant of infection outcomes in metazoan hosts.
Fig. 7.
Impairment of oyster cellular immunity as a common strategy of immune evasion in vibrios. Virulent strains of the species V. tasmaniensis and V. crassostreae exert cytotoxic activity toward immune cells. They thereby repress key cellular defenses, gain access to the conjunctive tissues, and cause systemic infection. In V. crassostreae, cytotoxicity is a rapid, contact-dependent phenotype that requires the R5.7 virulence factor, which is strongly induced inside the host. In V. tasmaniensis, which behaves as an intracellular pathogen, cytotoxicity is phagocytosis-dependent and requires the intracellular action of a constitutively expressed type 6 secretion system present on chromosome I. In contrast to virulent strains, nonvirulent strains of both species are controlled by hemocyte clumps in the hemolymph compartment, preventing them from causing systemic infection. We conclude that the outcome of the infection depends on independently evolved molecular determinants that impair oyster cellular defenses as a strategy of immune evasion.
Materials and Methods
Strains, Plasmids, and Primers.
All strains, plasmids and primers used or constructed in the present study are described in SI Appendix, SI Materials and Methods and Tables S6–S8.
Animals.
C. gigas diploid oyster spat were used from 3 families of siblings (Decipher 1, 14, and 15) (10), and a batch of standardized Ifremer spats (NSI) were produced from a pool of 120 genitors (51). Spats were used for experimental infections and dual RNA-seq. Adults produced from a pool of 120 genitors (ASI) were used to collect hemolymph for cytotoxicity assays (SI Appendix, SI Materials and Methods).
Experimental Infections.
Experimental infections were performed at 20 °C as in ref. 52 by injection at 7.5 × 107 CFU into oyster adductor muscle (details in SI Appendix, SI Materials and Methods). Statistical analyses were performed by using a nonparametric Kaplan–Meier test to estimate log-rank values for comparing conditions (GraphPad Prism 6).
Molecular Microbiology.
Vibrio isolates were cultured as described in SI Appendix, SI Materials and Methods. Conjugation between Escherichia coli and Vibrio were performed at 30 °C as described previously (53). To label the strains with a fluorochrome, gfp gene was cloned in Apa1/Xho1 sites of the pMRB plasmid known to be stable in Vibrio spp. (54), resulting in a constitutive expression from a Plac promoter. Gene inactivation was performed by cloning approximately 500 bp of the target gene in pSW23T (55) and selecting on Cm the suicide plasmid integration obtained by a single recombination (56). Mutants were screened for insertion of the suicide vector by PCR by using external primers flanking the different targeted genes (SI Appendix, Table S2).
Histology.
Spats (1.5–2 cm) were injected with 7.5 × 107 CFU of GFP-expressing LGP32, J2-9, LMG20 012T, or J2-8. Noninjected oysters were used as controls. Spats were kept in seawater tanks at 20 °C, sampled, and fixed 8 h after injection. Histological sectioning was performed at HISTALIM (Montpellier, France). Longitudinal sections were immunostained with an anti-GFP primary antibody coupled to alkaline phosphatase (Abcam). Sections and hemocytes were observed as described in SI Appendix, SI Materials and Methods.
In Vitro Cytotoxicity Assays.
Cytotoxicity of vibrios was measured on hemocyte monolayers according to procedures described in SI Appendix, SI Materials and Methods. Statistical analysis was performed by using RM-ANOVA.
Confocal Microscopy of Interaction Between Hemocytes and Vibrios in Vivo.
Oysters were injected with 1.5 × 108 CFU of GFP-expressing vibrios (J2-8 or J2-9) a priori washed and resuspended in sterile seawater. Control oysters received an injection of an equal volume of sterile seawater. For each experimental condition, hemolymph was collected from 5 oysters at 2 h, 4 h, and 8 h after injection. Samples were fixed with 4% paraformaldehyde and stained with DAPI and Phalloidin-TRITC and observed by confocal microscopy as described in SI Appendix, SI Materials and Methods.
Dual RNA-Seq Experiments.
3 independent experimental infections were performed. For every experiment, 150 oysters (30 animals per condition) were injected with bacteria (107 CFU of LGP32, LMG20012T, J2-9, or J2-8) or sterile seawater (mock-infected oysters). In addition, both bacterial suspensions and 30 nontreated oysters were collected at time 0. For every experimental condition, 30 oysters were then sampled 8 h after injection (i.e., before mortality occurred) before RNA extraction (SI Appendix, SI Materials and Methods). For oyster RNA-seq, library construction and sequencing were performed on polyadenylated mRNAs by Fasteris (Geneva, Switzerland; https://www.fasteris.com). Directional cDNA libraries were constructed and sequenced on an Illumina HiSeq system in paired-end reads of 2 × 75 bp. For bacterial RNA-seq, total RNA obtained from oyster tissues was used to prepare non–rRNA-bacterial RNA enriched libraries. After polyadenylated mRNAs (i.e., host mRNAs) were removed by using a MICROBEnrich Kit (Ambion), cDNA-oriented sequencing libraries were prepared starting from 100 ng enriched RNA by using an Ovation Universal RNA-Seq System kit (NuGEN). Library construction included a depletion step of abundant, nondesired sequences by using a Nugen-customized mixture of primers complementary to RNA sequences to be depleted, viz. oyster nuclear, mitochondrial, and ribosomal and bacterial ribosomal RNA sequences. The libraries were quantified by DeNovix dsDNA Broad Range Assay (DeNovix), and the quality was monitored by using an Agilent High Sensitivity DNA kit (Agilent Technologies). For each condition, 3 biological replicate libraries were prepared. Libraries were sequenced in paired-end mode (2 × 150 bp) by Fasteris on an Illumina HiSeq 4000 system to obtain ∼100 million reads per sample per run.
Oyster Transcriptome Analysis.
Data were treated using the galaxy instance of the IHPE laboratory (57). Phred quality scores were checked by using the FastX toolkit galaxy interface (http://hannonlab.cshl.edu/fastx_toolkit/) and were greater than 26 for more than 90% of the read length for all of the sequences. All of the reads were thus kept for subsequent analyses. Mapping to the C. gigas reference genome [assembly version V9 (58)] was performed by using RNA STAR (Galaxy version 2.4.0d-2; ref. 59). HTSeq-count was used to count the number of reads overlapping annotated genes (mode Union; Galaxy Version v0.6.1) (60). The resulting files were used as input to determine differential expression for each feature between oysters injected with vibrios and mock-infected oysters by using DESeq2 (61). Fold changes in gene expression between test animals and controls were considered significant when the adjusted P value (Padj) for multiple testing with the Benjamini–Hochberg procedure, which controls FDR, was <0.05. The same threshold was applied to fold changes in gene expression between mock-infected oysters and naïve controls. As genes for C. gigas antimicrobial peptides (AMPs) are not annotated in the C. gigas reference genome (assembly version V9; ref. 58), read counts for all AMPs were obtained by alignment to an in-house database of AMP protein sequences by using DIAMOND 0.7.9 (62) as described in SI Appendix, SI Materials and Methods. Oyster RNA-seq data have been made available through the SRA database (BioProject accession no. PRJNA515169) under SRA accession nos. SRR8551076–SRR8551093 (63).
Vibrio Transcriptome Analysis.
Raw Illumina sequencing reads were trimmed by Trimmomatic in paired-end mode (64) to remove sequences corresponding to the NuGEN-provided adapters, clipping part of reads having a quality score of below 20, and dropping reads of less than 36 nt. The 2 resulting files were used as input for Bowtie2 (65) to align the reads on V. tasmaniensis LGP32 or V. crassostreae J2-9 genome, respectively (SI Appendix, SI Materials and Methods). As the rate of read alignment on the bacterial genome was ∼2% for vibrios in oyster tissues, as opposed to >98% for the in vitro grown bacteria, we introduced a random down-sampling step of the aligned reads (http://broadinstitute.github.io/picard/command-line-overview.html#DownsampleSam; SI Appendix, SI Materials and Methods). Counts for each annotated feature (CDS + sRNAs) of V. tasmaniensis LGP32 or V. crassostreae J2-9 were then computed by using featureCounts (66). Differential expression was determined for each feature between oysters injected with vibrios and vibrios grown in vitro by DESeq2 (61). Genes were considered as differentially expressed between tested conditions and controls when |log2(fold change)| was ≥1 and Padj for multiple testing with the Benjamini–Hochberg procedure was <0.05. Bacterial RNA-seq data have been made available through the SRA database: BioProject accession no. PRJNA521688 under SRA accession nos. SRR8567597–SRR8567602 (V. crassostreae J2-9) (67) and BioProject accession no. PRJNA521693 under SRA accession nos. SRR8573808–SRR8573813 (V. tasmaniensis LGP32) (68).
Functional Gene Ontology Annotations.
For oyster genes, Blastx (69) comparison against NR database was performed for each set of genes with a maximum number of target hits of 20 and a minimal e-value of 0.001. XML blast result files were loaded onto Blast2GO (70) for GO mapping and annotation of the B2G database. Functional categories corresponding to significantly modulated genes (331 genes) were then determined manually; this was supported by background literature review. A heat map of the mean of log2 fold change in 3 independent experiments was then generated by using cluster 3.0 (http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm). For bacterial genes, annotations of differentially expressed genes (fold change ≥ 2 and Padj ≤ 0.05) were manually curated by using the MicroScope platform (71) and the MetaCyc database (72). Based on these curated annotations, each gene was assigned to 1 of 32 functional categories (the list of categories is provided in Fig. 5 and SI Appendix, Fig. S6). The MicroScope tools were used to compare genomes based on sequence and synteny conservation (SI Appendix, SI Materials and Methods).
RT-qPCR.
RT-qPCR was performed on the same RNA samples as those used for deep sequencing to validate RNA-seq data (SI Appendix, Fig. S4) or on specific RNA samples to monitor the time course of bacterial gene expression in oysters (SI Appendix, Fig. S7). cDNA synthesis and real-time qPCR were performed as described in SI Appendix, SI Materials and Methods.
Specific Vibrio Quantification by qPCR.
Genomic DNA was extracted with the DNA kit Promega (Wizard SV Genomic DNA Purification System) on the same oyster samples used for RNA-seq. Real-time qPCR was used to amplify monocopy genes from specific vibrio genomes (SI Appendix, Table S2). To quantify oyster genomes, Cg-BPI (GenBank accession no. AY165040) primers were used (SI Appendix, Table S2). The vibrio charge in oyster tissues was calculated as a number of vibrio genome copies per number of oyster genome copy (SI Appendix, SI Material and Methods).
Vibrio Genome Sequencing and Assembly.
Genomic DNA for J0-13, J5-13, and J5-9 was sequenced at the Bioenvironment platform (University of Perpignan, Cemeb) by using a NextSeq 550 instrument resulting in ∼100-fold coverage (SI Appendix, SI Material and Methods). Reads were assembled de novo by using Spades software. Computational prediction of coding sequences together with functional assignments was performed by using the automated annotation pipeline implemented in the MicroScope platform (73). The genome sequences of V. tasmaniensis J0-13, J5-13, and J5-9 were deposited in GenBank (BioProject accession no. PRJNA520819, nucleotide accession nos. SEOP00000000 (J0-13), SESM00000000 (J5-13), and SESL00000000 (J5-9) (74). The genome sequence of LMG20012T was given the BioProject accession no. PRJNA542263, nucleotide accession no. VARQ00000000 (75). Other Vibrio genomes presented in this study were previously deposited under the following accession numbers: J2-8 (PRJEB5890), J2-9 (PRJEB5876), J5-4 (PRJEB5877), LGP8 (PRJEB5884), J5-20 (PRJEB5882), and LGP32 (PRJEA32815).
Supplementary Material
Acknowledgments
We thank Drs. François Bonhomme and Evelyne Bachère for fruitful discussions, Marc Leroy and Agnès Vergnes for precious technical assistance, and Jean-François Allienne at the Bioenvironment platform at University Perpignan Via Domitia for support in NGS library preparation and sequencing, and the qPHD platform/Montpellier genomix for access to qPCR. We thank Montpellier RIO Imaging (https://www.mri.cnrs.fr). A.J. thanks Emilie Drouineau and Claire Toffano-Nioche for advice on the use of bioinformatics tools. This work was supported by Agence Nationale de la Recherche (ANR) Grants Vibriogen ANR-11-BSV7-0023, Decipher ANR-14-CE19-0023, and Revenge ANR-16-CE32-0008-01; European Union’s Horizon 2020 Research and Innovation Program Grant Vivaldi 678589; the University of Montpellier (Doctoral School Gaia; T.R.); Conicyt Pfcha/Doctorado En El Extranjero Becas Chile Grant 2016-72170430 (to D.O.); the Spanish Ministry of Science and Education Fellowship FPU13-0466 (to M.T.); the University of Granada Campus de Excelencia Internacional en Biosalud y Tecnologías de Información y Comunicación Grant CEI1-2015 (to M.T.); and the Chinese Scholarship Council (to X.L.). This study is set within the framework of the “Laboratoires d’Excellence (LABEX)” Tulip (ANR‐10‐LABX‐41).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Genbank database (nucleotide accession nos. SEOP00000000, SESM00000000, SESL00000000, and VARQ00000000) and SRA database (BioProject accession no. PRJNA515169, SRA accession nos. SRR8551076–SRR8551093; BioProject accession no. PRJNA521688, SRA accession nos. SRR8567597–SRR8567602; BioProject accession no. PRJNA521693; SRA accession nos. SRR8573808–SRR8573813).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905747116/-/DCSupplemental.
References
- 1.Takemura A. F., Chien D. M., Polz M. F., Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 5, 38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kremer N., et al. , Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 14, 183–194 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McFall-Ngai M., Divining the essence of symbiosis: Insights from the squid-vibrio model. PLoS Biol. 12, e1001783 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborty S., Nair G. B., Shinoda S., Pathogenic vibrios in the natural aquatic environment. Rev. Environ. Health 12, 63–80 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Dubert J., Barja J. L., Romalde J. L., New insights into pathogenic vibrios affecting bivalves in hatcheries: Present and future prospects. Front. Microbiol. 8, 762 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Roux F., Blokesch M., Eco-evolutionary dynamics linked to horizontal gene transfer in vibrios. Annu. Rev. Microbiol. 72, 89–110 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Thompson F. L., Iida T., Swings J., Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68, 403–431 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Roux F., Wegner K. M., Polz M. F., Oysters and vibrios as a model for disease dynamics in wild animals. Trends Microbiol. 24, 568–580 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Schmitt P., et al. , The antimicrobial defense of the pacific oyster, Crassostrea gigas. How diversity may compensate for scarcity in the regulation of resident/pathogenic microflora. Front. Microbiol. 3, 160 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lorgeril J., et al. , Immune-suppression by OsHV-1 viral infection causes fatal bacteraemia in Pacific oysters. Nat. Commun. 9, 4215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruto M., et al. , Vibrio crassostreae, a benign oyster colonizer turned into a pathogen after plasmid acquisition. ISME J. 11, 1043–1052 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruto M., et al. , Ancestral gene acquisition as the key to virulence potential in environmental Vibrio populations. ISME J. 12, 2954–2966 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gay M., Berthe F. C., Le Roux F., Screening of Vibrio isolates to develop an experimental infection model in the Pacific oyster Crassostrea gigas. Dis. Aquat. Organ. 59, 49–56 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Lemire A., et al. , Populations, not clones, are the unit of vibrio pathogenesis in naturally infected oysters. ISME J. 9, 1523–1531 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duperthuy M., et al. , Use of OmpU porins for attachment and invasion of Crassostrea gigas immune cells by the oyster pathogen Vibrio splendidus. Proc. Natl. Acad. Sci. U.S.A. 108, 2993–2998 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanhove A. S., et al. , Copper homeostasis at the host vibrio interface: Lessons from intracellular vibrio transcriptomics. Environ. Microbiol. 18, 875–888 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Vanhove A. S., et al. , Outer membrane vesicles are vehicles for the delivery of Vibrio tasmaniensis virulence factors to oyster immune cells. Environ. Microbiol. 17, 1152–1165 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Westermann A. J., Barquist L., Vogel J., Resolving host-pathogen interactions by dual RNA-seq. PLoS Pathog. 13, e1006033 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robb C. T., Dyrynda E. A., Gray R. D., Rossi A. G., Smith V. J., Invertebrate extracellular phagocyte traps show that chromatin is an ancient defence weapon. Nat. Commun. 5, 4627 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poirier A. C., et al. , Antimicrobial histones and DNA traps in invertebrate immunity: Evidences in Crassostrea gigas. J. Biol. Chem. 289, 24821–24831 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hornef M. W., Wick M. J., Rhen M., Normark S., Bacterial strategies for overcoming host innate and adaptive immune responses. Nat. Immunol. 3, 1033–1040 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Reddick L. E., Alto N. M., Bacteria fighting back: How pathogens target and subvert the host innate immune system. Mol. Cell 54, 321–328 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beutler B., Tlr4: Central component of the sole mammalian LPS sensor. Curr. Opin. Immunol. 12, 20–26 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Oldenburg M., et al. , TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science 337, 1111–1115 (2012). [DOI] [PubMed] [Google Scholar]
- 25.McDowell I. C., et al. , Transcriptome of American oysters, Crassostrea virginica, in response to bacterial challenge: Insights into potential mechanisms of disease resistance. PLoS One 9, e105097 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosa R. D., et al. , Big defensins, a diverse family of antimicrobial peptides that follows different patterns of expression in hemocytes of the oyster Crassostrea gigas. PLoS One 6, e25594 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bae Y. S., Choi M. K., Lee W. J., Dual oxidase in mucosal immunity and host-microbe homeostasis. Trends Immunol. 31, 278–287 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Bevins C. L., Salzman N. H., Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356–368 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Jia Z., et al. , An integrin from oyster Crassostrea gigas mediates the phagocytosis toward Vibrio splendidus through LPS binding activity. Dev. Comp. Immunol. 53, 253–264 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Pang X., et al. , Mosquito C-type lectins maintain gut microbiome homeostasis. Nat. Microbiol. 1, 16023 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Wang X. W., Xu J. D., Zhao X. F., Vasta G. R., Wang J. X., A shrimp C-type lectin inhibits proliferation of the hemolymph microbiota by maintaining the expression of antimicrobial peptides. J. Biol. Chem. 289, 11779–11790 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenstiel P., et al. , Regulation of DMBT1 via NOD2 and TLR4 in intestinal epithelial cells modulates bacterial recognition and invasion. J. Immunol. 178, 8203–8211 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Li J., Metruccio M. M. E., Evans D. J., Fleiszig S. M. J., Mucosal fluid glycoprotein DMBT1 suppresses twitching motility and virulence of the opportunistic pathogen Pseudomonas aeruginosa. PLoS Pathog. 13, e1006392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lv Z., et al. , Comparative study of three C1q domain containing proteins from pacific oyster Crassostrea gigas. Dev. Comp. Immunol. 78, 42–51 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Takemura A. F., et al. Natural resource landscapes of a marine bacterium reveal distinct fitness-determining genes across the genome. Environ. Microbiol. 19, 2422–2433 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Basler M., Type VI secretion system: Secretion by a contractile nanomachine. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho B. T., Dong T. G., Mekalanos J. J., A view to a kill: The bacterial type VI secretion system. Cell Host Microbe 15, 9–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joshi A., et al. , Rules of engagement: The type VI secretion system in Vibrio cholerae. Trends Microbiol. 25, 267–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen V. S., et al. , Towards a complete structural deciphering of Type VI secretion system. Curr. Opin. Struct. Biol. 49, 77–84 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Russell A. B., Peterson S. B., Mougous J. D., Type VI secretion system effectors: Poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X., et al. , Edwardsiella tarda T6SS component evpP is regulated by esrB and iron, and plays essential roles in the invasion of fish. Fish Shellfish Immunol. 27, 469–477 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Chen H., et al. , The bacterial T6SS effector EvpP prevents NLRP3 inflammasome activation by inhibiting the Ca2+-dependent MAPK-jnk pathway. Cell Host Microbe 21, 47–58 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Lennings J., West T. E., Schwarz S., The Burkholderia type VI secretion system 5: Composition, regulation and role in virulence. Front. Microbiol. 9, 3339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma A. T., Mekalanos J. J., In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc. Natl. Acad. Sci. U.S.A. 107, 4365–4370 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma A. T., McAuley S., Pukatzki S., Mekalanos J. J., Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe 5, 234–243 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Speare L., et al. , Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc. Natl. Acad. Sci. U.S.A. 115, E8528–E8537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Labreuche Y., et al. , Cellular and molecular hemocyte responses of the Pacific oyster, Crassostrea gigas, following bacterial infection with Vibrio aestuarianus strain 01/32. Microbes Infect. 8, 2715–2724 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Labreuche Y., et al. , Vibrio aestuarianus zinc metalloprotease causes lethality in the Pacific oyster Crassostrea gigas and impairs the host cellular immune defenses. Fish Shellfish Immunol. 29, 753–758 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Parizadeh L., et al. , Ecologically realistic model of infection for exploring the host damage caused by Vibrio aestuarianus. Environ. Microbiol. 20, 4343–4355 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Bailey S. F., Rodrigue N., Kassen R., The effect of selection environment on the probability of parallel evolution. Mol. Biol. Evol. 32, 1436–1448 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Petton B., Pernet F., Robert R., Boudry P., Temperature influence on pathogen transmission and subsequent mortalities in juvenile Pacific oysters Crassostrea gigas. (Translated from English). Aquacult. Environ. Interact. 3, 257–273 (2013). [Google Scholar]
- 52.Duperthuy M., et al. , The major outer membrane protein OmpU of Vibrio splendidus contributes to host antimicrobial peptide resistance and is required for virulence in the oyster Crassostrea gigas. Environ. Microbiol. 12, 951–963 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Le Roux F., Binesse J., Saulnier D., Mazel D., Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl. Environ. Microbiol. 73, 777–784 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le Roux F., Davis B. M., Waldor M. K., Conserved small RNAs govern replication and incompatibility of a diverse new plasmid family from marine bacteria. Nucleic Acids Res. 39, 1004–1013 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Demarre G., et al. , A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 156, 245–255 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Le Roux F., et al. , Genome sequence of Vibrio splendidus: An abundant planctonic marine species with a large genotypic diversity. Environ. Microbiol. 11, 1959–1970 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Afgan E., et al. , The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang G., et al. , The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anders S., Pyl P. T., Huber W., HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buchfink B., Xie C., Huson D. H., Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Rubio T., Toulza E., Chaparro C., Charrière G. M., Destoumieux-Garzón D., Crassostrea gigas response to Vibrio strains J2-9, LGP32, J2-8 and LMG20012T, strain:Decipher Family 14 (Pacific oyster). BioProject. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA515169. Deposited 15 January 2019.
- 64.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao Y., Smyth G. K., Shi W., FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Rubio T., Luo X., Charrière G. M., Destoumieux-Garzón D., Jacq A., Vibrio crassostreae strain:J2-9 response to oyster colonization. BioProject. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA521688. Deposited 12 February 2019.
- 68.Rubio T., Luo X., Charrière G. M., Destoumieux-Garzón D., Jacq A., Vibrio tasmaniensis strain: LGP32 response to oyster colonization. BioProject. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA521693. Deposited 11 February 2019.
- 69.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 70.Conesa A., et al. , Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Médigue C., et al. , MicroScope-an integrated resource for community expertise of gene functions and comparative analysis of microbial genomic and metabolic data. Brief. Bioinform., 10.1093/bib/bbx113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Caspi R., et al. , The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 46, D633–D639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vallenet D., et al. , MicroScope–An integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 41, D636–D647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bruto M., Lagorce A., Toulza E., Le Roux F., Destoumieux-Garzón D., Vibrio tasmaniensis, strains: J0-13, J5-13 and J5-9. BioProject. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA520819. Deposited 4 February 2019.
- 75.Bruto M., Charrière G. M., Le Roux F., Destoumieux-Garzón D., Vibrio tasmaniensis strain:LMG 20012T. BioProject. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA542263. Deposited 5 May 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.