Significance
In clinical in vitro fertilization (IVF), the prevailing method for preimplantation genetic testing for aneuploidy (PGT-A) requires biopsying a few cells from the trophectoderm (TE), the lineage that forms the placenta. The test is invasive, requires specialized skills, and suffers from false positives and negatives because chromosome numbers in the TE and the inner cell mass (ICM), which develops into the fetus, are not always the same. Noninvasive PGT-A (niPGT-A), which is based on sequencing DNA released into the culture medium from both TE and ICM, may offer a solution to these problems but has previously had limited efficacy. The present results show improved sensitivity and reliability of niPGT-A, suggesting a potentially superior test for noninvasive and cost-effective PGT-A in clinical IVF.
Keywords: preimplantation genetic testing, noninvasive PGT-A, culture medium, ICSI, human blastocyst
Abstract
Preimplantation genetic testing for aneuploidy (PGT-A) with trophectoderm (TE) biopsy is widely applied in in vitro fertilization (IVF) to identify aneuploid embryos. However, potential safety concerns regarding biopsy and restrictions to only those embryos suitable for biopsy pose limitations. In addition, embryo mosaicism gives rise to false positives and false negatives in PGT-A because the inner cell mass (ICM) cells, which give rise to the fetus, are not tested. Here, we report a critical examination of the efficacy of noninvasive preimplantation genetic testing for aneuploidy (niPGT-A) in the spent culture media of human blastocysts by analyzing the cell-free DNA, which reflects ploidy of both the TE and ICM. Fifty-two frozen donated blastocysts with TE biopsy results were thawed; each of their spent culture medium was collected after 24-h culture and analyzed by next-generation sequencing (NGS). niPGT-A and TE-biopsy PGT-A results were compared with the sequencing results of the corresponding embryos, which were taken as true results for aneuploidy reporting. With removal of all corona-cumulus cells, the false-negative rate (FNR) for niPGT-A was found to be zero. By applying an appropriate threshold for mosaicism, both the positive predictive value (PPV) and specificity for niPGT-A were much higher than TE-biopsy PGT-A. Furthermore, the concordance rates for both embryo ploidy and chromosome copy numbers were higher for niPGT-A than TE-biopsy PGT-A. These results suggest that niPGT-A is less prone to errors associated with embryo mosaicism and is more reliable than TE-biopsy PGT-A.
Preimplantation genetic testing for aneuploidy (PGT-A) using trophectoderm (TE) biopsy is currently the most widely used genetic test for identification of de novo aneuploidies in embryos in clinical in vitro fertilization (IVF). Despite the overall high implantation rates achieved following transfer of euploid embryos (1–4), debate continues regarding the accuracy and the safety of this approach, as well as which patients and/or age groups may truly benefit with an increased probability of having a healthy live birth (5–8).
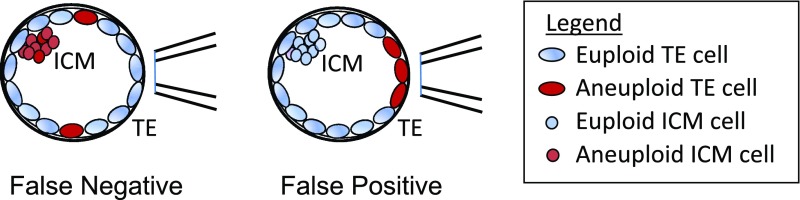
Accuracy of PGT-A with TE biopsy relates to embryonic mosaicism, a phenomenon characterized by the presence of two or more genetically distinct cell lineages (9). Mosaicism affects as many as 30–40% of human blastocysts (10, 11), with the reported incidence of euploid/aneuploid mosaicism varying (12) from as low as 2.0–2.9% (13, 14) to as high as 14.0–17.3% (10, 11). Development of the human blastocyst will therefore inevitably result in some TE test results failing to reflect the genome profile of the inner cell mass (ICM), which ultimately forms the fetus (Fig. 1). Although any risk of transferring a miscalled embryo is considered too great, false positives are particularly concerning as these may lead to discarding embryos that would otherwise lead to normal births. There are now several reports of healthy babies being born after transfer of embryos diagnosed as aneuploid or diploid/aneuploid mosaic (15–17), with some studies reporting a live-birth rate of over 40% (18, 19). False negatives are also of concern, because their transfer may lead to either no pregnancy or, worse yet, an abnormal fetus.
Fig. 1.
False positives and false negatives arise from embryo mosaicism in TE biopsy PGT-A.
In addition, there are safety concerns with TE biopsy. The removal of TE cells is inherently traumatic and may lead to a decrease in implantation potential. While one study showed equivalent implantation rates with and without TE biopsy (20), several other studies have not shown the expected improvement in live-birth rates following PGT-A with TE biopsy (7). To our knowledge, there is only one study investigating safety as measured by clinical outcomes (21). Although reassuring for the outcomes assessed (gestational age and birth weight), the study population was small. No long-term data on biosafety of TE biopsy in humans exist.
Given the above limitations of TE biopsy, recent attention has been given to another invasive approach, which involves aspiration of blastocoelic fluid. Although analysis of the cell-free DNA in this fluid contains detectable amounts of cell-free DNA, a wide range of ploidy concordance between this fluid and the whole embryo (48–97%) has been reported (22–24).
Notably, noninvasive preimplantation genetic testing for aneuploidy (niPGT-A) was developed in 2016 to analyze DNA leaked from human blastocysts into the spent culture medium (25). Xu et al. (25) compared sequencing results between the medium and the whole embryo and observed an 11.8% false-negative rate (FNR) and a 16.0% false-positive rate (FPR). These FNR and FPR were primarily attributed to contamination from maternal cumulus cells and embryo mosaicism, respectively. Considering that niPGT-A does not interfere with the embryos, the clinical live births with these initial results were encouraging (25).
At around the time of this initial publication, another paper was published, also reporting detection of amplified DNA in culture medium; however, concordance was low compared with TE biopsy (26). In the past 2 y, further studies have evaluated niPGT-A (27–30). While none of them provided encouraging results, two showed that TE-biopsy PGT-A was superior to niPGT-A (28, 30). However, one very recent study in women <30 y reported 27 normal births following transfer of 50 embryos identified as euploid by niPGT-A (31). Moreover, in another report, when combining niPGT-A with blastocoel fluid DNA analysis, concordance was higher than PGT-A with TE biopsy alone (29).
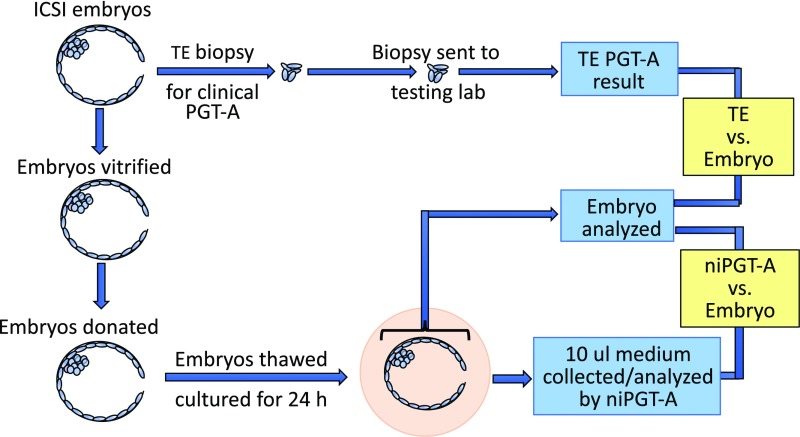
In the present study, we used donated human blastocysts that had previously undergone TE biopsy testing by an outside laboratory as part of clinical care of the IVF patients. The thawed blastocysts were cultured individually for 24 h in microdrops of culture medium, after which the medium and embryos were collected, amplified, and subjected to next-generation sequencing (NGS) (Fig. 2). We compared ploidy results obtained from the spent culture media and the TE biopsy samples, with the true result obtained from sequencing the whole embryo. We used a stringent procedure to set a mosaicism threshold so as to minimize the FPR and FNR and so identify true aneuploidy. In doing so, we have critically examined the accuracy and reliability of niPGT-A and report its improved efficacy compared with TE biopsy.
Fig. 2.
Workflow of sample processing for PGT-A analysis of TE biopsy, embryo, and spent culture media.
Results
Fifty-two embryos (average age, 35.9 y, based on maternal ages at retrieval) were included in this study; 41 were thawed after vitrification on day 5, and 11 were thawed after vitrification on day 6. Regardless of the day of thawing, all 52 embryos were cultured for exactly 24 h, and all embryos and their corresponding 52 spent media samples were successfully amplified. The sequencing data are shown in SI Appendix, Table S1. Forty-eight of the spent culture medium sequencing results gave interpretable copy number (CN) profiles, while four were not interpretable; these four were deemed “Noisy trace.” In contrast, two of the TE biopsy samples failed to provide informative PGT-A results (one reported as “No Result” and the other as “Degraded DNA”). There was no obvious relationship between embryo ploidy results and the morphological grade of the embryos. No DNA was detectable in any of five amplified blank culture media samples cultured under identical conditions.
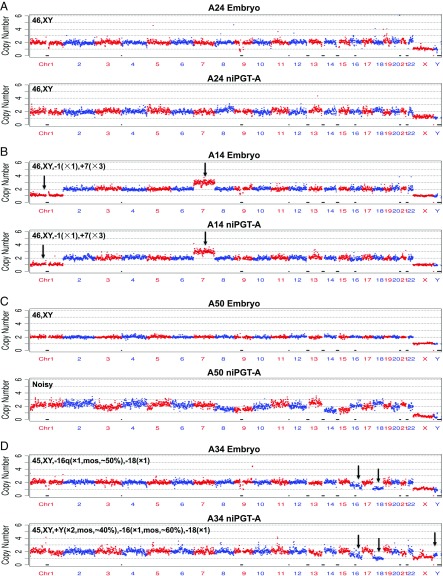
The purpose of niPGT-A is to distinguish euploid from aneuploid embryos by using the spent culture media, with minimum complications from contamination, measurement noise, and embryo mosaicism. Fig. 3A shows the CN profile of a euploid male embryo (A24) and its corresponding euploid niPGT-A CN profile. The CNs in both profiles are two across all autosomes, and one for each of the X and Y chromosome. This suggests the detected DNA in the spent culture medium comes primarily from the apoptosis of euploid cells. This provides the basis for niPGT-A to identify normal embryos. Fig. 3B shows the CN profile of an aneuploid male embryo (A14) with a CN of one for chromosome 1 and three for chromosome 7, both of which were consistently observed in the corresponding niPGT-A CN profile. This suggests that apoptosis of aneuploid cells allows abnormal embryos to be identified using the spent media.
Fig. 3.
The CN plots of embryo vs. spent culture medium at a sequencing depth of 0.02×. The chromosomes are shown in alternating red and blue colors. (A) A male euploid embryo with corresponding euploid niPGT-A profile. (B) A male aneuploid embryo with corresponding aneuploid niPGT-A profile. (C) A male euploid embryo with corresponding noisy niPGT-A profile. (D) A male aneuploid embryo with discordant mosaicism in the embryo compared with corresponding niPGT-A profile.
We note that the fact that the Y chromosomes in Fig. 3 A and B have a CN of one in the niPGT-A data indicates the lack of maternal contamination from the cumulus cells in the culture media, which is essential for the ploidy detection.
Among the 52 niPGT-A CN profiles, four showed numerous chromosomes with multiple gains and losses, and have high coefficients of variation (CVs) (mean, 0.23 vs. 0.16 for the remaining 48 samples). Such “noisy” profiles are likely due to the higher gain in whole-genome amplification (WGA) required for these samples of insufficient DNA (lack of apoptosis) in the spent media. We note all four noisy niPGT-A CN profiles were associated with euploid embryos based on their corresponding whole embryo results (Fig. 3C and SI Appendix, Fig. S1). In general, we recommend that those traces with a CV of ≥0.19 should not be used to identify embryo aneuploidy.
The CVs of each of the X and Y chromosomes in male embryos were higher than the CV of the autosomes. By contrast, the CVs of the X chromosomes in female embryos were almost identical to that of the autosomes, as expected, due to averaging of the two alleles (SI Appendix, Table S2).
Another major complication of aneuploidy identification beside the amplification noise and insufficient apoptosis is mosaicism. A mosaic embryo is defined as one having cells with different CNs in at least one chromosome. In the CN profile of such an embryo, the CN of a mosaic chromosome is a noninteger.
Fig. 3D shows an example of a mosaic embryo (A34) with a noninteger CN for chromosome 16 of 1.5 (50% diploid and 50% monosomy) in the embryo profile and of 1.4 (40% diploid and 60% monosomy) in the niPGT-A CN profile. In addition, there is a noninteger CN of 1.4 for the Y chromosome in niPGT-A. Importantly, mosaicism was not reported for the TE biopsy samples by any of the outside laboratories.
We define the percentage mosaicism “M” to quantify mosaicism according to the following formula, where CN is copy number:
For example, in the above case for A34 in Fig. 3D, “M” = 50% for chromosome 16 for the embryo; and “M” = 40% for the Y chromosome and 60% for chromosome 16 for niPGT-A.
The CN profiles for the embryo and spent culture medium in Fig. 3D are therefore labeled as follows:
In general, while experimental “M” values are complicated by amplification noise, the higher the “M” value the more likely it is due to true biological mosaicism of the embryo. In reporting aneuploidy, we needed to set a threshold for “M” above which aneuploidy would be identifiable beyond noise. Clearly, false-positive and false-negative aneuploidy callings are dependent on the threshold of “M.” Initially, we set this threshold at 30% to generate the CN profiles for niPGT-A, embryo, and TE-biopsy PGT-A from the raw data as shown in SI Appendix, Table S1.
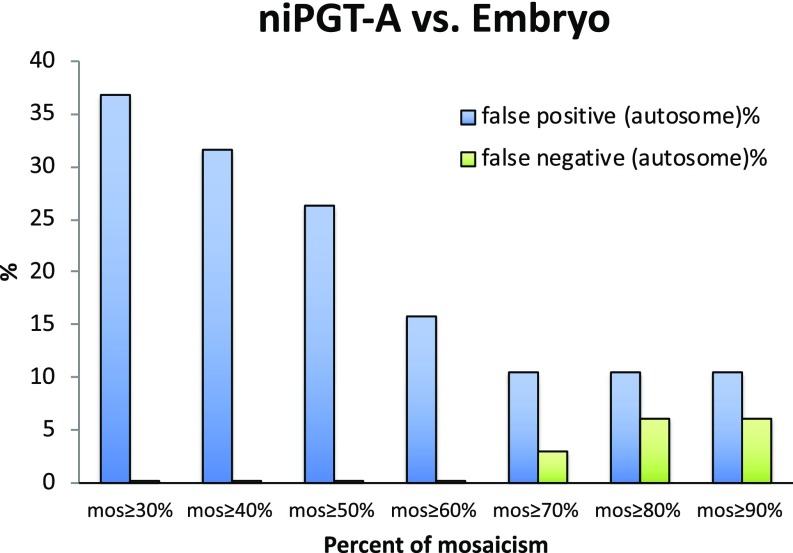
Fig. 4 shows the FPR and FNR detected by niPGT-A as a function of the “M” mosaicism threshold based on the data in SI Appendix, Table S1. Notably, false negatives are zero for the threshold ranging from ≥30% to ≥60%, which is due primarily to the lack of contamination by cumulus cells. The FPR decreases up to the “M” threshold of 70%, while the FNR rises above 70%. At a threshold of 60% mosaicism, the FPR was lowest while the false negative still remained at 0%. We therefore chose 60% as the threshold for distinguishing aneuploid from euploid embryos.
Fig. 4.
FPR and FNR as a function of the percent mosaicism in niPGT-A profiles. Sixty percent of mosaicism was set as the threshold for identifying aneuploidy.
To give an example, for an autosome in any embryo and for the two X chromosomes in a female embryo, a CN between 1.4 and 2.6 is considered diploid, and a CN <1.4 is considered monosomy, and >2.6 is considered trisomy. For each of the X and Y chromosomes of a male embryo, a CN between 0.4 and 1.6 is considered haploid.
Using the 60% “M” threshold for niPGT-A results, we determined the ploidy of each of the 48 spent culture media samples (Table 1). We then compared the FPR and FNR, as well as the concordance rates in chromosome CNs for niPGT-A and TE-biopsy PGT-A, with the embryo results as the true positives (Table 2). Being the same as the FNR for TE biopsy, the FNR for niPGT-A was zero, which is a significant improvement over our previous report by virtue of avoiding maternal contamination. The FPR of niPGT-A was 20.0% (3/15), much lower than that for TE-biopsy PGT-A, which was 50.0% (9/18).
Table 1.
Comparison of niPGT-A and TE-biopsy CN profiles with their corresponding embryos
| ID | niPGT-A | Embryo | TE-biopsy | Group |
| A21 | 46,XY | 46,XY | 46,XY | E/E/E |
| A23 | 46,XX | 46,XX | 46,XX | E/E/E |
| A24 | 46,XY | 46,XY | 46,XY | E/E/E |
| A42 | 46,XY | 46,XY | 46,XY | E/E/E |
| A48 | 46,XX | 46,XX | 46,XX | E/E/E |
| A49 | 46,XY | 46,XY | 46,XY | E/E/E |
| A51 | 46,XY | 46,XY | 46,XY | E/E/E |
| A01 | 46,XY,−5(pter→q31.3,∼141M,×1),+10(p12.31→qter,∼108M,×3) | 46,XY,−5(pter→q31.3,∼142M,×1),+10(p12.2→qter,∼111M,×3) | 46,XY,−5(pter→q33,×1),+10(p13→qter,×3) | A/A/A |
| A02 | 45,XX,−19(×1) | 46,XX,−19q(×1,mos,∼70%) | 45,XX,−19(×1) | A/A/A |
| A03 | 45,XY,−3p(pter→p12.2,∼84M,×1),−16(×1) | 46,XY,−16(×1) | 46,XY,−16(×1) | A/A/A |
| A05 | 45, XX, −4(×1) | 45,XX,−4(×1) | 45,XX,−4(×1) | A/A/A |
| A08 | 46,XX,−12p(p13.33→p12.1,∼25M,×1) | 46,XX,−12p(p13.33→p12.1,∼25M,×1) | 46,XX,+8p(pter-p23,×3),−12p(pter→p11.21,×1) | A/A/A |
| A09 | 46,XX,+12p(p13.33→p12.1,∼25M,×3) | 46,XX,+12p(pter→p12.1,∼24M,×3) | 46,XX,+8p(pter-p23 ×1),−12p(pter→p11.21,×3) | A/A/A |
| A10 | 46,XY,+12(p12.3→q24.31,∼106M,×3) | 46, XY,+12(p12.3→q24.31,∼108M,×3) | 46,XY,+8p(pter-p23,×3),+12(p11.2-qter,×3) | A/A/A |
| A11 | 46,XX, +12p(pter→p11.21,∼33M,×3) | 46, XX, +12p(pter→p12.1,∼24M,×3) | 46,XX,−8p(pter-p23 ×1),−12p(pter→p11.21,×3) | A/A/A |
| A12 | 47,XX,-X(×1,mos,∼60%),−1p(p31.1→p12,∼43M,×1),−1q(×1),−5(×1),+7(×3),+21(×3) | 46,XX, −1(p31.1→qter,∼179M,×1), +7(p21.2→qter,∼143M,×3), | 46,XX,+1(p31.2→qter,×1),+7(p15.2→qter,×3) | A/A/A |
| A13 | 46,XY,+22(×3,mos,∼60%) | 47,XY,+22(×3) | 47,XY,+22(×3) | A/A/A |
| A14 | 46,XY,−1(×1),+7(×3) | 46,XY,−1(×1),+7(×3) | 46,XY,−1(×1),+7(×3) | A/A/A |
| A15 | 46,XY,−1p(pter→p31.1,∼74M,×1), 7p(pter→p15.3 p15.3,∼23M,×3) | 46,XY,−1p(pter→p31.1,∼74M,×1), 7p(pter→p15.3,∼23M,×3) | 46,XY,−1p(pter→p31.1,×1),7p(pter→p15.3,×3) | A/A/A |
| A16 | 45,XX, −22(×1) | 45,XX, −22(×1) | 45,XX, −1(×1) | A/A/A |
| A17 | 46,XY,−1(p31.1→qter,∼173M,×1),+7(p15.3→qter,∼137M,×3) | 46,XY,−1(p31.2→qter,∼180M,×1), +7(p21.2→qter,∼143M,×3) | 46,XY,−1(p31.2→qter,×1),+7(p21.2→qter,×3) | A/A/A |
| A18 | 45,XY,−5(×1),+6(pter→q25.3,∼161M,×3),+19(×3),+22(×3,mos,∼60%) | 47,XY,−5(×1,mos,∼70%),+6(×3),+19(×3,mos,∼50%),+22(×3,mos,∼60%) | 48,XY,−5(×1),+6(×3),+19(×3),+22(×3) | A/A/A |
| A19 | 46,XX,+11(×3),−16(×1) | 47,XX,+11(×3),−16q(×1) | 47,XX,+11(×3),−16(×1) | A/A/A |
| A20 | 45,XX,−2(×1) | 45,XX,−2(×1) | 45,XX,−2(×1) | A/A/A |
| A25 | 45,XX,−21(×1) | 45,XX,−21(×1) | 45,XX,−21(×1) | A/A/A |
| A26 | 45,XY,+19(×3,mos,∼70%),−22(×1) | 46,XY,+19(×3),−22(×1) | 46,XY,+19(×3),−22(×1) | A/A/A |
| A27 | 47,XX,+16(×3) | 48,XX,+16(×4) | 48,XX,+16(×3) | A/A/A |
| A28 | 45,XX,−8(×1),+22(×3,mos,∼70%) | 45,XX,−8(×1) | 46,XX,−8(×1),+22(×3) | A/A/A |
| A29 | 46,XX,−10(×1),+11(×3),−12(×1),+21(×3) | 46,XX,−10(×1),+11(×3),−12(×1),+21(×3) | 45,XX,−10(×1),+11(×3),−12(×1) | A/A/A |
| A30 | 47,XX,+4(×3),+8(×3,mos,∼70%) | 46,XX,+4(×3,mos,∼70%),+8(×3,mos,∼70%) | 48,XX,+4(×3),+8(×3) | A/A/A |
| A32 | 46,XY,−22(×1,mos,∼60%) | 46,XY,+13(×3,mos,∼30%),+17q(×3,mos,∼40%),−22(×1,mos,∼60%) | 45,XY,−22(×1) | A/A/A |
| A34 | 45,XY,−16(×1,mos,∼60%),−18(×1) | 45,XY,−16q(×1,mos,∼50%),−18(×1) | 45,XY,−18(×1) | A/A/A |
| A35 | 47,XX, +16(×3), −18(×1), +22(×3) | 46, XX, +16(×3), −18(×1), +22(×3, mos,∼60%) | 47,XX,+16(×3)−18(×1),+22(×3) | A/A/A |
| A36 | 45,XY,−22(×1) | 45,XY,−22(×1) | 45,XY,−22(×1) | A/A/A |
| A37 | 47,XY,+21(×3) | 47,XY,+21(×3) | 47,XY,+21(×3) | A/A/A |
| A38 | 45,XX,−22(×1) | 45,XX,−22(×1) | 45,XX,−22(×1) | A/A/A |
| A41 | 46,XY,+3(×3,mos,∼70%) | 47,XY,+3(×3) | 47,XY,+3(×3) | A/A/A |
| A44 | 46,XX,+19q(×3,mos,∼70%) | 46,XX,+19(×3,mos,∼70%) | 47,XX,+19(×3) | A/A/A |
| A52 | 45,XY,−22(×1) | 45,XY,−22(×1) | 45,XY,−22(×1) | A/A/A |
| A50 | Noisy trace | 46,XY | 46,XY | NT/E/E |
| A04 | Noisy trace | 46,XY | 46,XY,−14q(q32.13-qter,×1) | NT/E/A |
| A07 | Noisy trace | 46,XX | 47,XX,+10(×3) | NT/E/A |
| A46 | Noisy trace | 46,XY | 47, XY,+10(×3) | NT/E/A |
| A06 | 46,XX | 46,XX | 45,XX,−13(×1) | E/E/A |
| A33 | 46,XY | 46,XY | 46,XY,−1(pter-p32.3,×1) | E/E/A |
| A43 | 46,XY | 46,XY | 47, XY,+20(×3) | E/E/A |
| A45 | 46,XX | 46,XX | 47,XX,+3(×3), +10(×3),−18(×1) | E/E/A |
| A40 | 46,XY,−16q(×1) | 46,XY | 47,XY,Tri/polysomy 10, Del/Dup 16 | A/E/A |
| A47 | 46,XX,−21(×1,mos,∼60%) | 46,XX | 47,XX,+22(×3) | A/E/A |
| A22 | 45,XX,−19(×1) | 46,XX | 46,XX | A/E/E |
| A39 | 46,XY | 46,XY | No Result | E/E/NR |
| A31 | 44,X,-X(×1),−16(×1) | 44,X,-X(×1),−16(×1) | Degraded DNA | A/A/DD |
The mosaicism threshold for niPGT-A was set at ≥60%. Different styles of font represent comparisons for niPGT-A with the embryo, and TE biopsy with the embryo; bold indicates concordance of chromosome CNs, and the underline indicates false positives. A, aneuploid; DD, degraded DNA; E, euploid; NR, no result; NT, noisy trace.
Table 2.
Comparison of the performance of niPGT-A versus TE biopsy for PGT-A
| Performance characteristic | niPGT-A (n = 48) | TE-biopsy (n = 50) |
| FPR | 20.0% (3/15) | 50.0% (9/18) |
| FNR | 0.0% (0/33) | 0.0% (0/32) |
| PPV | 91.7% (33/36) | 78.0% (32/41) |
| NPV | 100.0% (15/15) | 100.0% (18/18) |
| Sensitivity | 100.0% (33/33) | 100.0% (32/32) |
| Specificity | 80.0% (12/15) | 50.0% (9/18) |
| % Concordance for embryo ploidy | 93.8% (45/48) | 82.0% (41/50) |
| % Concordance for chromosome CNs | 83.3% (40/48) | 62.0% (31/50) |
niPGT-A and TE biopsy results were compared with those of the embryo. Sequencing threshold was set at 60% mosaicism.
We note that such a FPR of the TE biopsy is much higher than normal because of the high proportion of aneuploid embryos in our donated cohort (41/50 = 82% by TE biopsy testing), compared with the expected aneuploidy rate of ∼35% in embryos from women of the same age in clinical IVF (32). We note that the positive predictive value (PPV = [true positives]/[true positives + false positives]), is a better indicator than the FPR ([false positives]/[true negatives]), for assessing the efficacy as the former is less dependent on the aneuploidy rate of the cohort. The PPVs for niPGT-A and TE-biopsy PGT-A were 91.7% (33/36) and 78.0% (32/41), respectively.
For completeness, the sensitivities (Sensitivity = [true positives]/[true positives + false negatives]) and the negative predictive values (NPV = [true negatives]/[true negatives + false negatives]) for both niPGT-A and TE-biopsy PGT-A were 100%. The specificities (Specificity = [true negatives]/[true negatives +false positives]) of niPGT-A and TE-biopsy PGT-A were 80% (12/15) and 50% (9/18), respectively.
In addition to the comparison of the concordance for embryo ploidy (i.e., the aneuploidy/euploidy rate), we also assessed the concordance for chromosome CNs (with M ≥ 60%). As shown in Table 2, niPGT-A CN profiles had higher concordance to the embryo result compared with the TE biopsy CN profiles.
Discussion
The findings we report in this study extend and advance those of our previous work (21), in which we first reported the feasibility of niPGT-A. With the improved WGA method used in this study and by carefully setting a threshold for mosaicism to minimize the measurement noise, we show that niPGT-A outperformed PGT-A with TE biopsy.
niPGT-A relies on detection of DNA in the spent culture medium, which prompts the question as to the origin of the DNA. There are four sources that we can imagine: maternal DNA either from the polar bodies or from the cumulus cells, or embryonic DNA from either euploid or aneuploid apoptotic cells.
Contamination from either of the two polar bodies is very unlikely as they undergo apoptosis within 24 h of formation (33). Moreover, contaminating DNA from the polar bodies would mask the Y chromosome CN, if not fully degraded before the start time of the 24-h culture. Contamination by cumulus cells was minimized by our successful removal of all these cells in preparation for intracytoplasmic sperm injection (ICSI) as we were able to measure unity CN of the Y chromosome. Of note, in contrast to the report by Ho et al. (30) for a high male embryo misdiagnosis rate (63%), our result showed 0 of 24 male embryos were misdiagnosed.
The two remaining possible sources of DNA in the spent medium require careful consideration. In a developing embryo in culture, certain cells undergo apoptosis (34) and will expel DNA into the medium. As both the ICM and TE undergo apoptosis during preimplantation development (35, 36), the DNA in spent culture medium likely originates from both of these cell lineages. These apoptotic events increase in frequency as the total cell number increases exponentially in the ICM and TE at the blastocyst stage (37). It has recently been reported that a large number of cells exhibiting aneuploidy undergo apoptosis for clearance from the embryo (38). However, some euploid cells must also undergo apoptosis and be expelled into the spent culture medium. We suspect, at least for the euploid embryos, that leakage of DNA from the euploid cells outweighs that of the apoptotic aneuploid cells; otherwise, niPGT-A would not be able to report the euploid embryos successfully.
Aneuploid cells not cleared from the embryo will lead to aneuploidy in either the TE or the ICM, or in both lineages. When cells are not identical to each other, the embryo will be mosaic. Mosaicism has been the subject of much discussion in the interpretation of PGT-A errors when using TE biopsy (Fig. 1). In the present paper, we offer a solution to address this problem. Our basic assumption was that DNA in the spent culture medium will enable probing aneuploidy in both the ICM and TE, whereas the DNA in the TE biopsy will only probe aneuploidy in the TE.
Consistent with this assumption, Bolton et al. (39) showed in a mosaic mouse model that both aneuploid and euploid cells from the ICM, as well as from the TE, undergo apoptosis. Moreover, these investigators showed that a higher percentage of ICM cells compared with TE cells became apoptotic whether they were aneuploid (41.4% vs. 3.3%) or euploid (19.5% vs. 0.6%).
To accomplish our goal, we used an improved amplification method and NGS and, after setting an appropriate threshold of mosaicism, carried out niPGT-A to significantly reduce the chance of reporting errors due to mosaicism. We note that a good amplification method is critically important, and that the CV obtained after normalization with multiple annealing and looping based amplification cycles (MALBAC) technique is lower than those of other commercially available single-cell WGA methods (40). Technologies for DNA amplification are rapidly advancing with new preparation kits becoming commercially available that give even higher uniformity.
Even with the improved WGA, there can still be inadequate amplification or insufficient DNA leaked from aneuploid cells that are being cleared from euploid embryos (refer to the four embryos, A04, A07, A46, and A50, and their corresponding niPGT-A profiles in SI Appendix, Table S1). Because of the high CV of these four noisy niPGT-A profiles and the difficulty in interpreting their results, we chose to exclude these from our final analyses. Nevertheless, even when including all four noninformative samples, or when only excluding two of them, the performance characteristics of niPGT-A were still superior to those of TE biopsy (SI Appendix, Table S3). We recommend that only those embryos with CV < 0.19 are kept for further analyses. Assuming the WGA method used is sufficiently uniform, instead of discarding potentially euploid embryos with noisy niPGT-A profiles, we suggest that they should be cultured for several additional hours to allow more DNA to leak into the medium.
The variations along a CN profile arise from WGA noise, aneuploidy, and mosaicism. To distinguish true embryo aneuploidy from the noise associated with WGA and biological clearing of aneuploid cells, a threshold for mosaicism had to be carefully selected to minimize the false positives and false negatives. We chose M ≥ 60%, which enabled improved efficiency of niPGT-A compared with the previous reports.
Of significance, our FNR for niPGT-A was observed to be zero (Table 2), which is a substantial improvement over that recently reported by Ho et al. (42.1%) and that was previously reported by us (11.8%) (25). We assume that this improvement was due to the avoidance of maternal contamination resulting from stringent removal of all cumulus-corona radiata cells.
Our study is not without limitations. First, our sample size was relatively small, and most of the donated embryos were aneuploid by TE biopsy testing, and the incidence of false positives from TE-biopsy PGT-A was higher than expected, which was complicated by the fact that niPGT-A (by NGS) and TE-biopsy (by array comparative genomic hybridization) had different resolutions; and the TE-biopsy PGT-A results were from 4 companies that may have used different methods for data analysis. Nevertheless, comparison of niPGT-A and TE-biopsy PGT-A to their corresponding embryos has provided a robust procedure with which the efficacy of niPGT-A has been assessed. Second, the embryos had previously been biopsied, frozen, and then thawed on day 5 or 6, which is not the typical paradigm for PGT-A in clinical IVF. In reality, 2–3% of clinical TE biopsy samples fail to provide informative results (41), in which case the embryos often undergo a second biopsy. Our niPGT-A circumvents the risk of second or multiple biopsy, and hence offers a practical utility.
In summary, the findings reported here support the original report by Xu et al. (25) that cell-free DNA in medium used to culture human preimplantation embryos can be detected and used to determine ploidy of the embryos. In fact, compared with TE biopsy, the identical FNR of 0%, and the favorable FPR for niPGT-A we report here suggest that niPGT-A has the potential to be superior to TE biopsy for PGT-A, while avoiding the trauma associated with TE biopsy. However, further investigations are needed with embryos not previously biopsied and that also focus on development of new amplification techniques that minimize CVs and reduce requirements for the initial amount of DNA in spent culture medium. Testing of improved WGA methods with high CN variation (CNV) accuracy and resolution (42, 43) are underway, allowing further assessment as to whether niPGT-A would translate into a paradigm shift for PGT-A in clinical IVF.
Materials and Methods
Institutional Review Board Approval.
This study was approved by the Partners’ Healthcare Institutional Review Board. Full ethics committee approval was not required owing to the retrospective design of the study and the anonymized handling of the samples and data. All embryos were donated for research under informed consent by patients undergoing IVF for treatment of infertility at Brigham and Women’s Hospital (Boston, MA).
Study Subjects.
Patients (n = 13, mean age 35.0 y) using ICSI and PGT-A with TE biopsy for their infertility treatment at Brigham and Women’s Hospital during the period from October 2013 to January 2017 were included. Three of the 13 patients had balanced autosomal translocations, for whom the translocation breakpoints were available from the outside testing laboratory.
Laboratory Protocols.
Standard protocols were used for ICSI. Embryos were cultured individually in 25-µL microdrops of global total with human serum albumin (HSA) (LifeGlobal) overlain with mineral oil in Miri incubators (ESCO) at 37 °C in a dry atmosphere of 5% O2, 6–7% CO2 balanced with N2. On day 3, 64–68 h post-ICSI, embryos were evaluated and assisted hatched (AH) with a minimum of three laser pulses at 200 µs using a Zilos Tk laser (Hamilton Thorne Biosciences) and then moved to fresh drops of global total medium. Embryos were evaluated again on day 5, and again on day 6 if they failed to meet biopsy criteria on day 5. Blastocyst morphology was assessed according to developmental stage, blastocyst expansion, and quality of ICM and TE.
Blastocyst Biopsy and Vitrification.
TE biopsy was performed on good-quality expanded blastocysts with TE grades a or b, and a discernible ICM. The biopsy was performed in 25-µL drops global with Hepes medium under oil by holding the embryo under slight negative pressure with the ICM distant from the TE cells extruding through the AH hole created on day 3. On the rare occasion that the ICM was close to the extruding TE cells, great care was taken to avoid removing any ICM cells. Biopsy was performed with a biopsy pipette (30-µm inner diameter) and use of laser pulses at 300 µs to separate four to six extruded TE cells from the blastocyst. Immediately after biopsy, each biopsy sample was tubed per our routine protocol involving double identification before being sent to the outside testing laboratory for PGT-A. The biopsied blastocysts were returned to culture until vitrified shortly thereafter using Cryolocks (Irvine Scientific; one blastocyst per device) according to the manufacturer’s recommendations.
Outside Laboratory PGT-A Testing and Reporting.
PGT-A of the TE biopsies was performed by one of four outside testing laboratories (Reprogenetics; Reproductive Genetic Innovations; Genesis Genetics; Natera).
Noninformative results were reported as either “degraded DNA” or “no result.” A “degraded DNA” result may reflect cellular and/or DNA damage caused by the laser during the biopsy procedure itself; or it may be due to noisy profiles in embryo ploidy with numerous chromosomes showing multiple gains and losses. Such profiles are more common in arrested and morphologically abnormal embryos resulting in a high number of apoptotic and/or dying cells in the biopsy. By contrast, a report of “no result” indicates that the sample failed to amplify with the WGA process.
Culture of Donated Blastocysts and Sample Collection.
Immediately after warming, donated blastocysts were placed in 15-µL drops of equilibrated global total with HSA overlain with mineral oil. Exactly 24 h after onset of culture, blastocysts and their corresponding spent culture media samples were collected for analysis. Each blastocyst was gently moved with a pipette tip to the edge of its microdrop and then removed and transferred into a RNase-DNase–free PCR tube containing 2 µL of lysis buffer. For the media collections, 10 µL were removed from each of the residual spent medium drops and transferred into RNase-DNase–free PCR tubes. Pipette tips were changed between collections of each sample to avoid cross-sample contamination. Media drops incubated and collected under identical conditions to those used for blastocyst culture, but never containing embryos, served as negative controls. All samples were immediately frozen after collection and stored at −80 °C until analyzed. The workflow for sample processing is shown in Fig. 1.
WGA.
The frozen 10-µL spent culture medium samples were thawed and gently mixed, and 3.5 µL was removed for the niPGT-A assay. These and the embryo samples were lysed, and the DNA was amplified using the NICSwift Sample Preparation kit (Yikon Genomics), which is based on a modified MALBAC method (44). The DNA concentration of the product after amplification was measured using a Qubit 2.0 fluorometer (ThermoFisher Scientific) with the Qubit dsDNA HS Assay kit (Life Technologies). Sequencing library preparations were performed by the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs), following the manufacturer’s instructions.
Sequencing and Data Analysis.
The samples were subjected to NGS using the Illumina HiSeq 2500 system, yielding ∼1.5 million sequencing reads on each sample.
CNV detection for each sample was performed as previously described (25) after removal of sequencing library preparation adapters and low-quality bases from the sequencing results. High-quality reads were then aligned to the human reference genome hg19 (University of California, Santa Cruz Genome Browser; genome.ucsc.edu/). Mapped reads were subjected to correction of the GC bias resulting from DNA’s GC contents. After GC correction, the average read density of euploid samples was used to normalize the reads as reference (40). The normalized read counts for each bin of 1,000 kb were defined as the CN. A circular binary segmentation algorithm (45) was used to detect CNV segments to identify aneuploid segments larger than 10 Mb. The cytoband location of those aneuploid segments was calculated using the cytoband file for hg19 from the University of California, Santa Cruz, database (available at http://hgdownload.cse.ucsc.edu/downloads.html). The minimum resolution of CNV using this approach was 10 Mb.
A Perl script (available at https://www.perl.org/) was used to determine CNVs, and the CNVs were visualized by the R programming language (available at https://www.r-project.org/); detailed scripts have been previously submitted to GitHub.
The uniformity of amplification was characterized by the CV of the read density for each chromosome. The CV of the read density was calculated by the genome-wide SD divided by the mean. When niPGT-A results revealed noisy chromosome CN profiles in the spent culture medium, they were categorized as “noisy trace” and excluded from all comparisons.
Supplementary Material
Acknowledgments
We thank Shiping Bo and Shujie Ma (Yikon Genomics) for their assistance with the data analysis. We thank Dr. Lin Shao (Peking University) for helpful comments and suggestions. We thank the entire Embryology team at Brigham and Women’s Hospital for their assistance in sample collection. This work was supported by Beijing Advanced Innovation Center for Genomics at Peking University, by a generous gift grant from Xianhong Wu to Harvard University, and by a research grant from the Center of Infertility and Reproductive Surgery, Department of Obstetrics and Gynecology, Brigham and Women’s Hospital (Boston, MA).
Footnotes
Conflict of interest statement: S.L. and X.S.X. are cofounders and shareholders of Yikon Genomics. The other authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907472116/-/DCSupplemental.
References
- 1.Fiorentino F., et al. , Development and validation of a next-generation sequencing-based protocol for 24-chromosome aneuploidy screening of embryos. Fertil. Steril. 101, 1375–1382 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Lee H. L., et al. , In vitro fertilization with preimplantation genetic screening improves implantation and live birth in women age 40 through 43. J. Assist. Reprod. Genet. 32, 435–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simon A. L., et al. , Pregnancy outcomes from more than 1,800 in vitro fertilization cycles with the use of 24-chromosome single-nucleotide polymorphism-based preimplantation genetic testing for aneuploidy. Fertil. Steril. 110, 113–121 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Wells D., et al. , Clinical utilisation of a rapid low-pass whole genome sequencing technique for the diagnosis of aneuploidy in human embryos prior to implantation. J. Med. Genet. 51, 553–562 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barad D. H., Darmon S. K., Kushnir V. A., Albertini D. F., Gleicher N., Impact of preimplantation genetic screening on donor oocyte-recipient cycles in the United States. Am. J. Obstet. Gynecol. 217, 576.e1–576.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Kang H. J., Melnick A. P., Stewart J. D., Xu K., Rosenwaks Z., Preimplantation genetic screening: Who benefits? Fertil. Steril. 106, 597–602 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Paulson R. J., Preimplantation genetic screening: What is the clinical efficiency? Fertil. Steril. 108, 228–230 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Munne S., et al. , Global multicenter randomized controlled trial comparing single embryo transfer with embryo selected by preimplantation genetic screening using next-generation sequencing versus morphologic assessment. Fertil. Steril. 108, e19 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Taylor T. H., et al. , The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans. Hum. Reprod. Update 20, 571–581 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Fragouli E., et al. , Cytogenetic analysis of human blastocysts with the use of FISH, CGH and aCGH: Scientific data and technical evaluation. Hum. Reprod. 26, 480–490 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Northrop L. E., Treff N. R., Levy B., Scott R. T. Jr, SNP microarray-based 24 chromosome aneuploidy screening demonstrates that cleavage-stage FISH poorly predicts aneuploidy in embryos that develop to morphologically normal blastocysts. Mol. Hum. Reprod. 16, 590–600 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capalbo A., Ubaldi F. M., Rienzi L., Scott R., Treff N., Detecting mosaicism in trophectoderm biopsies: Current challenges and future possibilities. Hum. Reprod. 32, 492–498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capalbo A., et al. , Correlation between standard blastocyst morphology, euploidy and implantation: An observational study in two centers involving 956 screened blastocysts. Hum. Reprod. 29, 1173–1181 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Johnson D. S., et al. , Comprehensive analysis of karyotypic mosaicism between trophectoderm and inner cell mass. Mol. Hum. Reprod. 16, 944–949 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greco E., Minasi M. G., Fiorentino F., Healthy babies after intrauterine transfer of mosaic aneuploid blastocysts. N. Engl. J. Med. 373, 2089–2090 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Zore T., et al. , Transfer of embryos with segmental mosaicism is associated with a significant reduction in live-birth rate. Fertil. Steril. 111, 69–76 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Fragouli E., et al. , Analysis of implantation and ongoing pregnancy rates following the transfer of mosaic diploid-aneuploid blastocysts. Hum. Genet. 136, 805–819 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Maxwell S. M., et al. , Why do euploid embryos miscarry? A case-control study comparing the rate of aneuploidy within presumed euploid embryos that resulted in miscarriage or live birth using next-generation sequencing. Fertil. Steril. 106, 1414–1419.e5 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Zhang L., et al. , Rates of live birth after mosaic embryo transfer compared with euploid embryo transfer. J. Assist. Reprod. Genet. 36, 165–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott R. T. Jr, Upham K. M., Forman E. J., Zhao T., Treff N. R., Cleavage-stage biopsy significantly impairs human embryonic implantation potential while blastocyst biopsy does not: A randomized and paired clinical trial. Fertil. Steril. 100, 624–630 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Forman E. J., et al. , Single embryo transfer with comprehensive chromosome screening results in improved ongoing pregnancy rates and decreased miscarriage rates. Hum. Reprod. 27, 1217–1222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magli M. C., et al. , Deoxyribonucleic acid detection in blastocoelic fluid: A new predictor of embryo ploidy and viable pregnancy. Fertil. Steril. 111, 77–85 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Magli M. C., et al. , Preimplantation genetic testing: Polar bodies, blastomeres, trophectoderm cells, or blastocoelic fluid? Fertil. Steril. 105, 676–683.e5 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Tobler K. J., et al. , Blastocoel fluid from differentiated blastocysts harbors embryonic genomic material capable of a whole-genome deoxyribonucleic acid amplification and comprehensive chromosome microarray analysis. Fertil. Steril. 104, 418–425 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Xu J., et al. , Noninvasive chromosome screening of human embryos by genome sequencing of embryo culture medium for in vitro fertilization. Proc. Natl. Acad. Sci. U.S.A. 113, 11907–11912 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shamonki M. I., Jin H., Haimowitz Z., Liu L., Proof of concept: Preimplantation genetic screening without embryo biopsy through analysis of cell-free DNA in spent embryo culture media. Fertil. Steril. 106, 1312–1318 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Feichtinger M., et al. , Non-invasive preimplantation genetic screening using array comparative genomic hybridization on spent culture media: A proof-of-concept pilot study. Reprod. Biomed. Online 34, 583–589 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Vera-Rodriguez M., et al. , Origin and composition of cell-free DNA in spent medium from human embryo culture during preimplantation development. Hum. Reprod. 33, 745–756 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Kuznyetsov V., et al. , Evaluation of a novel non-invasive preimplantation genetic screening approach. PLoS One 13, e0197262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho J. R., et al. , Pushing the limits of detection: Investigation of cell-free DNA for aneuploidy screening in embryos. Fertil. Steril. 110, 467–475.e2. [DOI] [PubMed] [Google Scholar]
- 31.Fang R., et al. , Chromosome screening using culture medium of embryos fertilised in vitro: A pilot clinical study. J. Transl. Med. 17, 73 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franasiak J. M., et al. , The nature of aneuploidy with increasing age of the female partner: A review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil. Steril. 101, 656–663.e1 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Longo F. J., Fertilization (Chapman and Hall, New York, 1997). [Google Scholar]
- 34.Gahan P. B., Swaminathan R., Circulating nucleic acids in plasma and serum. Recent developments. Ann. N. Y. Acad. Sci. 1137, 1–6 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Hardy K., Cell death in the mammalian blastocyst. Mol. Hum. Reprod. 3, 919–925 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Hardy K., Apoptosis in the human embryo. Rev. Reprod. 4, 125–134 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Hardy K., Handyside A. H., Winston R. M., The human blastocyst: Cell number, death and allocation during late preimplantation development in vitro. Development 107, 597–604 (1989). [DOI] [PubMed] [Google Scholar]
- 38.Zhu P., et al. , Single-cell DNA methylome sequencing of human preimplantation embryos. Nat. Genet. 50, 12–19 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Bolton H., et al. , Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat. Commun. 7, 11165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang L., Ma F., Chapman A., Lu S., Xie X. S., Single-cell whole-genome amplification and sequencing: Methodology and applications. Annu. Rev. Genomics Hum. Genet. 16, 79–102 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Cimadomo D., et al. , Inconclusive chromosomal assessment after blastocyst biopsy: Prevalence, causative factors and outcomes after re-biopsy and re-vitrification. A multicenter experience. Hum. Reprod. 33, 1839–1846 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Chen C., et al. , Single-cell whole-genome analyses by linear amplification via transposon insertion (LIANTI). Science 356, 189–194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan L., Xing D., Chang C. H., Li H., Xie X. S., Three-dimensional genome structures of single diploid human cells. Science 361, 924–928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zong C., Lu S., Chapman A. R., Xie X. S., Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 338, 1622–1626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olshen A. B., Venkatraman E. S., Lucito R., Wigler M., Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5, 557–572 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.