Abstract
Psychrophilic green algae from independent phylogenetic lines thrive in the polar extreme environments, but the hypothesis that their psychrophilic characteristics appeared through parallel routes of molecular evolution remains untested. The recent surge of transcriptome data enables large-scale evolutionary analyses to investigate the genetic basis for the adaptations to the Antarctic extreme environment, and the identification of the selective forces that drive molecular evolution is the foundation to understand the strategies of cold adaptation. Here, we conducted transcriptome sequencing of two Antarctic psychrophilic green algae (Chlamydomonas sp. ICE-L and Tetrabaena socialis) and performed positive selection and convergent substitution analyses to investigate their molecular convergence and adaptive strategies against extreme cold conditions. Our results revealed considerable shared positively selected genes and significant evidence of molecular convergence in two Antarctic psychrophilic algae. Significant evidence of positive selection and convergent substitution were detected in genes associated with photosynthetic machinery, multiple antioxidant systems, and several crucial translation elements in Antarctic psychrophilic algae. Our study reveals that the psychrophilic algae possess more stable photosynthetic apparatus and multiple protective mechanisms and provides new clues of parallel adaptive evolution in Antarctic psychrophilic green algae.
Keywords: Antarctic, psychrophilic green algae, positive selection, molecular convergence, adaptation
Introduction
The Antarctic sea ice is considered to be one of the most extreme environments for living creatures on the earth. It comprised a system of brine channels which is characterized by temperatures under 0 °C, high salinity, low light, limited gas exchange, and highly oxic conditions, owing to the semienclosed pore system within the ice (Thomas and Dieckmann 2002). Although the environment of sea ice is extremely harsh, there are diverse and productive communities of organisms living in it (Kang and Fryxell 1992; Eddie et al. 2008). Unicellular eukaryotic green algae are well adapted to the Antarctic sea ice environment and become the dominant producer in the extreme environmental conditions (Arrigo et al. 1997).
Currently, several genome-wide studies have revealed various molecular adaptations (positive selection and convergent substitution) that may contribute to extreme environment adaptation in algae, including Fragilariopsis cylindrus (diatom) (Mock et al. 2017) and Trichormus sp. NMC-1 (cyanobacteria) (Qiao et al. 2016). However, the genomic and transcriptomic data of Antarctic psychrophilic green algae are limited to date, so that little is known regarding molecular adaptations. Psychrophilic green algae are distributed throughout different branches of the order Chlamydomonadales, and large-scale genomic data provide us opportunities to investigate the genetic basis for the adaptations to the Antarctic cold environment.
Both Chlamydomonas sp. ICE-L and Tetrabaena socialis of the Chlamydomonadales are psychrophilic green algae with an optimum growth temperature at 5 °C, and they have been separately isolated from the floating sea ice and snowfield in Antarctica (An et al. 2013). The Chlamydomonas sp. ICE-L (C. sp. ICE-L) is found to form large populations in the bottom layer of sea ice, and it may undergo lower light intensity as sea ice is an effective barrier to light transmission. The alga, T. socialis, has to experience higher light intensity and longer light exposure in the polar snowfield. The ambient light conditions of these two algae are different, but both of them have experienced and adapted to the same abiotic stress: cold temperatures below 0 °C. Psychrophilic and mesophilic algae are broadly defined as organisms that have optimal growth temperatures of <15 °C and >20 °C, respectively. Previous studies have revealed that C. sp. ICE-L is characterized as a psychrophilic and halophilic alga, and evidence of adaptive evolution on photosynthetic genes has been partially identified (Zhang et al. 2018). Photosynthesis is a temperature-sensitive energy transformation process that can convert light energy into chemical energy. Photosynthetic electron transport is driven by the absorption of light energy, and it flows through the photosystem II (PSII), Cytochrome b6f and photosystem I (PSI). Nicotinamide adenine dinucleotide phosphate (NADPH) is the product of electron transport, and ATP is generated by a chloroplastic Adenosine triphosphate (ATP) synthase using the proton gradient established across the thylakoid membrane during electron transport. Energy imbalance can occur as a consequence of exposure to low temperature at constant irradiance (Huner et al. 1998). The energy imbalance induces the generation of reactive oxygen species (ROS), and high level of ROS can attack the membrane and destroy the cell. Several solutions of stress-induced energy imbalance have been identified, such as a reduction in light-harvesting capacity, an increased capacity to dissipate excess energy as heat and an efficient ROS scavenging system (Maxwell et al. 1995). Maintenance of proper membrane fluidity is essential to membranes at low temperature condition. The photosynthetic apparatus of psychrophilic plants exhibited higher levels of unsaturated fatty acids in their membranes (Morgan-Kiss et al. 2002). Increases in fatty acid unsaturation and decreases in fatty acid length could increase membrane fluidity and decrease the temperature at which the transition from liquid-crystalline to gel phase takes place (Morgan-Kiss et al. 2006; Valledor et al. 2013). Cold stress may also cause the denaturation and subsequent aggregation of proteins, and many organisms have employed heat shock proteins to avoid such situation. The heat shock proteins act either alone or together to fold and assemble newly synthesized proteins, prevent the aggregation of unfolded proteins, and facilitate refolding of malfolded proteins (Liu et al. 2010; Storey and Storey 2013).
In this study, we conducted transcriptome sequencing of two Antarctic psychrophilic green algae and performed evolutionary analyses together with other available algal genomes and transcriptomes to investigate potential adaptive strategies of cold adaptation in Antarctica. We searched for the presence of positive selection on amino acid composition in two Antarctic psychrophilic algae and tested whether they have experienced adaptive evolution in the same sets of loci. The positive selection and convergent substitution analyses identified positively selected genes (PSGs) with convergent amino acid changes in the two algae, and functional analyses demonstrated that these adaptive modifications were associated with their psychrophilic characteristic. Our study revealed evolutionary pattern of adaptive evolution and provide insights into the molecular adaptations in Antarctic psychrophilic algae.
Materials and Methods
Algal Species and Culture
Two Antarctic psychrophilic green algae C. sp. ICE-L and T. socialis (NIES-691) were used in this study. C. sp. ICE-L was isolated from the floating ice near the Zhongshan Research Station of Antarctica, and T. socialis was obtained from the Microbial Culture Collection at the National Institute for Environmental Studies, Japan. The algae C. sp. ICE-L and T. socialis were separately cultured in Provasoli seawater (Provasoli 1966) and AF-6 medium (Provasoli and Pintner 1960; Kato 1982), at 5 °C under a 14-h light/10-h dark photoperiod (photon flux density of 40 μmol photons m−2 s−1).
Cold Tolerance Assay
We tested the psychrophilic characteristic of two Antarctic algae (C. sp. ICE-L and T. socialis; optimum growth temperature at 5 °C), and used a model mesophilic alga (Chlamydomonas reinhardtii; optimal growth temperature 21 °C) as a “control” in the assay. The logarithmic phase algal cells were placed in a low temperature chamber set to −5 °C for 24 h, and then transferred them to their optimal growth temperatures for 24 h. The photosynthetic efficiency (Fv/Fm) was measured at two time points: 1) 0 h—before cold treatment for 24 h and 2) 48 h—back to optimal temperature for 24 h after cold treatment.
RNA Extraction, Library Preparation, and Sequencing
To obtain as many expressed genes as possible, the logarithmic phase cells were harvested at three time points (9:00, 15:00, and 21:00) by centrifugation (8 °C, 5 min, 8,000 rpm). Collected cells were immediately flash frozen in liquid nitrogen until further processed for RNA extraction. Total RNA was extracted using RNAprep Pure Plant Kit (DP441) (Tiangen Biotech Co., Beijing, China) according to the manufacturer’s instructions. The eluted RNA was measured using the NanoPhotometer spectrophotometer (IMPLEN, CA) and the quality of RNA evaluated by gel electrophoresis. A total amount of 1.5 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Pair-end sequencing (2× 150 bp) was carried out on the Illumina HiSeq 4000 platform (Illumina, San Diego, CA) at Novogene Bioinformatics Technology Co., Ltd (Beijing, China).
Data Processing, Assembly, and Annotation
The raw data were first filtered by removing reads containing adapters, reads with more than 10% ambiguous bases (N) and low quality reads (more than 50% bases with small Qphred ≤20) through in-house perl scripts. All the downstream analyses were based on filtered reads. Transcriptome de novo assembly was performed using Trinity with default parameters (Grabherr et al. 2011). The Corset program was used with default parameter (distance thresholds = 0.3; minimum aligned reads number = 10) to cluster transcripts by sequence similarity and redundant transcripts were removed (Davidson and Oshlack 2014). All the assembled unigenes of the two algae were annotated by searching for sequence homology against the Nr (nonredundant protein sequences) and Nt (nonredundant nucleotide sequences) database from NCBI and Swiss-Prot (a manually annotated and reviewed protein sequence database) using the Blast algorithm with a E-value cutoff of 10−5, and KOG (Eukaryotic Orthologous Groups of proteins) database with a E-value cutoff of 10−3 (Altschul et al. 1997). PFAM protein family alignments were performed using the HMMER 3.0 package (Finn et al. 2008). The gene ontology (GO) classification of each gene model was carried out using Blast2GOv2.5 (Götz et al. 2008), and the KEGG classification was performed using the KEGG Automatic Annotation Server (Moriya et al. 2007). The BUSCO (Benchmarking Universal Single-Copy Orthologs) was employed to assess the completeness of transcriptome assemblies based on the Chlorophyta_odb10 (2168 putative universal single-copy genes of Chlorophyta) (Simão et al. 2015).
Ortholog Identification and Alignment
Total of 12 algae in Chlamydomonadales were used for comparative analyses (three whole genomes and nine transcriptomes) (supplementary table S1, Supplementary Material online). The genome sequences of C.reinhardtii (Merchant et al. 2007), Volvox carteri f. nagariensis (Prochnik et al. 2010), and Dunaliella salina (Polle et al. 2017) were obtained from the Joint Genome Institute. The transcriptome sequences were obtained from 1KP project (Wickett et al. 2014) (https://db.cngb.org/onekp/). The single-copy orthologous genes were identified from three algal genomes (C. reinhardtii, V.carteri f. nagariensis, and D.salina) using OrthoMCL (version_2.0.9) (Li et al. 2003). The OrthoMCL program provides a scalable method for constructing orthologous groups across multiple eukaryotes with well-annotated whole genome data. The orthologous transcripts of other algal transcriptomes were assigned to the identified single-copy orthologous genes using Orthograph (Petersen et al. 2017). Orthograph employs a best reciprocal hit search strategy using profile hidden Markov models and maps nucleotide sequences to the globally best matching cluster of reference orthologous genes. Each corresponding orthologous group of 12 species was extracted to generate multiple sequence alignments. All orthologous genes were aligned at the codon level with the option “-codon” using the PRANK program (Loytynoja and Goldman 2005). Stop codons were removed from the sequences prior to alignment. Several quality controls were performed: 1) each aligned nucleotide sequence was trimmed to exclude poorly aligned positions using Gblocks 0.91 b with default parameters (Castresana 2000), 2) all positions that had gaps (“-”) and “N” in the alignments were deleted to eliminate the effect of uncertain bases on the test of positive selection, and 3) alignments shorter than 120 bp were further discarded after the trimming process (Yang et al. 2015). The 2,182 putative single-copy orthologous genes were advanced to the subsequent evolutionary analyses after quality control.
Phylogenetic Inference
The maximum-likelihood phylogenetic trees are reconstructed based on the nucleotide data with GTRGAMMA model and amino acid data with PROTGAMMAAUTO model using RAxML v8.2 with a rapid bootstrap search of 1,000 replicates (Stamatakis 2014). The third codon positions are problematic to phylogenetic inference due to high saturation; therefore only first and second codon positions of nucleotide data are used for phylogenetic analyses to minimize negative effects of saturation (Nei and Kumar 2000). We also employed the coalescent approach to reconstruct the species tree, and the single maximum-likelihood gene trees were inferred using IQTREE (Nguyen et al. 2015) with the GTR + G model and 1,000 rapid bootstrap replicates. The coalescent-based species tree was inferred using ASTRAL-III v5.5.9 (Zhang et al. 2017) (hereafter ASTRAL) with node support estimated by local posterior probability.
Analyses of Evolutionary Selective Pressure in Antarctic Psychrophilic Algae
Codon Model Fitting
Based on the alignments from 12 Chlamydomonadales algae, the positive selection analyses were conducted using PAML 4.9 (Yang 2007). We first fitted the basic model (model = 0, Nsites = 0, which assumes no site-wise or branch-wise dN/dS variation) to estimate rates of synonymous (dS) and nonsynonymous substitutions (dN). The nonsynonymous to synonymous rate ratio ω (dN/dS) among branches indicates changes in patterns of natural selection, where ω = 1, ω < 1, and ω > 1 correspond to neutral evolution, purifying and positive selection (Yang 2007). Genes with high dS values were discarded to accurately estimate dN/dS across the phylogeny (Escalona et al. 2017).
Identification of PSGs
Two psychrophilic algae (C. sp. ICE-L and T. socialis) were separately assigned as foreground and performed branch-site model test. In order to determine the impacts of different characteristics (psychrophilic and halophilic) on selective pressure, we further set halophilic alga (D.salina) as foreground. The branch-site model was used to test positive selection, and model A (allows sites to be under positive selection; model = 2, NSsites = 2, and fix_omega = 0) was compared with the null model A1 (sites may evolve neutrally or under purifying selection; model = 2, NSsites = 2, fix_omega = 1, and omega = 1) (Yang 2007). The likelihood ratio test with a χ2 distribution was used to determine which models were statistically different from the null model at a threshold of P values <0.05. The QVALUE in R was used to correct for multiple testing with a false discovery rate cutoff of 0.05 (Storey and Tibshirani 2003). Bayes empirical Bayes method was used to statistically identify sites under positive selection with posterior probabilities ≥0.95 (Zhang et al. 2005).
To uncover the broad evolutionary pattern of parallelism in the psychrophilic algae, we used PSGs with uncorrected P < 0.05 in three algae. The overlapped PSGs were visualized with UpSet plots (Lex et al. 2014). Additionally, multiple test correction aims to remove false positives, but it will likely have led to the removal of some genuine cases of positive selection. Significance of observed intersections, calculated using expected intersections given the background of genes tested (the size of background population from which the intersections are sampled), was examined with the supertest function from SuperExactTest v.0.99.4 (Wang et al. 2015).
Inference of Molecular Convergence in Antarctic Psychrophilic Algae
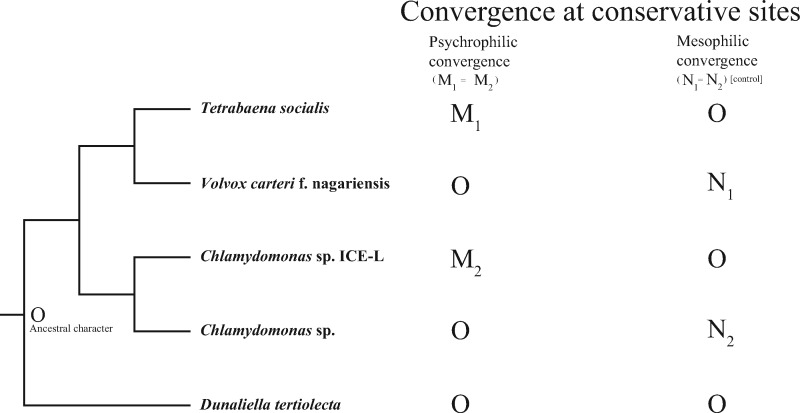
To identify genes underlying convergent substitution in two Antarctic psychrophilic algae (C. sp. ICE-L and T. socialis), the Convergence at Conservative Sites (CCS) method was applied in this study (Xu et al. 2017). The CCS method only infers convergence at conservative sites, and it has advantages in stringently filtering out noises in large-scale analyses. The symmetric design with two pairs of Antarctic psychrophilic and mesophilic algae (Chlamydomonas sp. and V. carteri f. nagariensis, ingroup control) provides a mean for evaluating the background level of convergence. Considering the living environment of these algae and their phylogenetic relationship, Dunaliella tertiolecta was selected as outgroup. The criteria for inferring convergence in CCS method are shown in figure 1 : 1) O indicates the character state of outgroup, and the ancestral states of both two branch pairs are assumed to be the same as outgroup in CCS method; 2) convergence is inferred only at conservative sites where two psychrophilic or two mesophilic algae shared the same character as outgroup (i.e., N1 = N2 = O or M1 = M2 = O); and 3) convergence is inferred when two psychrophilic or mesophilic algae share the same derived character.
Fig. 1.
—Convergence at conservative sites. The phylogeny consists of two psychrophilic and mesophilic pairs as ingroup species, with Dunaliella tertiolecta as outgroup. (M1/M2) and (N1/N2) indicate the observed character states in psychrophilic and mesophilic algae, respectively.
Functional Enrichment Analyses
Molecular evolution contributing to adaptation may act on multiple genes sharing similar functions or belonged to common pathways. We assessed enrichment in GO terms using BiNGO software in Cytoscape and in KEGG pathways using the KOBAS web server (http://kobas.org) (Mao et al. 2005). GO terms can be divided into three GO categories: molecular function (MF), cellular component (CC), and biological process (BP). Both the PSGs with uncorrected and corrected P value <0.05 were used to perform GO enrichment analyses, as small sets of genes after correction would preclude meaningful tests of enrichment. We implemented hypergeometric test to identify significantly overrepresented gene sets, and the Benjamini–Hochberg false discovery rate correction (corrected P < 0.05) was used for multiple testing (Benjamini and Hochberg 1995).
Results and Discussion
Summary of Sequencing, Assembly, and Functional Annotation
In total, 44.7 and 20.2-Gb high-quality sequences of C. sp. ICE-L and T. socialis were obtained from the transcriptomic sequencing (table 1). The de novo assemblies of clean reads produced 128,872 and 73,444 unigenes, respectively. The 55,448 and 33,810 unigenes (43.0% and 46.0% of the total unigenes) were separately annotated in at least one of the databases used in our study (supplementary table S2, Supplementary Material online). The top-hit species of the annotated unigenes against the NR database showed the highest similarity to C. reinhardtii and V.carteri f. nagariensis (supplementary table S3, Supplementary Material online). The BUSCO analyses showed that 87.2% and 77.9% of the conserved Chlorophyta genes (Chlorophyta_odb10) were annotated to be complete in our transcriptome data (table 1). The comparative analyses with other seven transcriptomes used also confirmed the high quality of our transcriptome assembly (supplementary fig. S1, Supplementary Material online).
Table 1.
Summary of Transcriptome Data in C. sp. ICE-L and Tetrabaena socialis
| C. sp. ICE-L | T. socialis | |
|---|---|---|
| No. of raw reads | 320,390,718 | 137,627,072 |
| No. of clean reads | 297,949,660 | 134,764,690 |
| Clean bases (Gb) | 44.7 | 20.2 |
| Number of unigenes | 128,872 | 73,444 |
| N50 | 821 | 1,531 |
| Percentage of BUSCO (Chlorophyta_odb10) (%) | 87.2 | 77.9 |
| Max unigene length (bp) | 16,998 | 16,030 |
| Mean unigene length (bp) | 570 | 938 |
| Number of unigenes (≥1 kb) | 16,753 | 20,589 |
The Phylogenetic Position of Antarctic Psychrophilic Algae
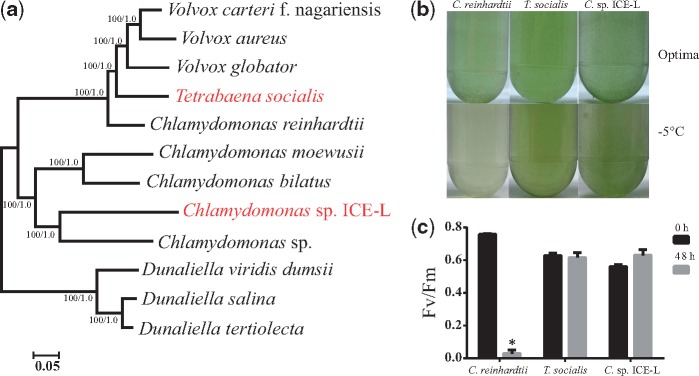
We constructed phylogenetic relationships of 12 Chlamydomonadales algae based on 2,182 orthologous genes using concatenation and coalescent approaches (fig. 2a). Consistent with previous studies (Nozaki et al. 2003; Liu et al. 2006; Lemieux et al. 2015; Featherston et al. 2018; Zhang et al. 2018), our phylogenetic analyses support that C. sp ICE-L, which belongs to the Monadinia clade, is close to Chlamydomonas sp., and T. socialis and V.carteri are sister group in the Reinhardtinia clade. Both sister lineages of the two psychrophilic algae (C. sp. ICE-L and T. socialis) are mesophilic algae, implying the independent origins of the psychrophilic characteristic. In the cold tolerance assay, the psychrophilic characteristic was confirmed in C. sp. ICE-L and T. socialis. Both of them grow well at −5 °C, but the mesophilic alga C. reinhardtii turns yellow and its photosynthetic efficiency decreases to zero in low temperature condition (fig. 2b and c).
Fig. 2.
—Phylogenetic position and physiological feature of psychrophilic algae. (a) Phylogenetic tree of 12 green algae inferred from 2,182 orthologous genes using concatenation and coalescent methods. The bootstrap support values and posterior probability scores are shown on the nodes from left to right; the two psychrophilic algae were colored in red. (b) C. reinhardtii, T. socialis, and C. sp. ICE-L were cultured in their optima temperature (up) and low temperature condition (−5 °C) (down). (c) Photosynthetic efficiency (Fv/Fm) of C. reinhardtii, T. socialis, and C. sp. ICE-L at two time points: 0 h (before cold treatment for 24 h) and 48 h (back to optimal temperature for 24 h after cold treatment); *The significant difference between control and treatment (P < 0.05). Abbreviation: C. sp. ICE-L, Chlamydomonas sp. ICE-L; T. socialis, Tetrabaena socialis; and C. reinhardtii, Chlamydomonas reinhardtii.
Parallel Positive Selection in Psychrophilic Algae
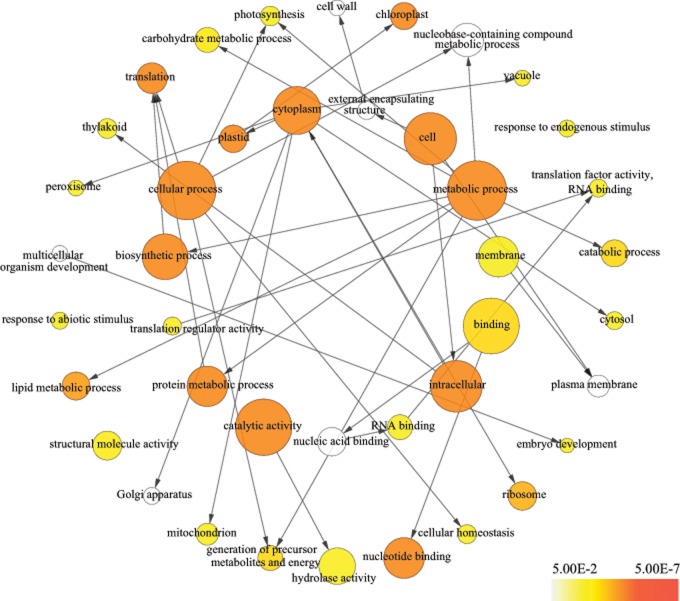
Gene families for three algal genomes (C. reinhardtii, V.carteri f. nagariensis, and D.salina) were delineated using OrthoMCL (4,738 families), and Orthograph was used to complete the identified single-copy orthologous gene families with genes from 9 species with transcriptomic data. Further analysis dealt with 2,182 families for which a single-copy gene was found for each species. The 913 genes with high dS value were discarded, and the dS values of the remaining 1,269 genes were below 1 for nearly all branches. The branch-site model was employed to test for positive selection in individual codons for these codon-aligned single-copy genes (1,269 genes), and 202 PSGs (corrected P < 0.05; 543 PSGs with uncorrected P < 0.05) were identified in two Antarctic psychrophilic algae. GO enrichment tests based on the 202 PSGs revealed that significantly enriched CC terms contains “thylakoid,” “peroxisome,” and “ribosome.” Several significantly enriched biological process terms can be grouped into three broad categories: photosynthesis, protein synthetic, and stress related process (e.g., “photosynthesis,” “translation,” “response to abiotic stimulus,” and “cellular homeostasis”) (fig. 3). We found that some PSGs involved in several protective mechanisms, such as photorespiration and antioxidant systems. Photorespiration is able to consume sufficient quantities of reducing equivalents to protect the photosynthetic apparatus, and antioxidant systems can protect cells from the toxic effects of ROS (Wise 1995; Allan and Fluhr 1997; Allen and Ort 2001; Yang et al. 2011). Moreover, evidence of positive selection were found in genes encoding ribosomal proteins, elongation factor-like proteins, and chaperonin family proteins associated with the protein synthesis process.
Fig. 3.
—GO enrichment results of the PSGs. The GO enrichment analyses were based on the 202 PSGs (corrected P < 0.05) identified in two Antarctic psychrophilic green algae. The GO terms with a significance level of 0.05 (corrected P < 0.05) were shown; P values were calculated and corrected by Benjamini–Hochberg approach; the size of the circles is proportional to the number of genes within the term; the color scale indicates the corrected P value corresponding to each term.
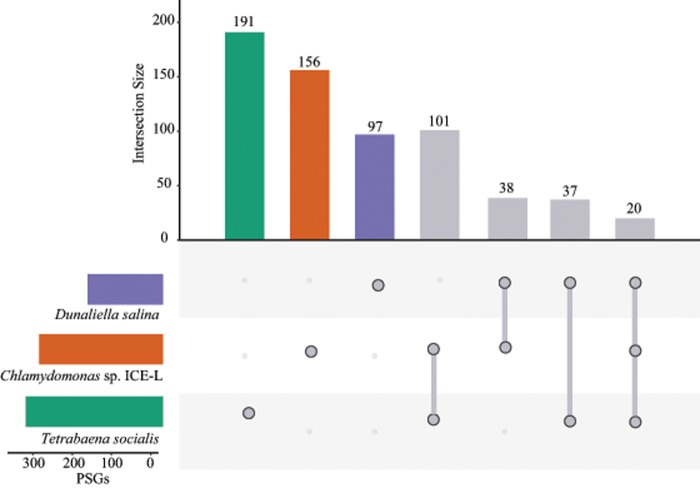
Comparative analyses were performed using 543 PSGs (P < 0.05) to uncover the broad evolutionary pattern of parallelism between psychrophilic and mesophilic algae. We found that 101 PSGs were specifically shared in the two psychrophilic algae, but the C. sp. ICE-L and T. socialis only separately shared 38 and 37 PSGs with D.salina (fig. 4). Significance of these gene intersections were tested by supertest, and only the number of observed gene intersection of two psychrophilic algae was significantly larger than expected (P < 0.01) (supplementary fig. S2, Supplementary Material online). These results provide hints that positive selection applies to similar set of genes in these two psychrophilic algae.
Fig. 4.
—Number and overlap of the PSGs in C. sp. ICE-L, Tetrabaena socialis, and Dunaliella salina. The specific PSGs in each alga are indicated by gray dots, and specific overlapping PSGs in respective sets are indicated by gray lines. Abbreviation: PSGs, positively selected genes.
Molecular Convergence in Antarctic Psychrophilic Algae
The 1,031 and 761 convergent amino acid substitutions were found in 493 genes of psychrophilic and 344 genes of mesophilic algae (table 2). The convergence level in psychrophilic algae is significantly higher than that of mesophilic algae (P = 1.69E-10 by χ2 test). The number of genes with convergent sites specifically in psychrophilic algae (493 genes) is larger than that of mesophilic algae (344 genes). For genes with two or more convergent sites, the psychrophilic/mesophilic ratio (convergence signal) is 1.77 (P = 3.63E-4, by the χ2 test), which may be driven by molecular convergence (table 2).
Table 2.
Results of Convergence Analyses between Psychrophilic and Mesophilic Algae
| No. of Convergent Sites per Gene | Psychrophilic Algae | Mesophilic Algae | Psychrophilic/Mesophilic Ratio |
|---|---|---|---|
| No. of genes with convergent sites in both psychrophilic and mesophilic algae | |||
| 255 | 255 | 1.00 | |
| No. of genes with convergent sites in psychrophilic and mesophilic algae only | |||
| ≥1 (=1) | 493 (392) | 344 (287) | 1.43 (1.36) |
| ≥2 (=2) | 101 (70) | 57 (43) | 1.77 (1.63) |
| ≥3 (=3) | 31 (25) | 14 | 2.21 (—) |
| Total number of sites | 1,031 | 761 | |
The noisy signals of molecular convergence should be considered in the large-scale analyses. The noisy signals might come from a variety of sources, including the possibility of differentially sorted ancestral variation (Storz 2016), random changes in parallel without being driven by the same selective pressure (Xu et al. 2017) and technical difficulties (obtaining accurate nucleotide substitution rates and patterns) (Zou and Zhang 2015). In addition, the determination of convergence in psychrophilic algae should be regarded as a probability statement, owing to the presence of convergence signal in mesophilic algae. To filter noisy signals, the genes with two or more convergent sites were used in subsequent analyses, and 33 genes containing convergent sites in both psychrophilic and mesophilic algae are excluded. In order to alleviate the possible false positive detection, we used a stringent criterion that the convergent amino acid substitutions in PSGs are a conservative signature of adaptive evolution. As a result, 54 genes with conservative signature of adaptive evolution were identified (supplementary table S4, Supplementary Material online). GO enrichment analyses of the genes under adaptive evolution revealed 22 significantly enriched GO terms. The enriched terms were related to biosynthesis, metabolic and stress response processes (“biosynthetic process,” “lipid/protein metabolic process,” “response to abiotic stimulus,” and “cellular homeostasis”) (supplementary fig. S3, Supplementary Material online), indicating that the adaptations to polar extreme environments is a complex process.
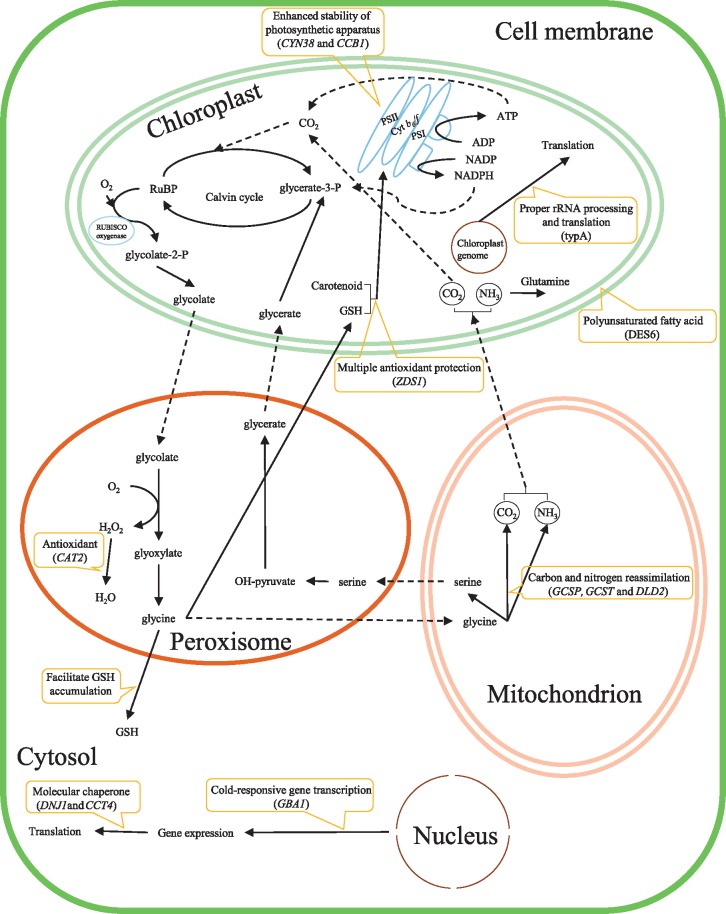
The maintenance of photostasis is essential for psychrophilic green algae ensuring adequate photosynthetic efficiency at cold condition in Antarctica (Huner et al. 2003; Ensminger et al. 2006). Several genes related to photosynthetic apparatus were identified to be under adaptive evolution, supporting that psychrophilic algae might evolve to maintain photostasis under polar extreme environments. The CYN38 gene was under adaptive evolution, and it encoded peptidyl-prolyl cis-trans isomerase CYP38 that can aid in the assembly and stabilization of PSII (fig. 5). Evidence of adaptive evolution was found in three genes (HemL, HemY, and CCB1) required for heme synthesis and accumulation of native cytochrome b6 in the thylakoid membrane (fig. 5). Cytochrome b6/f is an obligatory electron transfer and proton-translocating complex that can transfer electrons from PSII to PSI and pump protons into the thylakoid space contributing to create an electrochemical gradient. Moreover, the DES6 gene was evolving under adaptive evolution, and it encoded the chloroplast omega-6-fatty acids desaturase that introduces the second double bond in the biosynthesis of 16:3 and 18:3 fatty acids (fig. 5). It was reported that omega-6-fatty acids desaturase mutant could reduce levels of polyunsaturated fatty acids in the chloroplast galactolipids and slow the recovery rate of PSII complex at low temperature in Arabidopsis thaliana (Hugly and Somerville 1992; Vijayan and Browse 2002). There is a positive relationship between a higher level of polyunsaturated fatty acids and low temperature adaptation (Graham and Patterson 1982; Uemura and Steponkus 1994; Morgan-Kiss et al. 2002). Our evolutionary results revealed that maintaining the proper membrane fluidity is one adaptive strategy associated with adaptation to Antarctic cold environment in these two psychrophilic algae. Besides, the efficiency of the photosynthetic electron transport can be affected by low temperature resulting in changes in the redox state of the chloroplasts and in a rapid increase of free radicals and ROS, and high level of ROS attacks plant cells membrane (Baek and Skinner 2003; Plazek and Zur 2003). The signatures of adaptive evolution were identified in several genes related to antioxidant systems that can protect algae from oxidative stress, such as carotenoid, photorespiration and antioxidant enzymes. 1) The ZDS1 gene encoding zeta-carotene desaturase associated with carotenoids synthesis was under adaptive evolution. Carotenoids are crucial in photoprotection and stress respond process, it can quench the singlet oxygen and inhibit lipid peroxidation (Demmig-Adams and Adams 1996). 2) We found that several genes (GCSP, GCST, GCSL, and DLD2) associated with photorespiration were under adaptive evolution. Photorespiration is an important mechanism that can aid in the dissipation of excess light energy and protection of cells from photooxidative damage due to its characteristic of energy demanding and oxygen consuming. The A. thaliana mutant lacking photorespiration function showed hypersensitivity to abiotic stress, confirming that photorespiration is a key metabolic process required for efficient resistance to abiotic stress (Moreno et al. 2005). The photorespiratory glycine can facilitate the accumulation of glutathione that can also protect the photosynthetic components (Foyer et al. 1995; Noctor et al. 1999). These results suggest that photorespiration may be enhanced in Antarctic psychrophilic algae to minimize production of ROS at the chloroplast and mitigate oxidative damage induced by low temperature. 3) Evidence of adaptive evolution was also identified in an antioxidant defenses CAT2 gene, and it encodes Catalase 2 that can aid in the antioxidative response induced by cold stress in algae (Soitamo et al. 2008). It may serve to protect cells from the toxic effects of hydrogen peroxide during the enhanced photorespiration in psychrophilic algae. Furthermore, photorespiration shares enzymes and products with both the Calvin cycle and nitrate utilization, linking these major pathways of carbon and nitrogen assimilation. For example, photorespiration converts fixed carbon and nitrogen into CO2 and NH3, and these CO2 and NH3 can be reassimilated in the chloroplast (fig. 5) (Schuller and Randall 1989; Igamberdiev and Rodionova 1992). Together with these results, we deduced that the mechanisms involved in stabilization and protection of the photosynthetic machinery in psychrophilic algae are the crucial adaptation to Antarctic extreme environment.
Fig. 5.
—Summary of putative adaptive mechanisms in Antarctic psychrophilic algae. This scheme shows the principal cell components and pathways associated with cold adaptation. The solid arrows indicate the reactions; the dashed arrows represent the transport; and rounded orange rectangles denote the putative adaptive strategies.
Transcription and translation are temperature-sensitive steps, and it is important for the process of protein synthesis in psychrophilic green algae to adapt to low temperatures. The signatures of adaptive evolution were found in several genes (CSP41a, CPR, EIF3B, and EFG1) involved in rRNA processing and translation initiation, providing the clues for adaptive evolution associated with protein synthesis in the psychrophilic algae. It has been proved that ribosomes are essential for protein synthesis in all living cells and play a distinct role in plant cold tolerance (Zhang et al. 2016). Furthermore, we found that the GBA1 gene encoding GTP binding protein typA was under adaptive evolution, and the mutant with a deficient function can result in a pronounced chlorosis accompanying abnormal chloroplast rRNA processing and chloroplast protein accumulation under cold stress in A. thaliana (Liu et al. 2010). The PGH1 gene was under adaptive evolution, and it encoded a multifunctional enzyme (enolase) that acts a positive regulator of cold-responsive gene transcription in A. thaliana (fig. 5) (Lee et al. 2014). It was proved that the enolase abundance was associated with cold stress in C. reinhardtii (Valledor et al. 2013). Our analyses also present CCT4 and DNJ1 genes encoding T-complex protein delta subunit and chloroplastic chaperone DnaJ-like protein to be under adaptive evolution in Antarctic psychrophilic algae (fig. 5). Previous study has shown that DnaJ-like protein was localized to the soluble stroma fraction and thylakoids, and it can complement the temperature-sensitive phenotype of an Escherichia coli with DnaJ mutant (Willmund et al. 2008). Overall, our findings provide evidence of adaptive evolution in the translation elements and chaperones supporting that they aid in the protein synthesis during exposure to low temperature and freeze–thaw cycles in Antarctica algae.
In summary, multiple genes that account for adaptation to polar extreme environments were identified in the conjoint analyses of positive selection and convergent substitution. Functional analyses revealed that several adaptive modifications in photosynthesis, protein synthesis and stress related processes might underpin the adaptation to Antarctica extreme environment. The adaptations in psychrophilic algae present integral adaptive modifications and include multiple pathways involved in numerous cell components. It should be noted that our study only focused on detecting adaptive evolution in part of the coding regions of genomes. The adaptive mechanisms, such as expansion of the multiple copy genes, proliferation of noncoding regulatory regions, the gains and losses of genes, whole genome duplication, and horizontal gene transfer, will also offer considerable opportunities for future comprehensive research on the molecular adaptation of algae to polar environment.
Conclusion
Our analytical approach combining positive selection, convergent substitution, and functional enrichment analyses provides compelling evidence for molecular adaptations underpinning the evolution of the psychrophilic green algae in Antarctica. Our results revealed that positive selection applied to similar set of genes in these two psychrophilic green algae, indicating that their adaptation may arise via parallel molecular evolution. The adaptive evolution in two Antarctic psychrophilic algae is evident in several basic metabolic processes, implying that the adaptation to polar extreme environments is a complex process involved in several cellular components (CCs). The psychrophilic algae lived in polar habitats have evolved complex adaptive modifications in photosynthesis, including the enhanced stability of photosynthetic apparatus, increased membrane fluidity and multiple antioxidant strategies. The Antarctic psychrophilic algae also exhibit adaptations in protein synthesis (transcription regulator, protein translation initiation factor, and chaperone), which may aid in the cold response process and prevent the aggregation of unfolded proteins.
Data Availability
Unigenes and multiple sequence alignments used in this study are available in the FigShare repository: https://figshare.com/s/2f3a9ced53e87fd3cedf.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to Shaohua Xu and Suhua Shi for helpful discussion. The associate editor and anonymous reviewers provided constructive comments on the manuscript. This study was supported by the National Key Research and Development Program of China (2018YFD0900705 and 2018YFD0901103), the National Natural Science Foundation of China (31570219 and 41576187), the Jiangsu Province Key Project for Scientific Research (16KJA180002), Qing Lan Project, Young Elite Scientists Sponsorship Program and Six Talent Peaks Project of Jiangsu Province (16KJA180002), China Ocean Mineral Resources R&D Association (DY135-B2-14), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Data deposition: The sequencing reads have been deposited at Genome Sequence Archive (GSA; http://gsa.big.ac.cn/) under the BioProject accession PRJCA001235.
Literature Cited
- Allan AC, Fluhr R.. 1997. Two distinct sources of elicited reactive oxygen species in tobacco epidermal cells. Plant Cell 9(9):1559–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen DJ, Ort DR.. 2001. Impacts of chilling temperatures on photosynthesis in warm-climate plants. Trends Plant Sci. 6(1):36–42. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An M, Mou S, Zhang X, Ye N, Zheng Z.. 2013. Temperature regulates fatty acid desaturases at a transcriptional level and modulates the fatty acid profile in the Antarctic microalga Chlamydomonas sp. ICE-L. Bioresour Technol. 134:151–157. [DOI] [PubMed] [Google Scholar]
- Arrigo KR, Worthen DL, Lizotte MP, Dixon P, Dieckmann G.. 1997. Primary production in Antarctic sea ice. Science 276(5311):394–397. [DOI] [PubMed] [Google Scholar]
- Baek KH, Skinner DZ.. 2003. Alteration of antioxidant enzyme gene expression during cold acclimation of near-isogenic wheat lines. Plant Sci. 165(6):1221–1227. [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300. [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. [DOI] [PubMed] [Google Scholar]
- Davidson NM, Oshlack A.. 2014. Corset: enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 15:410.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WW.. 1996. The role of xanthophyll cycle carotenoids in the protection of photosynthesis. Trends Plant Sci. 1(1):21–26. [Google Scholar]
- Eddie B, Krembs C, Neuer S.. 2008. Characterization and growth response to temperature and salinity of psychrophilic, halotolerant Chlamydomonas sp. ARC isolated from Chukchi Sea ice. Mar Ecol Prog Ser. 354:107–117. [Google Scholar]
- Ensminger I, Busch FA, Huner NP.. 2006. Photostasis and cold acclimation: sensing low temperature through photosynthesis. Physiol Plant. 126(1):28–44. [Google Scholar]
- Escalona T, Weadick CJ, Antunes A.. 2017. Adaptive patterns of mitogenome evolution are associated with the loss of shell scutes in turtles. Mol Biol Evol. 34(10):2522–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherston J, et al. 2018. The 4-celled Tetrabaena socialis nuclear genome reveals the essential components for genetic control of cell number at the origin of multicellularity in the Volvocine lineage. Mol Biol Evol. 35(4):855–870. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. 2008. The Pfam protein families database. Nucleic Acids Res. 36(Database issue):D281–D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, et al. 1995. Overexpression of glutathione reductase but not glutathione synthetase leads to increases in antioxidant capacity and resistance to photoinhibition in poplar trees. Plant Physiol. 109(3):1047–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S, et al. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36(10):3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AD, Patterson BD.. 1982. Responses of plants to low, nonfreezing temperatures: proteins, metabolism, and acclimation. Annu Rev Plant Physiol. 33(1):347–372. [Google Scholar]
- Hugly S, Somerville C.. 1992. A role for membrane lipid polyunsaturation in chloroplast biogenesis at low temperature. Plant Physiol. 99(1):197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huner NPA, Öquist G, Melis A.. 2003. Photostasis in plants, green algae and cyanobacteria: the role of light harvesting antenna complexes. Dordrecht (the Netherlands: ): Springer. [Google Scholar]
- Huner NPA, Öquist G, Sarhan F.. 1998. Energy balance and acclimation to light and cold. Trends Plant Sci. 3(6):224–230. [Google Scholar]
- Igamberdiev AU, Rodionova MI.. 1992. Effect of glycolate pathway intermediates on succinate conversion in dark-incubated corn and wheat leaves. Soviet Plant Physiol. 39:87–92. [Google Scholar]
- Kang S, Fryxell GA.. 1992. Fragilariopsis cylindrus (Grunow) Krieger: the most abundant diatom in water column assemblages of Antarctic marginal ice-edge zones. Polar Biol. 12:609–627. [Google Scholar]
- Kato S. 1982. Laboratory culture and morphology of Colaclum vesiculosum Ehrb. (Euglenophyceae). Jpn J Phycol. 30:63–67. [Google Scholar]
- Lee H, et al. 2014. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J. 21(11):2692–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux C, Vincent AT, Labarre A, Otis C, Turmel M.. 2015. Chloroplast phylogenomic analysis of chlorophyte green algae identifies a novel lineage sister to the Sphaeropleales (Chlorophyceae). BMC Evol Biol. 15:264.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H.. 2014. UpSet: visualization of intersecting sets. IEEE Trans Vis Comput Graph. 20(12):1983–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Huang X, Wang X, Zhang X, Li G.. 2006. Phylogenetic studies on two strains of Antarctic ice algae based on morphological and molecular characteristics. Phycologia 45(2):190–198. [Google Scholar]
- Liu S, et al. 2010. Molecular cloning and expression analysis of a cytosolic Hsp70 gene from Antarctic ice algae Chlamydomonas sp. ICE-L. Extremophiles 14(3):329–337. [DOI] [PubMed] [Google Scholar]
- Liu X, Rodermel SR, Yu F.. 2010. A var2 leaf variegation suppressor locus, SUPPRESSOR OF VARIEGATION3, encodes a putative chloroplast translation elongation factor that is important for chloroplast development in the cold. BMC Plant Biol. 10:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N.. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A. 102(30):10557–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Cai T, Olyarchuk JG, Wei L.. 2005. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21(19):3787–3793. [DOI] [PubMed] [Google Scholar]
- Maxwell DP, Falk S, Huner N.. 1995. Photosystem II excitation pressure and development of resistance to photoinhibition (I. Light-harvesting complex II abundance and zeaxanthin content in Chlorella vulgaris). Plant Physiol. 107(3):687.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318(5848):245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock T, et al. 2017. Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature 541(7638):536. [DOI] [PubMed] [Google Scholar]
- Moreno JI, Martín R, Castresana C.. 2005. Arabidopsis SHMT1, a serine hydroxymethyltransferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. Plant J. 41(3):451–463. [DOI] [PubMed] [Google Scholar]
- Morgan-Kiss R, Ivanov AG, Williams J, Mobashsher K, Huner NP.. 2002. Differential thermal effects on the energy distribution between photosystem II and photosystem I in thylakoid membranes of a psychrophilic and a mesophilic alga. Biochim Biophys Acta. 1561:251–265. [DOI] [PubMed] [Google Scholar]
- Morgan-Kiss RM, Priscu JC, Pocock T, Gudynaite-Savitch L, Huner NPA.. 2006. Adaptation and acclimation of photosynthetic microorganisms to permanently cold environments. Microbiol Mol Biol Rev. 70(1):222–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M.. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35(Web Server):W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S.. 2000. Molecular evolution and phylogenetics. New York: Oxford University Press. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Arisi AM, Jouanin L, Foyer CH.. 1999. Photorespiratory glycine enhances glutathione accumulation in both the chloroplastic and cytosolic compartments. J Exp Bot. 50(336):1157–1167. [Google Scholar]
- Nozaki H, Misumi O, Kuroiwa T.. 2003. Phylogeny of the quadriflagellate Volvocales (Chlorophyceae) based on chloroplast multigene sequences. Mol Phylogenet Evol. 29(1):58–66. [DOI] [PubMed] [Google Scholar]
- Petersen M, et al. 2017. Orthograph: a versatile tool for mapping coding nucleotide sequences to clusters of orthologous genes. BMC Bioinformatics 18(1):111.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plazek A, Zur I.. 2003. Cold-induced plant resistance to necrotrophic pathogens and antioxidant enzyme activities and cell membrane permeability. Plant Sci. 164(6):1019–1028. [Google Scholar]
- Polle J, Barry K, Cushman J, Schmutz J, Tran D.. 2017. Draft nuclear genome sequence of the halophilic and beta-carotene-accumulating green alga Dunaliella salina strain CCAP19/18. Genome Announc. 5(43):e01105-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik SE, et al. 2010. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329(5988):223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provasoli L. 1966. Media and prospects for the cultivation of marine algae. In: Watanabe A, Hattori R, editors. Culture and collections of algae. Proc U.S.–Japan Conference Hakone. p. 63–75. Japanese Society of Plant Physiology, Tokyo.
- Provasoli L, Pintner IJ.. 1960. Artificial media for freshwater algae: problems and suggestions In: Tryon CA, Hartman RT, editors. The ecology of algae. Pittsburg: Pymatuning Lab Field Biology Univ Pittsburg; p. 84–96. [Google Scholar]
- Qiao Q, et al. 2016. The genome and transcriptome of Trichormus sp. NMC-1: insights into adaptation to extreme environments on the Qinghai-Tibet Plateau. Sci Rep. 6:29404.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller KA, Randall DD.. 1989. Regulation of pea mitochondrial pyruvate dehydrogenase complex: does photorespiratory ammonium influence mitochondrial carbon metabolism? Plant Physiol. 89(4):1207–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Soitamo AJ, Piippo M, Allahverdiyeva Y, Battchikova N, Aro EM.. 2008. Light has a specific role in modulating Arabidopsis gene expression at low temperature. BMC Plant Biol. 8:13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R.. 2003. Statistical significance for genome wide studies. Proc Natl Acad Sci U S A. 100(16):9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey KB, Storey JM.. 2013. Molecular biology of freezing tolerance. Compr Physiol. 3(3):1283–1308. [DOI] [PubMed] [Google Scholar]
- Storz JF. 2016. Causes of molecular convergence and parallelism in protein evolution. Nat Rev Genet. 17(4):239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DN, Dieckmann GS.. 2002. Antarctic sea ice: a habitat for extremophiles. Science 295(5555):641–644. [DOI] [PubMed] [Google Scholar]
- Uemura M, Steponkus PL.. 1994. A contrast of the plasma membrane lipid composition of oat and rye leaves in relation to freezing tolerance. Plant Physiol. 104(2):479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valledor L, Furuhashi T, Hanak AM, Weckwerth W.. 2013. Systemic cold stress adaptation of Chlamydomonas reinhardtii. Mol Cell Proteomics 12(8):2032–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan P, Browse J.. 2002. Photoinhibition in mutants of Arabidopsis deficient in thylakoid unsaturation. Plant Physiol. 129(2):876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zhao Y, Zhang B.. 2015. Efficient test and visualization of multi-set intersections. Sci Rep. 5:16923.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, et al. 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci U S A. 111(45):E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmund F, Dorn KV, Schulz-Raffelt M, Schroda M.. 2008. The chloroplast DnaJ homolog CDJ1 of Chlamydomonas reinhardtii is part of a multichaperone complex containing HSP70B, CGE1, and HSP90C. Plant Physiol. 148(4):2070–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RR. 1995. Chilling-enhanced photooxidation: the production, action and study of reactive oxygen species produced during chilling in the light. Photosyn Res. 45(2):79–97. [DOI] [PubMed] [Google Scholar]
- Xu S, et al. 2017. Genome-wide convergence during evolution of mangroves from woody plants. Mol Biol Evol. 34(4):1008–1015. [DOI] [PubMed] [Google Scholar]
- Yang H, et al. 2011. Profiling of the transcriptome of Porphyra yezoensis with Solexa sequencing technology. Chin Sci Bull. 56(20):2119–2130. [Google Scholar]
- Yang L, Wang Y, Zhang Z, He S.. 2015. Comprehensive transcriptome analysis reveals accelerated genic evolution in a Tibet fish, Gymnodiptychus pachycheilus. Genome Biol Evol. 7(1):251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang C, Sayyari E, Mirarab S.. 2017. ASTRAL-III: increased scalability and impacts of contracting low support branches In: Meidanis J, Nakhleh L, editors. Comparative genomics. RECOMB-CG. Lecture notes in computer science. Vol. 10562 Cham (Switzerland: ): Springer. [Google Scholar]
- Zhang J, Nielsen R, Yang Z.. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22(12):2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhang J, et al. 2016. Plastid ribosomal protein S5 is involved in photosynthesis, plant development, and cold stress tolerance in Arabidopsis. J Exp Bot. 67(9):2731–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, et al. 2018. The Antarctic sea ice alga Chlamydomonas sp. ICE-L provides insights into adaptive patterns of chloroplast evolution. BMC Plant Biol. 18(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Zhang J.. 2015. Are convergent and parallel amino acid substitutions in protein evolution more prevalent than neutral expectations? Mol Biol Evol. 32(8):2085–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Unigenes and multiple sequence alignments used in this study are available in the FigShare repository: https://figshare.com/s/2f3a9ced53e87fd3cedf.