Abstract
Introduction
In Japan, there are approximately 300 projects conducting research on rare diseases supported by the Ministry of Health, Labour and Welfare of Japan (MHLW) and the Japan Agency for Medical Research and Development (AMED). Diverse data, including clinical, genomic, and sample‐related data, are generated by these projects. However, at present, such data are managed individually by each project. This makes it difficult for third parties to ascertain the data generated by projects.
Methods
Again this background, at the beginning of 2017, the AMED started the National Platform for Rare Diseases Data Registry of Japan (RADDAR‐J), whose mission is to construct a cross‐sectional data integration platform incorporating projects supported by the AMED and MHLW. RADDAR‐J promotes data sharing by the projects in accordance with the data‐sharing policy established by the AMED, which classifies data sharing into three categories based on the strategies used to protect the rights of researchers while promoting data sharing. RADDAR‐J integrates and analyzes data shared by each project to add value to the resources and promote secondary use by third parties while protecting the rights of the researchers who shared their data. The platform is designed to provide incentives to projects that shared their data by supporting registry construction or genomic analysis to promote data sharing. RADDAR‐J also has the function of data identification to securely integrate data originating from the same person. RADDAR‐J accelerates clinical research by encouraging each project to utilize a central ethics committee.
Results/Conclusion
The use of the platform by projects is expected to lead to streamlined data collection, improved quality assurance, improved access to data, and promotion of joint research and the secondary use of shared data. These benefits will accelerate research into diagnosis and treatment technologies and will hopefully lead to improved quality of life for patients with rare diseases.
Keywords: clinical information, data integration platform, data sharing, genomic information, rare disease, registry
1. INTRODUCTION
Japan has a long history of researching rare diseases. The Ministry of Health, Labour and Welfare (MHLW) of Japan has played an essential role in supporting research on rare diseases for about four decades. Approximately 100 projects targeting rare diseases are involved in the Research Project on Rare/Intractable Disease organized by the MHLW every year, whose mission is to establish diagnostic criteria and clinical guidelines, perform cohort studies, promote public awareness, and support medical practice in this field.
The Japan Agency for Medical Research and Development (AMED), initiated in 2015 as a funding agency, has been committed to supporting research in this area as one of the nine major prioritized fields.1 In addition to research supported by the MHLW, the approximately 200 projects presently under the Practical Research Project for Rare/Intractable Diseases of the AMED2 run the gamut from basic research to clinical trials, genomic studies, and to those contributing to the development of patient registries and biorepositories in this specific field.
Among the long‐desired research infrastructure is a platform enabling data sharing, data integration, and secondary use under a predetermined policy. All data obtained by individual projects are indispensable to current and future research; especially in rare diseases, this need must be met to address the scarcity of patient populations. Therefore, the AMED launched a project in February 2017 to establish a national platform named the “National platform for Rare Diseases Data Registry of Japan (RADDAR‐J),3” whose aim is to construct an integrated data platform for projects conducting research on rare diseases managed by the AMED and MHLW.
This paper reviews the concept and missions of this project and the overall structure of the platform.
2. DATA SHARING
The AMED established the data sharing policy for facilitation of genomic medicine4 and is promoting data sharing. The platform has formulated a data‐sharing guideline in line with this policy and conducts appropriate data sharing with third parties.
The data classification and the content of the data sharing are as follows (see also Figure 1).
-
Group sharing data
Data are only shared on the platform with the projects that shared their data. This type of sharing is the basic form of data sharing on the platform. When projects share their data in this form with the platform, they can receive a variety of types of support from RADDAR‐J.
Such data can be shared with third parties upon request only with the consent of the data‐sharing project.
-
Controlled‐access data
Data for which provision to third parties has been approved in advance by the data‐sharing project will be shared who request access based on a review and the subsequent approval by the platform.
-
Open data
Data for which publication has been permitted in advance by the data‐sharing project will be published on the platform as data available for viewing and use by anyone. Target information includes topics by research3 and statistical value (prevalence, etc.) and by disease (group). Data on causative gene mutation in Mendelian genetic diseases will be published on MGeND.5
Figure 1.
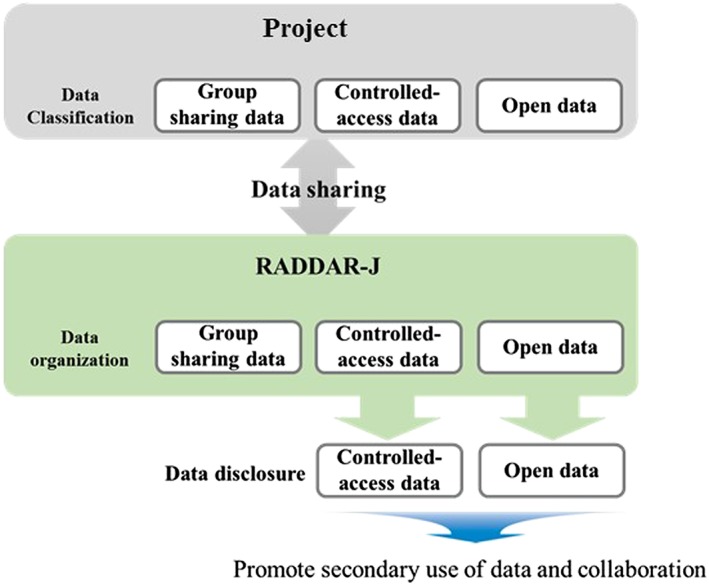
Classification of data sharing on Rare Diseases Data Registry of Japan (RADDAR‐J)
3. WHAT IS RADDAR‐J?
The mission of RADDAR‐J is to construct a cross‐sectional data integration platform for projects funded by the AMED and MHLW. This involves the construction of a data‐sharing platform among projects. This will also promote the secondary use of data for other research.
The platform is comprised of three units (Figure 2). An outline of each unit is provided below.
Clinical information integration unit
Figure 2.
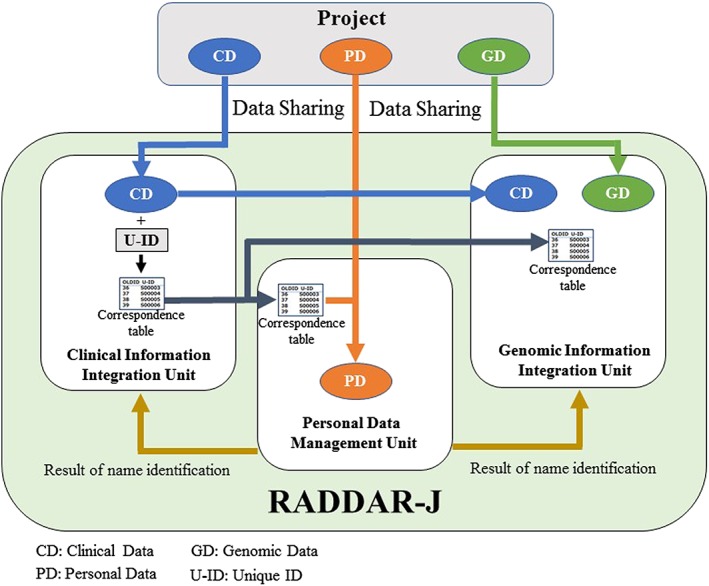
Overview and data flow of Rare Diseases Data Registry of Japan (RADDAR‐J)
This unit collects and stores clinical data and biological sample‐related data originating from patients with specific diagnosis and shared by each project. It then integrates them for advanced analysis, contributing back to the data‐sharing projects. Controlled‐access data will be shared with a public database. Open data will be made available on a RADDAR‐J website, listed by research topic.3 This is expected to promote secondary use of the data and joint research. Another mission of this unit is to support projects to construct and manage patient registries (see Section 7. Incentives for data sharing).
-
2.
Genomic information integration unit
The projects conducting genomic studies are handling a large amount of sequencing data on specific rare diseases. The mission of this unit is to collect and store genomic data shared by projects and integrate them for advanced analysis, contributing back to the data‐sharing projects. Controlled‐access data will be shared with a public database. Open data will be made available on a RADDAR‐J website, listed by research topic3 or on other public database, eg, Medical Genomics Japan Variant Database (MGeND),5 which is constructed by the Program for an Integrated Database of Clinical and Genomic Information6 of the AMED. Another mission of this unit is to support projects by suppling genetic analysis tools and genomic control information that can be used for analysis (see Section 7. Incentives for data sharing).
-
3.
Personal information management unit
In epidemiological research in Japan, it has been required that researchers de‐identify the collected information from patients and the information which is necessary for reidentification such as patient names is stored strictly and separately from other information by the persons in charge of personal information management. So‐called “name‐based aggregation” by collating the privacy information of the patients such as real name, gender, and date of birth will be necessary for (1) gathering patient information from multiple researches and (2) collecting additional information about the patients of the research from other databases, which is because there is no national identification number system available for medical research in Japan.
In order to achieve such purposes, the primary role of this unit is to control and manage the privacy information of the patients such as name, gender, date of birth, and to perform name‐based aggregation as necessary.
4. PERSONAL INFORMATION MANAGEMENT SYSTEM
Based on the role of personal information management unit described above, a personal information management system has been designed and developed for our platform, which stores patients' privacy information for personal identification in encrypted form and can perform name‐based aggregation of the patients without decryption.
4.1. Primary functions of the system
Real name entry function
It encrypts and stores patients' personal information such as name, gender, date of birth, address, and telephone number in the database.
-
2.
Name‐based aggregation of patients
By comparing phonetic spellings of names, genders, and dates of birth, it extracts identifiers of “possibly identical patients.” The results will be provided to the personal information management officer and shared with the clinical information integration unit and the genomic information integration unit.
4.2. Database encryption
An original searchable encryption technology has been developed and is used in the database of the personal information management system. The special characteristics of the system are as follows:
Search without decryption
Encrypted data to be stored in the database are limited to text data, and the data are encrypted character‐by‐character so that partial match retrieval can be performed without decryption.
-
2.
Blocking unauthorized decryption
Encrypted personal information is combined with dummy information when stored in the database.
Encrypted personal information is stored dispersively in multiple tables.
The personal information table stores not the personal information itself but the links to the information in encrypted form.
Encryption keys will be generated based on device‐specific information of the database server. Thus, even if the database were copied into a different environment, decryption would prove to be difficult.
-
3.
Security countermeasures
Login into the personal information management system requires, in addition to normal password authentication, client authentication via a client certificate.
A reverse proxy server protects the personal information management system against direct access from the outside.
5. DATA FLOW AND INTERACTION BETWEEN EACH UNIT (Figure 2)
5.1. Clinical information data sharing
Clinical information collected from each project is shared, in addition to data element information, with the clinical information integration unit in accordance with sharing permissions (group sharing data, controlled‐access data, open data) defined by the project themselves. In the clinical information integration unit, based on provided data element information, data element organization and definition of data structures are carried out. Afterwards, data is stored in the clinical information integration system. In the clinical information integration system, unique IDs are affixed to data based on the cases involved, and correspondence tables created using these IDs are shared with the genomic information integration unit and the personal information management unit.
5.2. Genomic information data sharing
Genomic information such as genome Variant Call File (gVCF) handled by projects is shared directly with the genomic information integration unit. Clinical information accompanying with genomic information will be shared with the clinical information integration unit first, and a unique IDs are affixed to it. Afterwards, correspondence tables and necessary clinical information (gender, date of birth, diagnosis, hereditary information) will be shared from the clinical information integration unit to the genomic information integration unit; then genomic information is combined with accompanied clinical information based on the correspondence tables.
5.3. Personal information data sharing
With regard to projects that possess personal information, such information is partitioned off from clinical information by the projects themselves. Next, clinical information is shared with the clinical information integration unit, and a unique IDs are affixed to it. Projects fill out correspondence tables generated using these unique IDs with the relevant personal information. That list then is shared with the personal information management unit.
The personal information management unit will use personal information provided by projects to collate records. Collated records then are shared with the clinical information integration unit and the genomic information integration unit. Finally, collated, organized records are provided back to the projects that initially collected it.
6. ETHICAL MEASURES FOR DATA SHARING
Data sharing with this platform requires that informed consent has been appropriately obtained from the registrant regarding sharing the data with the platform and secondary use of the data.
Template documents outlining the ethical items required for data sharing have been prepared on the platform for each project (Table 1). Modification guide documents have also been prepared describing the required items to be added to existing documents when research is already underway.
Table 1.
Standard documents supplied by Rare Diseases Data Registry of Japan
| ① Protocol |
| ② Written consent form |
| ③ Database construction definitions |
| ④ Database construction definition guideline |
| ⑤ Case report form |
| ⑥ Statistical analysis plan |
| ⑦ Examples of form templates |
| ⑧ Standard operating procedures for implementing research |
| ⑨ Version management procedures for research‐related documents |
| ⑩ Standard operating procedures for data management |
| ⑪ Standard operating procedures for system construction and management |
| ⑫ Standard operating procedures for monitoring and auditing procedures |
| ⑬ Manual for collection of samples and data |
| ⑭ Manual for data input to electric data capture |
7. INCENTIVES FOR DATA SHARING
The platform is designed to give some incentives to each project to promote data sharing.
7.1. Registry construction and operation support
The following is provided on the platform to support registry construction and operation for each project:
Standard documents
Standard data registry system
“Registry” on the platform refers to “research aiming to discover new findings by collecting and analyzing data on patients with rare diseases.”
-
Supply of standard documents
The standard documents supplied by the platform are template documents compiled based on considerations of the ethical items required for implementing research (Table 1). The specifications can be adapted to suit the content of research with a variety of patterns.
The documents contain ethical items in line with the Ethical Guidelines for Medical and Health Research Involving Human Subjects,7 Ethical Guidelines for Human Genome/Gene Analysis Research,8 and the Revised Personal Information Protection Act9; information on methods for collecting personal data; and information required for sharing data with the platform.
-
Provision of standard data registry system
A standard data registry system is provided on the platform to ensure efficient registry data input as well as data quality and security.
7.1.1. Characteristics of the standard data registry system provided by the platform
By providing the user with an environment that enables them to connect to the Internet, they can input case data without installing computer applications.
It is also possible to store and manage personal information. Personal and other information will be stored in quarantine in separate databases, and the independent storage of each database is assured.
Data collection elements can be customized for each individual project.
The system satisfies the conditions of using electromagnetic records and electronic signatures for application for approval or licensing of drugs (ER/ES guideline).10
The database server that stores the input data is secure and highly robust.
The system is equipped with a system to backup data to remote servers as a countermeasure against disaster and a recovery measure.
7.2. Genetic analysis support for research
The following genetic analysis services are provided by the platform to support genetic analysis for each project:
Provision of population control
Provision of a standard complete genome sequence using DNA samples selected by considering the genetic diversity of the Japanese population, which is essential for searching disease‐related genes using complete genome sequence (initially, there will be 3000 samples, which will be increased as the platform project progresses). Implementation of a joint call with the above reference sequence to improve the accuracy of complete data on patients with rare diseases in each project.
-
2.
Provision of a bioinformatics pipeline
Provision of a bioinformatics pipeline that enables rapid and very accurate extraction of a candidate's disease‐related genetic mutations from a patient's complete genome sequence.
8. ACTIVITIES OF THE CENTRAL ETHICS COMMITTEE
The AMED recommends the use of the Central Ethics Committee with the aim of streamlining ethical reviews and ensuring the uniform quality of the reviews.11
The standard documents for the protocol and written consent form provided by the platform (Table 1) have been approved by the Kyoto University Graduate School and Faculty of Medicine, Ethics Committee,12 which is accredited as the Central Ethics Committee. Therefore, each project that makes use of the Ethics Committee of Kyoto University can expect a smooth review process.
When utilizing the Ethics Committee of Kyoto University, the exchange of documents will be conducted by the representative research facility (main medical institution). The institutions engaged in joint research (subordinate institutions) only need to submit their Ethics Review Request Form to the research representative.
9. DISCUSSION
RADDAR‐J was started in February 2017 as a project to develop an information integration infrastructure for activities of the Practical Research Project for Rare/Intractable Diseases,2 which was run by the AMED, and the Research Project on Rare/Intractable Disease, which was run by the MHLW. As each project uses the platform, it is expected to lead to more efficient information gathering, quality assurance, improved information access, and the promotion of joint research and cooperation.
9.1. Data sharing
In Japan, the “5th Science and Technology Basic Plan” (a cabinet decision13 made on January 22, 2016) aimed at cooperation beyond researchers' affiliated organizations, expert fields, and national borders to accelerate the creation of knowledge and new values, through the promotion of open science and wide use of research results by diverse users. In addition, it is important to acknowledge the difference in the methods used in the storage and sharing of research data between research fields and be aware of ensuring intellectual properties and strategies to balance data sharing by protecting the rights of researchers while promoting data sharing in a way that considers national interests.
Against this background, the AMED established a data‐sharing policy for genomic information.4 A characteristic of this policy is that data sharing is classified into three types. Specifically, by creating classifications of group sharing data, controlled‐access data, and open data, it aims to protect researchers who provide data and information and to promote research in related areas.
According to the policy, RADDAR‐J promotes data sharing. In the classification of data sharing, RADDAR‐J uses group sharing as a base. Group sharing data is fundamentally sharing among each project and RADDAR‐J; the judgment on the secondary use of shared data can be controlled by the data sharing project. Using group sharing as the base lowers the physical and emotional hurdles for researchers and ultimately promotes data sharing with RADDAR‐J.
To promote data sharing, it is important to consider incentives for projects who share their data.14 RADDAR‐J supports registry development and genetic analysis. These are incentives to projects; improved research quality and data standardization are expected through such support. Ultimately, the quality of data provided to RADDAR‐J can be ensured, and data integration will become possible.
9.2. Registry development operation support
In the field of rare diseases, disease registries are used in various fields, such as epidemiology, natural history, and drug discovery; their importance is also increasing.15 Improving the quality of registries is important not only for a specific study but also for other studies that use knowledge gained from such research. However, it is not easy for all researchers to develop a high‐quality registry. In particular, the use of a registration system that ensures safety and quality is important for registry development, but the introduction of such a system incurs cost. Presently, the burden on researchers is high.
RADDAR‐J provides standard documents and a standardized system to support registry development and operation. With such support, projects can use documents and systems with guaranteed quality for their studies; in this way, sharing of high‐quality data with RADDAR‐J is expected. In addition, with such support, the cost burden on projects, as well as the required effort, can be minimized, allowing researchers to focus on the promotion of the original study.
9.3. Ethical response
In Japan, the Law on the Protection of Personal Information was revised on May 30, 2017.9 The characteristics of this revision were that “individual identification codes,” such as physical characteristics, were also defined as personal information, and personal information requiring special care was newly defined. Certain types of genomic information are handled by RADDAR‐J with individual identification codes. Clinical information is also considered to be personal information requiring special care. Thus, both types of information need appropriate handling as personal information. In addition, with this protection law, the provision of personal information to third parties requires informed consent of the said person in principle.
As such, to share data with third parties, it is necessary that each obtains the appropriate consent. RADDAR‐J provides consent explanation documents and a consent template to each project. In this manner, data with consent obtained using unified contents are shared with RADDAR‐J. As a result, data can have ethical consideration secured for secondary use.
9.4. Reidentification
In a typical study in Japan, each study deidentifies and uses IDs for their subjects; thus, whether data obtained from another study are from the same patient cannot be determined. In cases of patients with rare diseases, they sometimes happen to be registered with multiple projects at the same time or at different times. When performing a cross‐sectional analysis or a unification of study resources, such as registry, reidentification is necessary.16 In previous cases, reidentification was performed by using unique IDs.17 In Japan, there is the My Number System,18 in which each citizen is provided with a unique number. However, it has not been used for research. RADDAR‐J established a personal information management unit to manage the information provided by each project independently, which can directly identify an individual in a safe environment, and perform reidentification. By using this function, unification and cross‐sectional analysis of data obtained from different projects has become possible.
9.5. International collaboration
RADDAR‐J has been promoting collaboration with the Europe‐centered rare diseases platform RD = Connect19 with regard to mutual information exchange and sharing of genomic analysis tools. Many other similar undertakings on rare diseases exist around the world. In order to encourage research on rare diseases, we must promote further international collaboration through mutual sharing of information and translation of information on the platform into mutually understood languages.
10. CONCLUSIONS
RADDAR‐J is a project that has started with a new concept. It aims to share data, integrate analysis of the gathered data, promote secondary use, and develop studies of rare diseases.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
RADDAR‐J RESEARCH AND DEVELOPMENT GROUP
Ryo Yamada2, Yasuharu Tabara2, Yoichiro Kamatani2, Syuji Kawaguchi2, Shinji Kosugi5, Koichiro Higasa6, Yoichi Matsubara7, Naomichi Matsumoto8, Yoshihiro Aasano9, Ichizo Nishino10, Harumasa Nakamura11, Atsushi Takano12, Motoi Ito4, Shinya Sakai4
5 Department of Medical Ethics and Medical Genetics, Kyoto University Graduate School of Medicine, Kyoto, Japan
6 Human Disease Genomics, Center for Genomic Medicine, Kyoto University Graduate School of Medicine, Kyoto, Japan
7 National Center for Child Health and Development, Tokyo, Japan
8 Department of Human Genetics, Yokohama City University Graduate School of Medicine, Yokohama, Japan
9 Department of Cardiovascular Medicine, Osaka University Graduate School of Medicine, Osaka, Japan
10 Department of Neuromuscular Research, National Institute of Neuroscience, National Center of Neurology and Psychiatry, Tokyo, Japan
11 Translational Medical Center, National Center of Neurology and Psychiatry, Tokyo, Japan
12 Healthcare & Life Sciences, Public and Communication Sector, Global Business Services, IBM Japan
ACKNOWLEDGMENTS
This study was supported by the Japan Agency for Medical Research and Development (AMED) under Grant Number JP18ek0109348.
Furusawa Y, Yamaguchi I, Yagishita N, et al. National platform for Rare Diseases Data Registry of Japan. Learn Health Sys. 2019;3:e10080 10.1002/lrh2.10080
REFERENCES
- 1. Japan Agency for Medical Research and Development. Available from: https://www.amed.go.jp/en/
- 2. Practical research project for rare/intractable diseases of the AMED. Available from: https://www.amed.go.jp/en/program/list/01/05/001.html
- 3. Rare Disease Data Registry of Japan. Available from: https://www.raddarj.org/en/
- 4. Data Sharing Policy for the Realization of Genomic Medicine. Available from: https://www.amed.go.jp/content/000017356.pdf
- 5. Medical genomic Japan variant database. Available from: https://mgend.med.kyoto‐u.ac.jp/
- 6. Program for an integrated database of clinical and genomic information. Available from: https://www.amed.go.jp/en/program/list/04/01/006.html
- 7. Ethical guidelines for medical and health research involving human subjects. Available from: http://www.mhlw.go.jp/english/wp/wp‐hw10/dl/13e.pdf [DOI] [PubMed]
- 8. Ethical guidelines for human genome/gene analysis research. Available from: http://www.mhlw.go.jp/english/wp/wp‐hw10/dl/13e.pdf
- 9. Revised Personal Information Protection Act. Available from: https://www.ppc.go.jp/en/
- 10. Japanese ERES Guidelines. Available from: http://ecompliance.co.jp/english/Japanese%20ERES%20Guideline.html
- 11. Project for development of central institutional review board. Available from: https://www.amed.go.jp/en/program/list/05/01/010.html
- 12. Kyoto University Graduate School and Faculty of Medicine, Ethics Committee . Available from: http://www.ec.med.kyoto‐u.ac.jp/
- 13. 5th Science and Technology Basic Plan. Available from: http://www.mext.go.jp/en/policy/science_technology/lawandplan/title01/detail01/1375311.htm
- 14. Thompson R, Johnston L, Taruscio D, et al. RD‐connect: an integrated platform connecting databases, registries, biobanks and clinical bioinformatics for rare disease research. J Gen Intern Med. 2014;29(Supple 3):780‐787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kodra Y, Posada PM, Coi A, et al. Data quality in rare diseases registries. Adv Exp Med Biol. 2017;1031:149‐164. [DOI] [PubMed] [Google Scholar]
- 16. Hansson MG, Lochmüller H, Riess O, et al. The risk of re‐identification versus the need to identify individuals in rare disease research. Eur J Hum Genet. 2016;24(11):1553‐1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mohammed EA, Slack JC, Naugler CT. Generating unique IDs from patient identification data using security models. J Pathol Inform. 2016;7:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. My Number System. Available from: http://www.soumu.go.jp/main_content/000510925.pdf
- 19. RD‐Connect. Available from: https://rd‐connect.eu/


