Abstract
Approximately 40% of patients with stage I-III triple-negative breast cancer (TNBC) recur after standard treatment, while the remaining 60% experience long-term disease-free survival (DFS). There are currently no clinical tests to assess the risk of recurrence in TNBC patients. We previously determined that TNBC patients with major histocompatibility complex class II (MHCII) pathway expression in their tumors experienced significantly longer DFS. To translate this discovery into a clinical test, we developed an MHCII Immune Activation assay, which measures expression of 36 genes using NanoString technology. Preanalytical testing confirmed that the assay is accurate and reproducible in formalin-fixed paraffin-embedded (FFPE) tumor specimens. The assay measurements were concordant with RNA-seq, MHCII protein expression, and tumor infiltrating lymphocyte counts. In a training set of 44 primary TNBC tumors, the MHCII Immune Activation Score was significantly associated with longer DFS (HR = 0.17, P = 0.015). In an independent validation cohort of 56 primary FFPE TNBC tumors, the Immune Activation Score was significantly associated with longer DFS (HR = 0.19; P = 0.011) independent of clinical stage. An Immune Activation Score threshold for identifying patients with very low risk of relapse in the training set provided 100% specificity in the validation cohort. The assay format enables adoption as a standardized clinical prognostic test for identifying TNBC patients with a low risk of recurrence. Correlative data support future studies to determine if the assay can identify patients in whom chemotherapy can be safely de-escalated and patients likely to respond to immunotherapy.
Keywords: Biomarker, prognosis, breast cancer, gene expression, immunology
Precis:
The MHCII Immune Activation assay identifies TNBC patients with a low risk of recurrence, addressing a critical need for prognostic biomarker tests that enable precision medicine for TNBC patients.
Introduction
Triple negative breast cancer (TNBC) is a clinical subtype of invasive breast cancer that is defined by the absence of standard markers used for prognosis and treatment decisions (Estrogen receptor (ER), Progesterone receptor (PR), and HER2). TNBC is notable for its aggressive behavior and high rates of local and distant recurrence (1). TNBC patients are treated with local therapy and cytotoxic chemotherapy. Patient outcomes are disparate. Approximately 42% of patients experience rapid relapses with a peak at three years from diagnosis, while the remaining 58% of patients have long-term disease-free survival (DFS) (2). Physicians cannot currently predict which patients will relapse, even after intensive chemotherapy, and which patients will have long-term disease-free survival and might do equally well with de-escalation of their chemotherapy regimen. Currently, most TNBC patients are treated with aggressive chemotherapy, which can result in serious long-term toxicity including permanent peripheral neuropathy, cardiac toxicity, and secondary malignancies (3–8). A current goal of the TNBC biomarker field is to develop clinical tools that can be used to identify patients who do not require aggressive treatment and can be spared the associated toxicities.
We recently reported that expression of the major histocompatibility complex Class II antigen presentation pathway (MHCII) in TNBC tumor cells is significantly associated with long-term DFS (9). Further, high MHCII expression in tumor cells was associated with the presence of tumor-infiltrating lymphocytes (TILs) (9), which are known to be associated with good prognosis in patients with TNBC (10–14). An independent research team performed IHC on 681 TNBC patient tumors and confirmed that high expression of MHCII in tumor cells was associated with large amounts of tumor infiltrating CD4 and CD8 positive T cells, and longer disease-free survival (15). Mouse studies have shown that MHCII expression on tumor cells triggers T cell recruitment and inhibits tumor progression (16–23). A standardized method for the morphologic evaluation of TILs in patient tumor samples has been developed, but has not entered routine clinical practice (24,25). Although promising, broad clinical implementation of this method may be limited by pathologist training, inter-observer variability, and time required for assessment (26). Furthermore, this approach does not discern lymphocyte subsets or T cell activation states (24,25).
Although histologic assays for several MHCII proteins and TIL counting could be combined to develop diagnostic criteria, the process would be complex. Historically, multiplexed immunohistochemistry (IHC) assays (e.g. IHC4) have not performed as well as multiplexed gene expression assays (27,28). Compared to traditional pathologic scoring systems, a multiplexed gene expression test can measure the expression of many genes in the MHCII pathway, quantify TIL markers simultaneously, and has a larger dynamic range of measurements with finer resolution.
In routine clinical practice patients’ tumors are collected and processed as formalin-fixed, paraffin-embedded (FFPE) tissues, which results in significant degradation of mRNA (29). PCR was the first technology used to demonstrate that small fragmented RNA transcripts could be recovered from FFPE tissue and used to accurately quantify gene expression in breast tumors (30). This enabled the development of the first gene expression prognostic assay for patients with hormone receptor positive (HR+) breast cancer (Oncotype Dx) (31). There are now several gene expression assays that are indicated for use in patients with HR+ breast cancer (32–37); however, there are no clinically validated assays available for patients with TNBC.
The NanoString nCounter platform is an alternative method for measuring gene expression in clinical FFPE specimens. NanoString nCounter technology is unique in that it measures RNA directly without amplification or cloning, which eliminates the biases that can be introduced by other PCR or sequencing-based methodologies (38,39). One clinical prognostic test for HR+ breast cancer (Prosigna) utilizes NanoString technology (32,37,40). NanoString obtained a CE Mark for its Prosigna assay in 2012, followed by U.S. Food and Drug Administration (FDA) clearance in September 2013. Prosigna is now included in clinical oncology guidelines for the management of HR+ breast cancer (41) and is performed in qualified clinical laboratories around the world. In this study, we leveraged this previous success in clinical assay development on the NanoString nCounter platform to develop an assay for MHCII and TIL gene expression that could be used to assess prognosis in TNBC patients.
Materials and Methods
NanoString Probe Design
A custom panel of probes for measuring expression of 36 genes on the NanoString nCounter platform was designed. Probe sequences were compared to RNA-seq data from TNBC tumors (9) to confirm that mRNA isoforms in TNBC would be detected by the probe sequences, and redesigned as necessary. The probe sequences were then synthesized by Integrated DNA Technologies, Inc. The probe A oligos were HPLC purified and the Probe B oligos were PAGE purified. The full sequence of the probes is provided in Supplementary Table 1.
NanoString nCounter Assay
We used NanoString nCounter Elements™ TagSets and Master Kits to develop the assay. Custom gene-specific oligonucleotide probes (Probe Sequence in Supplementary Table 1) were produced by IDT (Integrated DNA Technologies, Coralville, IA). Hybridization and counting were performed according to the manufacturer’s specifications. Briefly, gene-specific probes were hybridized with NanoString Elements™ TagSets and RNA at 67°C for 24 hours. After hybridization, samples were transferred to the automated nCounter Prep Station for purification and immobilization onto the sample cartridge. After sample preparation was complete, the sample cartridge was transferred to the nCounter Digital Analyzer for imaging and analysis. All samples were analyzed using the maximum resolution setting (555 images per sample).
Approval for use of patient specimens
Approval for the use of archival tissue specimens was granted by Institutional Review Boards at the University of Utah and the University of Kentucky. The research was conducted in accordance with recognized ethical guidelines including the U.S. Common Rule. Written informed consent was obtained for fresh frozen tissue collections. For previously collected archival FFPE blocks, the IRBs waived the requirement for informed consent.
RNA from frozen tissues
RNA remaining from frozen tissue collected for previous studies was used (9,42). The RNA-seq data from these samples are publicly available through GEO Accession GSE58135. For the comparison of frozen and FFPE sections from the same tumor, frozen breast cancer specimens were obtained from the University of Kentucky Markey Cancer Center Biospecimen Procurement and Translational Pathology Shared Resource Facility (BPTP SRF). These tissues were collected from breast surgical specimens under IRB protocols # 04–0454 and 11–0750. Fresh frozen breast tissues were embedded in Tissue-Tek O.C.T. Compound (Sakura Finetek, Torrance, CA) and sectioned at −20° C on a cryostat. An initial 4 μm tissue section was cut and stained using H&E (hematoxylin and eosin) so that tumor cellularity could be assessed by a pathologist. Only cases with ≥10% tumor cellularity were included. After assessing the H&E slide, a pathologist cut an additional 10 unstained sections at 10 μm each. Unstained sections were collected in lysis buffer and homogenized in a Bullet Blender (NextAdvance, Troy, NY); RNA was then isolated using an E.Z.N.A RNA Isolation Kit (Omega Bio-tek, Norcross, GA). After frozen sections had been taken for RNA isolation, the remnant block was taken off the cryostat, placed in a tissue cassette, and submitted for routine processing and embedding (creation of an FFPE block) in a pathology laboratory.
FFPE sample identification
This project was performed under an approved University of Utah IRB protocol (#24487). Natural language searches were used to identify surgical pathology cases with a diagnosis of invasive carcinoma of the breast. Only breast tumors from patients with primary stage I-III breast cancer were included in the study. Surgical pathology reports were reviewed by a pathologist to determine ER, PR, and HER2 status. Only TNBC cases with pre-treatment tumor material available in the archives were included. Detailed clinicopathologic, stage and outcome data were obtained through review of the pathology report and medical record. DFS was defined as the length of time that the patient survived after a primary diagnosis of breast cancer without any evidence of local disease recurrence or distant metastases. Events included ipsilateral breast recurrence and distant metastases.
Slide review, macrodissection, and RNA Isolation from FFPE tissue
A pathologist reviewed all cases and selected the best FFPE block from each case for analysis, taking care to avoid blocks with low tumor cellularity, or with large areas of necrosis, calcification, or fibrosis. For each block, a fresh hematoxylin and eosin (H&E) stained slide and adjacent unstained sections (10 μm) were obtained. A board-certified pathologist reviewed each H&E section and confirmed the presence of invasive breast cancer. Tumors were required to be ≥ 4mm in size and to have at least 10% tumor cellularity. Using these requirements, only a single case was initially deemed inadequate due to low tumor cellularity (<10%). In this case, an alternate block was selected from the same surgical pathology specimen; the alternate block had 60% tumor cellularity and was therefore included in the study. After assessing tumor cellularity, the pathologist circled tumor on the H&E slide for macrodissection, taking care to exclude large areas of necrosis, hemorrhage, calcification, and ductal carcinoma in situ. The pathologist also measured the tumor surface area to determine the number of unstained slides required for the assay. Prior to macrodissection, unstained slides (10 μm) were de-paraffinized using Hemo-De (Scientific Safety Solvents, Keller TX), washed in 100% Ethanol, air-dried for 10 minutes, and then briefly rinsed in 3% glycerol. Tumor macrodissection was performed with a scalpel in order to isolate tumor-rich regions from unstained FFPE sections. Macrodissected tissue was subject to RNA isolation using a Roche column-based kit (HighPure FFPET RNA Isolation Kit, Roche Diagnostics, Indianapolis, IN). Briefly, macrodissected tissue from FFPE unstained slides was digested overnight in Proteinase K, RNA was bound to a silica column, treated with DNase, then eluted in 30 μL of buffer according to the manufacturer’s instructions. Isolated RNA was quantified using the Qubit 3.0 and the RNA-BR (Broad-Range) assay kit (ThermoFisher Scientific, Waltham, MA). RNA quality was assessed on the 2200 TapeStation (Agilent Technologies, Santa Clara, CA) using the Agilent RNA ScreenTape Assay. RIN (RNA integrity number) values for each specimen were recorded.
Normalization of gene expression values
The gene expression count values for each sample were normalized to correct for differences in background signal intensity across runs, and to correct for differences in RNA template quality and quantity between samples. The first step of normalization is background subtraction. In each Nanostring nCounter run a “no template” control sample was analyzed. The count values for each probe in this control were subtracted from the count values for each of the patient samples in the run. This is called “Blank lane background subtraction” in the NanoString nSolver analysis software. Next, the geometric mean of the Housekeeping genes was used as a normalization factor for each sample. Notably, the probe for the Housekeeping gene PSMC4 exhibited a very high percent coefficient of variation (101%) (Supplementary Figure 1) and was excluded from the normalization factor calculation. The normalized counts for each sample were then analyzed as described below.
Statistical Analyses
Statistical analyses were performed in R version 3.5.0 and GraphPad Prism Version 7.0C.
The geometric mean is often used in the literature, as well as the NanoString nSolver software, to calculate a composite score of multiple internal control housekeeping genes for normalization of gene expression assays (43,44). In this study the geometric mean was used to calculate composite scores for Basal-like gene expression, MHCII gene expression, TIL gene expression and Immune Activation gene expression. This ensures that each gene in the score has similar weight, regardless of its baseline expression levels and dynamic range. This was particularly important when incorporating TIL genes into the same score as the MHCII genes expressed in tumor cells, since TIL genes inherently have lower mRNA counts because they are derived from a smaller fraction of cells in the sample. Thus, higher scores represent higher expression of all of the genes in the signature, and avoids the risk that a single extremely high or low expressed gene in the signature will have uneven influence on the score.
The Basal-like Subtype score, MHCII Score, TIL Score, and Immune Activation Score were calculated using the geometric mean of normalized counts for each gene as noted in following formulas:
Heatmaps of log normalized gene counts were created using the R package ‘pheatmap’ version 1.0.10. Survival analysis (Kaplan Meier plots and cox regression) was performed using the R package ‘survival’ version 2.42–3 and the R package ‘survminer’ version 0.4.2. Receiver Operator Characteristic curve analysis was performed using the R package ‘pROC’ version 1.12.1. The linear model of Risk of Recurrence was created using the glm package in R.
Analysis of Public Microarray Data
The Kaplan-Meier Plotter tool (http://kmplot.com) (45) was used to perform correlative analysis of publicly available gene expression datasets. The intrinsic subtype classification provided by the Kaplan-Meier Plotter tool was used to select cases for analysis (46). The following selections were applied to all analyses: only one JetSet best probe (47) for each gene was used in the multigene classifier that calculates the mean expression of the selected probes, relapse free survival was selected for the analysis, patients were censored at the follow-up threshold (60 months), biased arrays were excluded, and redundant samples were removed. The most significant cutpoint was used to split patients into two groups.
Immunohistochemistry (IHC)
IHC staining was performed on 4 μm thick sections of FFPE tissue. The following antibodies were used: HLA-DR (Santa Cruz Biotechnology (sc-53319)) and HLA-DR/DP/DQ/DX (Santa Cruz Biotechnology (sc-53302)). FFPE sections were air-dried and then melted in a 60°C oven for 30 minutes. Slides were loaded onto the Ventana BenchMark™ ULTRA automated staining instrument (Ventana Medical Systems, Tucson, AZ) and de-paraffinized with the EZ Prep solution. Antigen retrieval was perform using Cell Conditioning 1 (CC1, pH 8.5) for 64 minutes at 95°C (HLA-DR and HLA-DR/DP/DQ/DX). The primary antibody (Concentration of 1:1000 for HLA-DR/DP/DQ/DX; 1:2000 for HLA-DR) was applied for 1 hour at 37°C. Signal amplification was performed with the Amplification kit (HLA-DR and HLA-DR/DP/DQ/DX). Positive signal was visualized using the UltraView DAB detection kit, which is a Universal HRP Multimer that contains a cocktail of HRP labeled antibodies (goat anti-mouse IgG, goat anti-mouse IgM, and goat anti-rabbit), utilizing DAB (3–3’ diaminobenzidine) as the chromogen. Tissue sections were counterstained with hematoxylin for 8 minutes. The slides were removed from the immunostainer and placed in a dH2O/DAWN ™ mixture. The sections were gently washed in a mixture of de-ionized water and DAWN™ solution to remove any coverslip oil applied by the automated instrument. The slides were gently rinsed in deionized water until all of the wash mixture was removed. The slides were de-hydrated in graded ethanol, cleared in xylene and then coverslipped. For all staining runs, positive and negative controls were included and stained appropriately in all cases. Benign human tonsil was used as a positive control, while skeletal muscle was used as a negative control. In addition, positive staining in macrophages and infiltrating lymphocytes served as internal positive controls for all cases. Scoring for HLA-DR and HLA-DR/DP/DQ was performed by a board-certified pathologist who was blinded to clinical variables. Expression of HLA-DR and HLA-DRDPDQ was assessed in tumor epithelial cells using a standard semi-quantitative system: negative (0), weak (1), moderate (2), and strong (3).
Results
A diagrammatic outline of this study’s design and analyses is provided in Supplementary Figure 2.
Design of the MHCII Immune Activation assay
The major goal of this study was to develop a multiplexed gene expression assay on the NanoString nCounter platform that could accurately measure the expression of MHCII and TIL genes in FFPE TNBC tumor specimens. We have named this the “MHCII Immune Activation” assay.
The MHCII Immune Activation assay uses custom gene-specific oligo probes designed to 36 genes including MHCII Signature genes, TIL genes, Subtype Verification genes, and Housekeeping Control genes (Figure 1A) (Probe Sequences in Supplementary Table 1). The MHCII genes were selected based on significant association with longer DFS in the previous study (9). CIITA is the master transcriptional transactivator of the MHCII pathway and is required to induce expression of the other genes in the pathway(48,49). Candidate TIL genes were selected based on high spearman correlation (R > 0.5) with CIITA expression in the TNBC tumors in the previous study (9) and membership in the Gene Otology classification “Positive regulation of T cell activation” (50–52). Nine candidate genes that were identified as TIL markers in recent publications were selected for the assay (53–55). The selected TIL genes include markers of T cell types, as well as markers of T cell activation, T cell memory and T cell interactions with tumor cells. The Subtype Verification genes were previously determined to be the best distinguishers of Basal-like TNBC from other subtypes using the PAM50 gene set (56). During the analytical/technical development of the PAM50 signature, statistical algorithms to identify the best housekeeping control gene sets for normalization in breast cancer were developed by our group (57). The 5 best housekeeping control genes for normalizing classifier genes across all types of breast cancer and across different ages of FFPE procurement were selected for this assay (57).
Figure 1. Pre-analytical testing of the MHCII Immune Activation assay.
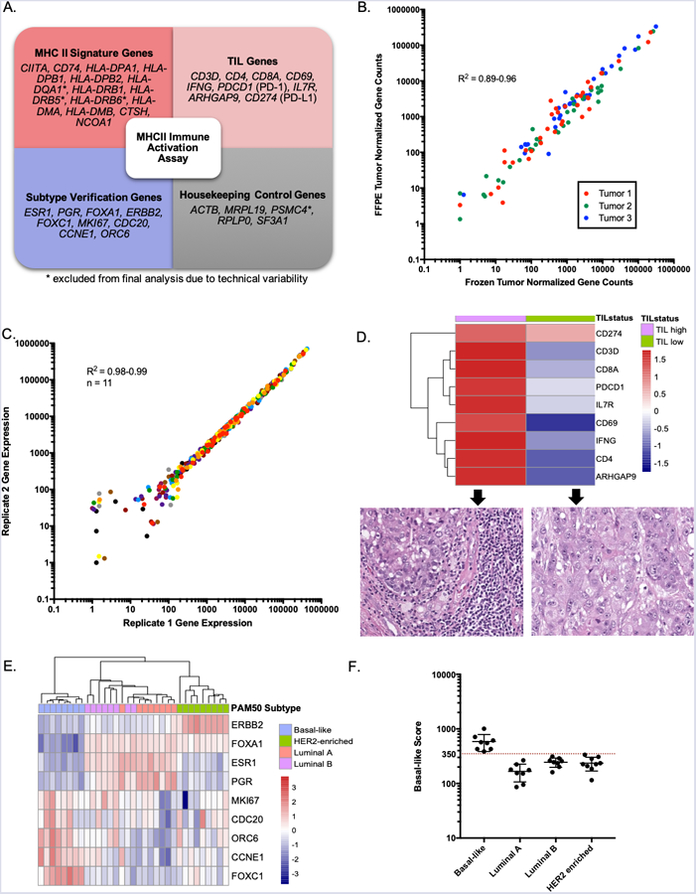
(A) Gene sets measured by the assay. (B) The assay provided similar measurements of gene expression in Frozen and FFPE sections from the same tumor (n= 3). Each point in the scatter-plot represents the expression values for one of 36 genes. (C) The assay provided highly similar gene expression measurements between two replicates of each of 11 different FFPE breast tumor RNA samples. Each point in the scatter-plot represents the expression values for one of 36 genes in one of 11 samples. Each of the 11 samples is depicted in a different color. (D) The TIL genes in the assay were differentially expressed between histologically confirmed TIL high and TIL low TNBC tumors. (E) The Subtype Verification genes in the MHCII Immune Activation assay were differentially expressed between FFPE tumor specimens previously classified by the PAM50 assay as Basal-like (n = 8), Luminal A (n = 8), Luminal B (n = 8), and HER2-enriched (n = 9). (F) A threshold chosen for the Basal-like score distinguishes Basal-like tumors from other subtypes.
Pre-analytical testing of the MHCII Immune Activation assay
We chose to develop the assay on the NanoString nCounter platform because previous studies reported that the platform provides accurate gene expression measurements even in degraded RNA from FFPE specimens (39). To ensure that the MHCII Immune Activation assay accurately measures gene expression in FFPE specimens, the MHCII Immune Activation Assay was performed on three pairs of matched frozen and FFPE breast tumor specimens. Measurements were highly correlated (Spearman R2 = 0.89–0.96; P < 0.0001) between the high-quality RNA from frozen tumor sections (RNA Integrity Number (RIN) = 9.0–9.7) and the degraded RNA from matched FFPE tumor sections (RIN =1.0–4.5) (Figure 1B). Thus, the MHCII Immune Activation assay on the NanoString platform can accurately quantify gene expression in FFPE specimens.
To evaluate the reproducibility of the MHCII Immune Activation assay, 11 pairs of replicate FFPE breast tumor RNA samples were analyzed on the NanoString nCounter instrument. The two sets of replicate samples were processed by two different technical teams at our institution. The normalized counts were highly correlated between the pairs of replicates for each of the 11 samples (Figure 1C; Spearman R2 = 0.98–0.99; P < 0.0001). Genes whose normalized counts were below 10 in both replicates have higher variation between replicates, reflecting natural variation in counting rare molecules. Therefore, the MHCII Immune Activation assay provides highly reproducible results on RNA isolated from FFPE tissue.
To confirm that the MHCII Immune Activation assay accurately measures TIL genes, the assay was performed on FFPE specimens from histologically confirmed TIL-high and TIL-low TNBC tumors. The TIL genes were differentially expressed between TIL high and TIL low TNBC tumors, as expected (Figure 1D).
The MHCII signature is associated with improved DFS in patients with Basal-like TNBC, but not in patients with HR+ breast cancer (Supplementary Figure 3). This observation is consistent with previous studies that have investigated immune and TIL signatures across breast cancer subtypes. In one of the largest studies, the presence of tumor infiltrating lymphocytes (TILs) was identified as an adverse prognostic factor in patients with luminal breast cancer, potentially reflecting the unique immunobiology of this HR+ subtype (58). Subtype Verification genes are included in the MHCII Immune Activation assay to exclude tumors that are not Basal-like TNBCs from analysis. To confirm that the Subtype Verification genes in the assay are able to discern Basal-like TNBC from other subtypes of breast cancer, the MHCII Immune Activation Assay was performed on 33 FFPE breast tumor RNA samples that had been previously classified into intrinsic subtypes (Basal-like (n = 8), Luminal A (n = 8), Luminal B (n = 8), and HER2-enriched (n = 9) using the PAM50 assay (59). The Subtype Verification genes were differentially expressed between these subtypes of breast cancer, as expected (Figure 1E). To develop an inclusion criterion threshold for Basal-like TNBC tumors a “Basal-like score” was calculated for each sample, defined as the geometric mean of the Subtype Verification genes that are highly expressed in Basal-like tumors (FOXC1, MKI67, CDC20, CCNE1, ORC6). A threshold for the Basal-like score that perfectly distinguished Basal-like tumors from other subtypes was selected (Figure 1F).
Performance of the MHCII Immune Activation assay in a training set of TNBC tumors
To evaluate the accuracy of the MHCII Immune Activation assay in TNBC tumor specimens, we analyzed RNA from fresh frozen tissue samples (n = 44) that had been previously analyzed using RNA-seq (9). From each sample, 50–250 ng of RNA was hybridized with the custom gene-specific probes and Elements TagSets and analyzed on the NanoString nCounter Analysis System. The gene expression counts in each sample were background subtracted and normalized to Housekeeping genes, as described in the Materials and Methods section. Five samples were excluded from analysis because they did not meet the Basal-like score threshold defined in the pre-analytical testing. The remaining 39 samples were analyzed for MHCII and TIL gene expression.
Three gene probes (HLA-DQA1, HLA-DRB5, and HLA-DRB6) were excluded from further analysis due to poor concordance between the RNA-seq and NanoString data (Supplementary Figure 4). The remaining MHCII gene expression measurements obtained from the MHCII Immune Activation assay and from RNA-seq on the same samples were highly correlated (mean Spearman R2 = 0.88, mean P = 0.008, Figure 2A). This result confirms the accuracy of this new MHCII Immune Activation assay on the NanoString nCounter instrument.
Figure 2. MHCII Immune Activation assay in a training set of TNBC tumors.
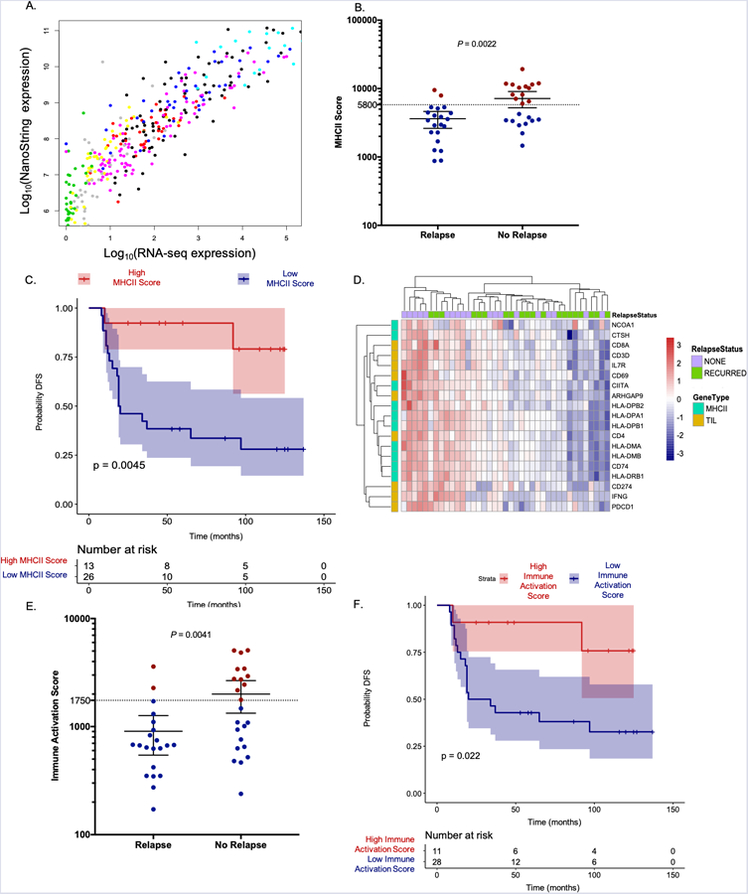
(A) MHCII gene expression measurements from the MHCII Immune Activation assay and RNA-seq on the same TNBC tumor samples were highly correlated. Each of the 10 genes is a different color. (B) MHCII Scores were significantly higher in patients who did not relapse. Mean and 95% confidence interval shown. Threshold is a dashed line, red circle classified as high, blue circle classified as low. (C) A Kaplan Meier curve and log rank p-value show significantly longer DFS in patients with high MHCII Scores. (D) Expression of MHCII and TIL genes are highly correlated within TNBC patient tumors in the training set. (E) Immune Activation Scores calculated using MHCII and TIL genes were significantly higher in patients who did not relapse. Mean and 95% confidence interval shown. Threshold is dashed line, red circles classified as high, blue circles classified as low. (F) A Kaplan Meier curve and log rank p-value show significantly longer DFS in patients with high Immune Activation Scores using the threshold depicted in 2E.
To determine if the MHCII Immune Activation assay could detect differential expression of MHCII genes between TNBC patients who relapsed and those who did not, an “MHCII Score” for each sample was calculated, defined as the geometric mean of the MHCII gene expression values. MHCII Scores were significantly higher (one-sided Mann Whitney P = 0.0022) in TNBC patients who did not relapse compared to those who did relapse (Figure 2B). A Kaplan Meier curve using a threshold for MHCII Score that provides the most significant log rank p-value demonstrated that the MHCII Immune Activation assay reproduced the significant prognostic difference between tumors with high and low MHCII expression (log rank P = 0.0045, Figure 2C, threshold depicted in Figure 2B). This result confirms that the MHCII gene expression signature maintains its prognostic significance on the Nanostring nCounter platform.
A heatmap of the MHCII and TIL genes in TNBC patient tumors demonstrated that expression of MHCII and TIL genes is highly correlated within a tumor (Figure 2D). Similarly, MHCII and TIL scores were correlated across samples (Spearman R2= 0.71, Supplementary Figure 5). To determine whether expression of the MHCII and TIL genes could be combined into score that could be used to assess prognosis, an Immune Activation Score for each sample was calculated using the geometric mean of the MHCII and TIL gene expression values. Immune Activation Scores were significantly higher (one-sided Mann Whitney P = 0.0041) in TNBC patients who did not relapse compared to those who did relapse (Figure 2E). A Kaplan Meier curve using a threshold for the Immune Activation Score that provides the same Specificity (90%) as the MHCII score demonstrated that patients with high Immune Activation Scores have a significantly higher probability of disease-free survival than those with low Immune Activation Scores (log rank P = 0.022, Figure 2F, threshold=1750 depicted in Figure 2E). This result confirms the prognostic power of the Immune Activation Score generated by the MHCII Immune Activation assay.
Validation of the MHCII Immune Activation assay in an independent cohort
The second major goal of this study was to validate that the MHCII Immune Activation assay could be used to assess prognosis in an independent institutional cohort of TNBC patients. Chart review was used to select cases that generally represent the diverse presentation and outcomes that are seen in TNBC patients in clinical practice at the University of Utah (n = 56). Selected cases included age 35–70 (median 55), Stage I-III disease (majority Stage II), tumor size T1–T4 (majority T2), Histologic Grade 2–3 (majority Grade 3), and patients with positive and negative lymph nodes (Supplementary Table 2). Overall, these demographics, and the number of cases, is similar to the cohort used in the previous study and the training set (Supplementary Table 2) (9).
A board-certified anatomic pathologist selected clinical FFPE tissue blocks in which there was adequate tumor tissue for macrodissection. All specimens were collected prior to chemotherapy. The MHCII Immune Activation assay was performed on RNA isolated from the TNBC FFPE specimens using a protocol similar to the Prosigna test, as described in detail in the Materials and Methods section.
Eleven samples were excluded from analysis because they did not meet the Basal-like score threshold defined in the pre-analytical testing. The observation that not all TNBC tumors will be classified into the Basal-like subtype based on gene expression is consistent with prior studies that report the presence of Luminal Androgen Receptor subtype tumors and HER2-enriched subtype tumors among TNBCs (59,60). The remaining 45 samples were analyzed for MHCII and TIL gene expression.
The expression of MHCII and TIL genes were correlated within each tumor, similar to the training set (Figure 3A). MHCII and TIL scores were also correlated across samples (Spearman R2 = 0.58, Supplementary Figure 5). The geometric mean of the MHCII and TIL gene expression values was used to calculate an Immune Activation Score for each sample. Immune Activation Scores were significantly higher (one-sided Mann Whitney P = 0.0278) in TNBC patients who did not relapse compared to those who did relapse (Figure 3B). A Kaplan Meier curve using the same Immune Activation Score threshold as the training set demonstrated a significant prognostic difference between tumors with high and low Immune Activation Scores (log rank P = 0.021, Figure 3C, threshold=1750 depicted as a dashed line in Figure 3B). This result confirms the prognostic significance of the MHCII Immune Activation assay in this independent cohort.
Figure 3. MHCII Immune Activation assay in an independent validation cohort of FFPE TNBC tumors.

(A) Expression of MHCII and TIL genes are highly correlated within TNBC patient tumors in the independent validation cohort. (B) Immune Activation Scores calculated using MHCII and TIL genes in the MHCII Immune Activation assay were significantly higher in patients who did not relapse. Mean and 95% confidence interval shown. Threshold is a dashed line, red circle classified as high, blue circle classified as low. (C) A Kaplan Meier curve and log rank p-value show significantly longer DFS in patients with high Immune Activation Scores using the threshold depicted in 3B.
Assessing Risk of Recurrence using the MHCII Immune Activation assay
The most likely clinical use of the MHCII Immune Activation assay would be to identify patients that have a very low risk of relapse, and distinguish them from patients who have an average risk of relapse. To determine if the MHCII Immune Activation Assay could be used to identify patients that have a very low risk of relapse, a Receiver Operator Characteristic (ROC) curve was calculated for the Immune Activation Scores in the training set and validation cohort (Figure 4A, ROC statistics are provided in Supplementary Figure 6). This clinical application of the assay requires high specificity to correctly identify patients who have a low risk of recurrence, and avoid misclassifying patients that may recur. To evaluate the specificity of the assay, threshold analysis of the ROC curve was used to calculate the Immune Activation Score that results in 95% specificity for identifying patients who do not relapse in the training set (threshold = 2400). The 95% confidence intervals for the threshold that provides 95% specificity are depicted in the ROC curve in Figure 4A. When this Immune Activation Score threshold was applied to the validation cohort, the specificity for identifying patients who did not relapse was 100%, i.e. zero patients with Immune Activation Scores above the threshold relapsed (Figure 4B). Kaplan Meier curves were created using this Immune Activation Score threshold to stratify patients, which demonstrates the difference in probability of disease-free survival in both the training set (Figure 4C) and the validation cohort (Figure 4D).
Figure 4. Using Immune Activation Scores to identify patients with a low risk of recurrence.
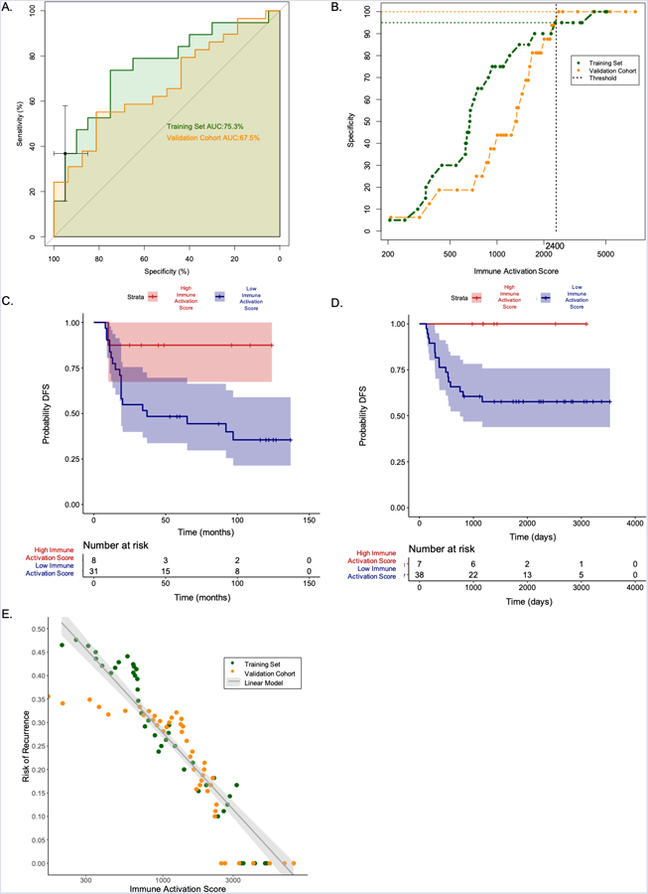
(A) ROC curve analysis of the training set was used to select an Immune Activation Score threshold that results in 95% specificity for identifying patients who do not relapse. Training set ROC curve in green. Validation cohort ROC curve in orange. 95% confidence intervals for the threshold that provides 95% specificity in Training set shown as black error bars. (B) When this Immune Activation Score threshold was applied to the independent validation cohort, the specificity for identifying patients who did not relapse was 100%. (C) Kaplan Meier curve that stratifies patients in the training set based on the Immune Activation Score threshold that provides 95% specificity. (D) Kaplan Meier curve of the same threshold applied to the independent validation cohort demonstrates longer DFS in patients with Immune Activation Scores above the threshold. (E) Risk of recurrence can be modeled as a linear function of the log10 Immune Activation score in both the training set and validation cohort.
In multi-gene clinical tests used to assess prognosis in HR+ breast cancer (e.g. Prosigna and Oncotype Dx), the results are continuous variables that are linearly related to a patient’s risk of recurrence (27,61). Currently, the quantitative results of these tests are used to classify patients into groups of low, intermediate and high risk of recurrence for clinical management. The Immune Activation Score produced by this assay is also a continuous variable. To determine if the Immune Activation Score produced by this assay is linearly related to a patient’s risk of recurrence, the cumulative risk of recurrence was calculated for patients across the range of Immune Activation Scores observed in the training set and validation cohort. The Risk of Recurrence in both the training set and validation cohort is a linear function of the log10 Immune Activation Score (Figure 4E). This result confirms that a patient’s risk of recurrence is monotonically related to the Immune Activation Score. In the future, larger studies could be used to define thresholds to classify TNBC patients into groups with low, intermediate or high risk of recurrence.
Cox proportional hazards regression models were generated to test the association between DFS, clinical variables, and Immune Activation Score in the training set and validation cohort. In univariate cox regression, Immune Activation Score and stage at diagnosis were significantly associated with DFS in both the training set and validation cohort (Table 1). The Immune Activation Score Hazard Ratio was 0.1430 (95% CI = 0.03683–0.5555) in the training set and 0.2111 (95% CI = 0.06075–0.7335) in the validation cohort, indicating a good prognostic factor. The Hazard Ratio for stage was 2.1227 (95% CI = 1.439–3.131) in the training set and 1.628 (95% CI = 1.204–2.201) in the validation cohort, indicating a poor prognostic factor. The other clinical parameters were not significantly associated with DFS, including age at diagnosis, and whether the patient received chemotherapy (Table 1). In the multivariable Cox proportional hazards regression model for both the training set and the validation cohort, Immune Activation Score and stage at diagnosis both remained significant, and their hazard ratios were similar to those in the univariate analysis (Table 1). This result indicates that the Immune Activation Score is an independent predictor of DFS, even when accounting for the differences in DFS associated with a patient’s disease stage at diagnosis.
Table 1.
Cox regression models of DFS
| Univariate | Multivariate | ||||
|---|---|---|---|---|---|
| Variable | Hazard Ratio (95% Confidence Interval) | P-value | Hazard Ratio (95% Confidence Interval) | P-value | |
| Training Set | Immune Activation Score (log10 transformed) | 0.1430 (0.03683– 0.5555) | 0.00496* | 0.1688 (0.04039–0.7054) | 0.014758* |
| Stage at Diagnosis | 2.1227 (1.439– 3.131) | 0.000147* | 2.0310 (1.33617–3.0871) | 0.000911* | |
| Age at Diagnosis | 1.006 (0.9661– 1.048) | 0.765 | 1.0363 (0.99081–1.0840) | 0.119459 | |
| Received Chemotherapy | 0.4879 (0.1752– 1.359) | 0.17 | 0.4660 (0.14512–1.4965) | 0.199589 | |
| Validation Cohort | Immune Activation Score (log10 transformed) | 0.2111 (0.06075–0.7335) | 0.01440* | 0.1939 (0.05451–0.6896) | 0.011280* |
| Stage at Diagnosis | 1.628 (1.204–2.201) | 0.00154* | 1.6363 (1.18309–2.2632) | 0.002920* | |
| Age at Diagnosis | 1.013 (0.9812–1.045) | 0.43200 | 1.0198(0.96152–1.0605) | 0.696870 | |
| Received Chemotherapy | 0.6166 (0.1986–1.915) | 0.40300 | 0.7696 (0.14986–3.9519) | 0.753700 | |
significant P-values
A cox proportional hazards model of the effect of stage alone in the validation cohort predicts that a patient diagnosed with Stage IIB disease has a 59% probability of 5-year disease free survival. A cox proportional hazards model including both stage and Immune Activation score predicts that a Stage IIB patient with a high Immune Activation Score of 4000 has an 79% probability of 5-year disease free survival, while a patient with the same disease stage and a low Immune Activation Score of 400 has a 32% probability of 5-year disease free survival. This suggests that a clinical decision-making tool that incorporated the Immune Activation Score in addition to the patient’s disease stage could provide improved assessment of a patient’s risk of recurrence. Further studies in larger cohorts will be needed to train and evaluate a predictive model that incorporates Immune Activation score.
Comparison of MHCII Immune Activation assay to IHC and histologic TIL counting
The results from the MHCII Immune Activation assay confirm that elevated expression of MHCII and TIL genes is associated with a significantly reduced risk of recurrence in TNBC patients. To determine if these gene expression measurements correlate with traditional histologic assessment of MHCII expression and TIL counting, IHC and H&E staining was performed on FFPE sections from the specimens analyzed in the validation cohort, which was reviewed by a board-certified anatomic pathologist who specializes in breast pathology.
In tumors with the highest Immune Activation Scores, MHCII protein was strongly expressed in a membranous pattern within infiltrating carcinoma cells and in associated TILs (Figure 5A). Tumors with an intermediate Immune Activation Score showed variable MHCII expression; in these cases, staining was often heterogeneous and of moderate intensity (Figure 5A). In tumors with the lowest Immune Activation Scores, MHCII protein expression was absent in invasive carcinoma cells and present only in rare tumor-associated inflammatory cells (Figure 5A).
Figure 5. Comparison of the MHCII Immune Activation assay to IHC and histologic TIL counting.
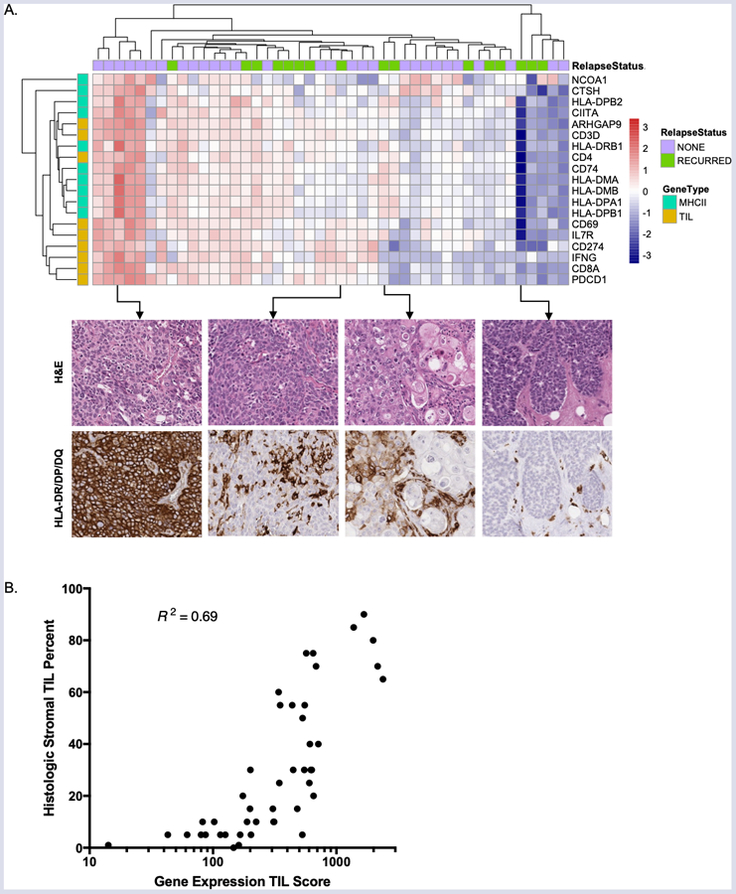
(A) Immunohistochemical analysis of MHCII expression in patients with high, intermediate, and low Immune Activation Scores. (B) The TIL Score calculated from TIL gene expression using the MHCII Immune Activation assay is correlated with histologic assessment of stromal TIL percentage.
TIL quantification was performed using a histologic “gold standard” protocol developed by a consensus committee on Tumor Infiltrating Lymphocytes (TILs) in breast cancer (24,25). The TIL Score measured by the MHCII Immune Activation assay was highly correlated with morphologic assessment of stromal TIL percentage (Spearman R2 = 0.69, P < 0.0001, Figure 5B). These results confirm that the MHCII Immune Activation assay on the Nanostring nCounter provides a standardized and multiplexed procedure for measuring MHCII expression and TILs in FFPE tumor specimens that is highly correlated with histologic assessments.
Discussion
The purpose of this study was to develop and validate a multiplexed assay for MHCII and TIL gene expression that could be used on FFPE tissue to assess a TNBC patient’s risk of recurrence. The results of this study demonstrate that performing the MHCII Immune Activation assay on FFPE tumor specimens using the Nanostring nCounter instrument provides accurate measurements of MHCII and TIL gene expression that are highly correlated with reduced risk of recurrence in TNBC patients with primary Stage I-III breast cancer.
The most likely clinical use of the MHCII Immune Activation assay would be to distinguish TNBC patients who have a very low risk of relapse from those who have an average risk of relapse. We demonstrate that an Immune Activation Score threshold can be established to identify patients who have a very low risk of recurrence (Figure 4) and may not require systemic therapy. Both the training set and validation cohort in this study included patients who did not receive systemic chemotherapy for a variety of reasons including advanced age, comorbidities, and patient preference (Supplementary Table 2). Excitingly, we found that patients with high Immune Activation Scores who did not receive systemic chemotherapy did not relapse (Supplementary Figure 7A). To investigate this preliminary association further, we analyzed public microarray data from a larger cohort of patients with primary Stage I-III Basal-like breast cancer who did not receive systemic chemotherapy. We found that patients with higher expression of MHCII and TIL genes had significantly longer relapse-free survival, even without systemic treatment (Supplementary Figure 7B). Future clinical studies are warranted to evaluate whether this assay could be used routinely to identify TNBC patients who inherently have a good prognosis and can safely be treated with local therapy alone. The MHCII Immune Activation assay enables precision medicine for TNBC patients and could help reduce the burden of chemotherapy-induced side effects in TNBC survivors.
Another potential clinical application of the MHCII Immune Activation assay is predicting response to immunotherapy. Recent studies have shown that expression of MHC Class II molecules in melanoma cells is associated with improved response to anti-PD-1 immunotherapy in melanoma patients (62–64). Data presented at the American Society of Clinical Oncology 2017 annual meeting from the Phase 2 randomized, controlled, multi-center I-SPY 2 trial ( NCT01042379) demonstrated that 60% of newly diagnosed TNBC patients achieved pathologic complete response (pCR) when treated with the immune checkpoint inhibitor pembrolizumab in combination with standard neoadjuvant chemotherapy. This was a significant improvement compared to the 20% of patients who achieved pCR with standard neoadjuvant chemotherapy alone (65). While this result is promising, it also indicates that 40% of TNBC patients in the pembrolizumab arm did not achieve pCR but were exposed to the significant risks associated with immunotherapy, which in this trial included autoimmune mediated adrenal insufficiency, hepatitis, colitis, and hypothyroidism. Future studies are needed to determine whether the MHCII Immune Activation assay can be used to identify patients that are most likely to benefit from immunotherapy.
The MHCII Immune Activation assay produces similar measurements as histologic assays for MHCII expression and TIL counting (Figure 5), but provides standardized methodology, a larger dynamic range of measurements, and multiplexed analysis of small specimens. The development of the Prosigna test for HR+ breast cancer has demonstrated that one key strength of assays developed on the NanoString nCounter is the ability to implement them as Laboratory Developed Tests (LDTs) in clinical laboratory sites across the world while maintaining standardized protocols and data analysis. Following demonstration of its clinical utility, the format of the MHCII Immune Activation assay will enable similar broad adoption as a clinical test for prognosis in TNBC patients, for which there are currently no clinical tests available.
Supplementary Material
Acknowledgements
We acknowledge the direct financial support for the research reported in this publication provided by the Huntsman Cancer Foundation and the Women’s Cancer Disease Oriented Team at Huntsman Cancer Institute; we also acknowledge support by the National Cancer Institute of the National Institutes of Health under Award Number P30CA042014. The project described was also supported by the NIH National Center for Advancing Translational Sciences through grant number KL2TR001996. Research reported in this publication utilized the University of Utah Huntsman Cancer Institute Biorepository and Molecular Pathology Shared Resource and the High-Throughput Genomics Shared Resource which are supported by the National Cancer Institute of the National Institutes of Health under Award Number P30CA042014. This research was also supported by the Biospecimen Procurement and Translational Pathology and Oncogenomics Shared Resource Facilities of the University of Kentucky Markey Cancer Center (P30CA177558). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Special thanks to Sheryl Tripp at ARUP Laboratories for her histologic expertise, and special thanks to Darah Johnson, S. Emily Bachert, and Donna Wall at the University of Kentucky for their technical assistance.
Financial Support: National Cancer Institute Award Number P30CA042014 (KEV, REF, NLH, KMB, & PSB)
NIH National Center for Advancing Translational Sciences through grant number KL2TR001996 (RLS)
N.L. Henry reports receiving other commercial research support from Pfizer, AbbVie, Innocrin Pharmaceutical, and H3 Biomedicine. P.S. Bernard has an ownership interest (including stock, patents, etc.) in Bioclassifier LLC and PhenoTx LLC. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Newman LA, Reis-Filho JS, Morrow M, Carey LA, King TA. The 2014 Society of Surgical Oncology Susan G. Komen for the Cure Symposium: triple-negative breast cancer. Ann Surg Oncol 2015;22(3):874–82 doi 10.1245/s10434-014-4279-0. [DOI] [PubMed] [Google Scholar]
- 2.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 2007;13(15 Pt 1):4429–34 doi 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 3.Addington J, Freimer M. Chemotherapy-induced peripheral neuropathy: an update on the current understanding. F1000Res 2016;5 doi 10.12688/f1000research.8053.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seretny M, Currie GL, Sena ES, Ramnarine S, Grant R, MacLeod MR, et al. Incidence, prevalence, and predictors of chemotherapy-induced peripheral neuropathy: A systematic review and meta-analysis. Pain 2014;155(12):2461–70 doi 10.1016/j.pain.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 5.Winters-Stone KM, Horak F, Jacobs PG, Trubowitz P, Dieckmann NF, Stoyles S, et al. Falls, Functioning, and Disability Among Women With Persistent Symptoms of Chemotherapy-Induced Peripheral Neuropathy. J Clin Oncol 2017;35(23):2604–12 doi 10.1200/JCO.2016.71.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowles EJ, Wellman R, Feigelson HS, Onitilo AA, Freedman AN, Delate T, et al. Risk of heart failure in breast cancer patients after anthracycline and trastuzumab treatment: a retrospective cohort study. J Natl Cancer Inst 2012;104(17):1293–305 doi 10.1093/jnci/djs317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirova YM, De Rycke Y, Gambotti L, Pierga JY, Asselain B, Fourquet A, et al. Second malignancies after breast cancer: the impact of different treatment modalities. Br J Cancer 2008;98(5):870–4 doi 10.1038/sj.bjc.6604241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahles TA, Root JC, Ryan EL. Cancer- and cancer treatment-associated cognitive change: an update on the state of the science. J Clin Oncol 2012;30(30):3675–86 doi 10.1200/JCO.2012.43.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forero A, Li Y, Chen D, Grizzle WE, Updike KL, Merz ND, et al. Expression of the MHC Class II Pathway in Triple-Negative Breast Cancer Tumor Cells Is Associated with a Good Prognosis and Infiltrating Lymphocytes. Cancer Immunol Res 2016;4(5):390–9 doi 10.1158/2326-6066.CIR-15-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Teijido P, Cabal ML, Fernandez IP, Perez YF. Tumor-Infiltrating Lymphocytes in Triple Negative Breast Cancer: The Future of Immune Targeting. Clin Med Insights Oncol 2016;10(Suppl 1):31–9 doi 10.4137/CMO.S34540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao Y, Qu Q, Chen X, Huang O, Wu J, Shen K. The Prognostic Value of Tumor-Infiltrating Lymphocytes in Breast Cancer: A Systematic Review and Meta-Analysis. PLoS One 2016;11(4):e0152500 doi 10.1371/journal.pone.0152500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stovgaard ES, Nielsen D, Hogdall E, Balslev E. Triple negative breast cancer - prognostic role of immune-related factors: a systematic review. Acta Oncol 2018;57(1):74–82 doi 10.1080/0284186X.2017.1400180. [DOI] [PubMed] [Google Scholar]
- 13.Wang K, Shen T, Siegal GP, Wei S. The CD4/CD8 ratio of tumor-infiltrating lymphocytes at the tumor-host interface has prognostic value in triple-negative breast cancer. Hum Pathol 2017;69:110–7 doi 10.1016/j.humpath.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 14.Loi S, Drubay D, Adams S, Pruneri G, Francis PA, Lacroix-Triki M, et al. Tumor-Infiltrating Lymphocytes and Prognosis: A Pooled Individual Patient Analysis of Early-Stage Triple-Negative Breast Cancers. J Clin Oncol 2019:JCO1801010 doi 10.1200/JCO.18.01010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park IA, Hwang SH, Song IH, Heo SH, Kim YA, Bang WS, et al. Expression of the MHC class II in triple-negative breast cancer is associated with tumor-infiltrating lymphocytes and interferon signaling. PLoS One 2017;12(8):e0182786 doi 10.1371/journal.pone.0182786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armstrong TD, Clements VK, Ostrand-Rosenberg S. MHC class II-transfected tumor cells directly present antigen to tumor-specific CD4+ T lymphocytes. J Immunol 1998;160(2):661–6. [PubMed] [Google Scholar]
- 17.Frangione V, Mortara L, Castellani P, De Lerma Barbaro A, Accolla RS. CIITA-driven MHC-II positive tumor cells: preventive vaccines and superior generators of antitumor CD4+ T lymphocytes for immunotherapy. Int J Cancer 2010;127(7):1614–24 doi 10.1002/ijc.25183. [DOI] [PubMed] [Google Scholar]
- 18.Ilkovitch D, Ostrand-Rosenberg S. MHC class II and CD80 tumor cell-based vaccines are potent activators of type 1 CD4+ T lymphocytes provided they do not coexpress invariant chain. Cancer Immunol Immunother 2004;53(6):525–32 doi 10.1007/s00262-003-0486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meazza R, Comes A, Orengo AM, Ferrini S, Accolla RS. Tumor rejection by gene transfer of the MHC class II transactivator in murine mammary adenocarcinoma cells. Eur J Immunol 2003;33(5):1183–92 doi 10.1002/eji.200323712. [DOI] [PubMed] [Google Scholar]
- 20.Mortara L, Castellani P, Meazza R, Tosi G, De Lerma Barbaro A, Procopio FA, et al. CIITA-induced MHC class II expression in mammary adenocarcinoma leads to a Th1 polarization of the tumor microenvironment, tumor rejection, and specific antitumor memory. Clin Cancer Res 2006;12(11 Pt 1):3435–43 doi 10.1158/1078-0432.CCR-06-0165. [DOI] [PubMed] [Google Scholar]
- 21.Pulaski BA, Ostrand-Rosenberg S. Reduction of established spontaneous mammary carcinoma metastases following immunotherapy with major histocompatibility complex class II and B7.1 cell-based tumor vaccines. Cancer Res 1998;58(7):1486–93. [PubMed] [Google Scholar]
- 22.Thompson JA, Dissanayake SK, Ksander BR, Knutson KL, Disis ML, Ostrand-Rosenberg S. Tumor cells transduced with the MHC class II Transactivator and CD80 activate tumor-specific CD4+ T cells whether or not they are silenced for invariant chain. Cancer Res 2006;66(2):1147–54 doi 10.1158/0008-5472.CAN-05-2289. [DOI] [PubMed] [Google Scholar]
- 23.Axelrod ML, Cook RS, Johnson DB, Balko JM. Biological Consequences of MHC-II Expression by Tumor Cells in Cancer. Clin Cancer Res 2018. doi 10.1158/1078-0432.CCR-18-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denkert C, Wienert S, Poterie A, Loibl S, Budczies J, Badve S, et al. Standardized evaluation of tumor-infiltrating lymphocytes in breast cancer: results of the ring studies of the international immuno-oncology biomarker working group. Mod Pathol 2016;29(10):1155–64 doi 10.1038/modpathol.2016.109. [DOI] [PubMed] [Google Scholar]
- 25.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol 2015;26(2):259–71 doi 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Loughlin M, Andreu X, Bianchi S, Chemielik E, Cordoba A, Cserni G, et al. Reproducibility and predictive value of scoring stromal tumour infiltrating lymphocytes in triple-negative breast cancer: a multi-institutional study. Breast Cancer Res Treat 2018;171(1):1–9 doi 10.1007/s10549-018-4825-8. [DOI] [PubMed] [Google Scholar]
- 27.Dowsett M, Sestak I, Lopez-Knowles E, Sidhu K, Dunbier AK, Cowens JW, et al. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol 2013;31(22):2783–90 doi 10.1200/JCO.2012.46.1558. [DOI] [PubMed] [Google Scholar]
- 28.Sestak I, Buus R, Cuzick J, Dubsky P, Kronenwett R, Denkert C, et al. Comparison of the Performance of 6 Prognostic Signatures for Estrogen Receptor-Positive Breast Cancer: A Secondary Analysis of a Randomized Clinical Trial. JAMA Oncol 2018;4(4):545–53 doi 10.1001/jamaoncol.2017.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Ahlfen S, Missel A, Bendrat K, Schlumpberger M. Determinants of RNA quality from FFPE samples. PLoS One 2007;2(12):e1261 doi 10.1371/journal.pone.0001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cronin M, Pho M, Dutta D, Stephans JC, Shak S, Kiefer MC, et al. Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol 2004;164(1):35–42 doi 10.1016/S0002-9440(10)63093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol 2006;24(23):3726–34 doi 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res 2010;16(21):5222–32 doi 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N Engl J Med 2018;379(2):111–21 doi 10.1056/NEJMoa1804710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sparano JA, Paik S. Development of the 21-gene assay and its application in clinical practice and clinical trials. J Clin Oncol 2008;26(5):721–8 doi 10.1200/JCO.2007.15.1068. [DOI] [PubMed] [Google Scholar]
- 35.van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002;415(6871):530–6 doi 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 36.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 2002;347(25):1999–2009 doi 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 37.Wallden B, Storhoff J, Nielsen T, Dowidar N, Schaper C, Ferree S, et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genomics 2015;8:54 doi 10.1186/s12920-015-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 2008;26(3):317–25 doi 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 39.Reis PP, Waldron L, Goswami RS, Xu W, Xuan Y, Perez-Ordonez B, et al. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol 2011;11:46 doi 10.1186/1472-6750-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen T, Wallden B, Schaper C, Ferree S, Liu S, Gao D, et al. Analytical validation of the PAM50-based Prosigna Breast Cancer Prognostic Gene Signature Assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer 2014;14:177 doi 10.1186/1471-2407-14-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duffy MJ, Harbeck N, Nap M, Molina R, Nicolini A, Senkus E, et al. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur J Cancer 2017;75:284–98 doi 10.1016/j.ejca.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 42.Varley KE, Gertz J, Roberts BS, Davis NS, Bowling KM, Kirby MK, et al. Recurrent read-through fusion transcripts in breast cancer. Breast Cancer Res Treat 2014;146(2):287–97 doi 10.1007/s10549-014-3019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3(7):RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bengtsson M, Stahlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res 2005;15(10):1388–92 doi 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123(3):725–31 doi 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 46.Mihaly Z, Gyorffy B. Improving Pathological Assessment of Breast Cancer by Employing Array-Based Transcriptome Analysis. Microarrays (Basel) 2013;2(3):228–42 doi 10.3390/microarrays2030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q, Birkbak NJ, Gyorffy B, Szallasi Z, Eklund AC. Jetset: selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics 2011;12:474 doi 10.1186/1471-2105-12-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LeibundGut-Landmann S, Waldburger JM, Krawczyk M, Otten LA, Suter T, Fontana A, et al. Mini-review: Specificity and expression of CIITA, the master regulator of MHC class II genes. Eur J Immunol 2004;34(6):1513–25 doi 10.1002/eji.200424964. [DOI] [PubMed] [Google Scholar]
- 49.Steimle V, Siegrist CA, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science 1994;265(5168):106–9. [DOI] [PubMed] [Google Scholar]
- 50.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 2017;45(D1):D183–D9 doi 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25(1):25–9 doi 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.The Gene Ontology C The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 2019;47(D1):D330–D8 doi 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med 2018;24(7):986–93 doi 10.1038/s41591-018-0078-7. [DOI] [PubMed] [Google Scholar]
- 54.Lee HJ, Lee JJ, Song IH, Park IA, Kang J, Yu JH, et al. Prognostic and predictive value of NanoString-based immune-related gene signatures in a neoadjuvant setting of triple-negative breast cancer: relationship to tumor-infiltrating lymphocytes. Breast Cancer Res Treat 2015;151(3):619–27 doi 10.1007/s10549-015-3438-8. [DOI] [PubMed] [Google Scholar]
- 55.Nirmal AJ, Regan T, Shih BB, Hume DA, Sims AH, Freeman TC. Immune Cell Gene Signatures for Profiling the Microenvironment of Solid Tumors. Cancer Immunol Res 2018;6(11):1388–400 doi 10.1158/2326-6066.CIR-18-0342. [DOI] [PubMed] [Google Scholar]
- 56.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27(8):1160–7 doi 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szabo A, Perou CM, Karaca M, Perreard L, Palais R, Quackenbush JF, et al. Statistical modeling for selecting housekeeper genes. Genome Biol 2004;5(8):R59 doi 10.1186/gb-2004-5-8-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Denkert C, von Minckwitz G, Darb-Esfahani S, Lederer B, Heppner BI, Weber KE, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol 2018;19(1):40–50 doi 10.1016/S1470-2045(17)30904-X. [DOI] [PubMed] [Google Scholar]
- 59.Bastien RR, Rodriguez-Lescure A, Ebbert MT, Prat A, Munarriz B, Rowe L, et al. PAM50 breast cancer subtyping by RT-qPCR and concordance with standard clinical molecular markers. BMC Med Genomics 2012;5:44 doi 10.1186/1755-8794-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hubalek M, Czech T, Muller H. Biological Subtypes of Triple-Negative Breast Cancer. Breast Care (Basel) 2017;12(1):8–14 doi 10.1159/000455820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gnant M, Filipits M, Greil R, Stoeger H, Rudas M, Bago-Horvath Z, et al. Predicting distant recurrence in receptor-positive breast cancer patients with limited clinicopathological risk: using the PAM50 Risk of Recurrence score in 1478 postmenopausal patients of the ABCSG-8 trial treated with adjuvant endocrine therapy alone. Ann Oncol 2014;25(2):339–45 doi 10.1093/annonc/mdt494. [DOI] [PubMed] [Google Scholar]
- 62.Johnson DB, Estrada MV, Salgado R, Sanchez V, Doxie DB, Opalenik SR, et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat Commun 2016;7:10582 doi 10.1038/ncomms10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yan H, Hou X, Li T, Zhao L, Yuan X, Fu H, et al. CD4+ T cell-mediated cytotoxicity eliminates primary tumor cells in metastatic melanoma through high MHC class II expression and can be enhanced by inhibitory receptor blockade. Tumour Biol 2016. doi 10.1007/s13277-016-5456-5. [DOI] [PubMed] [Google Scholar]
- 64.Johnson DB, Nixon MJ, Wang Y, Wang DY, Castellanos E, Estrada MV, et al. Tumor-specific MHC-II expression drives a unique pattern of resistance to immunotherapy via LAG-3/FCRL6 engagement. JCI Insight 2018;3(24) doi 10.1172/jci.insight.120360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nanda R, Liu MC, Yau C, Asare S, Hylton N,Veer LVt, et al. Pembrolizumab plus standard neoadjuvant therapy for high-risk breast cancer (BC): Results from I-SPY 2. Journal of Clinical Oncology 2017;35(15_suppl):506- doi 10.1200/JCO.2017.35.15_suppl.506.28029304 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


