Abstract
Autism spectrum disorder (ASD) affects up to 1 in 59 individuals1. Genome-wide association and large-scale sequencing studies strongly implicate both common variants2–4 and rare de novo variants5–10 in ASD. Recessive mutations have also been implicated11–14 but their contribution remains less well defined. Here we demonstrate an excess of biallelic loss-of-function and damaging missense mutations in a large ASD cohort, corresponding to ~5% of total cases, including 10% of females, consistent with a female protective effect. We document biallelic disruption of known or emerging recessive neurodevelopmental genes (CA2, DDHD1, NSUN2, PAH, RARB, ROGDI, SLC1A1, USH2A) as well as other genes not previously implicated in ASD including the transcription factor FEV, a key regulator of the serotonergic circuitry. Our data refine estimates of the contribution of recessive mutation to ASD and suggest new paths for illuminating novel biological pathways responsible for this condition.
Editorial Summary:
Analysis of whole exome sequencing data from 2,343 individuals with autism spectrum disorder (ASD) compared to 5,852 unaffected individuals demonstrates an excess of biallelic, autosomal mutations for both loss-of-function and damaging missense variants.
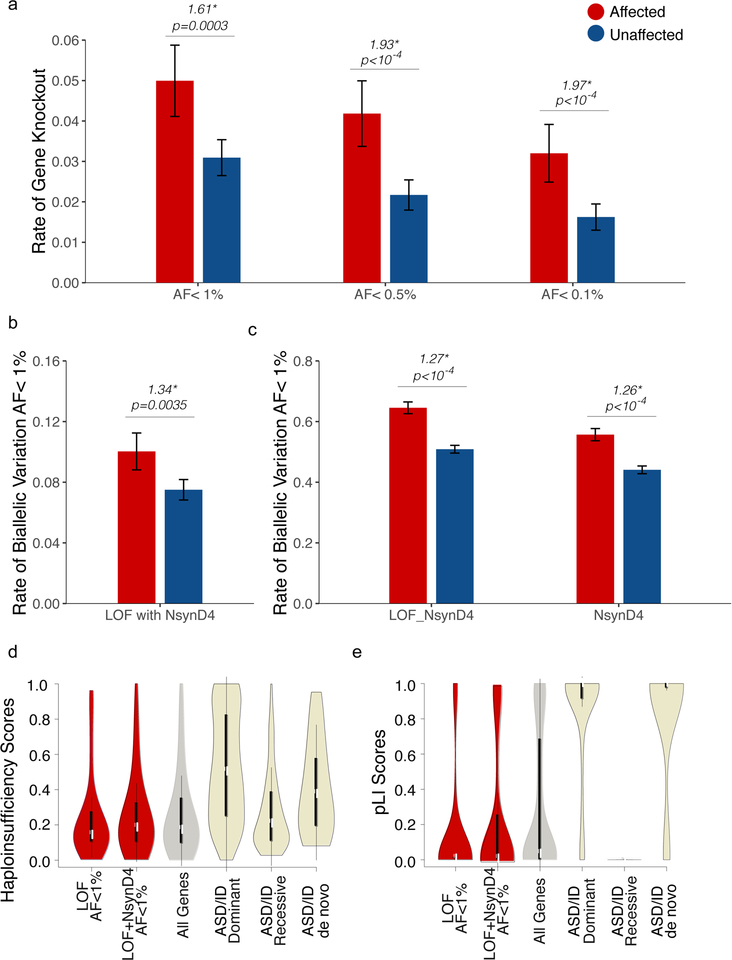
We performed a systematic analysis of exome data from the Autism Sequencing Consortium (ASC)15, representing 2,343 affected and 5,852 unaffected individuals (Supplementary Table 1, Supplementary Figure 1). We cataloged a total of 696,143 autosomal loss-of-function (LOF) events (representing 28,685 unique variants in 11,745 unique genes) that introduce a stop codon or disrupt a canonical splice site (Table 1). After excluding common variants (allele frequency (AF) >1%), there were 84,645 rare LOF events (27,648 unique alleles) for an average of ~10 LOF mutations per individual. After computational phasing, we found 298 events in the ASC (after filtering to exclude common polymorphisms) consistent with complete gene knockout (homozygous or compound heterozygous LOF mutations), affecting 266 individuals (Table 1). Affected individuals were disproportionately likely to harbor a gene knockout (62% more likely; 0.05 events per affected individual versus 0.031 per unaffected individual, p=0.0003 by random permutation testing), with the bulk of the excess arising from the rarest alleles (Table 1; Supplementary Table 2; Figure 1a). To control for possible differences in population and family structure (e.g., founder effects and/or consanguinity in the Finnish and Middle Eastern cohorts), we also normalized to background burdens of biallelic synonymous variants (see Methods and Supplementary Table 3–6). Individuals with ASD continued to exhibit higher knockout rates after normalization (0.042 biallelic LOF events per affected individual versus 0.031 per unaffected individual, p=0.008 by random permutation testing) (Supplementary Figure 2a). Based on the observed ascertainment differentials (AD) between affected and unaffected individuals (0.050 vs. 0.031, or 0.042 vs. 0.031 after normalization), these burdens predict a contribution of biallelic LOF alleles to ~1–2% of ASD cases (see Methods).
Table 1.
Patterns of biallelic LOF mutation in the Autism Sequencing Consortium
| Events per individual | ||||||
|---|---|---|---|---|---|---|
| Allele frequency | # of biallelic LOF events | # of unique genes involved | # of individuals harboring | Unaffected | Affected | p-value |
| All | 237,936 | 636 | 8195 | 28.997 | 29.128 | 0.2 |
| ≤10% | 1,646 | 414 | 1420 | 0.194 | 0.219 | 0.03 |
| ≤5% | 621 | 324 | 559 | 0.068 | 0.096 | 0.0005 |
| ≤1% | 298 | 237 | 266 | 0.031 | 0.050 | 0.003 |
| ≤0.5% | 225 | 182 | 201 | 0.022 | 0.042 | <0.0001 |
| ≤0.1% | 170 | 138 | 146 | 0.016 | 0.032 | <0.0001 |
Figure 1.
An excess of rare, damaging, biallelic mutation in ASD. (a) Rates of biallelic gene knockout (strict LOF) in the ASC, stratified by diagnosis and allele frequency. Rates of biallelic variation, considering (b) LOF variants paired with a damaging missense variant (NsynD4, predicted to be deleterious by at least 4 algorithms), (c) LOF or NsynD4 variants, or NsynD4 variants alone. Genes impacted by biallelic variation in cases exhibit (d) haploinsufficiency and (e) pLI score profiles consistent with recessive genes. Error bars represent the 95% confidence intervals.
We considered whether these findings might be extended to incorporate the impact of missense variation. Missense variation occurs much more frequently than truncating mutations, with a small yet significant subset likely to be damaging. We evaluated potential missense contributions of two different types: all missense/nonsynonymous variants (Nsyn), and damaging missense/nonsynonymous variants (NsynD4, defined as missense changes classified as damaging by 4 or more predictive algorithms, representing 4% of all Nsyn events). Individuals with ASD exhibited an excess of biallelic events specifically involving a LOF allele in trans with a NsynD4 allele (Figure 1b), an effect which persisted after normalization to synonymous rates (Supplementary Figure 2b). Similarly, considering biallelic events involving either rare LOF or NsynD4 alleles, individuals with ASD demonstrated an excess of biallelic variation (p<10−4) with or without normalization (Supplementary Figure 2c, Figure 1c). Biallelic NsynD4 alleles were also found in excess in cases (Figure 1c) but this did not persist after normalization (p=0.07, Supplementary Figure 2c).
Alleles contributing to biallelic LOF knockouts in cases were not in excess in the heterozygous state (Supplementary Figure 3). Genes impacted by biallelic mutation also scored low with respect to haploinsufficiency16 and gene constraint17 (Figure 1d–e, Supplementary Table 7), compared to known dominant and de novo ASD and ID genes18. These data support a recessive risk model, as opposed to the additive contributions of alleles acting co-dominantly.
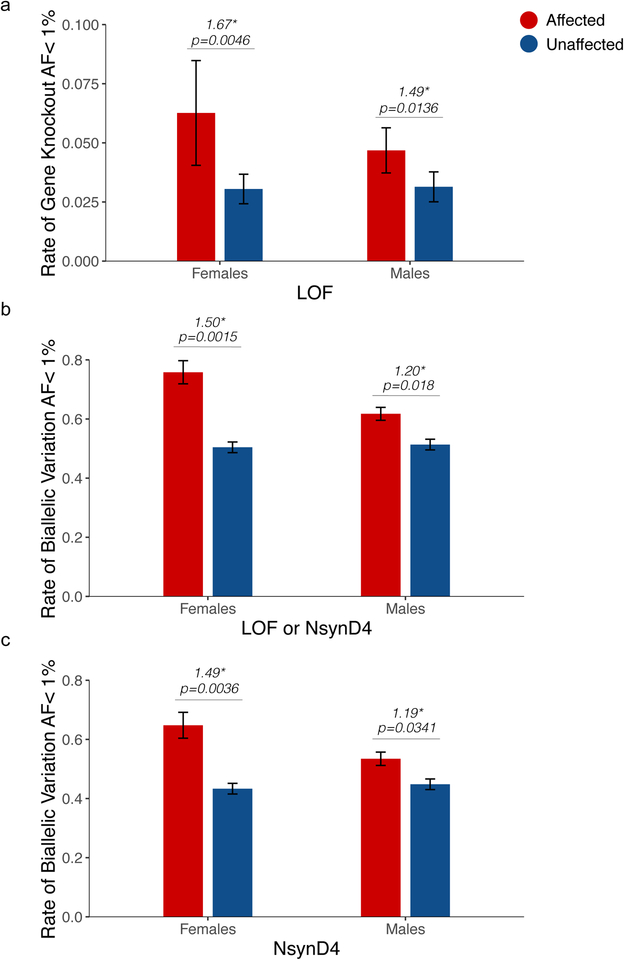
ASD exhibits a striking male bias, and females with ASD exhibit a disproportionate burden of de novo and inherited SNVs and CNVs, supporting a female protective/male susceptibility effect19,20. We asked whether this also applies to rare recessive mutations. Unaffected males and females did not differ in burdens of LOF knockout (Figure 2a, Supplementary Figure 4a). Females with ASD, however, exhibited a larger excess of biallelic LOF mutations (Figure 2a, Supplementary Figure 4a). Similar patterns were observed when the analysis was extended to incorporate damaging missense variants, either alone (NsynD4) (Figure 2b, Supplementary Figure 4b) or grouped with LOF mutation (LOF or NsynD4) (Figure 2c, Supplementary Figure 4c). As observed before, LOF alleles were not in excess in the heterozygous state (Supplementary Figure 3). From these patterns we conclude that the female protective/male susceptibility effect extends to recessive mutations as well.
Figure 2.
Biallelic mutations in ASD: effects of sex. (a) Rates of biallelic gene knockout (strict LOF) in the ASC, stratified by diagnosis and sex. (b, c) Rates of biallelic variation stratified by sex, considering (b) LOF or damaging missense (LOF or NsynD4) variants or (c) damaging missense variants alone (NsynD4). Error bars represent the 95% confidence intervals.
The ascertainment differentials observed led us to estimate the total contribution of biallelic mutation to ASD to be 3–5% (2% from LOF mutations, 1–3% from missense mutations). Furthermore, based on the ascertainment differentials, identifiable biallelic LOF or damaging missense alleles appeared to contribute to ~10% of female ASD cases in the ASC (Supplementary Figure 4b), suggesting that the category of biallelic mutation could be one of the largest genetic contributors to ASD in females.
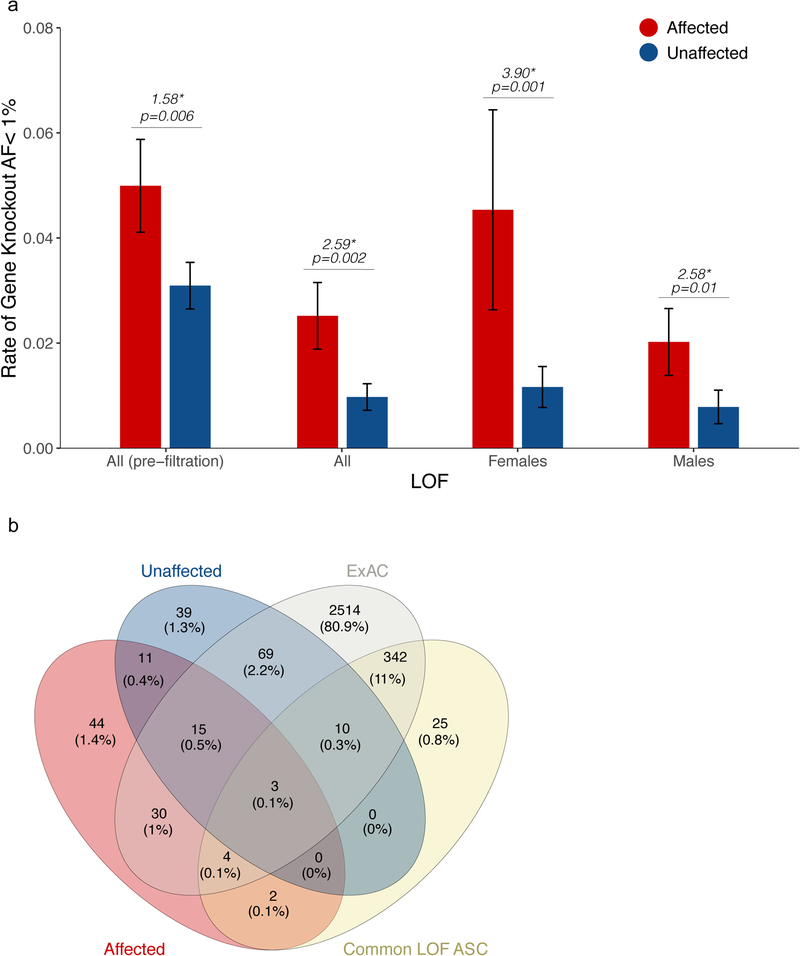
To address potential biological implications of these results, we examined the genes in which biallelic mutations were found. We first focused on the category of strict biallelic LOF mutations, estimating from the observed ascertainment differentials that ~25–40% of those found in ASD patients (0.019 out of 0.05; or 0.011 out of 0.042 after normalization to synonymous burdens) contribute to their condition. A total of 109 unique genes were knocked out (biallelic LOF) in affected individuals in our dataset. We reasoned that the most relevant of these genes would be those that are selected against biallelic inactivation in the general population. We therefore developed a filter to remove any gene that is also knocked out in one or more of 60,706 individuals sequenced as a part of ExAC (see Methods)17. To test its effectiveness, we examined its impact on observed patterns and burdens of gene knockouts in ASD. Overall, 131 of the 227 (58%) gene knockouts observed in the ASC were also observed in ExAC. Application of this filter reduced the overall burdens of observed biallelic knockout by half but left the ascertainment differentials intact (Figure 3a, Supplementary Figure 5). This significantly increased the observed proportional excess of biallelic LOF mutation: after filtering, on average, individuals with ASD were ~2.6 times as likely to have biallelic LOF mutations as individuals without ASD (p=0.002) (3.9X in females, p=0.001, and 2.6X in males, p=0.01) (Figure 3a).
Figure 3.
Biallelic mutations in ASD: ExAC filtration. (a) Rates of biallelic gene knockout (strict LOF) after filtration of commonly inactivated genes in ExAC. (b) Breakdown of candidate ASD genes excluded by presence of knockouts in ExAC, or by knockouts that are common and/or in unaffected individuals of the ASC. Error bars represent the 95% confidence intervals.
We applied a similar logic to biallelic missense mutations as well. We reasoned that missense alleles encountered in the homozygous state in the general population are not likely to contribute to recessive condition. ~50% of rare, biallelic damaging missense events in the ASC involved an allele encountered in the homozygous state in ExAC (Supplementary Figure 6a, Supplementary Table 8). Filtering these events allowed us to discern for the first time a significant excess of rare biallelic damaging missense variation (i.e. NsynD4) in cases compared to controls (Supplementary Figure 6a), with an ascertainment differential of ~4%. Biallelic Nsyn variants were also in slight excess (p=0.0037), which arose almost entirely from affected females (Supplementary Figure 6b).
These results indicate that the observed excess in ASD is driven by genes and alleles that are biallelically constrained in the control population, strongly supporting the biological relevance of these findings.
De novo gene disrupting mutations contribute strongly to ASD21. To examine possible interactions between de novo and biallelic gene disrupting events, we compared the burden of biallelic LOF events in cases with and cases without LOF de novo mutations (Supplementary Figure 10)21. An excess of rare biallelic LOF events is strongly evident in ASD cases lacking de novo LOF variants (Supplementary Figure 10, p=0.0006). Their contribution in ASD cases with de novo LOF variants is harder to distinguish, as the observed difference in burdens of biallelic LOF events did not achieve statistical significance (Supplementary Figure 7). These may represent orthogonal risk categories, although larger sample sizes will be required to clarify this in the future.
After ExAC filtering, a total of 57 biallelically constrained genes harbored biallelic LOF mutation in at least one ASD case. Of these, we can estimate that ~60% of these (based on the observed burdens, see Methods), and ~80% of the knockouts observed in females, likely confer ASD risk. To further enrich for relevant genes, we also set aside 13 genes knocked out in one or more unaffected individuals in our ASC dataset (Figure 3b, Supplementary Table 7), and three that failed Sanger confirmation, leaving a final set of 41 genes that were specifically knocked out in individuals diagnosed with ASD, but never in 66,557 controls (60,706 from ExAC and 5,852 unaffected individuals from ASC) (Supplementary Table 9,10).
Several of these represent genes already implicated in disease. Eight (CA2, DDHD1, NSUN2, PAH, RARB, ROGDI, SLC1A1, USH2A) were associated with recessive genetic conditions involving neurodevelopmental delay (Table 2 and Supplemental Information for clinical details). In at least six of these cases (CA2, NSUN2, PAH, RARB, ROGDI, USH2A), available medical and phenotypic records were concordant with the clinical features predicted from the genetic finding (Supplemental Information). Phenotypic data was either unavailable or incomplete, but not discordant, for other cases. In two cases, knockouts were encountered in genes that cause well-characterized autosomal recessive conditions without clear connection to autism (RFX5 and DNAI2, associated with autosomal recessive immunodeficiency and primary ciliary dyskinesia, respectively). In two other cases, knockouts were observed in genes associated with autosomal dominant disorders via recurrent gain-of-function mutations (IFITM5 and KIF22, causing osteogenesis imperfecta type V and spondyloepimetaphyseal dysplasia with joint laxity type 2, respectively). The phenotypic consequence of recessive loss-of-function mutations for these two genes is unknown. Interestingly, KIF22 is one of 25 genes in the 593kb core interval affected by 16p11.2 deletions.22 KIF22 was proposed as a one of two genes in the interval that demonstrates dosage sensitivity, with knockdown causing axon tract and behavioral defects in zebrafish,23 and disruption a KIF22 ortholog in Drosophila causes synapse development defects24.
Table 2.
Clinically relevant gene knockouts in the ASC
| Gene | Mutations | Disease relevance | Notes |
|---|---|---|---|
| CA2 | hom c.232+1G>A (NM_000067) | Carbonic anhydrase deficiency | Intellectual disability, osteopetrosis, renal tubular acidosis |
| DDHD1 | comp het c.1311–2A>T/c.2459–2A>T (NM_001160147) | Spastic paraplegia | Spastic paraplegia, mitochondrial abnormalities; only 3 cases reported |
| NSUN2 | hom c.C1708T:p.Q570X (NM_001193455) | Autosomal recessive intellectual disability | Syndromic intellectual disability; only 7 mutations reported |
| PAH | hom c.C703T:p.Q235X and hom c.592_613del:p.Y198fs (NM_000277) (two separate individuals) | Phenylketonuria | ~20% of individuals with PKU have autism (finding previously reported13) |
| SLC1A1 | hom c.G142T:p.E48X (NM_004170) | Dicarboxylic aminoaciduria | Elevated urinary glutamate and aspartate, variable neuropsychiatric symptoms; only 3 cases reported |
| RARB | hom c.C78A:p.C26X (NM_000965) | PDAC syndrome | Microphthalmia, pulmonary hypoplasia, diaphragmatic hernia, and cardiac defects; milder forms described |
| ROGDI | hom c.201–1G>T (NM_024589) | Kohlschütter-Tönz syndrome | Global developmental delay, epilepsy, spasticity, amelogenesis imperfecta |
| USH2A | comp het c.T12714G:p.Y4238X/ c.G6224A:p.W2075X (NM_206933) | Usher syndrome | Sensorineural hearing deficiencies, retinitis pigmentosa (finding previously reported14) |
Several of the remaining 33 genes are of strong neurobiological interest. One affected individual bore a homozygous LOF mutation (p.E48X) in SLC1A1, mutations in which have been linked to dicarboxylic aminoaciduria,25 an extremely rare disorder with prior associations with intellectual disability26,27 and obsessive compulsive disorder (OCD)25. SLC1A1 encodes a brain-expressed, activity-regulated glutamate transporter28,29 that modulates glutamatergic neurotransmission via a variety of mechanisms30,31,32,33,34,35 SLC1A1 knockout in mice results in altered locomotor behavior36 and learning and memory defects.37
Two brothers with ASD bore homozygous stop-gain mutations in FEV, a transcription factor that is required for both development and function of serotonergic neurons (Supplementary Figure 8a). These neurons send projections that ramify broadly throughout the CNS (Supplementary Figure 8b), and have wide-ranging neuromodulatory effects on many physiological states and behaviors, including mood, aggression, anxiety, sleep, and movement38. Serotonergic dysfunction has been strongly implicated in neuropsychiatric conditions including ASD39, schizophrenia and mood disorders38. In mice, Fev/Pet-1 has been shown to be highly expressed in the serotonergic raphe nuclei40, and inactivation of Fev during development41 or adulthood42 results in depletion of serotonergic neurons and neurobehavioral changes including anxiety and aggression.41,42 Human FEV is also expressed in the serotonergic raphe in adult brain43,44, and we found similar results via in situ hybridization analysis of human fetal brain as well (Supplementary Figure 8c–f). The older affected brother was diagnosed with autism and had a measured IQ of 69. He exhibited severe, stereotyped aggressive and self-injurious behaviors. He had a BMI of 25.6 (99%) and mildly dysmorphic facial features. EEG showed bilateral rolandic focus with generalization. The younger affected brother was diagnosed with PDD-NOS and intellectual disability (IQ not measured), had a BMI of 17.7 (91%), and no facial dysmorphisms (further details in Supplemental Information). We are not aware of any previous report of a human loss-of-function phenotype for FEV.
Gene knockouts were also identified in FCHSD2, a cytoskeletal adaptor protein that is highly expressed in human fetal brain and is the human homolog of Drosophila nervous wreck45, NCOA7, a brain-specific transcriptional coactivator for steroid and nuclear hormone receptors46, and MOB1B, a downstream effector in the Hippo signaling pathway, a pathway recently implicated in regulation of brain size47. Many additional genes in the candidate set have abundant brain expression (BRMS1L, ELOF1, GCN1L1, NUPR1, SLC22A6) but less clear neurobiological activity.
We also expanded our scope to incorporate biallelic missense mutation. There were 409 distinct genes that harbored biallelic, damaging missense mutations in cases but not in controls (Supplementary Figure 9, Supplementary Table 11). Only one gene was found that was independently hit in multiple families: AMT, an established cause of nonketotic hyperglycinemia (NKH). Hypomorphic mutations in AMT have previously been reported to cause ASD without NKH’s characteristic metabolic abnormalities. No other gene was hit more than once, so larger sample sizes would be necessary to establish any of these genes as ASD genes on this basis alone (Supplementary Table 11, Supplementary Figures 10 and 11). However, there were thirteen cases in which affected individuals bore biallelic mutations in alleles of established medical significance (in ClinVar as Pathogenic or Likely Pathogenic) (Table 3, Supplementary Table 12, Supplementary Information). All were diagnostic of genetic disorders with known neurodevelopmental consequences including classic metabolic diseases, mitochondrial depletion syndrome, and other syndromic conditions. All together, combined with the previously described eight LOF cases, biallelic mutations in known genes constituted ~1% of our cohort (21 out of 2,343 affected individuals), underscoring the importance of clinical screening for monogenic recessive conditions in this patient population.
Table 3.
Biallelic mutations identified in cases and controls previously reported as pathogenic or likely pathogenic in ClinVar.
| Gene | Condition | Sample identifier | Sex | Mutation | ClinVar (HGMD) | Variant pathogenicity | Damaging Predictions |
|---|---|---|---|---|---|---|---|
| Found in 13 cases | |||||||
| ASS1 | Citrullinemia type I | SKUSE5080161 | M | hom c.1168G>A:p.G390R (NM_000050.4) |
6329 (CM900037) | Pathogenic | 10 / 10 |
| ATP7B | Wilson disease | 80001103644 | M | het c.3207C>A:p.H1069Q (NM_000053.3) |
3848 (CM930059) | Pathogenic | 11 / 11 |
| het c.4087T>C:p.S1363P (NM_000053.3) |
Same site as p.S1363F (ClinVar 188908, HGMD CM992829) | (p.S1363F=Pathogenic) | 12 / 12 | ||||
| B3GALNT2 | Muscular dystrophy with brain malformations | 80001103921 | F | het c.1368+1G>A (NM_152490.4) |
na | (Likely Pathogenic) | na |
| het c.979G>A:p.D327N (NM_152490.4) |
424766 (CM144415) | Likely Pathogenic | 10 / 12 | ||||
| CA2 | Osteopetrosis, autosomal recessive 3 | AU-10501 | F | hom c.232+1G>A (NM_000067.2) |
288909 | Pathogenic | na |
| COQ8A | Coenzyme Q10 deficiency | AU-13901 | M | hom c.1805C>G:p.P602R (NM_020247.4) |
214050 (CM141297) | Pathogenic | 11 / 12 |
| GBE1 | Glycogen storage disease, type IV and Adult polyglucosan body disease | AU-24601 | F | hom c.986A>G:p.Y329C (NM_000158.3) |
371439 (CM128776) | Pathogenic | 11 / 11 |
| GLRA1 | Hereditary hyperekplexia | AU-12001 | M | hom c.277C>T:p.R93W (NM_000171.3) |
225379 (CM104312) | different missence LP in ClinVar (225379) | 10 / 11 |
| HADH | Deficiency of 3-hydroxyacyl-CoA dehydrogenase | 09C86928 | M | hom c.676T>C:p.Y226H (NM_005327.4) |
212734 (CM064046) | Likely pathogenic | 11 / 11 |
| MYO15A | Deafness, autosomal recessive 3 | SAGA-96 | F | het c.5515C>T:p.Q1839X (NM_016239.3) |
na | (Likely Pathogenic) | na |
| het c.8183G>A:p.R2728H (NM_016239.3) |
228276 (CM117958) | Pathogenic | 10 / 10 | ||||
| NAGLU | Sanfilippo syndrome B | AU-16201 | F | homc.934G>A:p.D312N (NM_000263.3) |
437446 (CM113463) | Pathogenic | 12 / 12 |
| PAH | Phenylketonuria | AU-16201 | F | het c.782G>A:p.R261Q (NM_000277.1) |
582 (CM910287) | Pathogenic | 12 / 12 |
| het c.842C>T:p.P281L (NM_000277.1) |
589 (CM910292) | Pathogenic | 11 / 11 | ||||
| POLG | Alpers-Huttenlocher syndrome, Childhood myocerebrohepatopathy spectrum | AU-16201 | F | hom c.3151G>A:p.G1051R (NM_002693.2) |
13501 (CM040472) | Pathogenic | 11 / 11 |
| USH2A | Usher Syndrome, Type 2A | 09C96107 | M | het c.12714T>G:p.Y4238X (NM_206933.2) |
48405 (CM134383) | Pathogenic | na |
| het c.6224G>A:p.W2075X (NM_206933.2) |
48554 (CM134384) | Pathogenic | |||||
| Found in 3 controls | |||||||
| MCCC2 | 3-methylcrotonyl-CoA carboxylase deficiency | AU-13303 | F | hom c.295G>C:p.E99Q(NM_022132.4) | 1920 (CM010910) | Pathogenic | 10 / 10 |
| PAH | Phenylketonuria | AU-16503 | F | hom c.898G>T:p.A300S(NM_000277.1) | 92751 (CM920555) | Pathogenic | 12 / 12 |
| AU-16502 | M | hom c.898G>T:p.A300S(NM_000277.1) | 92751 (CM920555) | Pathogenic | |||
Our study extends prior estimates of the contribution of recessive mutations to ASD13,14 and proves that the female protective / male susceptibility effect in ASD applies to biallelic mutations as well. Expanding these efforts to larger ASD cohorts should offer a path forward to delineate novel neurobiological mechanisms in autism.
METHODS
Whole exome data analyses
Whole-exome sequencing data were gathered as part of the Autism Sequencing Consortium48. We focused our analysis on nine datasets generated using the Illumina sequencing platform, Supplementary Table 1). Alignment and variant calling were performed as previously described. Low-quality variants were filtered with the following criteria: (1) SNPs with GQ<20, (2) SNPs not passing the standard PASS filter, and (3) all indels (due to excessive false positives leading to spurious frameshift calls). Variant annotation was performed using ANNOVAR49 to add allele frequencies, functional predictions, conservation, and gene annotations including OMIM and HGMD50 disease associations. Allele frequencies were also calculated for each contributing cohort and across the entire collection of ASC samples analyzed. Variants were categorized by their predicted impact into synonymous, nonsynonymous (missense), or loss-of-function (altering a canonical splice site or resulting in a stopgain). Additional classification of damaging missense variants (NsynD4) was performed for variants predicted to be deleterious by at least 4/6 algorithms (SIFT51, PolyPhen2_HDIV52, PolyPhen2_HVAR, MutationTaster53, MutationAssessor54, and LRT55).
Phasing was performed using Beagle 456 on the ASC dataset. VCFs were subdivided by chromosome and cohort prior to processing with Beagle employing default parameters.
Biallelic burden calculations
Allele frequency filtration was performed as described in the text using available public databases (1000G, EVS, ExAC). For rare variant calculations at maxAF 0.1%, 0.5%, 1%, 5%, and 10%, additional cohort-specific filters (cohort-specific AF of 5% or 10%) were applied to account for allelic variants that may be prevalent in a population but are not well represented in existing control databases. Burdens of homozygous and compound heterozygous variants were then counted for each cohort and for the entire ASC, stratifying by affected/unaffected status, gender, and mutational category (LOF, Nsyn, NsynD4, Syn).
To control for potential differences in underlying family and population structure of some of the constituent cohorts, we also measured rates of synonymous biallelic variation. In two cohorts (from the Middle East and Finland, respectively), rates of biallelic synonymous variation in affected individuals were higher than rates of biallelic synonymous variation in unaffected individuals (Supplementary Table 3). Therefore, for these cohorts, we normalized our LOF, NsynD4, and Nsyn burden calculations to the background rate of biallelic synonymous variation, to isolate the impact of biallelic LOF variation independent of background homozygosity in these cohorts.
The contribution of mutations to ASD risk was estimated using the following approach. First the ascertainment differential (AD) is calculated as the difference between the burden of biallelic mutations in cases and in controls and represents the estimated fraction of mutations that contribute to ASD (for example, an AD of 0.01 suggests that 1% of cases have a biallelic mutation contributing to ASD risk. The fraction of observed biallelic mutations contributing to disease is then calculated as AD divided by the total mutation burden in cases. For example, given a burden of 0.025 biallelic LOF events in cases and 0.01 biallelic LOF events in controls, the AD is 0.015 and the percentage of mutations identified which contribute to disease risk is 0.015 divided by 0.025, or 60%.
For burden comparisons, significance was assessed by permutation testing using 10,000 random assignments of diagnostic categorization to individuals within the study. A significance threshold of p<0.05 (i.e., without correction for multiple testing) was employed since the allele frequency cutoffs and variant annotation classes (LOF, LOF+NSynD4, LOF or NSynD4, NSynD4, NSyn) analyzed are not discrete. We do note that, all ExAC-filtered biallelic burden results (i.e., excess biallelic LOF, NSynD4, and NSyn mutations, summarized in Figure 3 and Supplementary Figures 5 and 6) were significant even with application of maximally conservative Bonferroni correction (e.g., p<0.01 assuming five fully independent hypotheses).
ExAC filtration
To generate a list of genes commonly inactivated by biallelic LOF mutation, we analyzed whole exome data from 60,706 individuals compiled by the Exome Aggregation Consortium (ExAC)17. We applied ANNOVAR to systematically re-annotate all variants from ExAC against RefSeq complete protein coding genes. Since phasing was unavailable, we focused on genes bearing putative homozygous LOF variants (any variant in stopgain, frameshift, or canonical splicing change). Homopolymer expansions / contractions in canonical splice regions were ignored, as expansions and contractions in these regions are common and have little impact on splicing. With these parameters, a total of 2986 genes were found to be inactivated by homozygous stopgain, frameshift, or canonical splice site mutation in ExAC. We also used this re-annotated dataset to generate a list of ExAC alleles encountered in the homozygous state, to identify missense and other mutations that are more likely to be clinically benign. For the final LOF candidate genes presented in Supplementary Tables 9 and 10, contributory alleles were also confirmed to be absent in the homozygous state in the gnomAD dataset17.
Variant confirmation
Sanger sequencing was employed to confirm the accuracy of exome variant calls for a subset of LOF and NsynD4 variants with an empirical validation rate of 96% (107 of 114 alleles tested). These included all LOF alleles reported here for which DNA samples were available. If DNA samples were not available, we note the presence of allelic variants in known databases and/or whether predicted genotypes follow Mendelian segregation patterns in parental exome data (where available) as additional support.
Human tissue collection and in situ hybridization for FEV
Human embryonic and fetal tissues were obtained from the MRC-Wellcome Trust Human Developmental Biology Resource (www.hdbr.org)57 following appropriate consent from the women involved and adhering to the relevant HTA guidelines with approval from Research Ethics Committee NRES Committee North East - Newcastle & North Tyneside 1.
RNA probes were produced by amplifying human embryonic cDNA using a forward primer to FEV (NM_017521, nt. 1286–1305) tagged with a T7 RNA promoter sequence and a reverse primer (NM_017521, nt. 1862–1843) tagged with a SP6 RNA promoter sequence (Supplementary Table 13). The resulting fragment was in vitro transcribed using the DIG RNA Labeling Kit (Roche).
The in situ hybridizations were performed as previously described48, except that 60ng RNA probe in 100ul ULTRAhyb (Ambion) per slide was used for the hybridization and the post-hybridization washes were modified: slides were washed for 10 minutes in 5 X SSC, twice in 2 X SSC, 50% formamide/1 X SSC, 1 X SSC and finally in 0.1X SSC. The hybridization incubation and all post-hybridization steps were performed at 63.5°C. Three human fetal brain samples were used: a Carnegie Stage (CS) 23 brain (12959), a CS23 head (12011), and a 10 post conception week hindbrain (sample 12721). Findings described were confirmed in at least two specimens. Expression patterns were analyzed relative to spatial models provided by the HuDSeN human gene expression spatial database project (http://www.hudsen.org).58
Reporting Summary
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Hossain and N. Hatem for their help with sample preparation, and Janet Kerwin for her help with analysis of in situ hybridization results. R.N.D. was supported by an NIH T32 fellowship from the Fundamental Neurobiology Training Grant (5 T32 NS007484–14) and the Nancy Lurie Marks Clinical and Research Fellowship Program in Autism. The ASC is supported by the National Institute of Mental Health (grants MH100233, MH100229, MH100209, MH100239, MH111661, MH111660, MH111662, and MH111658). Collection of the PAGES cohort is supported by the National Institute of Mental Health (grant MH097849). This work was supported in part through the computational resources provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai, and the Research Information Technology Group at Harvard Medical School, which is partially supported by NIH grant NCRR 1S10RR028832–01. Human embryonic and fetal material was provided by the joint MRC/Wellcome Trust (grant # MR/R006237/1) Human Developmental Biology Resource (www.hdbr.org). C.A.W. is an Investigator of the Howard Hughes Medical Institute. C.A.W. and T.W.Y. were supported by the National Institute of Mental Health (NIMH) (grants RC2MH089952 and RO1MH083565). T.W.Y. was supported by NIH/NIMH R01MH113761, NICHD/NHGRI/NIH U19HD077671, NIH/NICHD U24HD0938487 and a SFARI Pilot Research Award. S.D.R. and J.D.B. are supported by the Beatrice and Samuel A. Seaver Foundation.
Footnotes
DATA AVAILABILITY
Data included in this manuscript is deposited at dbGAP merged with published data under accession number phs000298.v4.p3.
COMPETING INTERESTS
The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to Timothy.yu@childrens.harvard.edu.
References
- 1.Prevention, C. f. D. C. a. & Baio J. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years. Morbidity and Mortality Weekly Report 63 (2014). [PubMed] [Google Scholar]
- 2.Robinson EB et al. Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat Genet 48, 552–555, doi: 10.1038/ng.3529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaugler T et al. Most genetic risk for autism resides with common variation. Nat Genet 46, 881–885, doi: 10.1038/ng.3039 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiner DJ et al. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat Genet 49, 978–985, doi: 10.1038/ng.3863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krumm N et al. Excess of rare, inherited truncating mutations in autism. Nat Genet 47, 582–588, doi: 10.1038/ng.3303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Rubeis S et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature News 515, 209–215, doi: 10.1038/nature13772 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy D et al. Rare De Novo and Transmitted Copy-Number Variation in Autistic Spectrum Disorders. 70, 886–897, doi: 10.1016/j.neuron.2011.05.015 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Sanders SJ et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241, doi: 10.1038/nature10945 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iossifov I et al. De novo gene disruptions in children on the autistic spectrum. 74, 285–299, doi: 10.1016/j.neuron.2012.04.009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronemus M, Iossifov I, Levy D & Wigler M The role of de novo mutations in the genetics of autism spectrum disorders. Nat Rev Genet 15, 133–141, doi: 10.1038/nrg3585 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Morrow EM et al. Identifying autism loci and genes by tracing recent shared ancestry. Science (New York, NY) 321, 218–223, doi: 10.1126/science.1157657 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chahrour MH et al. Whole-exome sequencing and homozygosity analysis implicate depolarization-regulated neuronal genes in autism. PLoS genetics 8, e1002635, doi: 10.1371/journal.pgen.1002635 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu TW et al. Using Whole-Exome Sequencing to Identify Inherited Causes of Autism. 77, 259–273, doi: 10.1016/j.neuron.2012.11.002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim ET et al. Rare Complete Knockouts in Humans: Population Distribution and Significant Role in Autism Spectrum Disorders. 77, 235–242, doi: 10.1016/j.neuron.2012.12.029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buxbaum JD et al. in Neuron Vol. 76 1052–1056 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang N, Lee I, Marcotte EM & Hurles ME Characterising and predicting haploinsufficiency in the human genome. PLoS genetics 6, e1001154, doi: 10.1371/journal.pgen.1001154 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291, doi: 10.1038/nature19057 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betancur C Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain research 1380, 42–77, doi: 10.1016/j.brainres.2010.11.078 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Jacquemont S et al. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. American journal of human genetics 94, 415–425, doi: 10.1016/j.ajhg.2014.02.001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson EB, Lichtenstein P, Anckarsäter H, Happé F & Ronald A Examining and interpreting the female protective effect against autistic behavior. Proceedings of the National Academy of Sciences of the United States of America 110, 5258–5262, doi: 10.1073/pnas.1211070110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Rubeis S et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215, doi: 10.1038/nature13772 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss LA et al. A recurrent genetic cause of autism: microdeletion at 16p11.2. (2007). [Google Scholar]
- 23.Blaker-Lee A, Gupta S, McCammon JM, De Rienzo G & Sive H Zebrafish homologs of genes within 16p11.2, a genomic region associated with brain disorders, are active during brain development, and include two deletion dosage sensor genes. Disease models & mechanisms 5, 834–851, doi: 10.1242/dmm.009944 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park SM, Littleton JT, Park HR & Lee JH Drosophila Homolog of Human KIF22 at the Autism-Linked 16p11.2 Loci Influences Synaptic Connectivity at Larval Neuromuscular Junctions. Experimental neurobiology 25, 33–39, doi: 10.5607/en.2016.25.1.33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey CG et al. Loss-of-function mutations in the glutamate transporter SLC1A1 cause human dicarboxylic aminoaciduria. The Journal of clinical investigation 121, 446–453, doi: 10.1172/JCI44474 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teijema HL, van Gelderen HH, Giesberts MA & Laurent de Angulo MS Dicarboxylic aminoaciduria: an inborn error of glutamate and aspartate transport with metabolic implications, in combination with a hyperprolinemia. Metabolism: clinical and experimental 23, 115–123 (1974). [DOI] [PubMed] [Google Scholar]
- 27.Swarna M, Rao DN & Reddy PP Dicarboxylic aminoaciduria associated with mental retardation. Human genetics 82, 299–300 (1989). [DOI] [PubMed] [Google Scholar]
- 28.Rothstein JD et al. Localization of neuronal and glial glutamate transporters. 13, 713–725 (1994). [DOI] [PubMed] [Google Scholar]
- 29.Ross JR, Porter BE, Buckley PT, Eberwine JH & Robinson MB mRNA for the EAAC1 subtype of glutamate transporter is present in neuronal dendrites in vitro and dramatically increases in vivo after a seizure. Neurochemistry International 58, 366–375, doi: 10.1016/j.neuint.2010.12.012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nieoullon A et al. The neuronal excitatory amino acid transporter EAAC1/EAAT3: does it represent a major actor at the brain excitatory synapse? Journal of neurochemistry 98, 1007–1018, doi: 10.1111/j.1471-4159.2006.03978.x (2006). [DOI] [PubMed] [Google Scholar]
- 31.Bianchi MG, Bardelli D, Chiu M & Bussolati O Changes in the expression of the glutamate transporter EAAT3/EAAC1 in health and disease. Cellular and molecular life sciences : CMLS 71, 2001–2015, doi: 10.1007/s00018-013-1484-0 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stafford MM, Brown MN, Mishra P, Stanwood GD & Mathews GC Glutamate spillover augments GABA synthesis and release from axodendritic synapses in rat hippocampus. Hippocampus 20, 134–144, doi: 10.1002/hipo.20600 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathews GC & Diamond JS Neuronal glutamate uptake Contributes to GABA synthesis and inhibitory synaptic strength. J Neurosci 23, 2040–2048 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scimemi A, Tian H & Diamond JS Neuronal Transporters Regulate Glutamate Clearance, NMDA Receptor Activation, and Synaptic Plasticity in the Hippocampus. Journal of Neuroscience 29, 14581–14595, doi: 10.1523/JNEUROSCI.4845-09.2009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diamond JS Neuronal glutamate transporters limit activation of NMDA receptors by neurotransmitter spillover on CA1 pyramidal cells. J Neurosci 21, 8328–8338 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peghini P, Janzen J & Stoffel W Glutamate transporter EAAC-1-deficient mice develop dicarboxylic aminoaciduria and behavioral abnormalities but no neurodegeneration. The EMBO journal 16, 3822–3832, doi: 10.1093/emboj/16.13.3822 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee S, Park SH & Zuo Z Effects of isoflurane on learning and memory functions of wild-type and glutamate transporter type 3 knockout mice. The Journal of pharmacy and pharmacology 64, 302–307, doi: 10.1111/j.2042-7158.2011.01404.x (2012). [DOI] [PubMed] [Google Scholar]
- 38.Lucki I The spectrum of behaviors influenced by serotonin. Biological psychiatry 44, 151–162 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Muller CL, Anacker AMJ & Veenstra-VanderWeele J The serotonin system in autism spectrum disorder: From biomarker to animal models. Neuroscience 321, 24–41, doi: 10.1016/j.neuroscience.2015.11.010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hendricks T, Francis N, Fyodorov D & Deneris ES The ETS domain factor Pet-1 is an early and precise marker of central serotonin neurons and interacts with a conserved element in serotonergic genes. The Journal of neuroscience : the official journal of the Society for Neuroscience 19, 10348–10356 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hendricks TJ et al. Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron 37, 233–247 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Liu C et al. Pet-1 is required across different stages of life to regulate serotonergic function. Nature Neuroscience 13, 1190–1198, doi: 10.1038/nn.2623 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iyo AH, Porter B, Deneris ES & Austin MC Regional distribution and cellular localization of the ETS-domain transcription factor, FEV, mRNA in the human postmortem brain. Synapse 57, 223–228, doi: 10.1002/syn.20178 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maurer P et al. The Ets transcription factor Fev is specifically expressed in the human central serotonergic neurons. Neuroscience letters 357, 215–218, doi: 10.1016/j.neulet.2003.12.086 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Cao H et al. FCHSD1 and FCHSD2 Are Expressed in Hair Cell Stereocilia and Cuticular Plate and Regulate Actin Polymerization In Vitro. PLoS ONE 8, e56516, doi: 10.1371/journal.pone.0056516 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shao W, Halachmi S & Brown M ERAP140, a conserved tissue-specific nuclear receptor coactivator. Molecular and cellular biology 22, 3358–3372 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poon CLC, Mitchell KA, Kondo S, Cheng LY & Harvey KF The Hippo Pathway Regulates Neuroblasts and Brain Size in Drosophila melanogaster. Current biology : CB 26, 1034–1042, doi: 10.1016/j.cub.2016.02.009 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Cheng YZ et al. Investigating embryonic expression patterns and evolution of AHI1 and CEP290 genes, implicated in Joubert syndrome. PLoS One 7, e44975, doi: 10.1371/journal.pone.0044975 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang K, Li M & Hakonarson H ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research 38, e164, doi: 10.1093/nar/gkq603 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stenson PD et al. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Human genetics 136, 665–677, doi: 10.1007/s00439-017-1779-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng PC & Henikoff S SIFT: Predicting amino acid changes that affect protein function. Nucleic acids research 31, 3812–3814 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adzhubei IA et al. A method and server for predicting damaging missense mutations. Nature methods 7, 248–249, doi: 10.1038/nmeth0410-248 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwarz JM, Rodelsperger C, Schuelke M & Seelow D MutationTaster evaluates disease-causing potential of sequence alterations. Nature methods 7, 575–576, doi: 10.1038/nmeth0810-575 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Reva B, Antipin Y & Sander C Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic acids research 39, e118, doi: 10.1093/nar/gkr407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chun S & Fay JC Identification of deleterious mutations within three human genomes. Genome research 19, 1553–1561, doi: 10.1101/gr.092619.109 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Browning SR & Browning BL Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81, 1084–1097, doi: 10.1086/521987 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerrelli D, Lisgo S, Copp AJ & Lindsay S Enabling research with human embryonic and fetal tissue resources. Development 142, 3073–3076, doi: 10.1242/dev.122820 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kerwin J et al. The HUDSEN Atlas: a three-dimensional (3D) spatial framework for studying gene expression in the developing human brain. Journal of anatomy 217, 289–299, doi: 10.1111/j.1469-7580.2010.01290.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.