Abstract
Despite considerable efforts to identify cancer metabolic alterations that might unveil druggable vulnerabilities, systematic characterizations of metabolism as it relates to functional genomic features and associated dependencies remain uncommon. To further understand the metabolic diversity in cancer, we profiled 225 metabolites in 928 cell lines from more than 20 cancer types in the Cancer Cell Line Encyclopedia (CCLE) using liquid chromatography-mass spectrometry (LC-MS). This resource enables unbiased association analysis linking cancer metabolome to genetic alterations, epigenetic features, and gene dependencies. Additionally, by screening barcoded cell lines, we demonstrated that aberrant ASNS hypermethylation sensitizes subsets of gastric and hepatic cancers to asparaginase therapy. Finally, our analysis revealed distinct synthesis and secretion patterns of kynurenine, an immune-suppressive metabolite, in model cancer cell lines. Together, these findings and related methodology provide comprehensive resources that will help to clarify the landscape of cancer metabolism.
Cell metabolism involves a highly coordinated set of activities in which multi-enzyme systems cooperate to convert nutrients into building blocks for macromolecules, energy currencies, and biomass1,2. In cancer, genetic or epigenetic changes can perturb the activity of key enzymes or rewire oncogenic pathways resulting in cell metabolism alterations3,4. Specific metabolic dependencies in cancer have also been the basis for effective therapeutics including inhibitors that target IDH1, as well as folate and thymidine metabolism5. The search for new drug targets, however, has been hampered, at least in part, by the fact that cancer metabolomic studies often draw conclusions from small numbers of cell lines from which generalizations are difficult. In contrast, there have been no systematic profiling efforts that encompass hundreds of cellular and genetic contexts. Furthermore, there is no high-throughput methodology that assesses cancer metabolic needs by perturbing related pathways across many cell lines. Consequently, the discovery of new anticancer metabolic targets might benefit from high-quality, comprehensive metabolomic data in addition to the current CCLE-related characterization that includes genomic, transcriptomic features as well as genetic dependency maps6-8.
Profiling metabolites from cultured CCLE cell lines
928 cancer cell lines from 20 major cancer types were cultured in vitro for metabolomic profiling of 124 polar and 101 lipid species (Fig. 1a). Extracted polar and lipid metabolites were analyzed using hydrophilic interaction chromatography (HILIC) and reversed phase (RP) chromatography (Fig. 1b). Sample measurements were obtained in four batches using pooled lysates as references to ensure consistent data quality. Trend normalization methods were applied before performing global comparisons. To assess experimental variation for each metabolite, we estimated a coefficient of variation (CV) based on 14 cell lines with biological replicates and found that 221 out of 225 metabolites had CV<0.1 (Extended Data Fig. 1a), indicating reproducible estimates for the majority of profiled metabolites. The complete metabolomic data and CV values are in Supplementary Table 1.
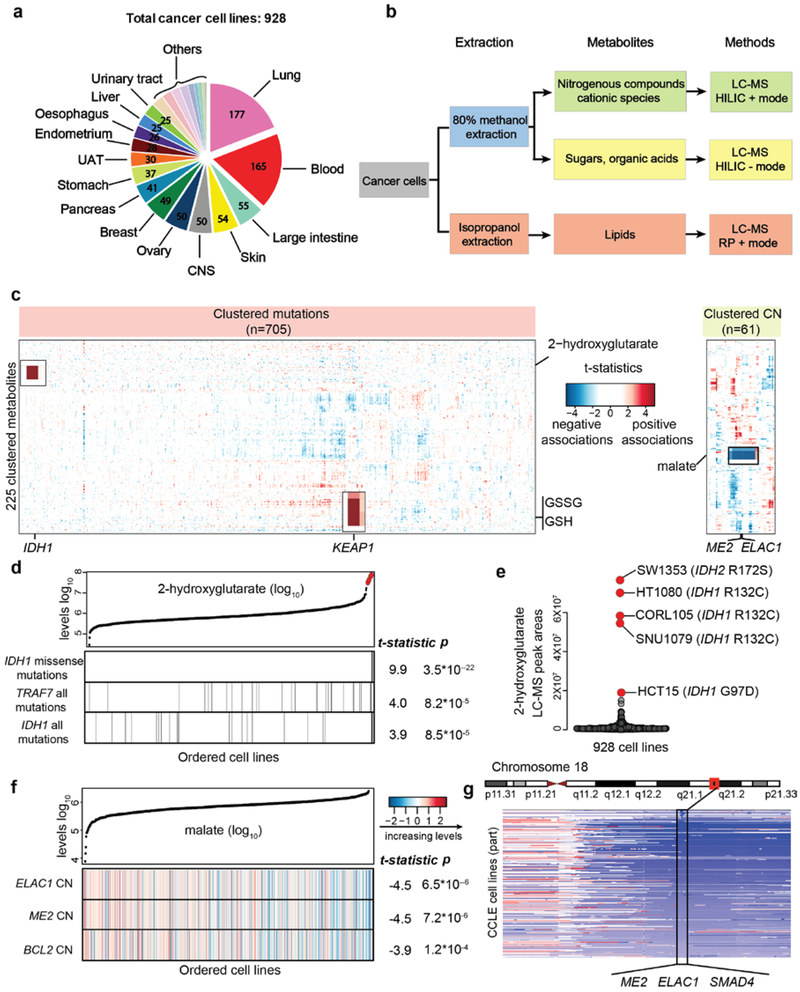
Fig.1 ∣. The CCLE database enables quantitative metabolomic modeling in relation to genetic features.
a, 928 cancer cell lines from more than 20 major tissues of origin were profiled for the abundance of 225 metabolites. The number of cell lines is annotated based on the tissues of origin. b, Schematic summarizing the workflow of metabolite profiling. c, Heatmap of 225 clustered metabolites (Y axis) and their associations with selected genetic features (X axis). T-statistics were calculated based on linear regression for each metabolite paired with each feature across all cell lines conditioned on the major lineages and were used to represent the regression coefficients scaled by standard deviations. Examples mentioned in the text are magnified and shown in black boxes. CN, copy number. d, 2HG and the top correlated mutations among all mutational features. Cell lines are shown as lines and ordered by increasing levels of 2HG. Those cell lines with corresponding mutations are labeled as black. The reported test statistics and p-values are based on the significance tests of genetic feature regression coefficients (cell line n=927, two-sided t-tests). e, Cancer cell lines with outlier levels of 2HG have specific IDH1/2 mutations. f, Malate levels and a heatmap representation of top correlated copy number alterations among all copy number features. The features are color-coded to indicate various copy number alterations (log2 scale) and are standardized for plotting. The reported test statistics and p-values are based on the significance tests of genetic feature regression coefficients (cell line n=912, two-sided t-tests). g, Schematic of the genomic locus containing ME2, ELAC1, and SMAD4.
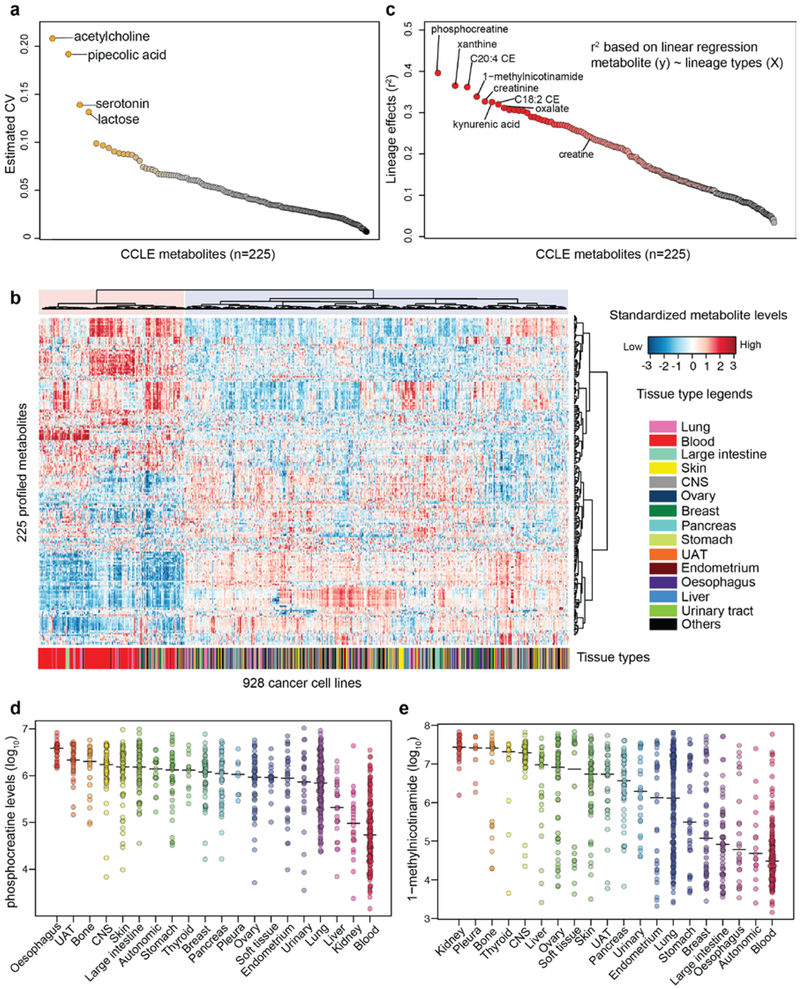
Next, we evaluated the metabolic similarities among cell lines using hierarchical clustering. The two main clusters strongly correlated with hematopoietic versus non-hematopoietic lineages, suggesting major metabolic differences in these two groups (p<1×10−10, Fisher’s exact test) (Extended Data Fig. 1b). To further quantify the degree that metabolite levels (y) associate with major cancer lineages (X), we adopted a linear regression model (see Methods). The result showed that 148 out of 225 metabolites had a lineage effect (defined as r2) greater than 0.1 (Extended Data Fig. 1c, Supplementary Table 1), suggesting that for these metabolites at least 10% of the cross-cell line metabolite level variances was related to lineage. To obtain lineage-specific distributions, we estimated the average abundance for each metabolite in major cancer lineages (Supplementary Table 1). This leads to a quantitative, cross-lineage view of metabolite levels in model cancer cell lines. For example, phosphocreatine accumulates in oesophagus cell lines but not in kidney or hematopoietic lineages (Extended Data Fig. 1d). In contrast, 1-methylnicotinamide is highly abundant in most kidney and pleura cell lines but is lower in many other lineages with multiple outliers (Extended Data Fig. 1e). These data suggest that lineage distinctions exist in model cancer cell lines and can influence cross-cell line metabolic comparisons.
Interrogating metabolite associations with genetic features
In addition to lineage, genetic or epigenetic events in cancer are likely to alter cellular metabolism. In order to identify metabolic variation that might be attributable to genetic differences, we curated a matrix of genetic features including 705 gene mutations and 61 amplifications or deletions. To look for associations between these genetic features and metabolite levels, linear regression models controlling for lineage effects were applied (Fig.1c, See Methods). The genetic features were scored by associations with each metabolite and can be compared in the order of statistical significance (Supplementary Table 2). Interestingly, we found that mechanistically relevant features often displayed strong correlations with aberrant metabolite levels. The selected examples are discussed below.
First, our unbiased comparison revealed the expected finding that for 2-hydroxyglutarate (2HG), the IDH1 hotspot missense mutation was a top predictive genetic feature (Fig. 1d). Cell lines with an aberrant accumulation of this metabolite are mostly IDH1/IDH2 mutants (Fig. 1e), recapitulating the known relationship9,10. Notably, although there are no known IDH1/IDH2 mutants in the CCLE renal cell carcinoma lines (RCC), additional lineage effect analysis revealed that on average RCC cells had a 3-fold higher level of 2HG than others (Supplementary Table 1). This is consistent with the observation of increased 2HG levels in RCC tumors11. Our analysis also showed KEAP1 mutation as the genetic feature most strongly associated with glutathione (GSH/GSSG) and NADP+ (Extended Data Fig. 2a, Supplementary Table 2). On average, cancer cell lines with mutated KEAP1 had ~2-fold higher levels of these redox metabolites (Extended Data Fig. 2b), suggesting a gain of buffering abilities against oxidative stresses12,13. Unlike the strong associations seen with KEAP1 and IDH1 alterations, it is noteworthy that a number of common oncogenic events including EGFR, KRAS, NRAS, TP53, PTEN, and TSC1/2 had weak to non-significant associations with the profiled metabolites when queried using unbiased analysis (Extended Data Fig. 2c-h, Supplementary Table 2).
In copy-number space, using malate as an example, we show that the most strongly associated features are deletions of ELAC1 and ME2 (Fig. 1f, Supplementary Table 2). These genes are co-localized in a 0.4 Mb region surrounding the tumor suppressor gene SMAD4 on chromosome 18 and are frequently co-deleted (Fig. 1g). ME2 (malic enzyme 2) catalyzes the oxidative decarboxylation of malate to pyruvate. Its mRNA levels were abolished in cells harboring homozygous deletion (Extended Data Fig. 2i), resulting in a correlation with higher malate levels (Extended Data Fig. 2j). While ME3 was previously found to be synthetic lethal with this deletion event14, we did not observe greater ME3 dependency in cancer cells having ME2 loss based on current CRISPR-Cas9 data in the cancer dependency map15 (Extended Data Fig. 2k).
To summarize, this resource enables unbiased association analysis between metabolites and various genetic features and confirms previous findings linking oncogenic changes (e.g., IDH1/KEAP1/ME2) to aberrant metabolite levels.
DNA methylation regulates metabolite abundances
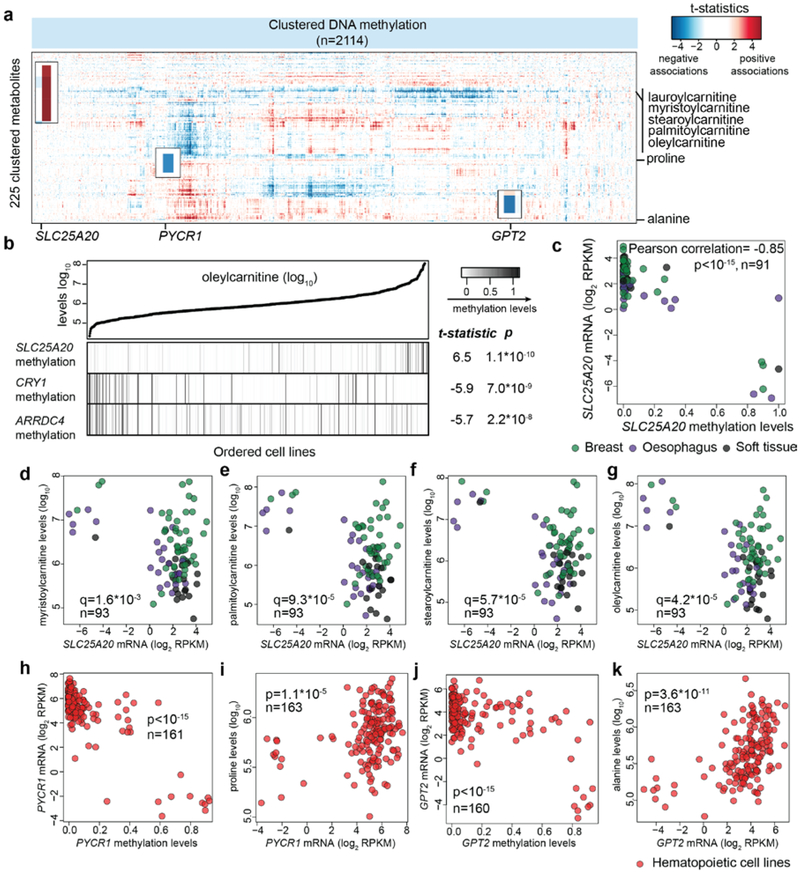
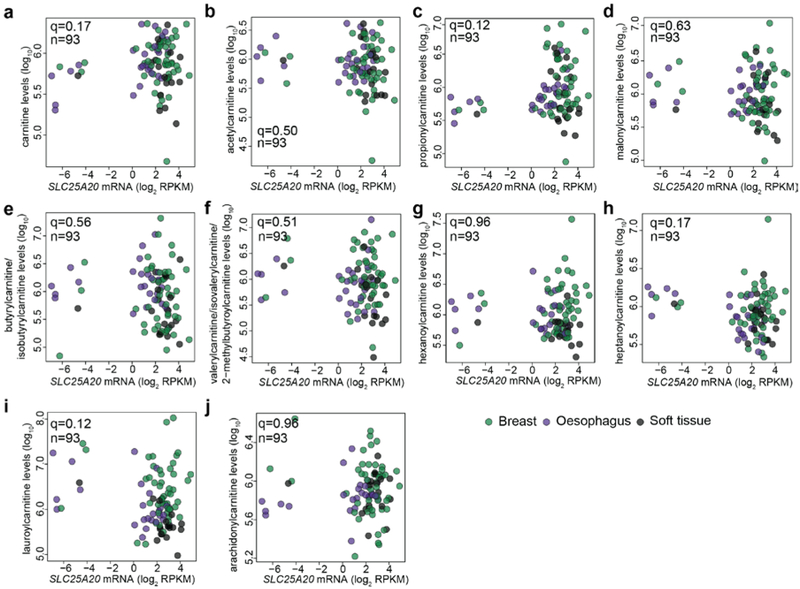
Next, we examined DNA methylation and assessed the associations with the metabolite levels. 2114 genes whose mRNA transcripts were significantly associated with their promoter CpG methylation levels were included in this analysis given that these selected genes were likely to be regulated via DNA methylation. Systematic analysis of the correlates revealed a surprising number of specific alterations related to potential metabolic dysregulation (Fig. 2a, Supplementary Table 2). These observations can be classified into two classes. First, DNA hypermethylation appears to influence metabolite levels via suppressing certain metabolite degradation pathways. For example, SLC25A20 methylation was strongly correlated with the accumulation of long-chain acylcarnitine species (e.g., oleylcarnitine) (Fig. 2b). SLC25A20, also known as carnitine/acylcarnitine translocase, shuttles acylcarnitines across the mitochondrial inner membrane for fatty acid oxidation16. SLC25A20 hypermethylation correlated with marked mRNA transcript reduction (Fig. 2c), which was associated with significantly elevated levels of acylcarnitine species having acyl chains of 14, 16 or 18 carbons (Fig. 2d-g), but not with other acylcarnitine species with shorter or longer acyl chains (Extended Data Fig. 3a-j), indicating an unusual specific fatty acid catabolism defects in these cell lines. Second, DNA hypermethylation appears to regulate metabolite levels by limiting components of biosynthetic pathways. For example, reduced proline levels were associated with the hypermethylation of PYCR1, an enzyme that converts pyrroline-5-carboxylate to proline (Fig. 2h, i). Additionally, decreased alanine levels were associated with the hypermethylation of GPT2, which can synthesize alanine via transamination (Fig. 2j, k). Both of these effects were particularly strong among hematopoietic cell lines. Taken together, this resource provides an unbiased way to assess the impact of DNA methylation events in regulating intracellular metabolite concentrations.
Fig.2 ∣. Systematic evaluations of metabolite associations with gene methylation patterns.
a, Heatmap of 225 clustered metabolites (Y axis) and their associations with selected gene methylation features (X axis). b, Oleylcarnitine (an example of long-chain acylcarnitines) and the top correlated features among all methylation features. The reported test statistics and p-values are based on the significance tests of DNA methylation feature regression coefficients (cell line n=811, two-sided t-tests). c, Scatter plot comparing SLC25A20 DNA methylation levels with its mRNA levels in selected lineages. d-g, Scatter plots comparing SLC25A20 mRNA levels with different acylcarnitines: myristoylcarnitine (d), palmitoylcarnitine (e), stearoylcarnitine (f), and oleycarnitine (g). The q-values were calculated based on the significance test of Pearson correlations (two-sided) with multiple hypothesis testing correction. h, Scatter plot comparing PYCR1 DNA methylation levels with its mRNA transcripts in hematopoietic cell lines. i, Scatter plot comparing PYCR1 mRNA transcripts with proline levels in hematopoietic cell lines. j, Scatter plot comparing GPT2 DNA methylation levels with its mRNA transcripts in hematopoietic cell lines. k, Scatter plot comparing GPT2 mRNA transcripts with alanine levels in hematopoietic cell lines. For h-k, the p-values were calculated based on the significance test of Pearson correlations (two-sided).
Metabolite-dependency association analysis
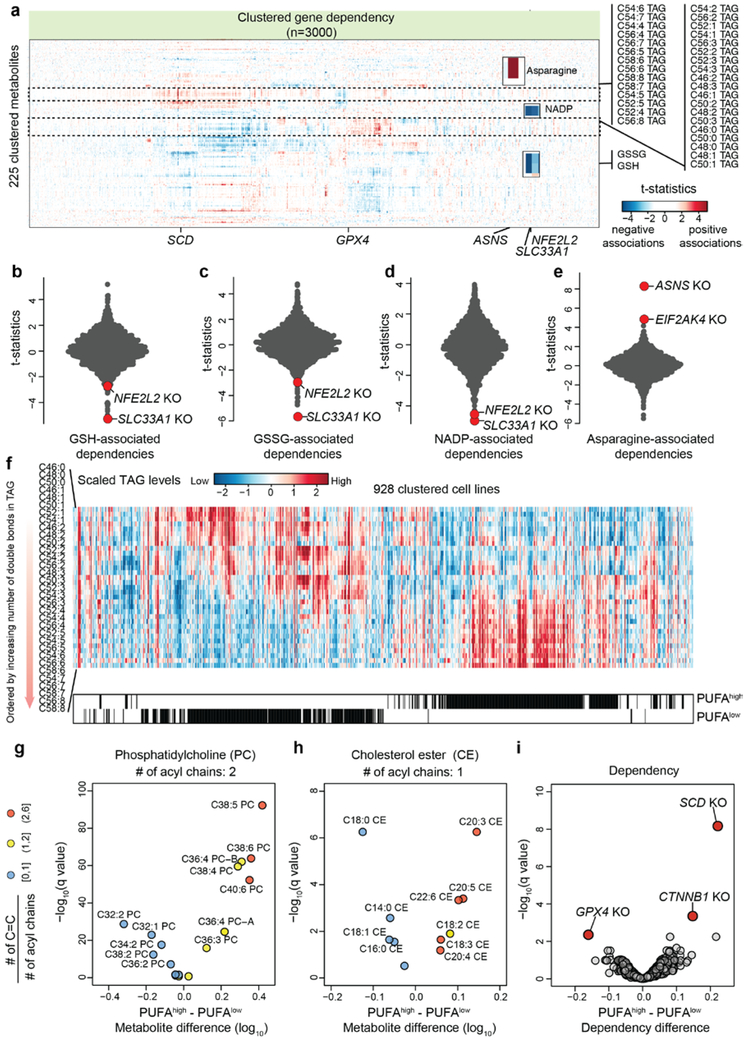
There has been a longstanding desire to take therapeutic advantage of dysregulated cancer metabolic states. To this end we sought to link metabolic alterations to cancer vulnerabilities unveiled in the DepMap CRISPR-Cas9 knockout dataset in which 483 CCLE cell lines have been screened with a library of ~74k sgRNAs targeting ~17,000 genes15. CERES scores were used to summarize gene-level dependency (small values indicate greater sensitivity to gene knockout)15 and then each gene level dependence was queried with respect to metabolite alterations. This unbiased metabolite-dependency association analysis shows that the dissimilar metabolic phenotypes observed in cancer cell lines are paired with distinct gene dependencies and therefore potential therapeutic targets (Fig.3a, Supplementary Table 3). Here, we focus on the top 3000 dependent genes (See Methods) and highlight representative examples in metabolism related to redox balance, amino acids, and lipids. First, as expected, aberrant accumulation of redox metabolites including GSH, GSSG, and NADP+ (partly attributed to KEAP1 mutation, vide supra) was associated with increased sensitivity to knockout of NFE2L2 (NRF2), a transcription activator involved in antioxidant response (Fig.3b-d). Notably, the most associated dependency was SLC33A1 (Fig.3b-d), an acetyl-CoA transporter whose role in redox homeostasis is currently unknown. As another example, we found that cells with lower asparagine levels were more dependent on its synthetase (ASNS) (explored further below) and EIF2AK4 (GCN2, involved in amino acid starvation response) (Fig.3e). Furthermore, we also observed an interesting association involving two distinct triacylglycerol (TAG) clusters (Fig.3a). One cluster consisted of polyunsaturated TAG species (at least 4 total C=C double bonds from 3 acyl chains) and the other cluster consisted of less unsaturated TAG species including monounsaturated fatty acyls (MUFA) (Fig.3a). To classify cancer cell lines enriched with either cluster, we labeled them as polyunsaturated fatty acyl high (PUFAhigh, n=315) or polyunsaturated fatty acyl low (PUFAlow, n=325) after excluding those with non-significant lipid unsaturation differences (Fig.3f). This unsaturation difference also existed in other lipid species such as phosphatidylcholines (PC, Fig.3g), and cholesterol esters (CE, Fig.3h). We asked whether this distinct lipid utilization pattern might link to targetable dependencies. By comparing CERES scores, we found that the PUFAhigh cell lines are sensitive to the knockout of GPX4 (Fig.3i), which mediates the detoxification of peroxidized PUFA17. In contrast, PUFAlow cell lines are sensitive to the loss of CTNNB1 or SCD (Fig.3i), which synthesizes MUFA. Together, these unbiased association analyses suggest that cancer cell lines cultured in vitro have significant lipidomic differences that can be selectively targeted based on PUFA classifications.
Fig.3 ∣. Systematic evaluations of metabolite-dependency associations.
a, Heatmap of 225 clustered metabolites (Y axis) and their associations with top 3000 gene dependencies (CERES scores) (X axis). The two distinct lipid groups revealed by clustering are highlighted. TAG, triacylglycerol. b-e, T-statistics based on selected metabolites (b, reduced glutathione, c, oxidized glutathione, d, NADP+, e, asparagine) and gene dependencies (CERES). Each point represents a gene knockout (KO). The statistical test was based on linear regression conditioned on major lineage types (cell line n=455). f, Heatmap showing relative levels of ordered TAG species in 928 cell lines. PUFAhigh and PUFAlow cell lines are selected by two-sample t-test (two-sided p<0.05) and are indicated by black lines below the heatmap. g-h, Volcano plots comparing the phosphatidylcholine (g) and cholesterol ester (h) species in the PUFAhigh (n=315) versus PUFAlow (n=325) cell lines. Each point represents a metabolite and is colored by the ratio of carbon-carbon double bonds to the acyl chain number. i, Volcano plot comparing the differential dependencies in the PUFAhigh (n=315) versus PUFAlow (n=325) cell lines. The dependency scores (CERES) used in comparison indicate cell line sensitivity in response to gene knockout (smaller values suggest greater sensitivity). For g-i, the q-values were calculated based on two-sample t-tests (two-sided) with multiple hypothesis testing correction.
Phenotypic profiling of barcoded CCLE lines
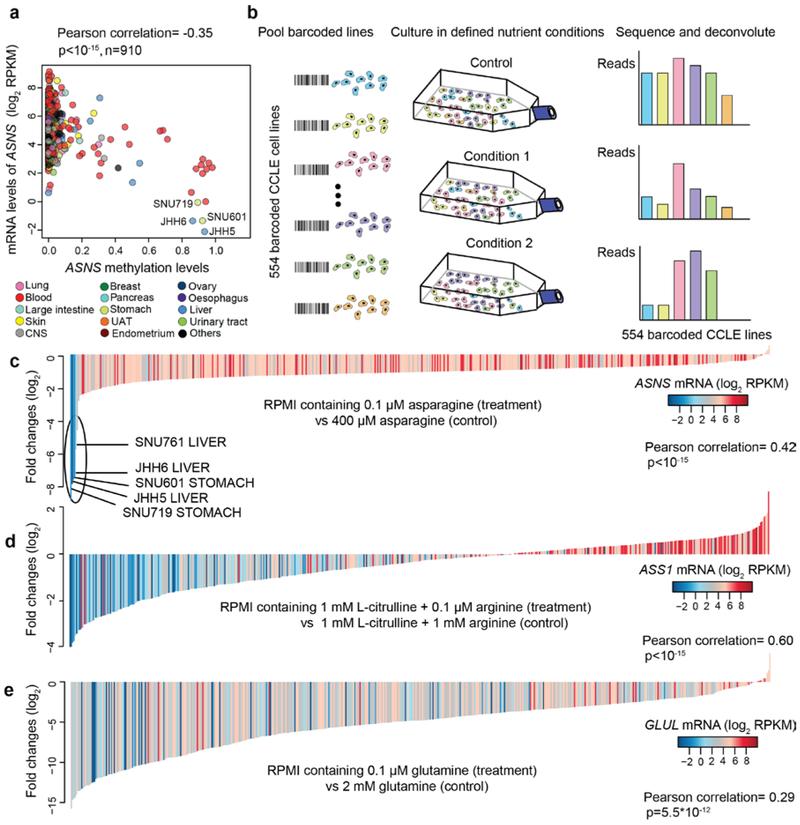
As is shown in the results above, lower asparagine levels strongly associated with increased sensitivity to loss of asparagine synthetase (ASNS) (Fig.3e). The non-essential amino acid asparagine is synthesized by ASNS but can also be imported directly from the media. We showed that ASNS knockdown significantly impeded cell proliferation when media asparagine was limiting (Extended Data Fig. 4a, b). Given that some CCLE cell lines with ASNS promoter hypermethylation have aberrantly low ASNS expression even in the presence of its transcriptional activator ATF4 (Fig.4a, Extended Data Fig. 4c, d), we hypothesized that intrinsic methylation-dependent gene suppression might be selectively targeted using specific nutrient deprivation. To explore this, we used a variation of the PRISM technology where 544 adherent CCLE lines labeled with 24-nucleotide barcodes were grown in a pooled format18 (Fig. 4b). The mixed cell pools were cultured under specific media conditions with defined amino acid concentrations and relative cell viability was then estimated by high-throughput sequencing of the barcode collected after 6 days of treatment (Supplementary Table 4). Here, we found that when the pooled cell populations were grown under limiting asparagine conditions, those with aberrantly low expression of ASNS were selectively depleted (Fig. 4c, Supplementary Table 4). In parallel, PRISM experiments were conducted in which arginine and glutamine (also non-essential amino acids, see Extended Data Fig. 4e) were set to different concentrations. Similarly, we found lower expressions of ASS1 and GLUL (influenced by promoter region methylation) marked cells that were more sensitive to reduced supplies of arginine and to a lesser degree, glutamine (Fig. 4d, e, Extended Data Fig. 4f, g, Supplementary Table 4). Notably, in the case of glutamine dependency, we also observed a strong correlation with the proliferative doubling rate (Extended Data Fig. 4h). Taken together, these examples suggest that DNA hypermethylation influences dependency on nutrient availability as exemplified by asparagine, arginine, and glutamine auxotrophy in subsets of cancer cell lines.
Fig.4 ∣. Revealing amino acid metabolism auxotrophs by pooled cancer cell line screens.
a, Scatter plot comparing ASNS DNA methylation levels with ASNS mRNA levels in all cell lines. b, Schematic summarizing the workflow of pooled cancer cell line screens. c-e, Waterfall plots showing the fold changes of pooled CCLE lines (n=554, median of 3 independent cell culture replicates) cultured in RPMI media containing 0.1 μM asparagine (c), 0.1 μM arginine + 1 mM L-citrulline (precursor required for arginine synthesis) (d), 0.1 μM glutamine (e) for 6 days (normalized to control). For c-e, the p-values were calculated based on the significance test of Pearson correlations (two-sided).
Expanding the therapeutic use of asparaginase
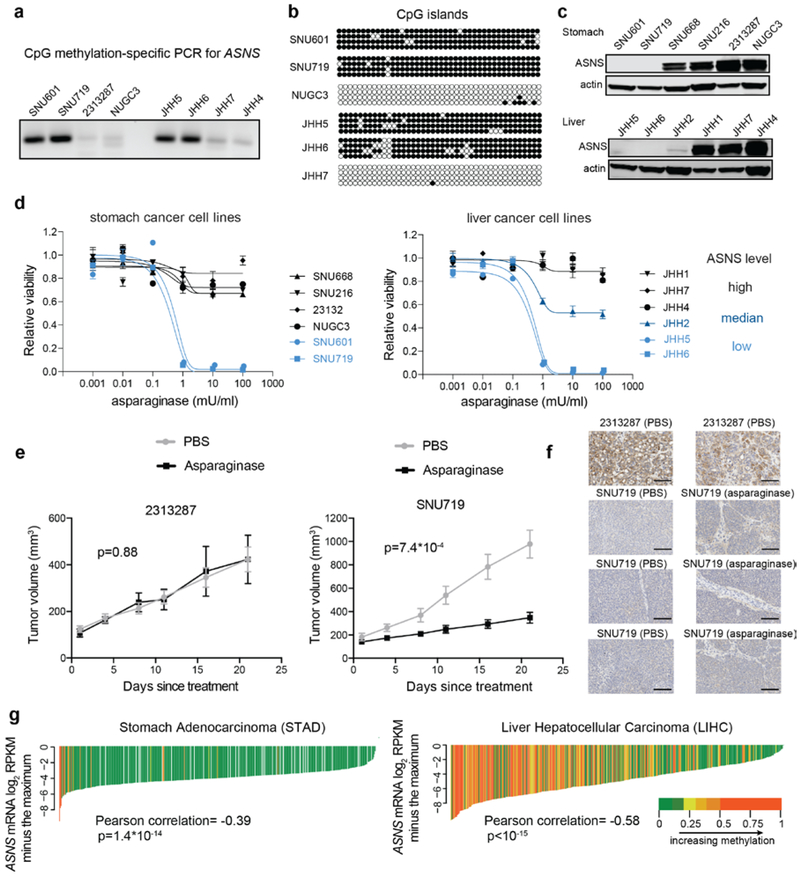
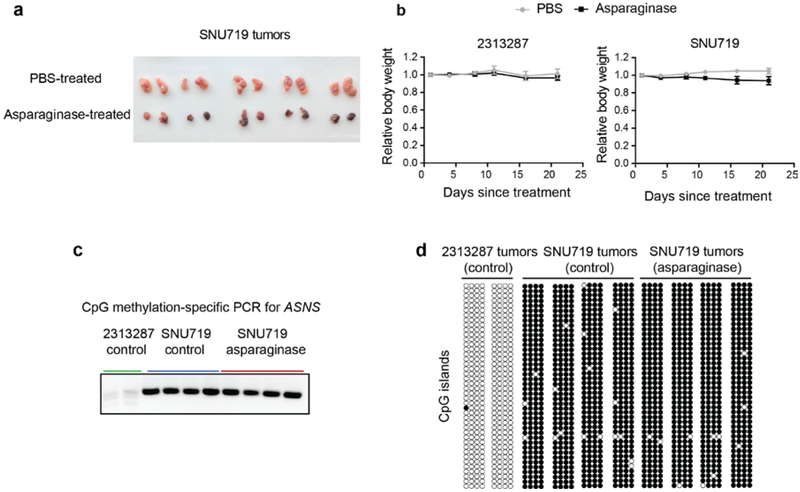
While arginine and glutamine dependence across cell lines was manifest as a graded response, the pronounced, nearly binary differences to asparagine depletion between cell lines with intrinsic lower expression of ASNS and the non-sensitive lines (Fig. 4c) prompted us to explore the potential therapeutic value of asparaginase beyond its use in treating acute lymphoblastic leukemia (ALL). We confirmed that cells with ASNS hypermethylation also lacked protein expression (Fig. 5a-c) and were profoundly sensitive to asparaginase in vitro (Fig. 5d). To determine whether this dependence could be reproduced in vivo, we then subcutaneously implanted 7*106 of 2313287 (ASNS high) or SNU719 (ASNS low) cells into both flanks of nude mice. After the tumors reached about 100–200 mm3 in volume, we then treated the mice with intraperitoneal injections of asparaginase (3000 units/kg/injection, 5 times a week) or vehicle control and monitored the tumor growth over a 3-week period. Here we observed a significant decrease of growth for SNU719 tumors but not 2313287 tumors with little body weight loss (Fig. 5e, Extended Data Fig. 5a, b). We also showed that ASNS hypermethylation and loss of expression was maintained during implantation and treatment of these xenografts (Fig. 5f, Extended Data Fig. 5c, d). These data also suggest that ASNS IHC might be applied to stratify and select patients for asparaginase trials. To define the relevant patient population based on data from human tumor samples, we examined DNA methylation among gastric and hepatic cancers in The Cancer Genome Atlas (TCGA) and found it significantly associated with reduced ASNS expression in tumor samples (Fig. 5g). Collectively, these results suggest that asparaginase can suppress the growth of defined subsets of cancer cell lines with loss of ASNS expression both in vitro and in vivo.
Fig.5 ∣. Therapeutic value of asparaginase in stomach and liver cancers.
a, Methylation-specific PCR for ASNS CpG islands (a cropped gel image is shown). This experiment was repeated once. b, Bisulfite sequencing for ASNS methylation status in different cell lines. Open circles indicate unmethylated CpG while solid circles indicate methylated CpG. This experiment was repeated once with 4 technical replicates for each cell line sample. c, Cropped immunoblot of ASNS in representative stomach and liver cancer cell lines. Actin was used as the loading control. This experiment was repeated independently twice with similar results. d, Evaluation of asparagine depletion on the viability of selected stomach and liver cancer cell lines. Viabilities were quantified by Cell-Titer Glo 6 days after treatment (mean ± SEM, n=3 cell culture replicates). e, Volume measurements for tumors resulting from subcutaneous injection of 2313287 cells and SNU719 cells with 3000 units/kg asparaginase treatment or vehicle control (10 tumors from 5 nude mice per condition, mean ± SEM). The p-values were calculated based on the tumor volume difference between Day 21 and Day 1 using two-sample t-tests (two-sided). f, Immunostaining of ASNS (brown) in xenograft tumors expressing high (2313287) or low (SNU719) levels of ASNS treated with vehicle control or 3000 units/kg asparaginase 5 times a week for 3 weeks. Each subplot is representative of a different tumor. The immunostaining was repeated independently twice with similar results. Scale bar, 100 μm. g, Waterfall plots showing the ASNS mRNA levels related to its DNA methylation (probe: cg08114476) in the STAD cohort (n=372) and the LIHC cohort (n=371) in TCGA. Each line represents a tumor sample. The p-values were calculated based on the significance test of Pearson correlations (two-sided).
The landscape of kynurenine metabolism in the CCLE
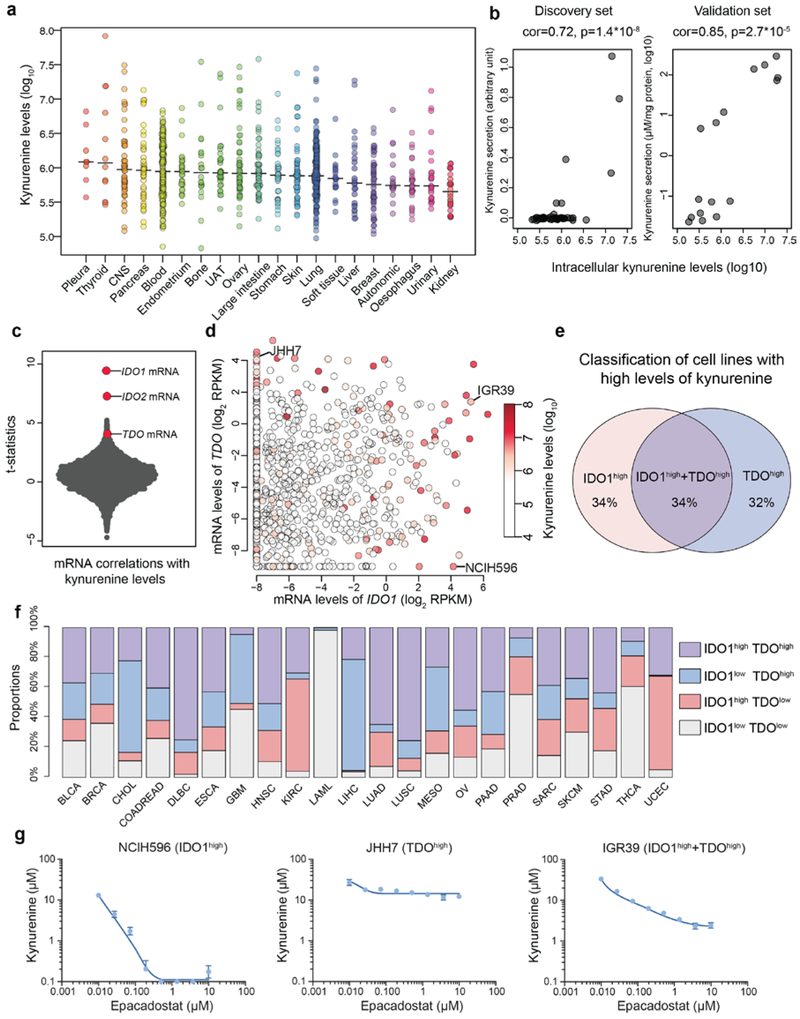
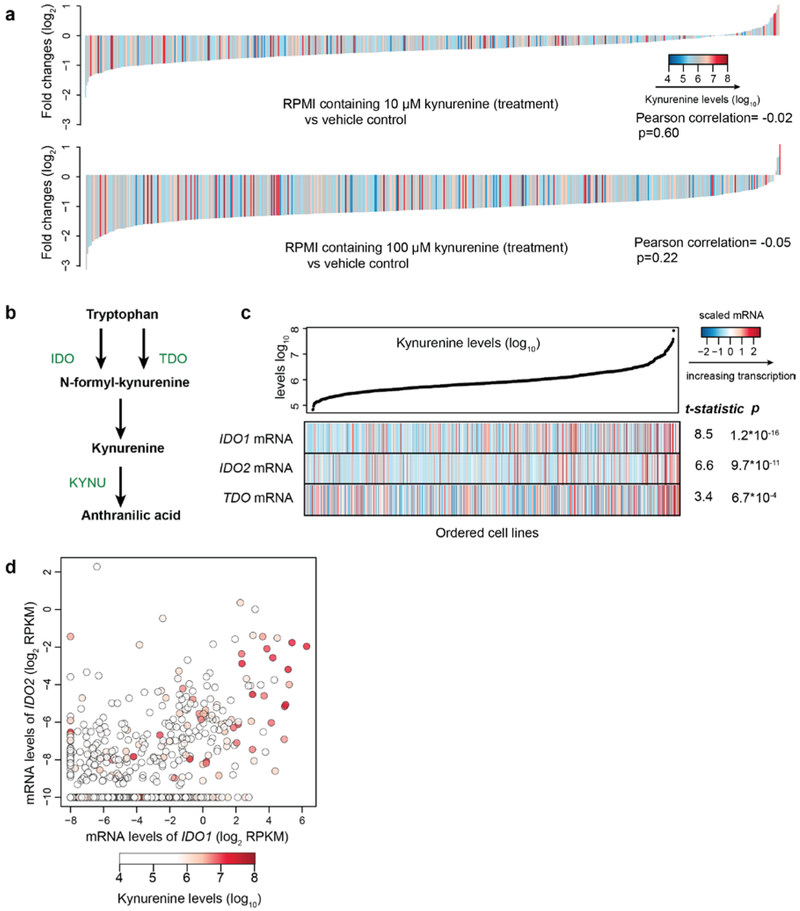
In addition to analyzing metabolites with nutritional value, we also explored the patterns of signaling metabolites including kynurenine. Kynurenine is produced from catabolism of tryptophan and can exert immunosuppressive effects through the aryl hydrocarbon receptor (AHR), leading to suppressed effector T cell proliferation and enhanced regulatory T cell generation19-21. These findings have led to clinical trials of IDO1 inhibitors targeting the kynurenine pathway in combination with PD1 blockade22. In the CCLE, we observed a wide range of intracellular kynurenine concentrations spanning three orders of magnitude across multiple cancer types (Fig. 6a). We found that higher intracellular kynurenine levels were strongly associated with greater secretion into the media in both a discovery (NCI6023) and validation set of cell lines (Fig. 6b). These data suggest that cell lines with higher intracellular kynurenine concentrations are also actively secreting it into the extracellular space.
Fig.6 ∣. The landscape of kynurenine metabolism in the CCLE.
a, Kynurenine levels with medians across the CCLE cell lines grouped by tissues of origin. b, Scatter plot showing intracellular kynurenine levels compared to secreted kynurenine levels in the selected cell lines (discovery set: n=49; validation set: n=16). Two cell culture replicates were used for each cell line in the validation set. The p-values were calculated based on the significance test of Pearson correlations (two-sided). c, T-statistics based on all gene transcripts and the basal levels of kynurenine conditioned on the major lineages. Each point represents a gene. The statistical test was based on linear regression conditioned on major lineage types (cell line n=913). d, mRNA levels of IDO1 and/or TDO correlate with kynurenine accumulation (Pearson correlation= 0.26 and 0.10 respectively; p<10−15 and p=0.002 respectively). Cell lines (n=913) are represented as points with color-coded kynurenine levels. e, Classification of cell lines with high kynurenine levels (the top 5% in the CCLE) based on their expression of IDO1 and TDO. Cell lines with IDO1/TDO mRNA transcripts lower than −2 (log2 RPKM) are considered to lack sufficient expression. f, Classification of TCGA tumor samples based on IDO1/TDO mRNA transcripts. The samples with above average IDO1/TDO levels were defined to be high in expression. The cancer type short names are based on standard TCGA notations. g, Kynurenine secretion to media in response to epacadostat in cell lines with expression of IDO1, TDO or both (mean ± SEM, n=3 cell culture replicates). The detection limit of the ELISA assay is 0.1 μM.
We next asked whether kynurenine levels have a direct impact on cell proliferation rates by applying increasing concentrations of kynurenine to the pooled, barcoded CCLE lines. A weak to moderate decrease of growth was observed in the majority of lines and was not correlated with the intrinsic synthesis capability (Extended Data Fig. 6a, Supplementary Table 4), suggesting that kynurenine does little to accelerate cell proliferation in vitro.
Notably, both indoleamine 2,3-dioxygenase (IDO) and tryptophan 2,3-dioxygenase (TDO) encode enzymes that catalyze the conversion of tryptophan to N-formylkynurenine, a precursor of kynurenine (Extended Data Fig. 6b). By correlating mRNA transcripts (n=18,205) with kynurenine levels, we found that IDO1, IDO2, and TDO were top hits that correlated significantly with kynurenine levels (Fig. 6c, Extended Data Fig. 6c). Interestingly, we observed abundant intracellular kynurenine in cell lines with either high IDO1/2 or TDO mRNA levels (Fig. 6d, Extended Data Fig. 6d). By classifying the cancer cell lines harboring high levels of kynurenine based on their expression of IDO (IDO1 as the major form) versus TDO, we found that about one third of cell lines have simultaneous IDO1 and TDO expression while the rest are driven by either IDO1 or TDO (Fig. 6e). This divergent expression pattern is also observed in different tumor samples in TCGA (Fig. 6f). This suggests that cancer cells can likely use either enzyme or potentially both to produce kynurenine. We next asked whether the IDO1 selective inhibitor epacadostat is effective in blocking kynurenine secretion in these distinct settings. To test this in vitro, we profiled epacadostat dose-dependent suppression of kynurenine secretion in three representative cell lines. We found that epacadostat completely inhibited kynurenine secretion in NCIH596 (IDO1high), caused a major decrease of kynurenine secretion in IGR39 (IDO1high + TDOhigh) but had little impact on JHH7 (TDOhigh) (Fig. 6g). Therefore, these data linking high kynurenine to IDO1 or TDO mRNA expression suggest that selective IDO1 inhibition might only work in a subset of these tumors and could be further subject to escape mediated by TDO. Recently, negative phase III data were reported for the combination of epacadostat with the anti-PD1 antibody pembrolizumab in unselected patients with metastatic melanoma24. Our data raise the possibility that selecting patients with increased tumor production of kynurenine coupled with an understanding of which enzymes are producing kynurenine might be critical to further IDO/TDO inhibitor development.
Discussion
Cancers are diverse in histology, in the pattern of underlying genetic alterations and in metabolic signatures. To date, there has been no systematic metabolomic profiling for hundreds of model cancer cell lines from multiple lineages with distinct genetic backgrounds. To bridge this gap, we profiled 225 metabolites in a collection of 928 cancer cell lines and intersected this data with other large-scale profiling datasets. This breadth and depth allows for various metabolic signatures to be probed in an unbiased manner and for metabolites with similar patterns to be identified. Beyond the diversity revealed in baseline metabolite levels, we also investigated the diverse proliferative responses to perturbations in the dynamic metabolic networks with pooled screens of 554 barcoded cell lines. Overall, the data and analyses suggest that distinct metabolic phenotypes exist in cancer cell lines both at the unperturbed and the perturbed states and that such phenotypes have direct implications for therapeutics targeting metabolism.
In particular, we delineated prevalent DNA methylation events in addition to somatic mutations and copy number alterations in various metabolic pathways and began to unveil their key regulatory roles both at the basal state and in the dynamics of cell growth. On one hand, gene hypermethylation events likely influence baseline metabolite abundance via reductions in key enzymes mediating metabolite degradation (e.g., SLC25A20 with long-chain acylcarnitines) or synthesis (PYCR1 with proline, GPT2 with alanine). Alternatively, methylation-dependent suppression of gene expression can have profound modulatory effects in cell proliferation under altered nutrient conditions (e.g., ASNS with asparagine, ASS1 with arginine).
In this resource, several observations relate to potential therapeutic applications. The observation of distinct associations between IDO/TDO enzymes and elevated levels of kynurenine suggests that clinical testing of either AHR inhibitors or dual IDO/TDO inhibitors might benefit from patient stratification based on kynurenine levels and the relative contribution of specific enzymes involved in its synthesis. Additionally, the suppressed ASNS expression in subsets of stomach and liver cancers raise the possibility of repurposing asparaginase as a therapeutic option for subpopulations in these diseases. Although asparaginase is an effective agent used in the regimen for ALL25, there has been no evidence for its potential efficacy for solid tumors in the clinic. This is consistent with our observation of abundant ASNS baseline expression in most lineages except the ALL where expression of ASNS is low (Extended Data Fig. 4c). This underlying intrinsic dependence sharply contrasts with the studies combining ASNS inhibition with asparagine depletion in solid tumors26,27. Consequently, our study of asparaginase use in treating solid tumors with intrinsic loss of ASNS may have therapeutic implications.
Several limitations of our study should be considered and improved in future large-scale metabolomic profiling efforts: First, only one biological replicate was used for most cell lines profiled except for quality control (QC) purposes. While QC measures have been implemented and multiple analyses demonstrated the rich information in this dataset, noise from multiple sources still exists and additional biological replicates would improve data quality; Second, we expect that a broader collection of metabolites would generate information about a larger array of biochemical pathways; Third, due to the scale of these studies we were unable to use dynamic measures of metabolic flux and hence we can only look at the steady-state levels of various metabolites profiled. Finally, the media used in tissue culture remain significantly different from in vivo environments. Consequently, large-scale metabolomic characterization of tumor samples will be an important next step.
METHODS
Cell lines and culture conditions
Human cancer cell lines were collected as described previously7. SNP genotyping was incorporated at each stage of cell culture to validate the identity of cell lines. The associated tissue type and gender information was annotated based on literature or vendor information when available. The cell culture media used in this study with catalog information were included in Supplementary Table 1. All cell lines were grown in T75 flasks with respective media using standard cell culture conditions (37°C, 5% CO2) and were free of microbial contamination including mycoplasma. For each actively growing cell line with a low passage number, two million cells were seeded per T75 flask, the metabolites were extracted after 2 days and before the cells reached a confluence of 90%. Separate flasks were used for polar metabolite or lipid extractions.
Polar metabolite extraction
LC-MS grade solvents were used for all of the metabolite extraction in this study. For adherent cells, the media were aspirated off as much as possible and the cells were washed with 4 mL cold Phosphate Buffered Saline (PBS, no Mg2+/Ca2+). After vacuum aspiration of PBS, the metabolites were extracted by adding 4 mL 80% methanol (−80°C) immediately and the samples were transferred to a −80°C freezer. The flasks were kept on dry ice during the transfer and were incubated at −80°C for 15 min. Then the lysate was collected by a cell scraper and transferred to a 15 mL conical tube on dry ice. The insoluble debris was removed by centrifuging at 3500 rpm for 10 min (4°C). The supernatant was transferred to a new 15 mL conical tube on dry ice and the tube with the pellet was kept for further extraction. Then, 500 μL 80% methanol (−80°C) was added to each pellet. The mixture was resuspended by vortexing or pipetting and transferred to a 1.5 ml centrifuge tube on dry ice. The cell debris was removed by centrifuging samples at 10,000 rpm for 5 min (4°C). The supernatant was transferred to the corresponding 15 mL conical tube on dry ice so that all extracts were combined. The pooled extracts were stored at −80°C before LC-MS analysis.
For cells growing in suspension, they were centrifuged to pellet at 300g for 5 min (4°C) and the supernatant was then aspirated off as much as possible. These cells were washed once with 4 mL cold PBS (no Mg2+/Ca2+) and they were pelleted at 300g for 5 min (4°C). After vacuum aspiration of PBS, the metabolites were extracted by adding 4 mL 80% methanol (−80°C) immediately and the samples were transferred to a −80°C freezer after brief vortexing. The samples were kept on dry ice during the transfer and were incubated at −80°C for 15 min. The insoluble debris was removed by centrifuging at 3500 rpm for 10 min (4°C). The subsequent steps were the same as those used for adherent cell lines.
Lipid extraction
For adherent cells, the medium was aspirated off as much as possible and the cells were washed with 4 mL cold PBS (no Mg2+/Ca2+). After vacuum aspiration of PBS, the lipid metabolites were extracted by adding 4 mL isopropanol (4°C) and the lysate was collected by a cell scraper and transferred to a 15 mL conical tube on ice. The samples were covered to avoid exposure to light and were allowed to sit for 1h at 4°C. Samples were then vortexed and the cell debris was removed by centrifuging at 3500 rpm for 10 min (4°C). The supernatant was transferred to a new 15 mL centrifuge tube on ice and stored at −20°C before LC-MS analysis.
For cells growing in suspension, they were centrifuged to pellet at 300g for 5 min (4°C) and the supernatant was then aspirated off as much as possible. These cells were washed once with 4 mL cold PBS (no Mg2+/Ca2+) and they were pelleted at 300g for 5 min (4°C). After vacuum aspiration of PBS, the lipid metabolites were extracted by adding 4 mL isopropanol (4°C) immediately. After brief vortexing, the samples were covered to avoid exposure to light and were allowed to sit for 1h at 4°C. The insoluble debris was removed by centrifuging at 3500 rpm for 10 min (4°C). The supernatant was transferred to a new 15 mL centrifuge tube on ice and stored at −20°C before LC-MS analysis.
LC-MS instrumentation and methods
A combination of two hydrophilic interaction liquid chromatography (HILIC) methods, either acidic HILIC method with positive-ionization-mode MS, or basic HILIC method with negative-ionization-mode MS was used to profile polar metabolites. Reversed Phase (RP) chromatography was used to profile lipid species. The LC-MS methods were based on a previous study28, where the metabolite retention time and the selected reaction monitoring parameters were also described. LC-MS related reagents were purchased from Sigma-Aldrich if not specified. Pooled samples composed of 11 cell lines from different lineages were used for trend and batch correction.
The LC-MS system for the first method consisted of a 4000 QTRAP triple quadrupole mass spectrometer (SCIEX) coupled to an 1100 series pump (Agilent) and an HTS PAL autosampler (Leap Technologies). Polar metabolite extracts were reconstituted with acetonitrile/methanol/formic acid (74.9:24.9:0.2 v/v/v) containing stable isotope-labeled internal standards (0.2 ng/μL valine-d8 (Isotec) and 0.2 ng/μL phenylalanine-d8 (Cambridge Isotope Laboratories)). The samples were centrifuged (10 min, 9,000g, 4 °C), and the supernatants (10 μL) were injected onto an Atlantis HILIC column (150 × 2.1 mm, 3 μm particle size; Waters Inc.). The column was eluted isocratically at a flow rate of 250 μL/min with 5% mobile phase A (10 mM ammonium formate and 0.1% formic acid in water) for 1 min followed by a linear gradient to 40% mobile phase B (acetonitrile with 0.1% formic acid) over 10 min. The ion spray voltage was set to be 4.5 kV and the source temperature was set to be 450 °C.
The second method using basic HILIC separation and negative ionization mode MS detection was established on an LC-MS system consisting of an ACQUITY UPLC (Waters Inc.) coupled to a 5500 QTRAP triple quadrupole mass spectrometer (SCIEX). Polar metabolite extracts spiked with the isotope labeled internal standards including 0.05 ng/μL inosine-15N4, 0.05 ng/μL thymine-d4, and 0.1 ng/μL glycocholate-d4 (Cambridge Isotope Laboratories) were centrifuged (10 min, 9,000g, 4 °C), and 10 μL supernatants were injected directly onto a Luna NH2 column (150 × 2.0 mm, 5 μm particle size; Phenomenex) that was eluted at a flow rate of 400 μL/min with initial conditions of 10% mobile phase A (20 mM ammonium acetate and 20 mM ammonium hydroxide in water (VWR) and 90% mobile phase B (10 mM ammonium hydroxide in 75:25 v/v acetonitrile/methanol (VWR)) followed by a 10-min linear gradient to 100% mobile phase A. The ion spray voltage was set to be −4.5 kV and the source temperature was set to be 500 °C.
Lipids were profiled using a 4000 QTRAP triple quadrupole mass spectrometer (SCIEX) coupled to a 1200 Series Pump (Agilent Technologies) and an HTS PAL autosampler (Leap Technologies). Lipid extracts in isopropanol, spiked with an internal standard (0.25 ng/μL 1-dodecanoyl-2-tridecanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids)), were centrifuged and 10 μL supernatants were injected directly to a 150 × 3.0 mm Prosphere HP C4 column (Grace) for reversed phase chromatography. Mobile phase A was 95:5:0.1 (v/v/v) 10 mM ammonium acetate/methanol/acetic acid. Mobile phase B was 99.9:0.1 (v/v) methanol/acetic acid. The column was eluted isocratically with 80% mobile phase A for 2 minutes, followed by a linear gradient to 80% mobile phase B over 1 minute, a linear gradient to 100% mobile phase B over 12 minutes, and then 10 minutes at 100% mobile phase B. MS analyses were carried out using electrospray ionization and performed in the positive-ion mode with Q1 scans. Ion spray voltage was set to be 5.0 kV, and the source temperature was set to be 400°C.
Generation of isogenic cell lines
A2058 cells were maintained in DMEM, supplemented with 10% FBS and 2 mM glutamine. 1% non-essential amino acids (NEAA, BioConcept, 5–13K00) was added if stated. This NEAA mix (100X) contained 10 mM of L-asparagine, L-alanine, L-aspartic acid, L-glutamic acid, L-proline, L-serine, and glycine. shRNA (Control_KD: AGAAGAAGAAATCCGTGTGAA, ASNS_KD1: GCATCCGTGGAAATGGTTAAA; ASNS_KD2: CATTCAGGCTCTGGATGAAGT; PLK1_KD: GGTATCAGCTCTGTGATAACA) were cloned in inducible pLKO-based lentiviral vectors (puromycin resistant). Wild type A2058 was infected with shRNA-expressing viruses respectively. After selection, the KD efficiency was evaluated by western blots upon 3 days of treatment with doxycycline (100 ng/mL).
Pooled screens of barcoded CCLE lines
The CCLE lines were barcoded and screened as described previously18. Briefly, cells were mixed as individual pools (~24 lines in each) and kept frozen in liquid nitrogen before use. On the day of experiment, the individual pools were mixed together in corresponding media conditions with equal numbers so that each line started from about 200 cells per T25 flask. After 6 days, the genomic DNA was extracted and the barcodes were amplified by PCR before high-throughput sequencing. Three biological replicates were used in each condition and the growth changes were calculated with the control conditions as reference.
Animal studies
The animal work was approved by the Institutional Animal Care and Use Committee (IACUC) at the Broad Institute. 4-week-old, female, athymic nude mice (CrTac:NCr-Foxn1nu, Taconic) were inoculated subcutaneously with 7*106 cancer cells in phenol red free RPMI media with 50% matrigel in both flanks. The mice were randomized into treatment or control group when tumors reached approximately 100–200 mm3 in size. Asparaginase (Abcam) was delivered with intraperitoneal injection at 3000 units/kg in 200 μl PBS 5 times per week (omitting Wednesday and Sunday) for 3 weeks. Tumor tissues were collected and processed for IHC staining by standard methods. All IHC staining was performed on the Leica Bond automated staining platform. Polyclonal Asparagine Synthetase (ASNS) antibody from Proteintech (#14861–1-AP) was run at 1:1500 dilution using the Leica Biosystems Refine Detection Kit with citrate antigen retrieval. Tumor sizes were calculated by 1/2*length*width*width.
Quantification of kynurenine secretion
To validate kynurenine secretion in the validation set of cell lines, cells were seeded on 6 well plates at a density of 4*105/2 ml RPMI (+10%FBS) per well. The 16 cell lines included SNU719, WM793, LOXIMVI, UACC257, U2OS, JHH5, JHH4, SNU601, SNU668, NUGC3, 2313287, NCIH2196, NCIH596, JHH7, IGR39, and NCIH2228. The supernatant was collected 3 days later and the cell protein mass was measured by BCA assay. To profile kynurenine secretion in response to epacadostat, cells were seeded at a density of 25,000/125μl/well in 96 well plates. Compounds were added at seeding and supernatants were collected 3 days after. Secreted kynurenine in the conditioned media was quantified using a kynurenine ELISA kit (Rocky Mountain Diagnostics, Inc) according to manufacturer’s protocol. RPMI (+10%FBS) was used as the negative control to determine background signal levels in the ELISA assay.
Analysis of DNA methylation
The CCLE reduced representation bisulfite sequencing (RRBS) data was used for gene methylation analysis. For independent validation and cell lines not covered (e.g., JHH5, JHH6), genomic DNA from cell line or tumor samples was isolated and bisulfite-converted using the EpiTect Fast LyseAll Bisulfite Kit (Qiagen) following manufacturer’s instructions. For methylation-specific PCR, the primer set consisted of 5’CGTATTGAGACGTAAGGCGT3’ and 5’CTAACTCCTATAACGCGTACGAAA3’. For bisulfite sequencing, the primer set consisted of 5’GTTAGAATAGTAGGTAGTTTGGG3’ and 5’AAAATACACATATAACATTTACAAAAACTC3’. Purified PCR products were cloned into the pCR™4-TOPO® TA vector using TOPO TA Cloning Kit (Invitrogen).
Statistical analysis
All statistical analyses used in this paper were done in R v 3.4.2 (downloaded from https://www.r-project.org/). Data visualization was done in R and Prism (GraphPad). Statistics and relevant information including the type and the number of replicates (n), the adopted statistical tests, and p-values are reported in the figures and associated legends. For Pearson correlations, we used the cor.test function in R to conduct significance test and obtain the p-values (two-sided). We used the Benjamini-Hochberg procedure to control for multiple hypothesis testing when applicable.
Metabolite data acquisition and quality control
Raw data were processed using MultiQuant 1.2 software (SCIEX) for automated LC-MS peak integration. All chromatographic peaks were also manually reviewed for the quality of integration and compared against known standards for each metabolite to confirm identities. Internal standard peak areas were monitored for quality control and to assess system performance over time. Additionally, pooled samples composed of mixed metabolites from 11 cell lines (NCIH446, DMS79, NCIH460, DMS53, NCIH69, HCC1954, CAMA1, KYSE180, NMCG1, UACC257, and AU565) were used after every set of 20 samples. This was an extra quality control measure of analytical performance and also served as a reference for scaling raw metabolomic data across samples. The peak area for each metabolite in each sample was standardized by computing the ratio between the value observed in the sample and the value observed in the “nearest neighbor” pooled sample. These ratios were then multiplied by the mean value of all reference samples for each analyte to obtain standardized peak areas.
To remove potential batch effects, the ratio between the mean standardized peak area for each metabolite in a given batch and the mean standardized peak area for that metabolite across all the batches was computed. Then the standardized peak areas for that metabolite in that given batch were divided by that ratio. These standardized peak areas are included in Supplementary Table 1. Note that the abundance of different metabolites cannot be compared given the nature of the LC-MS methods. Only for the same metabolite, the levels could be compared between different cell lines. The final batch-corrected standardized peak areas were then log10-transformed. Additionally, considering the cell line to cell line variation in biomass that could contribute to systematic differences in metabolite abundance detected by LC-MS, we processed the data by two steps. First, we calibrated each column of metabolites to have the same median. Then we calibrated each row (cell line) to have the same median. Empirically, this median normalization step effectively calibrated metabolomic datasets, adjusting artificial differences due to different sample biomass before metabolite extraction. The final metabolomic data matrix (928 rows of cell lines and 225 columns of metabolites) for downstream analyses or modeling is included in Supplementary Table 1.
Missing data handling
For the trend-corrected metabolomic dataset, a small fraction of values were missing. We first applied imputations using fully conditional specification implemented by the Multivariate Imputation via Chained Equations (MICE) algorithm from R package “mice”, which has the advantage of preserving intrinsic data matrix structure and information. The quality of predictive-mean-matching-based imputations was inspected using diagnostic tools in the package. We noticed that the generated multiple matrices had negligible differences for most downstream applications due to the small fraction (9%) of missing values and the strong signals from observed values. Therefore, we chose one representative imputed matrix for downstream regression analysis that required a complete data structure for efficient computation.
Other cancer cell line dataset acquisition
The CCLE datasets (e.g., mutation, copy number variation, RNAseq) were downloaded from the CCLE portal (https://portals.broadinstitute.org/ccle). The CRISPR-Cas9-based gene-essentiality data used (CERES scores, 2019Q1 release) were obtained from the Cancer Dependency Map project15 (https://depmap.org/portal/; https://doi.org/10.6084/m9.figshare.5319388.v2).
Clustering and heatmap plotting
Clustering was done in R with the function hclust. Note that each feature (e.g., metabolite) was scaled to have mean 0 and standard deviation 1 before hierarchical clustering analysis and heatmap plotting. The dissimilarity was defined as 1 minus the Pearson correlation between each pair of selected features. The resulting distance matrix was processed by the “centroid” method in the hclust function to get the clustering results. For heatmap plots, we used the heatmap.2 function in the R package gplots.
Metabolite lineage effect analysis
To evaluate the association between the metabolite levels and the lineage types, we applied a linear regression model. We coded the lineage types as binary covariates (X). Cell lines were represented by the rows, with 1 indicating presence of the corresponding feature. Each metabolite level (log10 scale) was used as the response variable Y. The calculated r2 was used to characterize the lineage effects quantitatively.
Genetic, epigenetic, and dependency feature collection
We curated genetic and epigenetic features in our association analysis with CCLE metabolites. These included all nonsynonymous mutations of 474 cancer-related genes, deleterious, loss-of-function mutations of 202 genes, and hotspot missense mutations of 29 genes (TCGA hotspot count >=10; https://portals.broadinstitute.org/ccle). Such discrete features were converted to binary indicators (1/0) in our analysis. We also selected 40 genes with frequent deletions and 21 genes with frequent amplifications. These copy number alteration events were validated to significantly associate with corresponding gene transcriptional levels (CCLE RNAseq data). Additionally, the methylation scores of 2,114 genes were included given their significant negative associations with the corresponding transcriptional levels (CCLE RNAseq data). To select dependencies, we focused on top 3,000 genes ordered by variance of CERES scores across the panel of cell lines. Genes with less cell-line-to-cell-line dependency difference (e.g., non-essential) were not prioritized for metabolite-dependency association analysis.
Linear regression analysis
We applied a linear regression model to evaluate associations between two different datasets of CCLE cell lines (e.g., genetic feature vs metabolite level). We included lineage variables to account for lineage-associated confounding effects when cell lines from different lineages were analyzed together.
First, we constructed a covariate matrix with cell lines as rows and features as columns for the linear regression. In addition to the intercept variable I, we also included binary variables indicating major lineages. Here, L1, L2,…, L17 represented the lineages of lung, large intestine, blood, urinary, bone, skin, breast, liver, ovary, oesophagus, endometrium, central nervous system, soft tissue, pancreas, stomach, kidney, and upper aerodigestive tract. We further added variable (X) to this covariate matrix: each mutation variable was binary-coded; each continuous variable (e.g., mRNA log2 RPKM) was rescaled to have mean 0 and standard deviation 1.
The dependent variable vector Y could be another type of cell features. The coefficient vector was represented as β and the model is shown below. For example, to answer the question that in a given cell line feature matrix (e.g., collections of genetic or epigenetic features) which feature was the most associated with a given metabolite vector under the condition of controlled lineage effects, this regression analysis was applied to individual features (e.g., individual genetic and epigenetic features) before comparisons. The calculated t-statistics, p-values, and estimated coefficients for X (βX) were reported to evaluate the associations.
Data availability
The CCLE metabolomic data and related analyses can be found in Supplementary Tables 1-4. Additional CCLE resources are available at: https://portals.broadinstitute.org/ccle. Source data of all the western blots and DNA gels in this work are provided in Source Data Figures.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 ∣. Quality control of the CCLE metabolomic dataset.
a, Metabolites ordered by estimated coefficients of variance (CV). b, Heatmap showing the hierarchical clustering of the CCLE metabolomic data with annotated tissues of origin. CNS: central nervous system; UAT: upper aerodigestive tract. For each metabolite (log10 scale), the abundance was standardized for plotting. c, Ordered lineage effects of the 225 profiled metabolites. Each dot represents a metabolite. d, Phosphocreatine levels with medians across the CCLE cell lines grouped by tissues of origin. e, 1-methylnicotinamide levels with medians across the CCLE cell lines grouped by tissues of origin.
Extended Data Fig. 2 ∣. Selected mutation features in relation to metabolite abundances.
a, Reduced glutathione (GSH) levels and the top correlated mutations among all mutational features. Cell lines are shown as lines and ordered by increasing levels of GSH. Those cell lines with corresponding mutations are labeled as black. The reported test statistics and p-values are based on the significance tests of genetic feature regression coefficients (cell line n=927, two-sided t-tests). b, Swarm plot comparing GSH, GSSG, and NADP+ levels in cell lines with or without KEAP1 mutations (n=72 and 855 respectively, mean ± SD). Each dot represents a cell line. The p-values were calculated based on two-sample t-tests (two-sided). c-h, Volcano plots comparing 225 profiled metabolites based on the mutational status of EGFR (c), KRAS (d), NRAS (e), TP53 (f), PTEN (g), TSC1/2 (h) in CCLE cell lines. Each point represents a metabolite and the red lines indicate a cutoff of q=0.05. The statistical significance and effect sizes were calculated using linear regression models conditioned on major lineage types (two sided t-test). Note that cell lines with either PTEN mutation or deletion were defined as PTEN deficient. i, Scatter plot comparing ME2 copy number alterations with its mRNA levels in all cell lines. j, Scatter plot comparing ME2 mRNA levels with malate levels in all cell lines. k, Scatter plot comparing ME2 mRNA levels with ME3 knockout CERES scores. For i-k, the p-values were calculated based on the significance test of Pearson correlations (two-sided). The number of cell lines used for analysis is shown above in each plot.
Extended Data Fig. 3 ∣. Influences of SLC25A20 methylation on acylcarnitine metabolism.
a-j, Scatter plots comparing SLC25A20 mRNA transcripts with different metabolites: carnitine (a), acetylcarnitine (b), propionylcarnitine (c), malonylcarnitine (d), butyrylcarnitine/isobutyrylcarnitine (e), valerylcarnitine/isovalerylcarnitine/2-methylbutyroylcarnitine (f), hexanoylcarnitine (g), heptanoylcarnitine (h), lauroylcarnitine (i), arachidonylcarnitine (j). The q-values were calculated based on significance test of Pearson correlations (two-sided) with multiple hypothesis testing correction.
Extended Data Fig. 4 ∣. Additional information regarding amino acid dependency.
a, Cropped immunoblot of ASNS in A2058 cells with or without dox-inducible ASNS knockdown (KD). Tubulin was used as the loading control. The experiment was repeated independently twice with similar results. b, Relative cell growth upon ASNS KD with or without rescue in the A2058 cell line grown in DMEM without asparagine (mean ± SEM, n=2 cell culture replicates, two-sample t-test, two sided). After 13 days, the relative growth was quantified by standard crystal violet staining. PLK1 KD was used as a control. NEAA, non-essential amino acids. c, ASNS mRNA levels with medians across the CCLE lines grouped by cancer types. DLBCL, diffuse large B-cell lymphoma; CML, chronic myeloid leukemia; AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia. d, Scatter plot comparing ATF4 mRNA levels with ASNS mRNA levels in all cell lines. e, Schematic depicting part of the metabolic pathway of asparagine, arginine, and glutamine. f, Scatter plot comparing ASS1 DNA methylation levels with ASS1 mRNA levels in all cell lines. g, Scatter plot comparing GLUL DNA methylation levels with GLUL mRNA levels in all cell lines. For f-g, the p-values were calculated based on the significance test of Pearson correlations (two-sided). The number of cell lines used for analysis is shown in the plot above. h, Waterfall plot showing the fold changes of pooled CCLE lines (n=554, median of 3 independent cell culture replicates) cultured in RPMI media containing 0.1 μM glutamine for 6 days (normalized to control). The colors show the associated cell doubling time. For h, the p-value was calculated based on the significance test of Pearson correlations (two-sided).
Extended Data Fig. 5 ∣. Evaluation of asparaginase therapeutic value in vivo.
a, Surgically removed SNU719 tumors after asparaginase treatment or vehicle control treatment (2 tumors per nude mouse). b, Relative mouse body weight changes in the duration of asparaginase treatment (3000 units/kg, 5 times a week) or vehicle control (n = 5 nude mice per condition, mean ± SEM). c, Methylation-specific PCR for ASNS CpG islands in different tumor samples (a cropped gel image is shown). This experiment was repeated once. d, Bisulfite sequencing for ASNS methylation status in different tumor samples. Open circles indicate unmethylated CpG while solid circles indicate methylated CpG. This experiment was repeated once with 4 technical replicates for each different tumor sample.
Extended Data Fig. 6 ∣. Kynurenine metabolism in the CCLE.
a, Waterfall plots showing the fold changes of pooled CCLE lines (n=554, median of 3 independent cell culture replicates) cultured in RPMI media containing 10 μM or 100 μM kynurenine for 6 days (normalized to control). The colors show the associated kynurenine levels in cells. The p-values were calculated based on the significance test of Pearson correlations (two-sided). b, Schematic depicting part of the tryptophan catabolism pathway. c, Kynurenine levels and a heatmap representation of top correlated mRNA transcripts in each cell line. The feature values (log2 RPKM) were standardized for plotting. The reported test statistics and p-values are based on the significance tests of mRNA feature regression coefficients (cell line n=913, two-sided t-tests). d, mRNA levels of IDO1 and/or IDO2 correlate with kynurenine accumulation (Pearson correlation= 0.26 and 0.22 respectively; two-sided p<10−15 and p=2.5*10−11 respectively). Cell lines (n=913) are represented as points with color-coded kynurenine levels.
Supplementary Material
Supplementary Table 1. The CCLE metabolomic data.
Supplementary Table 2. Metabolite associations with genetic and epigenetic features.
Supplementary Table 3. Metabolite-dependency association analysis.
Supplementary Table 4. Pooled screens of barcoded CCLE lines under various conditions.
Acknowledgements
The authors would like to thank E. Gelfand for assistance with project management. We acknowledge J. Jane-Valbuena, S. Gill, V. Viswanathan, L. Dailey and R. Almeida for helpful discussions. The PRISM team including J. Roth, S. Bender, K. Gore, C. Zhu, and J. Fonseka provided valuable help for the pooled CCLE cancer cell line screening at the Broad Institute. We thank Dana-Farber/Harvard Cancer Center in Boston, MA, for the use of the Specialized Histopathology Core, which provided histology and immunohistochemistry service. Dana-Farber/Harvard Cancer Center is supported in part by an NCI Cancer Center Support Grant # NIH 5 P30 CA06516. The CCLE project was supported by grants from Novartis and R35 Outstanding Investigator Award from the National Cancer Institute (L.A.G.) and U01 CA176058 (W.C.H.). This work was supported in part by the NCI’s Cancer Target Discovery and Development (CTD2) Network (grant number U01 CA217848, awarded to S.L.S.). Haoxin Li is a fellow in the Herchel Smith graduate fellowship program.
Footnotes
Competing interests
G.V.K is an employee of and has equity in KSQ Therapeutics. G.V.K is also a shareholder of Illumina and CRISPR Therapeutics. J.A, D.S, V.A, and G.G.G are current Novartis employees. J.B. is a shareholder and former employee of Novartis. R.A.P is a former employee of Novartis and a current employee of Celsius Therapeutics. W.C.H is a consultant for Thermo-Fisher, Seer, AjuIB, MPM Capital and KSQ Therapeutics and receives research funding from Deerfield Management. W.C.H is a founder and has equity in KSQ Therapeutics. L.A.G. is a current employee at Eli Lilly and Company. A.T is a consultant for Tango Therapeutics. Mar.G participated in an advisory board for Astrazeneca and receives research funding from Bristol-Myers Squibb. S.L.S. is a shareholder of Forma Therapeutics, a shareholder of and adviser to Jnana Therapeutics and Decibel Therapeutics, an adviser to Eisai, Inc. and F-Prime Capital Partners, and a Novartis Faculty Scholar. W.R.S is a shareholder and former employee of Novartis. W.R.S is a consultant and shareholder for Peloton Therapeutics and Ideaya Biosciences and is a consultant for Array, Astex, Ibsen, Servier and Sanofi Pharmaceuticals and Epidarex Capital and receives research funding from Deerfield Management. H.L and W.R.S have a patent application based on this work. The other authors declare no competing interests.
References
- 1.Vander Heiden MG et al. Understanding the Warburg effect : the metabolic requirements of cell proliferation. Science 324, 1029–1034 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pavlova NN & Thompson CB The emerging hallmarks of cancer metabolism. Cell Metabolism 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns RA, Harris IS & Mak TW Regulation of cancer cell metabolism. Nature Reviews Cancer 11, 85–95 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Vander Heiden MG & Deberardinis RJ Understanding the intersections between metabolism and cancer biology. Cell 168, 657–669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tennant DA, Durán RV & Gottlieb E Targeting metabolic transformation for cancer therapy. Nature Reviews Cancer 10, 267–277 (2010). [DOI] [PubMed] [Google Scholar]
- 6.McDonald ERI et al. Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-Scale, deep RNAi screening. Cell 170, 577–592 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Barretina J et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsherniak A et al. Defining a cancer dependency map. Cell 170, 564–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu W et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of a ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang L et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shim EH et al. L-2-hydroxyglutarate: An epigenetic modifier and putative oncometabolite in renal cancer. Cancer Discovery 4, 1290–1298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wakabayashi N et al. Protection against electrophile and oxidant stress by induction of the phase 2 response : Fate of cysteines of the Keap1 sensor modified by inducers. Proceedings of the National Academy of Sciences of the United States of America 101, 2040–2045 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohta T et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Research 68, 1303–1309 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Dey P et al. Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature 542, 119–123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyers RM et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nature genetics (2017). doi: 10.1038/ng.3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Indiveri C, Tonazzi A, Prezioso G & Palmieri F Kinetic characterization of the reconstituted carnitine carrier from rat liver mitochondria. Biochimica et Biophysica Acta 1065, 231–238 (1991). [DOI] [PubMed] [Google Scholar]
- 17.Yang WS et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu C et al. High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nature biotechnology 34, 419–423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mezrich JD et al. An Interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. The Journal of Immunology 185, 3190–3198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uyttenhove C et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nature Medicine 9, 1269–1274 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Munn DH & Mellor AL Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends in Immunology 34, 137–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brochez L, Chevolet I & Kruse V The rationale of indoleamine 2, 3-dioxygenase inhibition for cancer therapy. European Journal of Cancer 76, 167–182 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Jain M et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336, 1040–1044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long GV et al. Epacadostat (E) plus pembrolizumab (P) versus pembrolizumab alone in patients (pts) with unresectable or metastatic melanoma: results of the phase 3 ECHO-301/KEYNOTE-252 study. in J Clin Oncol 36, 2018 (suppl; abstr 108) (2018). [Google Scholar]
- 25.Narta UK, Kanwar SS & Azmi W Pharmacological and clinical evaluation of L-asparaginase in the treatment of leukemia. Critical Reviews in Oncology/Hematology 61, 208–221 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Hettmer S et al. Functional genomic screening reveals asparagine dependence as a metabolic vulnerability in sarcoma. eLife 4, 1–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knott SRV et al. Asparagine bioavailability governs metastasis in a model of breast cancer. Nature 554, 378–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Reference for methods
- 28.Townsend MK et al. Reproducibility of metabolomic profiles among men and women in 2 large cohort studies. Clinical Chemistry 59, 1657–1667 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. The CCLE metabolomic data.
Supplementary Table 2. Metabolite associations with genetic and epigenetic features.
Supplementary Table 3. Metabolite-dependency association analysis.
Supplementary Table 4. Pooled screens of barcoded CCLE lines under various conditions.
Data Availability Statement
The CCLE metabolomic data and related analyses can be found in Supplementary Tables 1-4. Additional CCLE resources are available at: https://portals.broadinstitute.org/ccle. Source data of all the western blots and DNA gels in this work are provided in Source Data Figures.