Abstract
BACKGROUND
Cerebral cavernous angiomas (CAs) with a symptomatic hemorrhage (CASH) have a high risk of recurrent hemorrhage and serious morbidity.
METHODS
Eighteen plasma molecules with mechanistic roles in CA pathobiology were investigated in 114 patients and 12 healthy subjects. The diagnostic biomarker of a CASH in the prior year was derived as that minimizing the Akaike information criterion and validated using machine learning, and was compared with the prognostic CASH biomarker predicting bleeding in the subsequent year. Biomarkers were longitudinally followed in a subset of cases. The biomarkers were queried in the lesional neurovascular unit (NVU) transcriptome and in plasma miRNAs from CASH and non-CASH patients.
RESULTS
The diagnostic CASH biomarker included a weighted combination of soluble CD14 (sCD14), VEGF, C-reactive protein (CRP), and IL-10 distinguishing CASH patients with 76% sensitivity and 80% specificity (P = 0.0003). The prognostic CASH biomarker (sCD14, VEGF, IL-1β, and sROBO-4) was confirmed to predict a bleed in the subsequent year with 83% sensitivity and 93% specificity (P = 0.001). Genes associated with diagnostic and prognostic CASH biomarkers were differentially expressed in CASH lesional NVUs. Thirteen plasma miRNAs were differentially expressed between CASH and non-CASH patients.
CONCLUSION
Shared and unique biomarkers of recent symptomatic hemorrhage and of future bleeding in CA are mechanistically linked to lesional transcriptome and miRNA. The biomarkers may be applied for risk stratification in clinical trials and developed as a tool in clinical practice.
FUNDING
NIH, William and Judith Davis Fund in Neurovascular Surgery Research, Be Brave for Life Foundation, Safadi Translational Fellowship, Pritzker School of Medicine, and Sigrid Jusélius Foundation.
Keywords: Inflammation, Vascular Biology
Keywords: Diagnostics, Stroke
A highly sensitive and specific plasma biomarker signature of symptomatic hemorrhage in cavernous angiomas is identified as are biomarkers predicting the likelihood of rebleeding events.
Introduction
Cavernous angiomas (CAs), also known as cerebral cavernous malformations or cavernomas, are a common neurovascular pathology affecting 0.3%–0.9% of the population, predisposing patients to a lifetime risk of stroke, seizures, and consequent disability related to lesional hemorrhage (1, 2). CA with recent symptomatic hemorrhage (CASH) is an important event defined by evidence of new bleeding on diagnostic imaging, with new attributable neurological symptoms (1). Based on a systematic review of studies with no selection criteria, the risk of first symptomatic hemorrhage has been estimated at 0.4%–2.4% per year, with the higher estimate in brainstem lesions (1, 2). Once a patient experiences an initial symptomatic hemorrhage, the risk of rebleeding increases more than 10-fold, to 3.8%–29.5% per year (1). It would be desirable to develop a therapy that stabilizes the CASH and prevent future hemorrhagic sequelae (1, 3). CASH lesions have been singularly targeted for therapeutic development and trial readiness (NIH 1-U01-NS104157-01) (3).
CAs are clusters of thin-walled vascular caverns lined by endothelium that lack mature vessel wall architecture (1). A sporadic form of the disease manifests a solitary lesion and accounts for almost two-thirds of cases (1). A familial form of the disease results from a germline mutation at one of 3 CCM gene loci (CCM1/KRIT1, CCM2/MGC4607, and CCM3/PDCD10), manifesting with multifocal lesion development throughout the brain. Lesions are histologically indistinguishable in both forms of the disease and harbor somatic mutations in the same CCM genes (4). Rarely, lesions develop after brain irradiation (5).
Biologic mechanisms associated with this disease hold the promise of characterizing the molecular signatures of the CASH lesion and potentially predicting future bleeding. In vivo and in vitro investigations in transgenic murine models support a complex CA pathobiology, with contribution of inflammatory and angiogenic processes during CA genesis, maturation, and hemorrhage (6). Proinflammatory gene variants have been associated with higher lesion counts in Hispanic American patients with a common CCM1 (Q455X) mutation, but not with the risk of bleeding from individual lesions (7, 8). A “proinflammatory cluster” of plasma cytokines, vitamin D, and non-HDL cholesterol plasma levels has been correlated with chronic CA disease severity, but not recent bleeding (9, 10). A systematic literature review of biologic mechanisms implicated in brain hemorrhage and CA identified a set of plasma molecules potentially playing a role in this disease (10, 11). We demonstrated that weighted combined plasma levels of 4 of these molecules could predict a symptomatic bleed in the subsequent year (11). However, it remains unclear whether a plasma biomarker can differentiate patients who have experienced CASH within the prior year, and if the same or different molecules can diagnose a recent bleed and predict future bleeding.
We hypothesized that levels of plasma molecules previously implicated in disease mechanisms would act as a diagnostic biomarker in patients who experienced a symptomatic hemorrhage within the prior year, thereby explaining the inflammatory and angiogenic processes occurring after a CASH (12). We conducted validations of this diagnostic CASH biomarker using adjunctive statistical and machine learning simulations, and examined it in healthy non-CA subjects. We identified the similarities between the diagnostic CASH biomarker and the previously identified prognostic biomarker of symptomatic hemorrhage (11). We queried the transcriptome of mRNA from lesional neurovascular units (NVUs) and circulating miRNA from plasma of CA patients in relation to the molecules in the diagnostic and prognostic CASH biomarkers. Finally, we analyzed changes in the diagnostic and prognostic CASH biomarkers during the follow-up of stable and unstable patients, and those recovering after a recent bleed.
Results
Demographics and lesion characterization of patients enrolled.
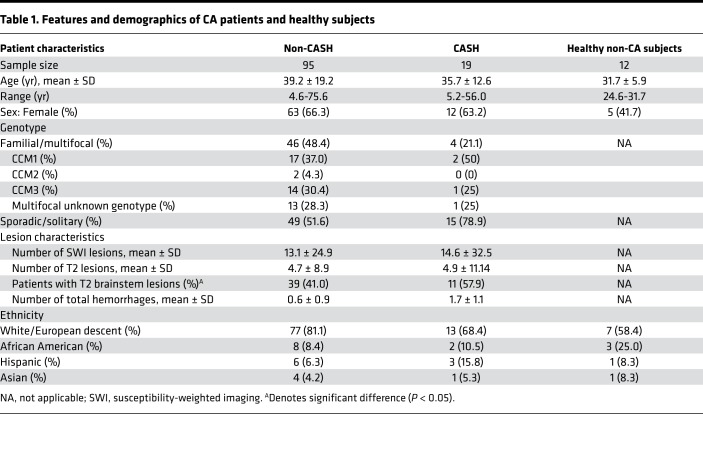
Among the 114 patients enrolled, 19 had experienced an adjudicated symptomatic hemorrhage in the prior year (±30 days) and were qualified as CASH patients (Figure 1). The remaining 95 patients did not experience CASH in the year prior to the plasma sample and were defined as non-CASH patients. There were no significant differences in patient age, sex, ethnicity, genotype, or lesion number between CASH and non-CASH cases (Table 1). We also did not find any difference among the cohorts with and without follow-up (Table 2 and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.128577DS1). As expected, there was a greater prevalence of brainstem lesions among CASH cases and in the follow-up cohort.
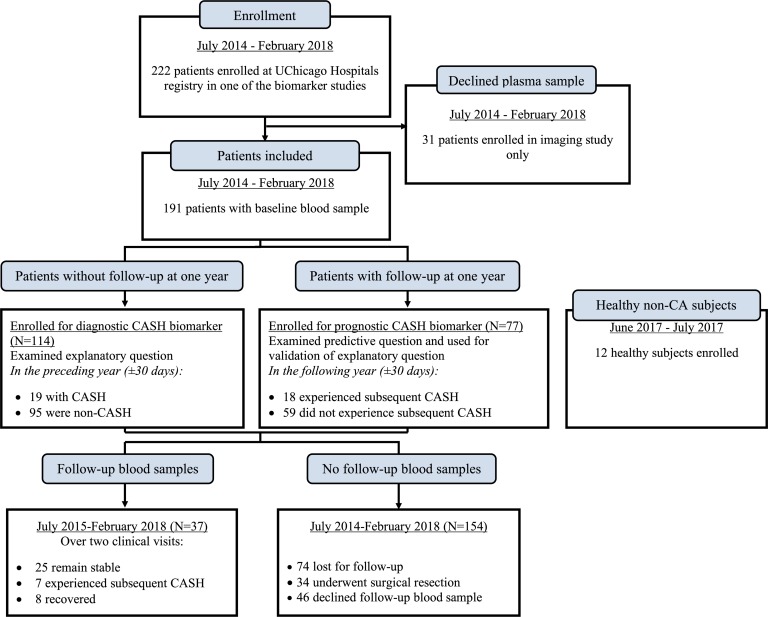
Figure 1. Consort diagram.
Two hundred and twenty-two patients were enrolled between July 2014 and February 2018 in one of the biomarker studies ongoing at the University of Chicago Neurovascular Research Center. In addition, 12 healthy subjects were enrolled between June 2017 and July 2017. Thirty-one patients were only enrolled in the imaging study. One hundred and fourteen patients were enrolled for the diagnostic CASH biomarker, while 77 constituted the prognostic CASH biomarker cohort. Of these 191 patients, 37 had a 1-year follow-up with a blood sample after the initial collection. One hundred and fifty-four patients were either lost to follow-up (n = 74), underwent surgical resection (n = 34), or declined follow-up blood samples (n = 46).
Table 1. Features and demographics of CA patients and healthy subjects.
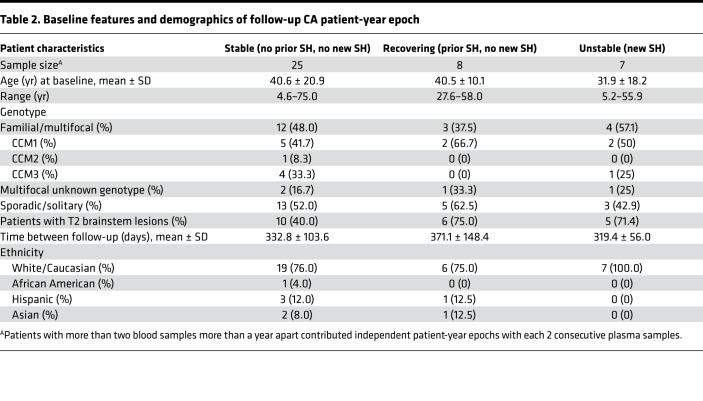
Table 2. Baseline features and demographics of follow-up CA patient-year epoch.
Soluble CD14, VEGF, IL-1β, IL-6, IL-8, IL-10, and TNF-α plasma levels were lower, while C-reactive protein and TNF receptor 1 were significantly increased in patients with CASH.
Plasma levels of 18 molecules previously identified in a systematic literature search were first compared between CASH and non-CASH patients (10, 11). Plasma levels of soluble CD14 (sCD14; P = 0.004, FDR corrected), C-reactive protein (CRP; P = 0.004, FDR corrected), IL-8 (P = 0.005, FDR corrected), IL-1β (P = 0.007, FDR corrected), IL-6 (P = 0.01, FDR corrected), IL-10 (P = 0.01, FDR corrected), VEGF (P = 0.004, FDR corrected), TNF-α (P = 0.01, FDR corrected), and TNF receptor 1 (TNFRI; P = 0.01, FDR corrected) were significantly different in CASH patients. Of these molecules, CRP and TNFRI were increased, while sCD14, VEGF, IL-1β, IL-6, IL-8, IL-10, and TNF-α were decreased. We did not observe any significant differences in the levels of these plasma molecules in relation to patient age, sex, phenotype (sporadic/solitary or familial/multifocal), or genotype (CCM1, -2, or -3), or in association with brainstem lesion location (Supplemental Table 2).
The best weighted linear combination of sCD14, VEGF, IL-10, and CRP is able to distinguish patients with CASH.
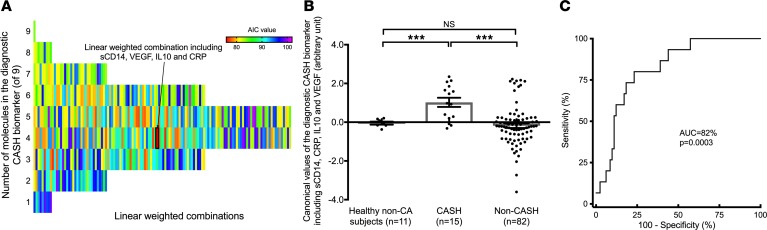
The best diagnostic biomarker to distinguish CASH patients (Akaike information criterion [AIC] = 75.9) among the 511 possible combinations included a weighted combination of sCD14, VEGF, IL-10, and CRP levels (Figure 2), as formulated: diagnostic biomarker = –3.37*[sCD14] +1.47*[CRP] –0.36*[VEGF] –0.57*[IL-10].
Figure 2. The best diagnostic biomarker for CASH included sCD14, VEGF, CRP, and IL-10.
(A) Among the 511 possible combinations, the best-weighted diagnostic biomarker to differentiate patients who experienced CASH included sCD14, VEGF, CRP, and IL-10 (Akaike information criterion [AIC] = 75.9). The colors are shaded in relation to the AIC of all the possible combination of molecules, which were significant individually. The deepest red color represents the lowest AIC, which determines the best predictive combination. (B) A 2-tailed 2-sample t test showed that the estimated values were increased in CASH (n = 15; mean estimated value ± SD, 1.02 ± 0.92) compared with healthy non-CA subjects (n = 11; mean estimated value ± SD, –0.05 ± 0.21) and non-CASH patients (n = 82; mean estimated value ± SD, –0.18 ± 1.05) (P = 0.0005 and P = 0.0002, respectively). (C) Receiver operating characteristic (ROC) analysis was able to differentiate CASH patients with 77% sensitivity and 80% specificity (AUC = 82%, P = 0.0003). ***P < 0.001. Data represent mean ± SEM.
The individual receiver operating characteristic (ROC) curves generated for sCD14, VEGF, CRP, and IL-10 showed “poor” or “fair” accuracy (Figure 3). However, the canonical value calculated using the diagnostic biomarker demonstrated sensitivity and specificity of 77% and 80%, respectively (AUC = 82%, P = 0.0003) to distinguish CASH patients (Figure 2). It was significantly increased in CASH patients compared with healthy non-CA subjects (mean estimated value ± SD = –0.05 ± 0.21) and non-CASH patients (P = 0.0005 and P = 0.0002, respectively), and was not significantly different between healthy non-CA subjects and non-CASH patients. The mean weighted combination value was 6-times increased (P = 0.0002) in CASH patients (mean estimated value ± SD = 1.02 ± 0.92) compared with non-CASH patients (mean estimated value ± SD = –0.18 ± 1.05). The plasma levels of the 4 individual components encompassed in the diagnostic CASH biomarker were not correlated between each other, and hence showed independent contribution (Supplemental Table 3).
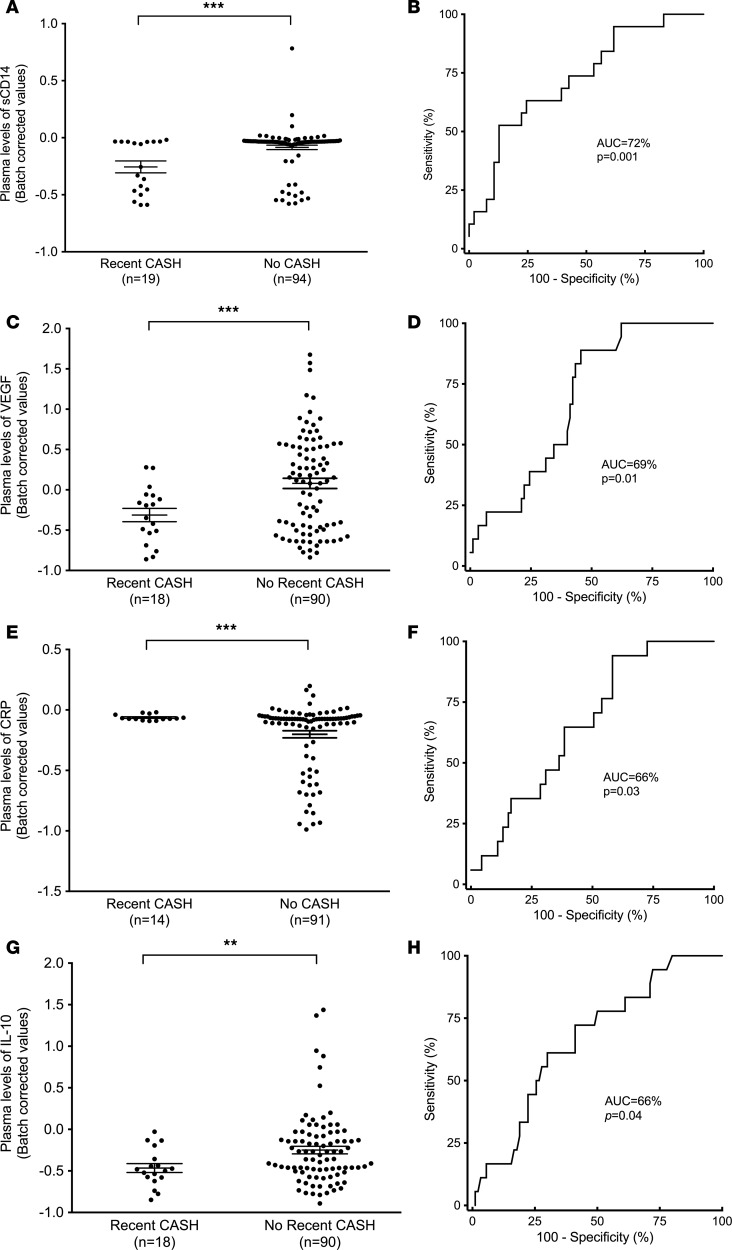
Figure 3. Differentially expressed inflammatory proteins in CASH patients.
Among the 18 plasma molecules, CASH patients showed lower plasma levels of sCD14 (AUC = 72%, P = 0.001, 52% sensitivity, 87% specificity) (A and B), VEGF (AUC = 69%, P = 0.01, 89% sensitivity, 54% specificity) (C and D), and IL-10 (AUC = 66%, P = 0.04, 61% sensitivity, 70% specificity) (G and H), along with higher CRP (AUC = 66%, P = 0.03, 94% sensitivity, 42% specificity) plasma levels (E and F). The plasma levels of the 18 plasma molecules were compared between CASH and non-CASH patients using a 2-tailed 2-sample t test with an FDR correction. **P < 0.01, ***P < 0.001. Data represent mean ± SEM.
Statistical and machine learning validation of the diagnostic CASH biomarker.
The derived diagnostic CASH biomarker (the weighted combined levels of sCD14, VEGF, IL-10, and CRP) was then statistically validated using a simulated population using Monte Carlo, random sampling (Supplemental Figure 1), and machine learning approaches. We first simulated the plasma levels of sCD14, VEGF, IL-10, and CRP, assuming normal probability distribution and batch effect–corrected values, for 1000 simulated non-CASH patients and 1000 CASH patients using a Monte Carlo approach. The simulated values of the diagnostic CASH biomarker resulted in sensitivity and specificity of 89% and 67% (AUC = 84%, P < 0.0001) (Supplemental Figure 1A).
The second statistical validation was performed using a random-sampling parsing approach whereby a simulated cohort was generated by selecting randomly 70% of the patients over the course of 1000 simulations. The diagnostic CASH biomarker was able to differentiate CASH patients in the simulated cohort with sensitivity and specificity of 80% and 77% (AUC = 82%, P < 0.0001) (Supplemental Figure 1B).
We also employed machine learning techniques to validate the diagnostic CASH biomarker. First, we used a 1000-times-repeated sampling with a 70:30 partition bootstrapping aggregation (bagging) that identified the following equation: diagnostic biomarker = –3.39*[sCD14] +1.48*[CRP] –0.38*[VEGF] –0.56*[IL-10], with sensitivity and specificity of 77% and 80% (P < 0.0001, AUC = 82%). This bagging approach showed high similarity to the equation previously identified through statistical methods using AIC. Finally, a 10-fold cross-validation support vector machine (SVM) hyperbolic tangent–based kernel validation resulted in a best sensitivity and best specificity of 75% and 77%, respectively, for the diagnostic CASH biomarker.
The diagnostic CASH biomarker showed shared and unique molecules with the prognostic CASH biomarker.
We then compared the diagnostic CASH biomarker with a recently published prognostic CASH biomarker using an extended cohort of 77 (42 sporadic/solitary, 35 familial/multifocal) patients, which included 18 patients who experienced CASH in the year (±30 days) following plasma collection and 59 who did not experience a subsequent CASH (11): prognostic biomarker= –0.135*[sCD14] +7.73*[IL-1β] –0.775*[VEGF] +0.658*[sROBO4].
This prognostic extended cohort was independent from the cohort used to develop the CASH biomarker. The mean of the canonical values estimated by the prognostic CASH biomarker was greater (P < 0.0001) in subjects who experienced a subsequent CASH in the following year (mean estimated value ± SD= –0.61 ± 0.68) compared with subjects who did not experience subsequent CASH (mean estimated value ± SD = –2.00 ± 0.55). Patients who did not experience a subsequent CASH over the following year were significantly different in comparison to healthy non-CA subjects (mean estimated value ± SD= –1.01 ± 0.32; P < 0.0001), as well as to patients who experienced CASH in the following year after the blood collection (P < 0.0001). In addition, healthy non-CA subjects showed a trend toward lower canonical values (P = 0.10) than patients who experienced a CASH in the following year (Supplemental Figure 2A). ROC analysis showed sensitivity and specificity of 83% and 93% (AUC = 94%, P < 0.0001) in distinguishing patients who would experience future hemorrhagic expansion within the following year (±30 days) from patients who did not experience future hemorrhagic expansion (Supplemental Figure 2B). The SVM hyperbolic tangent–based kernel validation of the prognostic CASH biomarker resulted in best sensitivity and best specificity of 100% and 86%.
Cross-comparison of the diagnostic and prognostic CASH biomarkers.
We then used the prognostic cohort as an independent validation cohort for the diagnostic CASH biomarker, which yielded sensitivity and specificity of 80% and 78%, respectively (AUC = 84%, P = 0.003). The diagnostic and prognostic CASH biomarkers were cross-compared to determine whether one biomarker could distinguish the others characteristic (Supplemental Figure 3). The prognostic biomarker was used on the cohort of CASH patients to test its ability to distinguish CASH rather than predicting a future CASH. This cross-comparison yielded a “poor” ROC curve with sensitivity and specificity of 50% and 86%, respectively (AUC = 62%, P = 0.03).
The opposite comparison was also performed to test whether the diagnostic CASH biomarker could predict future CASH in the extended prognostic cohort. This analysis also yielded a poor ROC curve, with sensitivity and specificity of 50% and 93% (AUC = 60%, P = 0.01). This cross-comparison analysis demonstrated that each biomarker cannot distinguish the other biomarker’s characteristic event. The canonical values estimated by the 2 biomarkers were not correlated. Finally, the canonical values calculated by each biomarker in the other defining characteristic demonstrated no significant differences between groups for the diagnostic biomarker.
3D hypersurface-under-manifold between healthy control and CA patients defined by CASH.
The 3D ROC analysis among the 3 groups enrolled including healthy non-CA subjects, CASH and non-CASH patients showed a hypersurface-under-manifold (HUM) of 52% and best differentiating surface point of (73%, 86%, 56%) to distinguish the 3 cohorts.
The diagnostic and prognostic CASH biomarkers performed as anticipated in recovering, unstable (new bleed), and stable cases over time.
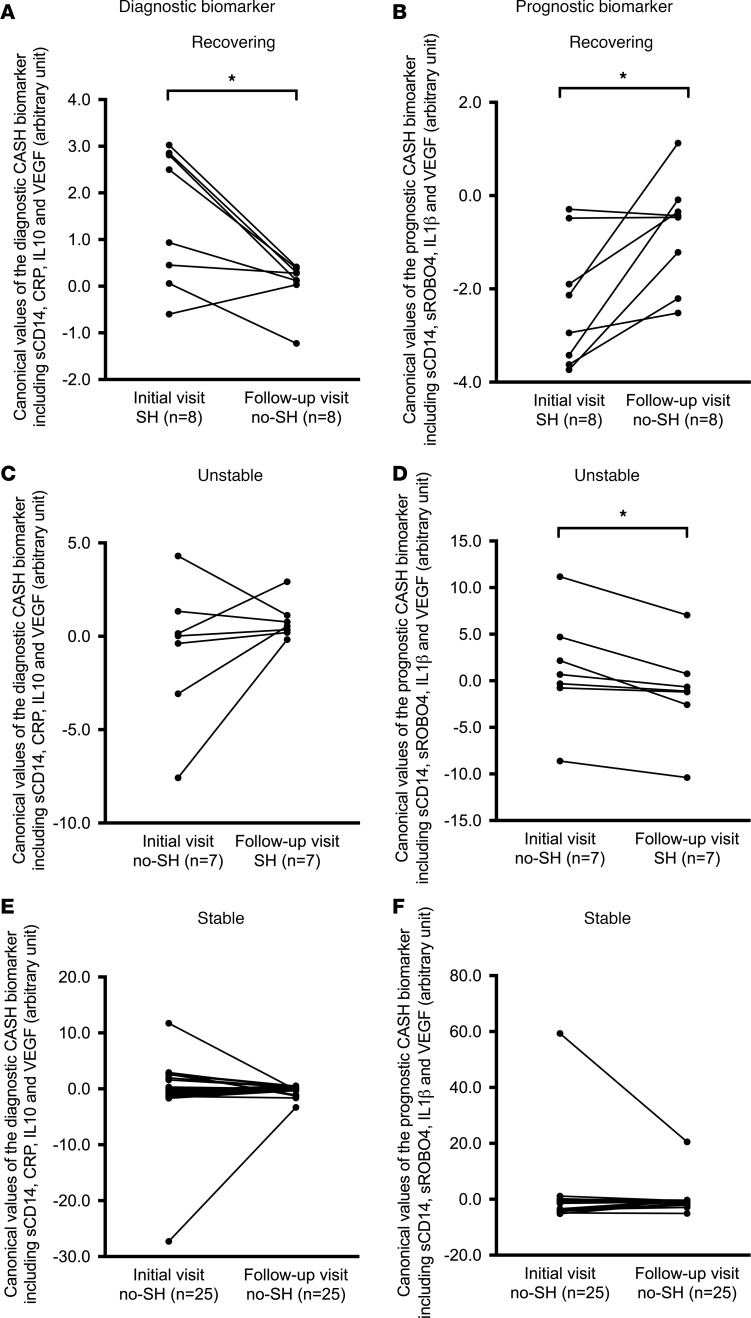
Thirty-seven patients had at least one follow-up blood sample within 1 year (±30 days) of their initial blood sample; 25 remained stable over time, with no symptomatic bleeding at either time point (stable); 7 experienced a new CASH (unstable); and 8 recovered after a CASH (recovering) (Table 2). As would be anticipated in cases recovering after a CASH, the canonical diagnostic biomarker decreased in all but 1 case (mean significantly decreased, P = 0.01) (Figure 4A). Conversely the prognostic biomarker increased in all but 2 recovering cases (mean significantly increased, P = 0.02), potentially heralding future rebleed risk (Figure 4B). In cases experiencing a new bleed during the epoch between plasma samples, the diagnostic biomarker increased in all but 2 cases (means not significantly different), while the prognostic biomarker started higher as expected in predicting the bleed, and decreased in all cases following the new bleed (mean significantly decreased, P = 0.01) (Figure 4, C and D). Finally, the follow-up values of stable patients did not change over time in all but 2 cases in the diagnostic and all but 1 case in prognostic CASH biomarker (both means nonsignificantly different) (Figure 4, E and F).
Figure 4. Diagnostic biomarker decreased and the prognostic biomarker increased in recovering patients, while diagnostic biomarker increased and prognostic biomarker decreased in unstable patients.
In recovering patients (n = 8), (A) the diagnostic biomarker showed decreased (P = 0.01) while (B) the prognostic biomarker showed increased (P = 0.02) canonical followed-up plasma values (1 year ± 30 days). In unstable patients (n = 7), the followed-up plasma values (C) increased for the diagnostic biomarker (NS) and (D) decreased for the prognostic biomarker (P = 0.01). (E and F) No significant change over time was observed in diagnostic or prognostic CASH biomarker canonical values in stable patients (n = 25). Patients with more than 2 longitudinal blood samples were considered as independent epochs between each 2 consecutive blood samples. The difference over time were assessed using Wilcoxon’s tests. *P < 0.05. SH, symptomatic hemorrhage.
Transcriptome analyses of CASH and non-CASH lesional NVUs.
Transcriptomic analyses were performed in order to both validate the current biomarkers and identify new candidate biomarkers for future study (Supplemental Figure 4 and Supplemental Table 4). A total of 1027 and 866 differently expressed genes (DEGs) were identified (P < 0.05, FDR corrected) in, respectively, the CASH and non-CASH compared with healthy NVUs harvested from brains at autopsy brain (13). Among the 1027 DEGs, 398 were only dysregulated in the CASH transcriptome. Network analysis of the 398 DEGs (Supplemental Table 5) showed 17 highly interconnected genes (5 or more connections), including RHOF, RAC2, CD3G, GNAI1, GNA15, PRKACB, GLI2, BTRC, PLK4, CKAP5, PCM1, CEP290, SDCCAG8, CLASP1, ITGAL, ITGB5, and COL6A1 (Supplemental Figure 5). In addition, gene ontology (GO) enrichment analysis identified 28 enriched GO terms (P < 0.01, FDR corrected) (Supplemental Figure 6).
Among the 866 DEGs identified in non-CASH NVUs, 237 were only found in the non-CASH transcriptome (Supplemental Table 6). GO analysis of these 237 DEGs showed 44 enriched GO terms (P < 0.01, FDR corrected). Finally, 629 DEGs were commonly differently expressed in both CASH and non-CASH transcriptome (Supplemental Table 7), and were associated with 460 enriched GO terms (P < 0.01, FDR corrected) (Supplemental Figure 6).
The mRNA CASH transcriptome in CASH patients supports the established biomarker.
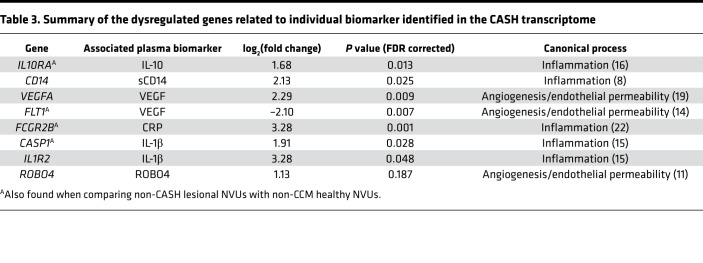
Specific transcriptomic changes supporting the diagnostic and prognostic biomarkers were identified in the 398 DEGs (P < 0.05, FDR corrected) in CASH but not non-CASH NVUs. Five of the 6 molecules composing the prognostic and diagnostic biomarkers were significantly related (P < 0.05, FDR corrected) to DEGs in CASH NVUs, including CASP1 (log2[fold change] [log2FC] = 1.91), IL1R2 (log2FC = 3.28), CD14 (log2FC = 2.13), FCGR2B (log2FC = 3.28), VEGFA (log2FC = 2.29), as well as FLT1 (log2FC = –2.10) (Table 3) (13–15). IL10RA (log2FC = 1.68) was also identified in the CASH transcriptome (as well as in the non-CASH transcriptome). No dysregulated genes related to ROBO4 reached significance (log2FC = 1.13; P = 0.18, FDR corrected).
Table 3. Summary of the dysregulated genes related to individual biomarker identified in the CASH transcriptome.
Plasma miRNAs and integration analyses from transcriptome of CASH and non-CASH lesional NVUs and dysregulated plasma miRNA.
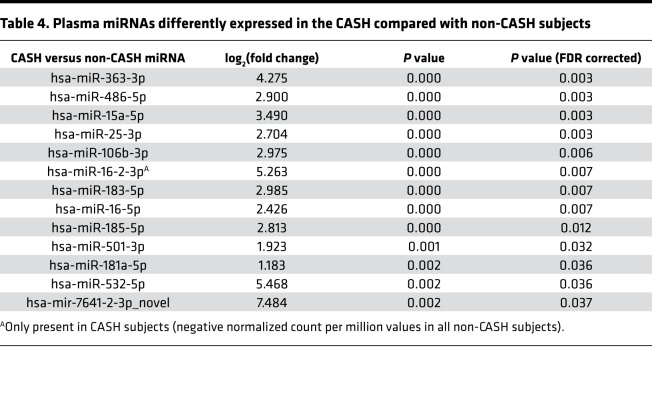
Thirteen plasma miRNAs were differently expressed (P < 0.05, FDR corrected) between CASH and non-CASH patients (Table 4). Demographic characteristics of the subset of patients included in miRNA analyses are summarized in Supplemental Table 8. The integration analyses of these 13 miRNAs found 16 putative target genes in the 3′ UTR, 3 in the 5′ UTR, and 19 in the coding sequence (CDS) region of mRNA found among the 629 DEGs common to both CASH and non-CASH NVUs (Supplemental Table 9). In addition, 14 putative target genes were also found in the 3′ UTR, 2 in the 5′ UTR, and 16 in the CDS region of mRNA within the 398 DEGs identified only in the CASH transcriptome (Supplemental Table 10).
Table 4. Plasma miRNAs differently expressed in the CASH compared with non-CASH subjects.
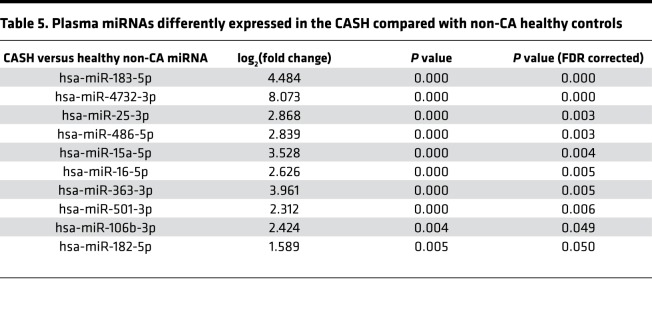
Finally, 10 plasma miRNAs were also differently expressed (P < 0.05, FDR corrected) between CASH patients and healthy non-CA subjects (Table 5). No difference in miRNA profiling was identified between non-CASH patients and non-CA healthy subjects. One of the miRNAs, hsa-miR-185-5p, putatively targets IL10RA (Supplemental Table 9).
Table 5. Plasma miRNAs differently expressed in the CASH compared with non-CA healthy controls.
Discussion
Herein we examined candidate inflammatory and angiogenic molecules as potential biomarkers of CASH (10, 11). Nine inflammatory plasma molecules were differentially detected in patients who experienced a symptomatic hemorrhage within the prior year (±30 days). The weighted combination using plasma levels of 4 molecules, sCD14, VEGF, CRP, and IL-10, was defined as the best diagnostic biomarker able to distinguish CASH patients who had experienced a symptomatic hemorrhage in the prior year. This combination was independently derived using traditional statistical methods and the AIC, and validated using 2 distinct machine learning approaches, as well as Monte Carlo random-sampling simulations. Comparison between the diagnostic CASH biomarker and our previously published prognostic CASH biomarker predicting a symptomatic bleed suggests the existence of shared and distinct components of the inflammatory and angiogenic changes occurring in CA patients after a hemorrhagic event and leading to a future bleed. In the context of the most recently published guidelines, this study begins to develop a method by which CA patients can be further classified into high-risk and low-risk groups in association with CASH, which is one of the most clinically relevant characteristics of this disease (1). We also provided transcriptomic evidence that these markers are related to hemorrhagic CAs
Neuroinflammation is increasingly a focus of research in cerebrovascular hemorrhagic events (10, 11). Although certain elements of inflammation are well documented, such as the involvement of IL-10 in recovery after an inflammatory response, there is still a significant gap in knowledge regarding inflammatory processes surrounding cerebral hemorrhage (16). Here we have shown significant changes in IL-10, CRP, VEGF, and sCD14, all in response to a clinically relevant hemorrhagic event in the prior year. The previously identified prognostic CASH biomarker also showed different plasma levels of VEGF and sCD14 in patients who would experience a subsequent hemorrhage within a year after the plasma collection. Furthermore, a lesional NVU transcriptome database identified dysregulated genes related to 5 of the 6 plasma molecules.
Similar to CD14 involvement in LPS signaling, IL-10 has been demonstrated to be involved in metabolic reprogramming of macrophages by inhibiting LPS uptake (16). Following acute brain injuries, IL-10 mediates a resolution of various inflammatory processes most commonly associated to M2 macrophage activation through STAT3, alongside a possible neuroprotective effect prior to traumatic events (17, 18). Of note, VEGF is known to activate STAT signaling, recapitulated here by the altered levels of VEGF in the plasma along with altered levels of IL10RA mRNA in the lesional transcriptome, as well the potential modulation by hsa-miR-185-5p found in the plasma of CASH patients (19). IL-10 signaling has been reported to be acutely elevated in peripheral plasma following an intracranial hemorrhage, but it is unclear how its levels evolve after the acute stage (20). Here we report decreased IL-10 levels in cases who had suffered a symptomatic CA hemorrhage in the prior year, but we were unable to assess whether IL-10 levels would have been elevated in the acute phase. Our group had previously reported an inverse relationship between quantitative susceptibility mapping, a measure of iron accumulation in CAs, and IL-10 after a symptomatic hemorrhage (21).
CRP is also a well-defined marker of the inflammatory response, typically associated with acute events, and is routinely used in clinic as a marker of the acute phase of general inflammation (22). We here observed an increase in CRP plasma levels for up to a year after a CASH, indicating a persistent response, not limited to the acute phase. We also noted changes at the transcriptomic level in CASH lesional NVUs of the CRP-related receptor FCGR2B (22). Interestingly, abnormal levels of both IL-10 and CRP have also been associated with increased risk for developing vascular dementia, although multiple studies have yielded conflicting results (23).
Two of 4 molecules (sCD14 and VEGF) were common to both the diagnostic and prognostic CASH biomarkers. This suggests a role for these molecules in both angiogenic and inflammatory processes following a hemorrhage, and susceptibility to subsequent CASH. This may explain, in part, by a consistent natural history observation that lesions that recently bled are more likely to rebleed (1, 2). sCD14 has been suggested to possibly promote either proinflammatory or antiinflammatory phenotypes via interaction with LPS-related pathways (8, 24). Conversely, gene polymorphisms expected to cause enhanced expression of CD14 receptors have been correlated with more severe CA disease (7, 8). CD14 receptors on brain endothelial cells were also found to mediate LPS-induced CA lesion development in mouse models (8). Here, lower plasma levels of sCD14 were detected in both diagnostic and prognostic CASH biomarkers (11). CD14 has been linked to STAT regulation involving IL-10, which was reflected here in the dysregulated IL10RA and CD14 genes found in the transcriptome data (25). These results suggest a role for STAT signaling and an interaction of these molecules in CASH.
VEGF has also been heavily implicated in CA genesis (26). Aberrant VEGF expression has been extensively studied in CA disease, suggesting a role in changing the permeability of the blood-brain barrier and lesion progression (27). Notably, decreased plasma levels of VEGF have been shown to occur in CA patients who bled in the prior year, and in cases who would experience a hemorrhage in the upcoming year. Altered levels of VEGFA and FLT1 (also known as VEGFR-1) mRNAs were also observed in the CASH transcriptome. This suggests a similar state of aberrant VEGF signaling, which predisposes patients to a lack of vascular integrity. We believe our results will generate hypotheses about the potential role of lower plasma VEGF levels in CASH cases to be examined in future studies.
Our 2 biomarkers may enable clinicians to adjudicate when patients have experienced a symptomatic hemorrhage, and whether they have increased risk of a future hemorrhagic event. Notably, the values of the diagnostic and prognostic CASH biomarkers did not change over time in patients who remained stable, supporting their potential clinical utility in managing CA patients. Their changes over time in recovering cases and in new bleeds were consistent with their hypothesized roles.
We did not document a correlation of CASH biomarkers with brainstem lesion location. Our study may have been underpowered to detect such an association. Or this may reflect a lack of true biologic risk with lesions in that location. The reported increased prevalence of symptomatic hemorrhage in brainstem lesions may reflect a sensitivity bias, due to the highly eloquent (sensitive) location, where the slightest bleed is more likely to cause symptoms. This will need to be clarified in future studies.
We validated the importance of the molecules involved in the diagnostic and prognostic CASH biomarkers by identifying associated dysregulated genes specifically in the transcriptome of CASH lesional NVUs. In addition, transcriptome analyses identified that hsa-miR-185-5p is dysregulated in the plasma of CASH patients and putatively targets IL10RA. This result suggests that the IL-10 and related processes may be critical to lesion pathology, and that differing regulation of IL-10 may play a critical role in hemorrhagic events.
Our transcriptomic lesional mRNA and plasma miRNA databanks may serve as an important resource for identifying new mechanistic targets involved in CASH pathogenesis, as well as possible biomarker exploration in the future (13). Network analysis of the DEGs identified only in the CASH transcriptome showed 17 highly interconnected genes, such as BTRC, COL6A1, and ITGB5 (Supplemental Figure 5), which may be considered as additional candidate biomarkers or undergo mechanistic studies in the future (Supplemental Figure 4). These molecules may be measured in plasma and have documented roles in TLR4 signaling, apoptosis pathways, and endothelial junction stability, respectively, which are mechanisms reported to be involved in CA pathogenesis (8, 13). RHOF was highly connected within the network and has been reported to be involved in lamellopodium formation, which may be an ongoing process in the endothelium of the CA following vascular injury (28). Furthermore, some of highly interconnected genes have protein-encoded products that are involved with extracellular matrix stability and cell-cell interactions, such as COL6A1, ITGB5, and ITGAL, suggesting a remodeling of the extracellular compartment as an important process following hemorrhagic damage (29). Finally, various cell signaling transduction molecules with recognized superfamilies, such as RHOF and RAC2, as well as smaller G-associated proteins such as GNAI1 and GNA15 may serve as an interesting starting point for future mechanistic studies exploring pathway dysregulation following injury. Although such G-associated proteins may initially appear as nonspecific markers of altered regulatory pathways, generous amounts of evidence exist for the role of G-associated proteins in vascular disease, such as in Sturge-Weber syndrome. In addition, GNAO1 has been reported to be differently expressed across multiple different CA model species (13, 30).
The GO analyses of the 398 DEGs identified only in the CASH transcriptome showed the importance of extracellular remodeling, as well as ongoing catabolic processes, as these processes were absent in the non-CASH transcriptome (Supplemental Figure 6). These catabolic GO terms included collagen and multicellular organismal catabolic processes, and the relevance of these pathways being activated following a hemorrhagic event may simply be related to the ongoing breakdown of dead cells and debris (31). Additional studies are needed to determine whether CCM1, CCM2, or CCM3 differ in their response following a hemorrhagic event, and whether or not these microlevel functions significantly differ in patients that will rebleed. The non-CASH transcriptome demonstrated a number of GO terms that had an overarching theme involving membrane function, including various types of channel activity, mechanisms involving synaptic transmission, and regulation of downstream and upstream events of synaptic transmission. It is unclear how these functions protect from a hemorrhagic event. We identified significant differences in the transcriptomes of CASH and non-CASH patients. There is notably some expected commonality between CASH and non-CASH lesions as well, which is likely due to underlying pathologic mechanisms of CA disease. Ultimately these transcriptomic studies provide different insights into the intralesional pathology occurring following CASH in CA disease. These results may also motivate novel biomarker discoveries and therapeutic approaches, including immunomodulation, small molecule inhibition, and mRNA modulation.
The limitations of this study include selection and follow-up biases inherent to a single referral center, despite our careful comparison of features in cases with and without follow-up. Future multicenter studies will mitigate these biases and allow further analytic validations. Another limitation is the use of batch correction, which provides arbitrary units instead of measurable units that may be directly translated into the clinical setting. This may be addressed by new ELISA methodology. We did not document a relationship of the diagnostic CASH biomarker with brainstem lesion location, or other potential confounders of age, sex, ethnicity, or genotype, but our sample was potentially underpowered to detect such correlations. However, the results herein allow us to postulate sample sizes sufficient to validate correlations or lack thereof in relevant subgroups, in the context of future larger confirmatory studies, needed for the accreditation of the biomarkers in the clinic (12). There is an opportunity to address these questions in a larger cohort and in different sexes, age groups, specific genotypes, and lesion locations, in conjunction with the recently launched Trial Readiness in Cavernous Angiomas with Symptomatic Hemorrhage Cash Trial Readiness project (NIH 1U01NS104157-01), enrolling CASH patients at multiple sites (3). We had too few cases to reliably assess the value of changes in the diagnostic and prognostic biomarkers during longitudinal follow-up; hence, their value as monitoring biomarkers remains speculative. The biomarker associations were queried in relation to symptomatic hemorrhage within 1 year preceding and following plasma sample collection. For optimal clinical context of use, future longitudinal studies should assess the biomarker performance at different time points after and before a bleed. Finally, the diagnostic and prognostic CASH biomarkers showed imperfect sensitivity and specificity. This may reflect inherent imprecisions in the diagnostic and/or the clinical definitions of CASH (i.e., asymptomatic hemorrhagic lesion). This imperfection in sensitivity and specificity of our biomarkers may also reflect biologic limitations of the queried candidate biomarkers. It is possible that the biomarker precision can be enhanced by integrating additional putative candidates (Supplemental Figure 4).
Notwithstanding these limitations, this is the largest such study in CA, a rare disease, and the first to our knowledge to demonstrate and mechanistically validate overlapping and distinct biomarkers explaining and predicting a cardinal symptomatic event. With these promising results, we can look toward refining these models by discarding noncorrelated biomarker candidates, and querying new candidate molecules emerging from ongoing mechanistic studies, miRNA sequencing, and transcriptomic discoveries (8, 32–34). The approach employed herein may lead to significant advances in patient care in this disease and risk stratification in clinical trials. It may be applicable in other hemorrhagic cerebrovascular diseases and age-related pathologies, such as hemorrhagic microangiopathy, vascular dementia, and Alzheimer’s disease, where similar pathophysiological processes have been implicated (35, 36).
Methods
Study design and participants.
This prospective observational cohort study included 114 consecutive CA (50 familial/multifocal, 64 sporadic/solitary) patients (mean age ± SD, 38.6 ± 18.3 years), evaluated clinically at a single referral center (Center of Excellence for Cerebral Cavernous Malformations, University of Chicago Medical Center) and who had a baseline blood sample collected between July 2014 to February 2018 (Table 1 and Figure 1).
CA diagnosis was confirmed via MRI. Cases were classified as sporadic/solitary if they harbored a single CA lesion on susceptibility-weighted imaging MRI. They were defined as familial/multifocal if they presented with multifocal lesions, a germline mutation in one of 3 CA gene loci, or a history of CA in a first-degree relative (1). Patients with partial or complete CA lesion resection or any prior brain irradiation were excluded from the study.
Among the 114 consecutive cases, 37 had at least one follow-up blood sample within a year (±30 days) after the baseline collection. The remaining 77 patients either did not have a clinical follow-up visit, declined a follow-up blood sample, or underwent surgical resection.
Patients with a follow-up clinical visit and plasma collection were classified as stable (no CASH in the year preceding the initial or follow-up clinical visit), unstable (CASH occurring between the initial and follow-up clinical visit), or recovering (CASH within the prior year [±30 days] of the initial sample, with no recurrent hemorrhage) (21). Patients who had more than 2 longitudinal blood samples greater than a year apart were considered for the same criteria of stable, recovering, or unstable in independent epochs between each 2 consecutive blood samples. CASH patients were reviewed and adjudicated by the senior author (IAA), who has extensive experience in treating this disease. All clinical assessments were performed blinded to the biomarker data.
In addition, blood plasma samples were collected from 12 healthy non-CA subjects (mean age ± SD, 31.7 ± 5.9 years). We excluded subjects who had (a) any medical or neurologic condition requiring ongoing follow-up or medical treatment in the preceding year, (b) been pregnant or lactating in the preceding year, (c) a history of concussion or brain trauma in the preceding year, (d) a history of prior brain irradiation at any time, (e) used recreational, psychoactive, or neuroleptic drugs in the prior year, or (f) MRI contraindications or known claustrophobia.
Plasma isolation and biomarker multiplex assessment.
Standard clinical 10-ml heparinized Vacutainer tubes (BD Vacutainer, Becton, Dickinson and Company) were used to draw blood samples. The plasma was isolated by centrifugation at 500 g at 4°C for 10 minutes (AllegraX-30R, Beckman Coulter). Subsequently 200 μl supernatant plasma was aliquoted into 1.7-ml microcentrifuge tubes for storage at –80°C.
Eighteen different plasma molecules were assessed according to the manufacturer’s instructions using 5 customized magnetic bead–based multiplex Luminex screening immunoassay kits (R&D Systems), including TNF-α, TNFRI, soluble MMP2 and MMP9, CCL2/MCP1, soluble endoglin/CD105 (sENG), soluble VCAM1 (sVCAM1), soluble ICAM1 (sICAM1/CD54), IL-2, IL-6, IL-8/CXCL-8, IL-10, IL-1β, soluble VEGF, soluble roundabout guidance receptor 4 (sROBO4), IFN-γ, sCD14, and CRP (R&D Systems) (37, 38). Measurements were performed with a Bio-Rad BioPlex-100 analyzer running BioPlex Manager Software version 5.0 or the Luminex 200 System (Luminex Corp.) running with xPONENT Software.
In each plate, the plasma samples were loaded in parallel duplicate wells, and then averaged. Fifty beads per region were collected for each well, and 5-parameter logistic regression analysis was performed to estimate the sample concentration. All assessments were performed at the Flow Cytometry Core Facility at the University of Chicago.
Statistics.
Nonbiological experimental variations are common when using multiplex or microarray analysis (39). A canonical discriminant analysis revealed significant variation between batches in the plasma molecules. A marked difference in first, third and fourth principal component values demonstrated that the second multiplexed immunoassay kits acted as a confounding factor. Seven plasma molecules levels, namely CRP (P = 0.02), IL-2 (P < 0.0001), IL-10 (P = 0.0004), sMMP2 (P < 0.0001), sROBO4 (P = 0.0069), sICAM1/CD54 (P = 0.012), and IFN-γ (P = 0.018), were affected by a batch effect; the other 11 plasma molecules were unaffected.
After batch effect correction, the plasma levels of the 18 plasma molecules were compared between CASH and non-CASH patients using a 2-tailed 2-sample t test with a FDR correction. For each plasma molecule, a corrected value greater than ± 2 SDs away from the mean was defined as an outlier (40, 41). The correlations between the relevant molecules were assessed using a linear Pearson’s correlation coefficient.
Following canonical discriminant function analysis, ROCs were generated along with computed AUC for each significant molecule as well as the combination models. The optimal cutoff point was generated from ROC curves utilizing the Youden index method (42). Finally, a 3D ROC curve analysis was performed among the healthy non-CA, CASH, and non-CASH subjects using the HUM package.
All the 511 possible linear combinations of the 9 plasma molecules showing significant associations (P < 0.05, FDR corrected) in CASH patients were processed using canonical discriminant function analysis (43, 44). ROC curves were generated and AUC was calculated for each molecule individually. The best diagnostic biomarker to differentiate CASH and non-CASH patients was selected according to the AIC, representing the best-fit parsimonious model to the data with the fewest number of predictors (45). The difference in mean canonical values calculated using the diagnostic and prognostic CASH biomarkers among CASH and non-CASH patients and healthy non-CA subjects was assessed using a 2-tailed 2-sample t test. Finally, the canonical values calculated using both the diagnostic and prognostic CASH biomarkers were analyzed to evaluate for difference over time in plasma levels in stable, unstable, and recovering patients separately using Wilcoxon’s tests.
Monte Carlo analysis was subsequently conducted to validate the ability of the diagnostic CASH biomarker to differentiate CASH and non-CASH patients in a simulated population (11, 46). This statistical validation was performed by simulating 1000 CASH and 1000 non-CASH patients. We also conducted a cross-validation analysis using the 2/3 to 1/3 random parsing methods with 1000 iterations in order to further validate the biomarker. ROC curves were generated, AUC was calculated, and the best sensitivity/specificity was estimated for each validation. We also employed a machine learning approach using an ensemble meta-algorithm through bootstrap aggregation (bagging) to generate a new weighted combination of the 4 molecules using 1000-repeated samples at a 70:30 partition with replacement. Finally, a 10-fold cross-validation utilizing an SVM based on a hyperbolic tangent–based kernel using an 80-20 partition of the data was conducted for both the diagnostic and prognostic CASH biomarker (47).
All statistical analyses were conducted using a combination of R (https://www.r-project.org/), SAS9.4 (SAS Institute Inc.), and Prism 4.0 (GraphPad).
RNA isolation from laser microdissected NVUs of human CA lesions and nonlesional brain capillaries.
Five CASH (3 familial/multifocal, 2 sporadic/solitary) and 5 non-CASH (1 familial/multifocal, 4 sporadic/solitary) surgically resected human lesions (Supplemental Table 4) as well as 3 control brain tissue samples from autopsy were used for the transcriptomic analyses. The samples were fixed in formalin and embedded in paraffin blocks. All autopsy patients were free of neurological disease.
Samples were snap-frozen and embedded in optimal cutting temperature compound for subsequent storage at –80°C. Frozen samples were mounted as 5-μm-thick sections onto Leica slides (Leica Biosystems), and stained utilizing HistoGene (Applied Biosystems). The NVUs from the CASH lesions, non-CASH lesions, and control brains were laser capture microdissected at ×40 using a Leica LMD 6500 system (Leica Biosystems) and then stored at –80°C as per the recently published protocol (13).
RNA was then isolated using an RNeasy Micro Kit (QIAGEN). The single-end 47-bp or 50-bp read sequencings of the 13 samples were performed on 3 batches (13).
Transcriptome bioinformatics methods.
RNA libraries were generated using a commercial low-input strand-specific RNA-Seq kit (Clontech) and sequenced using an Illumina HiSeq4000 platform (Illumina). The raw sequencing quality was analyzed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). RSeQC and Picard analysis tools were then used for evaluation of postalignment quality control (48). Ultimately, reads were mapped into the GENCODE human genome model (GRCh38 V28) using STAR (49). The count-based method featureCounts was used to quantify and assemble the gene transcripts (50).
DEG analyses were conducted with DESeq2, in combination with a batch effect correction when needed (51). The network analysis was performed using ReactomeFIViz in Cytoscape (release 2016, http://www.cytoscape.org/) based on a highly reliable Reactome function interaction network (52, 53). GO enrichment analyses were conducted using R bioconductor package clusterProfiler (http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html).
Plasma miRNA extraction and bioinformatics methods.
miRNA extractions from 12 plasma samples (9 CA patients and 3 healthy non-CA subjects) were performed using the miRNeasy Serum/Plasma Kit (QIAGEN). cDNA libraries were generated with commercially available Illumina small RNA-Seq kits, and sequenced with the Illumina HiSeq 4000 platform using single-end 50-bp reads. Raw sequencing quality was assessed using FastQC (v0.11.5). The small RNA adapter sequences were trimmed from small RNA sequencing data with cutadapt (54). The adapter trimmed reads were then mapped and quantified to the human mature miRNA database (miRBase 21) with sRNAbench (release 10/14) library mapping strategy, wrapped bowtie alignment (with alignment type = –n, seed length for alignment = 20, minimum read-count = 2, allowed number of mismatch = 0, minimum read count = 2, and maximum number of multiple mappings = 20) (55). Low-expressed miRNAs were then removed for further downstream analyses. Finally, the miRNA expression values were normalized following trimmed mean of M-values (TMM) normalization methods with library size correction (56).
The differently expressed miRNAs were identified between CASH and non-CASH patients with a correction factor for healthy non-CA subject effects using R bioconductor package DESeq2 (51). We also analyzed differently expressed miRNAs between healthy non-CA subjects and CASH, and non-CASH patients. The miRNA putative target genes were predicted on the 3 different targeting gene regions (3′ UTR, 5′ UTR, and CDS) based on the database platforms provided by miRWalk (release 3.0, http://mirwalk.umm.uni-heidelberg.de/) using a random forest tree algorithm with a bonding prediction probability higher than 95% (57). The exploration and integrative analyses of miRNA-mRNA gene regulations were assisted using R.
Study approval.
All subjects enrolled in the study gave written informed consent in compliance with the Declaration of Helsinki, and the study was approved by the University of Chicago IRB. The ethical principles guiding the IRB are consistent with the Belmont Report, and comply with the rules and regulations of the US Department of Health and Human Services Federal Policy for the Protection of Human Subjects (56 FR 28003).
Data and materials availability.
Anonymized data are available by request from any qualified investigator with experience handling sensitive patient information. The raw sequencing data used in this study are available in the NCBI’s Gene Expression Omnibus database (GEO) GSE130176.
Author contributions
SL created figures, collected, analyzed, and interpreted data, and wrote the manuscript. RG created figures, contributed to the study design, collected, analyzed, and interpreted data, and wrote the manuscript. JK created figures, collected, analyzed, and interpreted data, and wrote the manuscript; HAZ and DZ collected data. YC analyzed and interpreted data. YL created figures, and analyzed and interpreted data. AS collected data and wrote the manuscript. TM and RL collected data. CS collected data and designed the study. RS designed the study. JCP and SPP collected data. SR collected data and wrote the manuscript. AA and MLR collected data. KJW designed the study. MLK, MHG, and DAM collected data and designed the study. IAA designed the study, interpreted and collected data, and wrote the manuscript. All authors critically reviewed the manuscript and accepted the final version.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R21NS087328 to IAA and P01 NS092521 to IAA, MLK, MHG, and DAM, and P30 CA14599 to the University of Chicago Comprehensive Cancer Center; a core subsidy to RS from the National Center for Advancing Translational Sciences of the NIH (1UL1TR002389-01, which funds the Institute for Translational Medicine); the William and Judith Davis Fund in Neurovascular Surgery Research; the Be Brave for Life Foundation; the Safadi Translational Fellowship to RG; the Pritzker School of Medicine to SL; and the Sigrid Juselius Foundation to JK. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Version 1. 06/20/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(12):e128577. https://doi.org/10.1172/jci.insight.128577.
Contributor Information
Romuald Girard, Email: rgirard@surgery.bsd.uchicago.edu.
Janne Koskimäki, Email: koskimaki@uchicago.edu.
Hussein A. Zeineddine, Email: h.a.zeineddine@gmail.com.
Dongdong Zhang, Email: dzhang4@surgery.bsd.uchicago.edu.
Ying Cao, Email: ycao2@surgery.bsd.uchicago.edu.
Yan Li, Email: yli22@bsd.uchicago.edu.
Agnieszka Stadnik, Email: astadnik@surgery.bsd.uchicago.edu.
Thomas Moore, Email: tmoore10@surgery.bsd.uchicago.edu.
Rhonda Lightle, Email: rlightle@surgery.bsd.uchicago.edu.
Changbin Shi, Email: changbinshi@hotmail.com.
Robert Shenkar, Email: rshenkar@uchicago.edu.
Julián Carrión-Penagos, Email: jcarrion@surgery.bsd.uchicago.edu.
Sean P. Polster, Email: Sean.Polster@uchospitals.edu.
Sharbel Romanos, Email: sromanos@uchicago.edu.
Mark L. Kahn, Email: markkahn@mail.med.upenn.edu.
Douglas A. Marchuk, Email: douglas.marchuk@duke.edu.
Issam A. Awad, Email: iawad@surgery.bsd.uchicago.edu.
References
- 1.Akers A, et al. Synopsis of guidelines for the clinical management of cerebral cavernous malformations: consensus recommendations based on systematic literature review by the Angioma Alliance Scientific Advisory Board Clinical Experts Panel. Neurosurgery. 2017;80(5):665–680. doi: 10.1093/neuros/nyx091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Shahi Salman R, et al. Untreated clinical course of cerebral cavernous malformations: a prospective, population-based cohort study. Lancet Neurol. 2012;11(3):217–224. doi: 10.1016/S1474-4422(12)70004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polster SP, et al. Trial readiness in cavernous angiomas with symptomatic hemorrhage (CASH) Neurosurgery. 2019;84(4):954–964. doi: 10.1093/neuros/nyy108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDonald DA, et al. Lesions from patients with sporadic cerebral cavernous malformations harbor somatic mutations in the CCM genes: evidence for a common biochemical pathway for CCM pathogenesis. Hum Mol Genet. 2014;23(16):4357–4370. doi: 10.1093/hmg/ddu153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutsforth-Gregory JK, Lanzino G, Link MJ, Brown RD, Jr, Flemming KD. Characterization of radiation-induced cavernous malformations and comparison with a nonradiation cavernous malformation cohort. J Neurosurg. 2015;122(5):1214–1222. doi: 10.3171/2015.1.JNS141452. [DOI] [PubMed] [Google Scholar]
- 6.Shi C, et al. B-cell depletion reduces the maturation of cerebral cavernous malformations in murine models. J Neuroimmune Pharmacol. 2016;11(2):369–377. doi: 10.1007/s11481-016-9670-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choquet H, et al. Polymorphisms in inflammatory and immune response genes associated with cerebral cavernous malformation type 1 severity. Cerebrovasc Dis. 2014;38(6):433–440. doi: 10.1159/000369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang AT, et al. Endothelial TLR4 and the microbiome drive cerebral cavernous malformations. Nature. 2017;545(7654):305–310. doi: 10.1038/nature22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girard R, et al. Peripheral plasma vitamin D and non-HDL cholesterol reflect the severity of cerebral cavernous malformation disease. Biomark Med. 2016;10(3):255–264. doi: 10.2217/bmm.15.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girard R, et al. Plasma biomarkers of inflammation reflect seizures and hemorrhagic activity of cerebral cavernous malformations. Transl Stroke Res. 2018;9(1):34–43. doi: 10.1007/s12975-017-0561-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girard R, et al. Plasma biomarkers of inflammation and angiogenesis predict cerebral cavernous malformation symptomatic hemorrhage or lesional growth. Circ Res. 2018;122(12):1716–1721. doi: 10.1161/CIRCRESAHA.118.312680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amur S, LaVange L, Zineh I, Buckman-Garner S, Woodcock J. Biomarker qualification: toward a multiple stakeholder framework for biomarker development, regulatory acceptance, and utilization. Clin Pharmacol Ther. 2015;98(1):34–46. doi: 10.1002/cpt.136. [DOI] [PubMed] [Google Scholar]
- 13.Koskimäki J, et al. Comprehensive transcriptome analysis of cerebral cavernous malformation across multiple species and genotypes. JCI Insight. 2019;4(3):e126167. doi: 10.1172/jci.insight.126167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stefater JA, et al. Regulation of angiogenesis by a non-canonical Wnt-Flt1 pathway in myeloid cells. Nature. 2011;474(7352):511–515. doi: 10.1038/nature10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayer-Barber KD, Yan B. Clash of the cytokine titans: counter-regulation of interleukin-1 and type I interferon-mediated inflammatory responses. Cell Mol Immunol. 2017;14(1):22–35. doi: 10.1038/cmi.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ip WKE, Hoshi N, Shouval DS, Snapper S, Medzhitov R. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science. 2017;356(6337):513–519. doi: 10.1126/science.aal3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia JM, et al. Role of interleukin-10 in acute brain injuries. Front Neurol. 2017;8:244. doi: 10.3389/fneur.2017.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ugarte-Berzal E, et al. VEGF/VEGFR2 interaction down-regulates matrix metalloproteinase-9 via STAT1 activation and inhibits B chronic lymphocytic leukemia cell migration. Blood. 2010;115(4):846–849. doi: 10.1182/blood-2009-08-239426. [DOI] [PubMed] [Google Scholar]
- 20.Dziedzic T, Bartus S, Klimkowicz A, Motyl M, Slowik A, Szczudlik A. Intracerebral hemorrhage triggers interleukin-6 and interleukin-10 release in blood. Stroke. 2002;33(9):2334–2335. doi: 10.1161/01.STR.0000027211.73567.FA. [DOI] [PubMed] [Google Scholar]
- 21.Girard R, et al. Vascular permeability and iron deposition biomarkers in longitudinal follow-up of cerebral cavernous malformations. J Neurosurg. 2017;127(1):102–110. doi: 10.3171/2016.5.JNS16687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mineo C, et al. FcgammaRIIB mediates C-reactive protein inhibition of endothelial NO synthase. Circ Res. 2005;97(11):1124–1131. doi: 10.1161/01.RES.0000194323.77203.fe. [DOI] [PubMed] [Google Scholar]
- 23.Bettcher BM, Kramer JH. Longitudinal inflammation, cognitive decline, and Alzheimer’s disease: a mini-review. Clin Pharmacol Ther. 2014;96(4):464–469. doi: 10.1038/clpt.2014.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lévêque M, et al. Soluble CD14 acts as a DAMP in human macrophages: origin and involvement in inflammatory cytokine/chemokine production. FASEB J. 2017;31(5):1891–1902. doi: 10.1096/fj.201600772R. [DOI] [PubMed] [Google Scholar]
- 25.Rahimi AA, Gee K, Mishra S, Lim W, Kumar A. STAT-1 mediates the stimulatory effect of IL-10 on CD14 expression in human monocytic cells. J Immunol. 2005;174(12):7823–7832. doi: 10.4049/jimmunol.174.12.7823. [DOI] [PubMed] [Google Scholar]
- 26.Jung KH, Chu K, Jeong SW, Park HK, Bae HJ, Yoon BW. Cerebral cavernous malformations with dynamic and progressive course: correlation study with vascular endothelial growth factor. Arch Neurol. 2003;60(11):1613–1618. doi: 10.1001/archneur.60.11.1613. [DOI] [PubMed] [Google Scholar]
- 27.Whitehead KJ, et al. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nat Med. 2009;15(2):177–184. doi: 10.1038/nm.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ridley AJ. Rho GTPase signalling in cell migration. Curr Opin Cell Biol. 2015;36:103–112. doi: 10.1016/j.ceb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali G, et al. Differential expression of extracellular matrix constituents and cell adhesion molecules between malignant pleural mesothelioma and mesothelial hyperplasia. J Thorac Oncol. 2013;8(11):1389–1395. doi: 10.1097/JTO.0b013e3182a59f45. [DOI] [PubMed] [Google Scholar]
- 30.Shirley MD, et al. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med. 2013;368(21):1971–1979. doi: 10.1056/NEJMoa1213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lapchak PA, Wu Q. Vascular dysfunction in brain hemorrhage: translational pathways to developing new treatments from old targets. J Neurol Neurophysiol. 2011;2011:S1-e001. doi: 10.4172/2155-9562.S1-e001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Z, et al. Cerebral cavernous malformations arise from endothelial gain of MEKK3-KLF2/4 signalling. Nature. 2016;532(7597):122–126. doi: 10.1038/nature17178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Ramirez MA, et al. Thrombospondin1 (TSP1) replacement prevents cerebral cavernous malformations. J Exp Med. 2017;214(11):3331–3346. doi: 10.1084/jem.20171178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13(8):622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 35.Poggesi A, Pasi M, Pescini F, Pantoni L, Inzitari D. Circulating biologic markers of endothelial dysfunction in cerebral small vessel disease: a review. J Cereb Blood Flow Metab. 2016;36(1):72–94. doi: 10.1038/jcbfm.2015.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajani RM, et al. Reversal of endothelial dysfunction reduces white matter vulnerability in cerebral small vessel disease in rats. Sci Transl Med. 2018;10(448):eaam9507. doi: 10.1126/scitranslmed.aam9507. [DOI] [PubMed] [Google Scholar]
- 37.McKay HS, et al. Host factors associated with serologic inflammatory markers assessed using multiplex assays. Cytokine. 2016;85:71–79. doi: 10.1016/j.cyto.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fleming BD, et al. The generation of macrophages with anti-inflammatory activity in the absence of STAT6 signaling. J Leukoc Biol. 2015;98(3):395–407. doi: 10.1189/jlb.2A1114-560R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clarke DC, Morris MK, Lauffenburger DA. Normalization and statistical analysis of multiplexed bead-based immunoassay data using mixed-effects modeling. Mol Cell Proteomics. 2013;12(1):245–262. doi: 10.1074/mcp.M112.018655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selst MV, Jolicoeur P. A solution to the effect of sample size on outlier elimination. Q J Exp Psychol A. 1994;47(3):631–650. doi: 10.1080/14640749408401131. [DOI] [Google Scholar]
- 41.Davies L, Gather U. The identification of multiple outliers. J Am Stat Assoc. 1993;88(423):782–792. doi: 10.1080/01621459.1993.10476339. [DOI] [Google Scholar]
- 42.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3(1):32–35. doi: 10.1002/1097-0142(1950)3:1<32::AID-CNCR2820030106>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 43. Rencher AC. Methods of Multivariate Analysis. Hoboken, New Jersey, USA: Wiley; 2003. [Google Scholar]
- 44. Huberty CJ. Applied Discriminant Analysis. New York, New York, USA: Wiley; 1994. [Google Scholar]
- 45. Lovric M, ed. International Encyclopedia of Statistical Science. Berlin, Germany: Springer; 2010. [Google Scholar]
- 46. Manly BFJ. Randomization, Bootstrap, and Monte Carlo Methods in Biology. London, United Kingdom: Chapman & Hall; 1997. [Google Scholar]
- 47. Karatzoglou A, Smola A, Hornik K. kernlab: Kernel-Based Machine Learning Lab. https://cran.r-project.org/web/packages/kernlab/index.html Updated August 8, 2018. Accessed April 18, 2019.
- 48.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28(16):2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 49.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 51.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 2010;11(5):R53. doi: 10.1186/gb-2010-11-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu G, Dawson E, Duong A, Haw R, Stein L. ReactomeFIViz: a Cytoscape app for pathway and network-based data analysis. F1000Res. 2014;3:146. doi: 10.12688/f1000research.4431.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):3. doi: 10.14806/ej.17.1.202. [DOI] [Google Scholar]
- 55.Barturen G, et al. sRNAbench: profiling of small RNAs and its sequence variants in single or multi-species high-throughput experiments. Methods in Next Generation Sequencing. 2014;1(1):21–31. [Google Scholar]
- 56.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sticht C, De La Torre C, Parveen A, Gretz N. MiRWalk: an online resource for prediction of microRNA binding sites. PloS One. 2018;13(10):e0206239. doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.