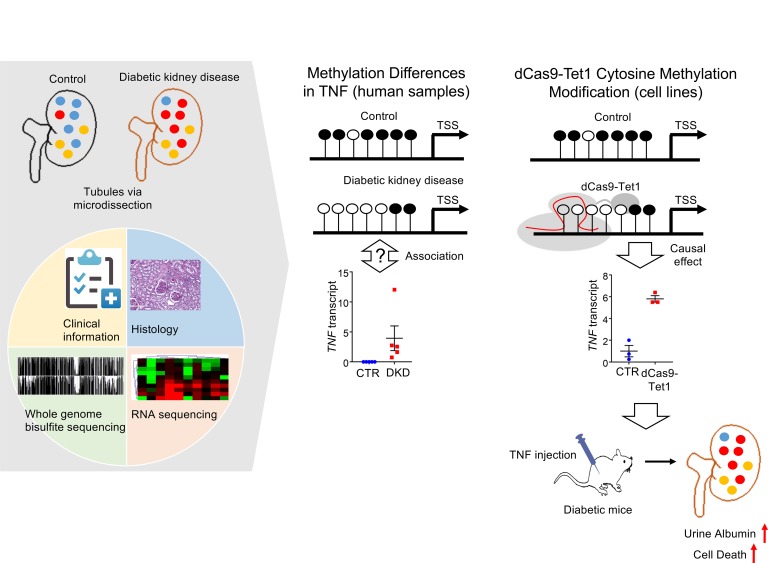
Abstract
In patients with diabetes mellitus, poor metabolic control has a long-lasting impact on kidney disease development. Epigenetic changes, including cytosine methylation, have been proposed as potential mediators of the long-lasting effect of adverse metabolic events. Our understanding of the presence and contribution of methylation changes to disease development is limited because of the lack of comprehensive base-resolution methylome information of human kidney tissue samples and site-specific methylation editing. Base resolution, whole-genome bisulfite sequencing methylome maps of human diabetic kidney disease (DKD) tubule samples, and associated gene expression measured by RNA sequencing highlighted widespread methylation changes in DKD. Pathway analysis highlighted coordinated (methylation and gene expression) changes in immune signaling, including tumor necrosis factor alpha (TNF). Changes in TNF methylation correlated with kidney function decline. dCas9-Tet1–based lowering of the cytosine methylation level of the TNF differentially methylated region resulted in an increase in the TNF transcript level, indicating that methylation of this locus plays an important role in controlling TNF expression. Increasing the TNF level in diabetic mice increased disease severity, such as albuminuria. In summary, our results indicate widespread methylation differences in DKD kidneys and highlights epigenetic changes in the TNF locus and its contribution to the development of nephropathy in patients with diabetes mellitus.
Keywords: Genetics
Keywords: Chronic kidney disease, Diabetes, Epigenetics
Whole genome bisulfate sequencing and methylome editing identifies the epigenetic role in TNF regulation in patients with diabetic kidney disease.
Introduction
Diabetic kidney disease (DKD) is the leading cause of end-stage renal disease (ESRD) (1). Most excess mortality of patients with diabetes mellitus can be explained by the presence of kidney disease (2). Although cardiovascular mortality in patients with diabetes mellitus has declined by 70% over the last 2 decades, DKD incidence has only marginally changed (3). About one-third to one-half of patients with diabetes mellitus develop progressive kidney disease. Patients with poor glycemic control exhibit an increased risk of nephropathy, but the correlation between metabolic control and kidney disease is relatively poor (4). Identification of patients who are at increased risk for rapidly progressive DKD is of great importance. Recent results indicate that circulating tumor necrosis factor receptor (TNFR) levels can predict kidney disease risk and kidney functional decline in patients with type 1 and type 2 diabetes mellitus (5, 6). Over the last few years, serum TNFR levels emerged as one of the best and most reproducible biomarker of DKD, and TNF activation has been observed in patients with diabetes mellitus (7). In our review of the literature, changes in TNF levels are not fully recapitulated in mouse DKD models. A recent paper by Gurley et al. (8) proposed that an immune/cytokine expression profile can also distinguish susceptible and resistant strains of mice, indicating that TNF activation might be an indicator of kidney disease susceptibility in mice as well. Although TNF is a key disease biomarker in patients with diabetes mellitus, the mechanism of TNF activation is unclear.
Metabolic abnormalities have a long-term effect on DKD development, such as patients who have poor glycemic control early on have an increased risk for the development of complications (including kidney disease) even after decades of adequate glycemic control (9, 10). This phenomenon is called metabolic memory. A similar phenomenon has been reported for fetal development, in which nutritional availability has a long-lasting influence on hypertension, kidney disease, and metabolic disease development (11, 12). Because metabolite availability can directly control the epigenome, the epigenome can potentially be viewed as a long-term environmental footprint (13, 14).
DNA cytosine methylation (5mC) is one of the best-studied epigenetic modifications that has been proposed to play a key role in the pathogenesis of disease conditions (15, 16). Several studies have attempted to describe methylation changes in DKD. Most of these studies have been carried out on mice (17) or surrogate cell types, such as blood or saliva samples (18, 19) and used relatively small sample sizes. These early studies highlighted potential sites with differential methylation in DKD; however, none of these studies reported changes that reached the genome-wide significance threshold or could be validated in external cohorts. A recently published large multi-ethnic study identified an association between the methylation level of a handful of loci and kidney function with genome-wide significant methylation changes in the whole blood of subjects with nondiabetic kidney disease (20). It is not clear whether changes observed in blood samples are also relevant for target tissue or cell type, such as the kidney. In addition, although studies have been able to define the association between DNA methylation, gene expression, and kidney disease development, none of the studies have been able to demonstrate a direct effect of DNA methylation on target gene expression and kidney disease development because of the lack of the target-specific epigenome editing techniques.
The CRISPR/Cas9 system is an RNA-guided adaptive immune system in bacteria that is able to recognize specific DNA sequences and, therefore, has been developed as an efficient tool for genome editing (21, 22). Mutations have been introduced into the Cas9 protein to diminish its nuclease activity (dead nuclease activity of Cas9 [dCas9]), thereby generating a unique tool that recognizes specific DNA sequences under sgRNA guidance (23). Fusing of dCas9 with epigenome-editing enzymes could provide a special opportunity for site-specific epigenome editing. For example, fusing of dCas9 with DNA methyltransferases can be used for site-specific methylation, whereas a dCas9-Tet fusion system can be used to hydroxymethylate and erase methylation (24). A recent study highlighted the potential clinical use of dCas9-Tet1 system for erasing the hypermethylation of the CGG expansion mutation of the FMR1 gene, which causes silencing of the FMR1 gene and fragile X syndrome (25).
In this study, we generated whole-genome DNA methylation maps for human kidney samples from healthy and DKD patients at base resolution and identified differentially methylated regions (DMRs). DNA methylation and RNA sequencing indicated marked changes in immune cell proportions in kidneys from DKD. DMRs were highly enriched in genes with metabolic functions and immune response, such as TNF. To define the contribution of DMRs to downstream gene expression and phenotype development, dCas9-Tet1 was used to lower the methylation of the target DMR regions of TNF. Reducing the methylation of the target DMR resulted in an increase in the TNF transcript level in kidney cells. Increased TNF led to increased epithelial injury and death and increased proteinuria development in mice, raising the possibility that methylation differences in TNF plays an important role in DKD development.
Results
Genome-wide base-resolution DNA methylation in DKD.
To define changes in cytosine methylation in DKD, we generated base-resolution cytosine methylation maps by whole-genome bisulfite sequencing (WGBS). Tubule cells play a key role in DKD. Tubule atrophy shows a strong correlation with kidney function and predicts future functional decline. We analyzed microdissected human kidney tubule samples from 5 controls (healthy) and 5 subjects with DKD as key target cells in DKD pathogenesis. The clinical characteristics of samples are detailed under Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.128886DS1 The presence of DKD was confirmed by histopathological analysis of the samples. We obtained 108–198G base-sequencing data that achieved 14- to 20-fold genome coverage. Control and DKD samples clearly separated based on their methylation patterns (Supplemental Figure 1). As expected, DKD samples showed greater heterogeneity than controls.
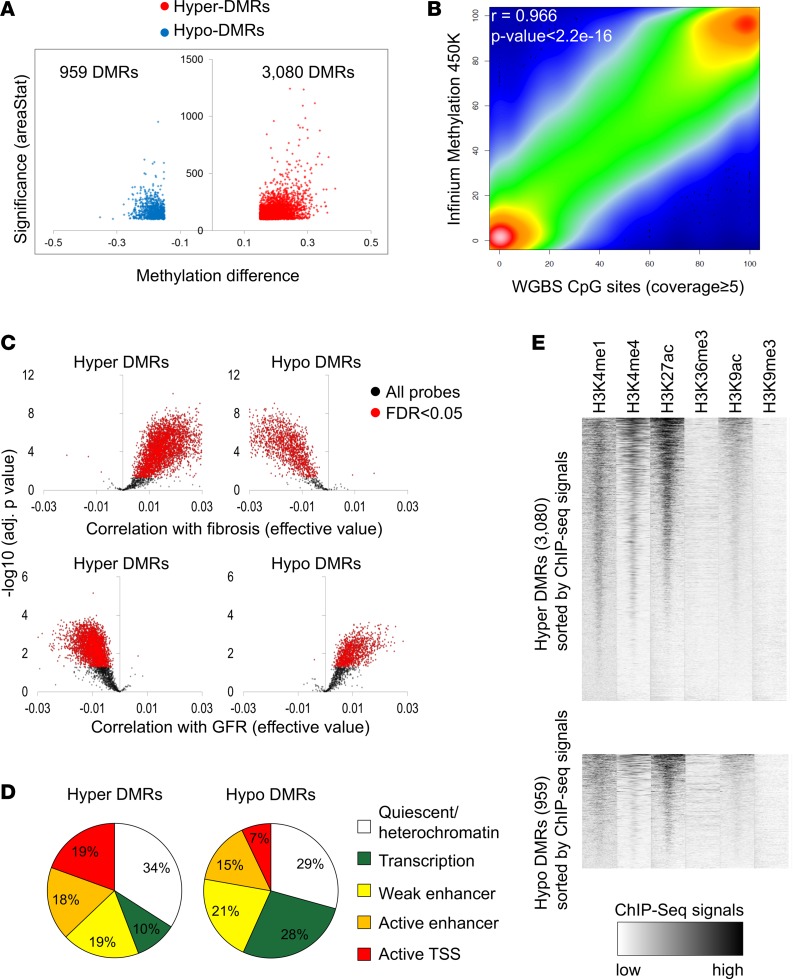
We next compared the methylation of control and DKD samples. DMRs were defined as at least 5 CpGs that showed greater than 15% methylation difference. Significance was defined as areaStat 100 and greater, which is the sum of the t-statistics for the individual CpGs in the DMRs. This analysis detected a total of 4,039 DMRs: 3,080 DMRs with higher methylation (hyper) and 959 DMRs with lower methylation (hypo) in DKD samples compared with controls (Figure 1A).
Figure 1. Whole-genome cytosine methylation analysis of control and DKD kidney tubule samples at base resolution.
(A) Volcano plot for the DMRs between control and DKD samples. The x axis represents methylation level difference. The y axis represents statistical significance of the DMRs shown by areaStat value from bsseq program. (B) The correlation between Infinium array and WGBS data. (C) Methylation patterns of the Infinium probes in the DMR regions identified by WGBS. The x axis represents correlation level (effective values calculated by linear regression) with interstitial fibrosis (upper panel) and GFR (lower panel). The y axis represents the significance of the correlation. The significant probes are colored in red (FDR < 0.05). (D) Genomic distribution of the DMRs in kidney-specific functional elements. Functional elements were defined by ChromHMM-based histone ChIP data. (E) Enrichment patterns of 6 different histone modifications in hyper- and hypo-DMRs. Normalized ChIP-seq read density was calculated for every 25-bp window in ±3 kb flanking DMR center.
Because the WGBS data set was obtained from a relatively small number of subjects, we sought to validate our results in a larger cohort. We used Infinium methylation 450K array data obtained from 91 microdissected human kidney tubule samples as our validation data set. The clinical and histological characteristics of the validation set were similar to the original cohort (Supplemental Table 2). The WGBS and array-based methylation data were highly concordant (Figure 1B) (r = 0.966, P < 2.2 × 10–16) and, as expected, showed a bimodal methylation pattern. Then, we specifically analyzed the methylation patterns of DMRs identified by WGBS on the Infinium arrays. Of the DMRs identified by WGBS, 58% had at least one overlapping probe on the Infinium methylation arrays, indicating that 42% of the identified DMRs were not present on the Infinium methylation arrays. Almost all of the probes (90.8%), localized in hyper-DMR and identified by WGBS analysis, showed significant and direction-consistent association with fibrosis level (adj. P < 0.05) (Figure 1C). In addition, 90.7% of the probes that were located within hypo-DMRs showed a significant negative association with the fibrosis level. DMRs identified by WGBS also correlated with probes associated with kidney function (eGFR) in the Infinium dataset (Figure 1C). In total, 79.2% of the probes in hyper-DMRs and 69.0% of the probes in hypo-DMRs showed the same direction of methylation changes with the WGBS. In summary, we identified base-resolution DNA methylation changes in human DKD tubule samples and validated changes in a larger cohort of patient samples.
Cytosine methylation correlates with gene expression.
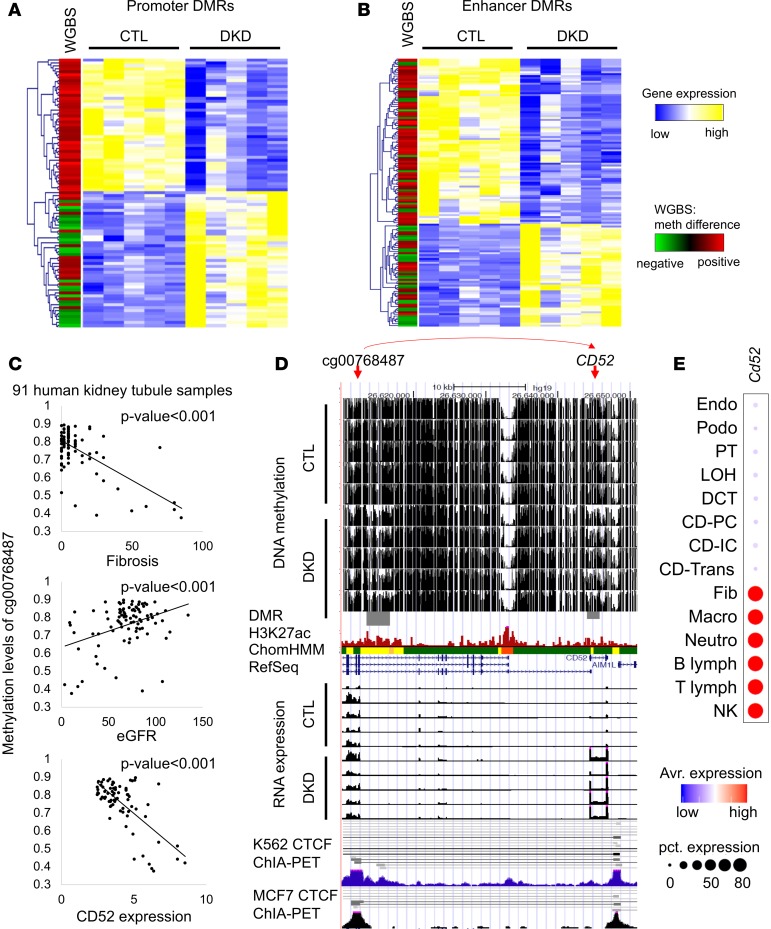
We hypothesized that cytosine methylation plays a role in disease development via interfering with transcription factor binding and downstream gene expression regulation. To assign DMRs to kidney-specific functional elements, we mapped the DMRs to chromatin states, which were identified by combining human kidney multiple histone modification chromatin immunoprecipitation (ChIP-seq) data using a Hidden Markov Model implemented in the ChromHMM method (26). We found that most hypo- and hyper-DMRs (close to 36% to 38%) were localized to kidney enhancer regions. Hyper-DMRs in DKD samples were highly enriched in active functional elements, particularly in active promoters (19%) (Figure 1D). There was a nice correlation of histone enrichment at the DMRs, specifically at the hyper-DMR sites (Figure 1E). In contrast, only 7% of hypo-DMRs were located in active promoters. Meanwhile, more transcription-associated regions showed lower methylation in DKD. Integrating functionally annotated DMRs with gene expression data, obtained via RNA sequencing of the same microdissected tubule samples, revealed a strong negative correlation between promoter methylation level and gene expression changes. Genes with methylated promoters had lower expression. We also observed a good correlation between enhancer methylation and gene expression, although this seemed less pronounced (Figure 2, A and B).
Figure 2. Association between DNA methylation and gene expression in human kidney tubules.
(A) Heatmap showing relative gene expression of control and DKD samples (yellow/blue); the differential methylation is shown in the first column (red/green). Association between gene expression and promoter DMRs. The genes that contain DMRs in their promoter region (TSS ± 3 kb) were clustered based on gene expression patterns. Each row is 1 DMR/gene and each column is 1 sample. (B) Heatmap showing relative gene expression of control and DKD samples (yellow/blue); the differential methylation is shown in the first column (red/green). Association between gene expression and the enhancer DMRs. Enhancers that are outside of promoter regions were linked to the nearest genes. Genes were clustered based on gene expression patterns. Each row is 1 DMR/gene and each column is 1 sample. (C) The methylation of cg00768487 was associated with interstitial fibrosis, kidney function (eGFR), and CD52 gene expression. (D) Screenshot of UCSC genome browser for the probe shown in C and the target gene CD52. The y axis shows WGBS DNA methylation level (control and DKD) followed by DMR highlight, human kidney H3K27ac ChIP-seq, ChromHMM data, and RNA expression (by RNA sequencing) and ChIA-PET data from K562 and MCF7 cells. ChromHMM color code as shown in A. (E) The expression of Cd52 in mouse kidney cells as identified by single-cell RNA sequencing (28).
The integrative analysis of methylation, histone modification, and gene expression changes highlighted multiple critical long-range interactions between promoter-enhancer–target gene regulatory units. For example, the methylation level of a CpG site, cg00768487, was strongly and significantly associated with kidney disease severity, such as interstitial fibrosis and eGFR (Figure 2C). WGBS showed substantial decrease of DNA methylation levels around the cg00768487 probe in DKD samples. Expression of CD52 was increased in kidney tubule samples as observed by RNA sequencing as well (Figure 2D). The methylation of cg00768487 was negatively correlated with the expression level of CD52, even though this region was 33 kb further away from the transcription start site of CD52 (Figure 2C). CTCF ChIA-PET data (from multiple cell lines) (27) indicated a direct interaction between the cg00768487 region and CD52 (Figure 2D). Single-cell RNA sequencing data indicated that Cd52 is highly expressed in immune cell types in mouse kidney (Figure 2E and ref. 28). In summary, the base-resolution methylation data not only highlighted important changes on regulatory regions of human kidneys and changes correlated with gene expression, but also with cell heterogeneity, potentially indicating that methylation of regulatory regions plays an important role in gene expression and phenotype development.
DNA methylation and RNA expression data indicate substantial cell composition changes in DKD.
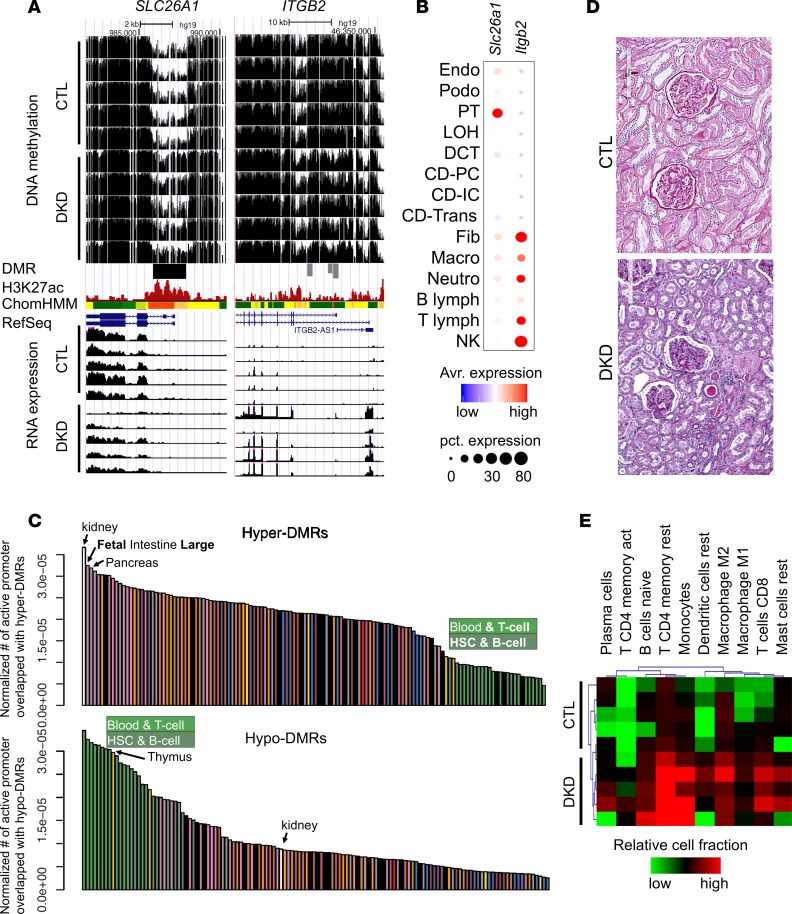
To understand potential differences between hypo- and hyper-DMRs, we performed integrative analysis of several loci. We observed an increase in the methylation at the promoter region of SLC26A1 in DKD samples (Figure 3A). This DMR was localized to the regulatory region and most likely to be an active promoter region based on the histone modification information. SLC26A1 is a sulfate transporter, which seems to be almost exclusively expressed by proximal tubule cells when analyzed by mouse single-cell RNA sequencing (Figure 3B and ref. 28). The higher methylation level of the SLC26A1 regulatory region was associated with a substantial decrease of mRNA expression of SLC26A1 in DKD samples (Figure 3A). These results are consistent with the observed tubule epithelial dedifferentiation observed in DKD (29). Conversely, dramatic decrease of DNA methylation and increase of mRNA expression of ITGB2 (integrin, β2) was observed in DKD tubule samples. Increase in ITGB2 transcript level in DKD tubules has already been reported earlier (29). In our kidney single-cell RNA sequencing atlas, ITGB2 was found to be almost exclusively expressed in immune cells (Figure 3B and ref. 28).
Figure 3. Differential methylation is driven by cell composition and cell type–specific changes.
(A) WGBS DNA methylation level (control and DKD) followed by DMR highlight, human kidney H3K27ac ChIP-seq, ChromHMM data, and RNA expression (by RNA sequencing). (B) The expression of Slc26a1 and Itgb2 in mouse kidney cells as identified by single-cell RNA sequencing. (C) Normalized enrichment of DMR by tissue-specific promoter regions. The y axis is the number of tissue-specific active promoters overlapping with hyper- and hypo-DMRs normalized by total length of tissue-specific active promoters. (D) Periodic acid–Schiff staining of kidney sections from control and DKD samples. (E) In silico deconvolution (by CIBERSORT) of bulk RNA sequencing data from normal and DKD human samples. Colors represent relative immune cell fractions in different samples.
Based on the differences we observed between hypo- and hyper-DMR regions, we next investigated whether unexpected cell heterogeneity potentially contributed to the observed methylation changes in our samples. Our samples were collected via careful manual microdissection and differential expression analysis confirmed the enrichment of key compartment-specific gene expression (29). However, some changes in cell composition cannot be excluded. We compared DMRs identified in DKD kidneys to active promoter regions identified in other tissue samples. As expected, hyper-DMRs showed the highest enrichment level for kidney-specific promoters (Figure 3C), indicating that DKD DMRs are kidney specific. To further substantiate their kidney specificity, hyper-DMRs also showed binding motif enrichment for renal tubule–specific transcription factors, such as HNF and LHX (Supplemental Figure 2). Conversely, hypo-DMRs were enriched on active promoters in blood, T cells, B cells, hematopoietic stem cells, and thymus rather than kidney tissue promoters (Figure 3C). We also found that hypo-DMRs showed enrichment for binding motifs of immune cell–specific transcription factors, such as ETS1, PU.1, and FLI1 (Supplemental Figures 2 and 3), indicating that hypo-DMR in DKD samples might potentially be the consequence of immune cell heterogeneity or immune cytokine activation in epithelial cells. Indeed, lymphocytic infiltrate scores measured by PAS staining were substantially higher in the DKD group than in the control group (Figure 3D and Supplemental Table 1). Markedly higher lymphocytic infiltrate scores in DKD (n = 22) than control (n = 69) were also observed in our replication cohort (Supplemental Table 2). To corroborate the contribution of cell heterogeneity, we performed in silico cell deconvolution analysis of the bulk tubule RNA sequencing data using kidney-specific single-cell data and the CIBERSORT software package (28, 30). The in silico deconvolution results further confirmed an increase in CD4+ and CD8+ T cells, monocytes, macrophages, mast cells, and dendritic cells in DKD samples compared with controls (Figure 3E). To summarize, gene expression, open chromatin, and cell heterogeneity data indicate that DMRs with higher methylation levels were mostly kidney epithelial specific and were associated with lower gene expression, whereas cell-specific or cell composition changes could contribute to loci with lower methylation.
Increased TNF signaling in DKD.
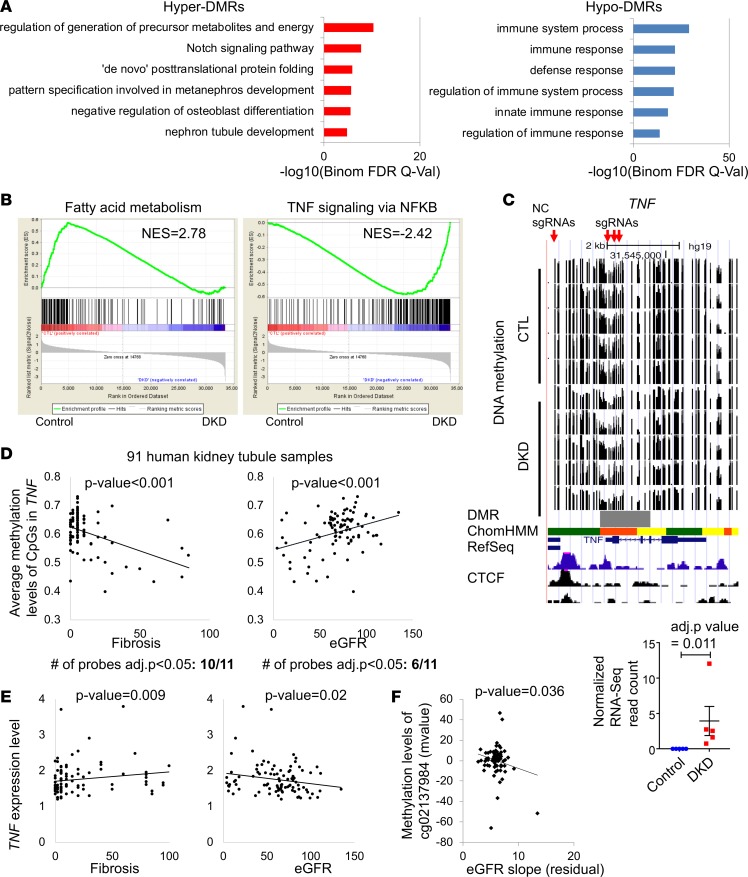
Next, we explored the functional annotation of genes that showed differential methylation in human DKD tubule samples. Gene ontology analysis indicated enrichment for developmental pathways, such as Notch signaling and nephron development in hyper-DMRs. Conversely, genes with lower methylation levels in DKD were enriched for immune function (Figure 4A and Supplemental Figure 4). Gene set enrichment analysis (GSEA) of RNA sequencing data revealed a lower level of fatty acid oxidation genes in DKD. Alternatively, immune pathway–associated genes, specifically TNF signaling, were enriched in DKD samples (Figure 4B and Supplemental Figures 5 and 6). RNA level of TNF was higher in DKD samples and was associated with lower methylation levels of the TNF promoter (Figure 4C). These changes in methylation and gene expression were not restricted to our WGBS samples but could be validated in the Infinium 450K data set that contained 91 microdissected human kidney samples as well. We observed significant correlation between the degree of methylation of TNF locus and fibrosis (negative) and GFR (positive) (Figure 4D). A correlation between kidney tubule TNF gene expression and fibrosis (positive) and GFR (negative) (Figure 4E) was also detected. In addition, the methylation of TNF probe cg0213984 in 91 human kidney tubule samples was significantly correlated with kidney function decline (eGFR slope P = 0.036) (ref. 31 and Figure 4F). Single-cell RNA sequencing data indicated that Tnf is mainly expressed in immune cell types in mouse kidney (Supplemental Figure 7 and ref. 28). In summary, GSEA indicated a concerted epigenetic dysregulation of the TNF pathway, which is one of the best-known early biomarkers to identify patients with progressive DKD.
Figure 4. Increased enrichment for TNF pathway in human DKD tubules.
(A) Functional annotation analysis by GREAT of the hyper- and hypo-DMRs. (B) Gene set enrichment analysis (GSEA) results of genes that are increased and decreased in DKD. (C) WGBS DNA methylation level (control and DKD) followed by DMR highlight (gray), human kidney H3K27ac ChIP-seq, ChromHMM data, and RNA expression (by RNA sequencing). ChromHMM color code is from Figure 1C. (D) Correlation between GFR or interstitial fibrosis and CpG methylation in TNF gene locus measured by Infinium array in microdissected tubule samples from 91 human kidneys. (E) Correlation between eGFR or interstitial fibrosis and human patient TNF gene expression. P value was calculated using linear regression models adjusted for age, sex, race, diabetes mellitus, and hypertension (36). (F) Correlation between eGFR slope and the methylation of cg02137984 (Infinium array) which is localized in the TNF gene locus (31). P value was calculated using linear regression models adjusted for age, sex, race, diabetes mellitus, and hypertension.
Site-specific methylation editing by dCas9-Tet1 increases TNF transcript level.
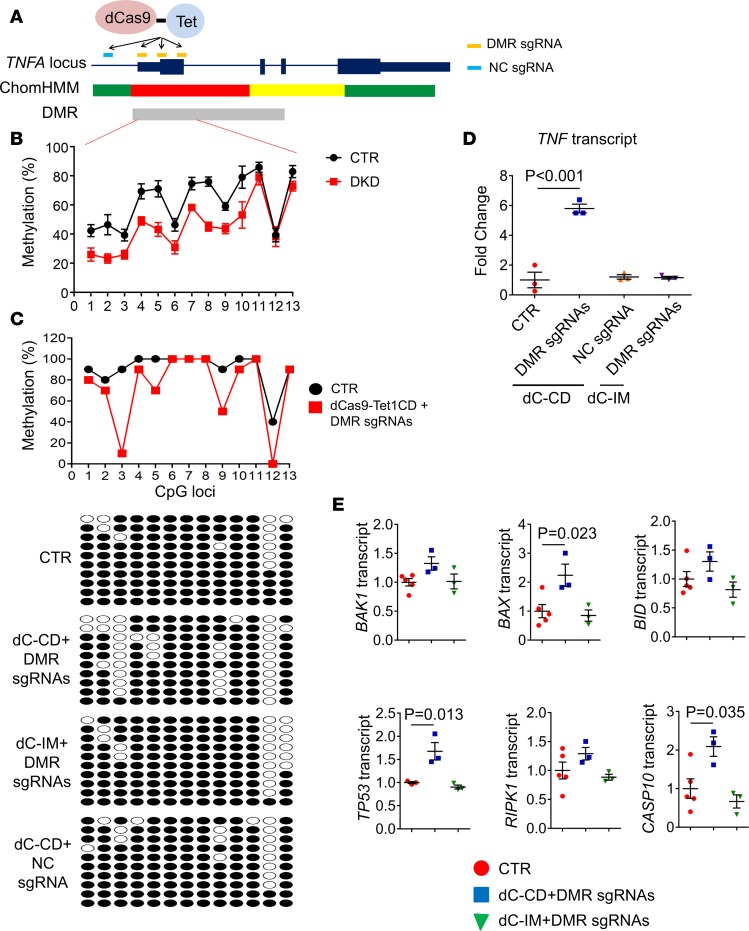
We identified a DMR on the promoter region of TNF that showed a lower methylation level in DKD. To test whether methylation of the TNF DMR, which was identified in DKD samples, plays a role in regulating the expression of TNF, we developed site-specific methylome editing via the dCas9-Tet1 system. We generated human embryonic kidney (HEK293) cells that stably express the dCas9-TET1, in which Cas9 has dead nuclease activity and is fused with Tet1 (the Tet eleven hydroxylase) that modifies methylated cytosine. Control cells contained an inactive mutant (IM) form of TET1 fused with dCas9 (dCas9-Tet1CD_IM) (Supplemental Figure 8). To determine whether the lower methylation of TNF DMR actually contributes to the increase of its transcript level, we designed 3 sgRNAs targeting 13 CpGs in the TNF DMR region (Figure 4C and Figure 5, A and B). Pooled DMR sgRNAs in HEK293 cells resulted in a substantial reduction of methylation in the third, fifth, and ninth CpGs (Figure 5C and Supplemental Figure 9), as evaluated by bisulfite PCR and sequencing. Lower methylation of the TNF promoter region led to an increase in expression of TNF in dCas9-Tet1CD cells with pooled DMR sgRNAs (Figure 5D). Single-guide RNA data showed a similar outcome to the pooled guide RNA (Supplemental Figure 10) and the results were consistent in multiple experimental repeats. No increase in expression of TNF was observed in the inactive mutant (dCas9-Tet1CD_IM) cells transfected with pooled DMR sgRNAs (Figure 5D). We also tested the effect of negative control (NC) guide RNA which is targeted outside of the TNF DMR region (Figure 4C and Figure 5A). TNF expression was not changed in dCas9-Tet1CD cells transfected with NC sgRNA (Figure 5D). In summary, direct manipulation of the DKD-specific DMR at the TNF regulatory region indicated that lowering the methylation of this region results in an increase in TNF transcript level.
Figure 5. Site-specific methylation editing of the TNF DMR by dCas9-Tet1 increases TNF transcript level.
(A) Schematic representation of methylome editing of the TNF DMR by dCas9-Tet1 and specific sgRNAs. ChromHMM color code is in Figure 1C. (B) Methylation level of 13 individual CpG sites at the TNF DMR of human kidney tubules from control and DKD patients. (C) Upper panel: Methylation level of individual CpGs in the TNF DMR from dCas9-Tet1CD cells (HEK293) with or without DMR-targeting sgRNAs. Bottom panel: Bisulfite sequencing analysis of specific cells. (D) Relative transcript level (qRT-PCR) of TNF in control, dCas9-Tet1CD cells with DMR sgRNAs or NC sgRNA, and dCas9-Tet1CD_IM cells with DMR sgRNAs. (E) Relative transcript levels of genes associated with cell death in control, dCas9-Tet1CD or dCas9-Tet1CD_IM cells with DMR sgRNAs. All data are presented as means ± SEM of 3 experiments and analyzed by 1-way ANOVA with Tukey’s post hoc test.
Higher TNF levels worsened DKD severity in diabetic mice.
To understand whether or not an increase in TNF level has a functional effect, we studied gene expression changes following epigenome editing. An increase in TNF level can induce cell death, including apoptosis (32). Consistent with the proapoptotic properties of TNF, we found that transcript expression of BAX, TP53, and CASP10 was significantly increased in dCas9-Tet1CD cells following transfection of pooled sgRNAs (Figure 5E) compared with dCas9-Tet1CD_IM controls. In summary, demethylation of TNF DMR leads to an increase in TNF expression with functional significance at the cellular level.
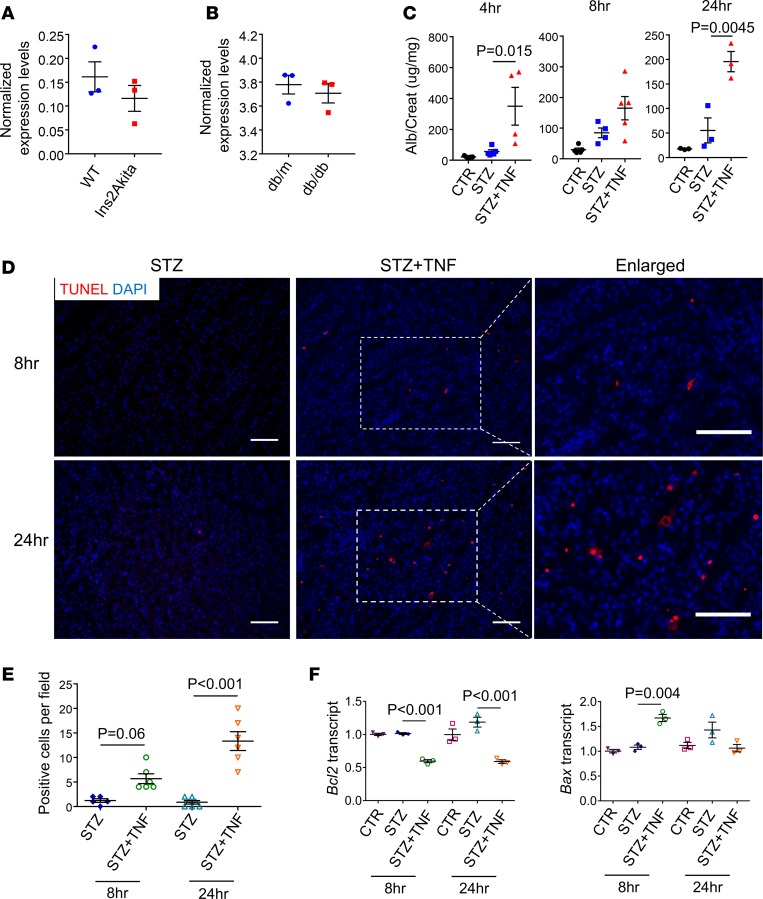
Next, we hypothesized that an increase in TNF levels in vivo is important for DKD development. Most strains of mice, when made diabetic, do not develop robust albuminuria and progressive kidney disease as evidenced by a decline in kidney function (33). We downloaded publicly available gene expression (microarray) data from Akita (type 1 diabetic) and db/db (type 2 diabetic) mice. We observed no significant change in TNF transcript level in kidneys of these diabetic mouse models (refs. 34, 35, and Figure 6, A and B). Conversely, we observed a modest increase in TNF and TNFRSF1B levels and signaling pathway–related gene expression in kidneys of patients with DKD (ref. 36; Figure 4E; and Supplemental Figures 5 and 11). To define the role of increased TNF levels in DKD development, we induced diabetes mellitus in mice by low-dose streptozotocin (STZ) injection (Supplemental Figure 12A). To increase circulating and renal TNF levels, we injected animals with recombinant TNF. Urine collected from TNF-injected diabetic mice showed 3-fold higher albuminuria (albumin-to-creatinine ratio) after TNF injection compared with the vehicle-injected STZ group (Figure 6C). The increase in albuminuria was sustained after TNF injection in the diabetic animals (Figure 6C). At the observation timeframe, we did not observe significant fibrosis in the diabetic or the TNF-injected diabetic animals (Supplemental Figure 12B).
Figure 6. Higher TNF levels augment diabetic albuminuria and cell death.
(A) Tnf expression in kidneys of wild-type and Ins2Akita mice (GSE87899) (34). (B) Tnf expression in kidneys of db/m and db/db mice (GSE87359) (35). (C) The urinary albumin-to-creatinine ratio using the urine collected from control, STZ-treated, and STZ/TNF–cotreated mice at 4, 8, and 24 hours following TNF injection. (D) Representative images of TUNEL assay of mouse kidney samples from STZ-treated and STZ/TNF–cotreated mice. (E) Quantification of positive cells per field as in D. (F) Relative transcript levels of Bcl2 and Bax in kidney samples dissected at 8 and 24 hours following TNF injection from control, STZ-treated, and STZ/TNF–cotreated mice. All data are presented as means ± SEM of 3 experiments and analyzed by 1-way ANOVA with Tukey’s post hoc test. Scale bars: 20 μm.
An increase in the TNF level was also associated with an increase in tubule epithelial cell death, as evident by an increase in TUNEL-positive cells (Figure 6, D and E). We also found an increase in expression of proapoptotic genes, such as Bax, and a decrease in expression of antiapoptotic genes, such as Bcl2, compared with STZ-diabetic mice (Figure 6F). These changes were consistent with our in vitro results. In summary, our results indicate that increasing the TNF level in diabetic mice results in a significant increase in albuminuria, via enhanced cell death, a key manifestation of DKD in patients, indicating a role for TNF in DKD.
Discussion
Clinical and observational data strongly imply the role of epigenetic changes in DKD. In support of this hypothesis, cytosine methylation differences have been described in blood and kidney samples when control and DKD samples have been compared. Most of these studies, however, used modest sample sizes and have not shown consistent results. Few, if any, studies reported results from kidney cells that are functionally linked to disease development. Kidney studies reported data from a mixed-CKD cohort that included patients with both DKD and non-DKD. Finally, no prior studies have reported base-resolution cytosine methylation analysis in kidney samples or site-specific methylation base editing.
Here we performed WGBS of microdissected human DKD kidney tubule samples. The current study only included a limited number of samples for whole-genome methylation analysis; however, we used array-based methylation data from a larger cohort of 91 human kidney tissue samples, in which patients with diabetes mellitus but no renal disease were also included. WGBS analysis showed a strong correlation with array-based analysis. Further integration of methylome data with histone modification analysis was highly consistent and reinforced the strong negative correlation between promoter methylation and transcript levels. Interestingly, a larger number of sites showed a higher methylation level in DKD. Sites that showed higher methylation were enriched on enhancer regions and for binding of kidney-specific transcription factors and correlated with the expression of tubule epithelial genes. These results are consistent with the decrease of expression of kidney tubule–specific genes in DKD, as it has been described as part of the epithelial dedifferentiation program. Further studies will need to analyze the potential causal role of increased methylation as part of the epithelial dedifferentiation in DKD.
A smaller number of sites showed lower methylation levels in DKD. Sites with lower methylation were also enriched on promoter regions; however, these regions showed enrichment for immune cell–specific transcription factor binding. Our histological analysis indicated an increase in immune cells in DKD samples. Changes in methylation of these sites strongly correlated with gene expression changes, indicating that although the changes might not be cell autonomous, they still likely play a role in gene expression regulation. These results suggest that DMRs with a lower methylation level in DKD samples probably resulted from cell composition changes (such as increased immune infiltrate), with a possible lower contribution from cell-autonomous changes.
We specifically analyzed the methylation of the TNF locus because TNF is a potent proinflammatory cytokine that has been shown to play a role in autoimmune renal diseases including lupus nephritis (37, 38). We found that TNF signaling genes, including TNF expression, were higher in human DKD samples compared with the control. The methylation of the TNF gene locus was consistently lower in diabetic samples. The DMR was localized to the promoter region of TNF and correlated with TNF expression. The change in methylation level was validated in an external cohort via methylation arrays and confirmed epigenetic changes in the TNF locus, indicating a potential regulatory mechanism. Site-specific methylation editing of the TNF region lead to increased TNF levels, further confirming the causal role of this DMR in regulating gene expression.
Blood TNFR1 and R2 levels are considered the best biomarker for progressive kidney disease in patients with diabetes mellitus (39). Consistent with these observations, in our data set, methylation of TNF was also remarkably associated with future kidney function decline, indicating that TNF is an important biomarker for progressive kidney disease. Our results indicate that the regulation of TNF likely occurs at the epigenetic level, potentially indicating a key role for epigenetic changes in DKD development. Given that epigenetic changes are under metabolic control, these changes could represent the biological basis of the long sought after “metabolic memory,” although further studies are needed to define such a connection.
The role of TNF in diabetes mellitus and DKD development has been proposed by earlier studies. For example, TNF-deficient mice show less severe nephropathy (40, 41) and TNF has been proposed as a potential therapeutic target for DKD. Our results are consistent with previous findings indicating the increase in TNF level is associated with increased epithelial injury and death both in vitro and in vivo, and administration of TNF to the diabetic mouse model exacerbates albuminuria, which is a key manifestation of DKD.
An important limitation of our study is that it is from a single center where both the primary and validation cohorts were collected at similar institutions. The samples are from an unaffected portion of tumor nephrectomy cases. Because both the cases and controls were obtained in the same manner, changes related to surgery or renal cell cancer should equally bias the cases and controls and therefore were eliminated during the analysis. In addition, although we show the critical role of TNF DMR in regulating TNF transcript levels, it is likely that the methylation change observed in DKD was also affected by cell heterogeneity, not only by cell-type specific changes. In summary, here we show what we believe is the first base-pair-resolution methylation atlas of control and diabetic human kidney tubules. Our site-specific methylome editing highlights the role of methylation in regulating the expression of TNF; the methylation of this locus correlates with kidney function changes and indicates that TNF likely plays a key role in the development of DKD in patients.
Methods
Sample procurement.
Human kidney tissues were collected from the unaffected portion of surgical nephrectomies. Samples were permanently deidentified, clinical information was provided by an honest broker, and no personal identifiable information was collected as part of the study. The collected kidney was immersed into RNAlater and stored at −80°C. Tubule compartments were manually microdissected from the tissue and subjected to RNA and DNA isolation. The rest of the tissue was formalin-fixed and paraffin-embedded for periodic acid–Schiff and hematoxylin and eosin staining and the diagnosis of DKD was established by histopathological analysis.
WGBS and mapping.
Genomic DNAs were isolated using DNeasy kits (QIAGEN) by following the manufacturer’s instructions. WGBS libraries were constructed from the purified genomic DNAs using the Illumina TruSeq DNA Methylation Library Preparation kit and subjected to high-throughput sequencing using an Illumina HiSeq X Ten system with 150-bp paired-end sequencing according to the manufacturer’s instructions. Low-quality and adaptor sequences were trimmed using the Trim Galore program. Bisulfite-converted sequencing reads were aligned to the human genome (hg19) using Bismark program version 0.7.0. Duplicate reads were removed using deduplicate_bismark. Methylation levels of every CpGs were calculated using bismark_methylation_extractor function with the non-default parameters --ignore 7 --ignore_r2 6 --ignore_3prime 0 --ignore_3prime_r2 1.
WGBS data analysis.
PCA plot was generated from methylation percentages of individual CpGs using RnBeads R package version 1.2.2. To identify DMRs, the BSmooth algorithm was first applied to the Bismark coverage files. After smoothing, DMRs were identified between 5 control and 5 DKD samples using bsseq R package version 1.6.0 with the following parameters: CpGs where 10 samples have at least 3× in coverage were used for computing t-statistics, estimate.var=”same” was used for BSmooth.tstat function, qcutoff=c(0.01,0.99) was used for dmrFinder function, number of CpGs included in DMRs 5 and greater, mean methylation difference 15% and greater, and area Stat 100 and greater. Human kidney ChIP-seq data for histone H3K4me1, H3K4me3, H3K27ac, H3K36me3, H3K9ac, and H3K9me3 (GSM621634, GSM621648, GSM621651, GSM670025, GSM772811, GSM1112806) were used to annotate chromatin states using ChromHMM (26). ChromHMM maps of other tissues were downloaded from ENCODE project. The genes that contain the DMRs in their promoter region were defined when the DMRs are localized within transcription start site ±3 kb and overlapped with active TSS chromatin states, which were identified by ChromHMM. The enhancer DMRs were defined when they are localized outside of transcription start site ±3 kb and overlapped with active enhancer chromatic states which were identified by ChromHMM. Significantly enriched transcription factor motifs were identified in the DMRs using the Homer program (42). The genomic regions enrichment of annotations (GREAT) version 3.0 was used for functional annotation analysis (43).
RNA sequencing analysis.
Total RNAs were isolated using the RNeasy Mini kit (QIAGEN). cDNA sequencing libraries were constructed using the Illumina TruSeq RNA Preparation Kit. High-throughput sequencing was performed using Illumina HiSeq 4000 with 100-bp single-end sequencing according to the manufacturer’s instruction. Low-quality and adaptor sequences were trimmed using the Trim Galore program. Sequencing reads were aligned to human genome (hg19) using STAR-2.4.1d. The expression level for each gene was quantified using HTSeq-0.6.1 (htseq-count) and GENCODE version 19 gtf file. Differentially expressed genes between control and DKD groups were identified using DESeq2 version 1.10.1. FPKM values were used for GSEA to identify the significantly enriched molecular signatures among MSigDB gene sets (44). In silico deconvolution analysis of RNA sequencing data was performed using CIBERSORT with FPKM values (30).
Illumina Infinium HumanMethylation450K BeadChip.
Infinium methylation 450k array data of 91 microdissected human kidney tubule samples were downloaded from GSE50874. DNA methylation values were extracted following our previous study (31). Pearson’s correlation coefficient was calculated between the methylation level of Infinium probes and corresponding CpG sites (coverage ≥ 5) in WGBS data. The correlation between methylation level of each probe, eGFR, and interstitial fibrosis score was calculated using linear regression models adjusted for age, sex, race, diabetes mellitus, hypertension, batch effect, and bisulfite conversion efficiency. Bonferroni’s correction was used for multiple comparisons. Linear regression models with the same covariates were applied to calculate the association between expression level of the genes and methylation level of probes that are located within 500 kb from their transcription start site. To validate DMRs identified by WGBS, less stringent criteria were applied (P < 0.01) to identify the associated probes with gene expression. Subject-specific unadjusted eGFR slopes were determined by linear regression across all available eGFR measures.
Data and materials availability.
RNA sequencing data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE115098. The authors are in the process of making the WGBS data publicly available in a manner that does not reveal personal identified information and the samples are not traceable back to tissue donors.
Plasmids and sgRNA design.
Fuw-dCas9-Tet1CD and Fuw-dCas9-Tet1CD_IM were gifts from Rudolf Jaenisch (Addgene plasmid 84475 and 84479). The sgRNA expression plasmid pLKO5.sgRNA.EFS.GFP was a gift from Zhaolan (Joe) Zhou (Addgene plasmid 57822). Annealed sgRNA oligos were subcloned into pLKO5.sgRNA.EFS.GFP with the Bsmb1 site. All constructs were verified by sequencing before transfection. Primers for sgRNA design are listed in Supplemental Table 3.
Bisulfite conversion, PCR, and sequencing.
Genomic DNA was isolated from cells 96 hours after transfection using DNeasy Blood and Tissue Kit (QIAGEN). Genomic DNA (500 ng) was bisulfite converted using an EpiTect Bisulfite Kit (QIAGEN) following the manufacturer’s instructions. The resulting modified DNA was PCR amplified using a PyroMark PCR kit (QIAGEN). The resulting amplified products were purified, subcloned using a TOPO TA Cloning Kit (Thermo Fisher Scientific), and sequenced.
Gene expression analysis by qPCR.
RNA was isolated using an RNeasy Mini Kit (QIAGEN). RNA was reverse transcribed using cDNA archival kit (Life Technologies), and quantitative real-time PCR was conducted in the ViiA 7 System (Life Technologies) machine using SYBR Green Master Mix. Primers used are listed in Supplemental Table 4.
Mouse studies.
Four-week-old wild-type mice were intraperitoneally injected with 50 mg/kg STZ for 5 days. One week later, STZ-treated mice were injected with 0.25 mg/kg murine recombinant TNF (Biolegend) at the first and eighth hour. Urine was collected at 4, 8, and 24 hours after TNF administration. Mice were sacrificed and kidneys were harvested 8 and 24 hours after TNF injection.
Immunofluorescence.
For immunohistochemistry, sections were deparaffinized, hydrated in 100%, 95%, and 70% alcohol, and washed in water. TUNEL assay was performed according to manufacturer’s instruction (Abcam).
Statistics.
The data are presented as means ± SEM. Unpaired 2-tailed Student’s t test was used for comparisons between 2 groups. We used 1-way ANOVA with Tukey’s post hoc test to compare multiple groups. P < 0.05 was considered significant.
Study approval.
The human subject study was approved by the institutional review boards of the Albert Einstein College of Medicine and Montefiore Medical Center (IRB 2002–202) and the University of Pennsylvania (IRB 815796). All experiments on animals were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania (Protocol 804250) and were performed in accordance with the institutional guidelines.
Author contributions
This study was conceived of and led by KS. J. Park performed computation analysis with assistance from XS, CG, CQ, and HL. YG performed experiments. MJS, AH, J. Pullman, and AV collected kidney tissues. MP performed histological analysis. J. Park, YG, and KS wrote the manuscript.
Supplementary Material
Acknowledgments
Work in the Susztak lab is supported by NIH National Institute of Diabetes and Digestive and Kidney Diseases grants R01DK076077, R01 DK087635, and DP3 DK108220. J. Park is supported by American Diabetes Association Training grant 1-17-PDF-036. We thank the University of Pennsylvania Diabetes Research Center (DRC) (P30-DK19525) and Center for Molecular Studies in Digestive and Liver Diseases (P30-DK050306).
Version 1. 06/06/2019
Electronic publication
Version 2. 06/06/2019
A. Ari Hakimi's name has been updated
Footnotes
J. Park’s present address is: School of Life Sciences, Gwangju Institute of Science and Technology, Gwangju, South Korea.
Conflict of interest: The Susztak lab is supported by Boehringer Ingelheim, Lilly, Regeneron, GSK, Merck, Pharma, Gilead, and Bayer.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(11):e128886. https://doi.org/10.1172/jci.insight.128886.
Contributor Information
Jihwan Park, Email: jihwan.park724@gmail.com.
Yuting Guan, Email: yutguan@pennmedicine.upenn.edu.
Xin Sheng, Email: xin.sheng@pennmedicine.upenn.edu.
Caroline Gluck, Email: caroline.gluck@gmail.com.
Matthew J. Seasock, Email: mseasock@pennmedicine.upenn.edu.
Chengxiang Qiu, Email: quorly@gmail.com.
Amit Verma, Email: amit.verma@einstein.yu.edu.
Hongzhe Li, Email: hongzhe@mail.med.upenn.edu.
Katalin Susztak, Email: ksusztak@pennmedicine.upenn.edu.
References
- 1.[No authors listed] USRDS: the United States Renal Data System. Am J Kidney Dis. 2003;42(6 suppl 5):1–230. [PubMed] [Google Scholar]
- 2.Groop PH, et al. The presence and severity of chronic kidney disease predicts all-cause mortality in type 1 diabetes. Diabetes. 2009;58(7):1651–1658. doi: 10.2337/db08-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregg EW, et al. Changes in diabetes-related complications in the United States, 1990–2010. N Engl J Med. 2014;370(16):1514–1523. doi: 10.1056/NEJMoa1310799. [DOI] [PubMed] [Google Scholar]
- 4.Thomas MC, et al. Diabetic kidney disease. Nat Rev Dis Primers. 2015;1:15018. doi: 10.1038/nrdp.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gohda T, et al. Circulating TNF receptors 1 and 2 predict stage 3 CKD in type 1 diabetes. J Am Soc Nephrol. 2012;23(3):516–524. doi: 10.1681/ASN.2011060628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niewczas MA, et al. Circulating TNF receptors 1 and 2 predict ESRD in type 2 diabetes. J Am Soc Nephrol. 2012;23(3):507–515. doi: 10.1681/ASN.2011060627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akash MSH, Rehman K, Liaqat A. Tumor necrosis factor-α: role in development of insulin resistance and pathogenesis of type 2 diabetes mellitus. J Cell Biochem. 2018;119(1):105–110. doi: 10.1002/jcb.26174. [DOI] [PubMed] [Google Scholar]
- 8.Gurley SB, et al. Inflammation and immunity pathways regulate genetic susceptibility to diabetic nephropathy. Diabetes. 2018;67(10):2096–2106. doi: 10.2337/db17-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Boer IH, et al. Long-term renal outcomes of patients with type 1 diabetes mellitus and microalbuminuria: an analysis of the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications cohort. Arch Intern Med. 2011;171(5):412–420. doi: 10.1001/archinternmed.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reidy K, Kang HM, Hostetter T, Susztak K. Molecular mechanisms of diabetic kidney disease. J Clin Invest. 2014;124(6):2333–2340. doi: 10.1172/JCI72271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vehaskari VM, Aviles DH, Manning J. Prenatal programming of adult hypertension in the rat. Kidney Int. 2001;59(1):238–245. doi: 10.1046/j.1523-1755.2001.00484.x. [DOI] [PubMed] [Google Scholar]
- 12.Hoppe CC, et al. Combined prenatal and postnatal protein restriction influences adult kidney structure, function, and arterial pressure. Am J Physiol Regul Integr Comp Physiol. 2007;292(1):R462–R469. doi: 10.1152/ajpregu.00079.2006. [DOI] [PubMed] [Google Scholar]
- 13.Susztak K. Understanding the epigenetic syntax for the genetic alphabet in the kidney. J Am Soc Nephrol. 2014;25(1):10–17. doi: 10.1681/ASN.2013050461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. doi: 10.1038/s41581-019-0135-6. Kato, M., Natarajan, R. Epigenetics and epigenomics in diabetic kidney disease and metabolic memory [published online ahead of print March 20, 2019]. Nat Rev Nephrol. doi: 10.1038/s41581.019.0135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beckerman P, Ko YA, Susztak K. Epigenetics: a new way to look at kidney diseases. Nephrol Dial Transplant. 2014;29(10):1821–1827. doi: 10.1093/ndt/gfu026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy MA, Natarajan R. Epigenetics in diabetic kidney disease. J Am Soc Nephrol. 2011;22(12):2182–2185. doi: 10.1681/ASN.2011060629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marumo T, et al. Diabetes induces aberrant DNA methylation in the proximal tubules of the kidney. J Am Soc Nephrol. 2015;26(10):2388–2397. doi: 10.1681/ASN.2014070665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z, et al. Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort. Proc Natl Acad Sci U S A. 2016;113(21):E3002–E3011. doi: 10.1073/pnas.1603712113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sapienza C, et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics. 2011;6(1):20–28. doi: 10.4161/epi.6.1.13362. [DOI] [PubMed] [Google Scholar]
- 20.Chu AY, et al. Epigenome-wide association studies identify DNA methylation associated with kidney function. Nat Commun. 2017;8(1):1286. doi: 10.1038/s41467-017-01297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li D, et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol. 2013;31(8):681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- 22.Shao Y, et al. CRISPR/Cas-mediated genome editing in the rat via direct injection of one-cell embryos. Nat Protoc. 2014;9(10):2493–2512. doi: 10.1038/nprot.2014.171. [DOI] [PubMed] [Google Scholar]
- 23.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu XS, et al. Editing DNA methylation in the mammalian genome. Cell. 2016;167(1):233–247.e17. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu XS, et al. Rescue of Fragile X syndrome neurons by DNA methylation editing of the FMR1 gene. Cell. 2018;172(5):979–992.e6. doi: 10.1016/j.cell.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ernst J, Kellis M. Chromatin-state discovery and genome annotation with ChromHMM. Nat Protoc. 2017;12(12):2478–2492. doi: 10.1038/nprot.2017.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462(7269):58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J, et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360(6390):758–763. doi: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woroniecka KI, Park AS, Mohtat D, Thomas DB, Pullman JM, Susztak K. Transcriptome analysis of human diabetic kidney disease. Diabetes. 2011;60(9):2354–2369. doi: 10.2337/db10-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ko YA, et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol. 2013;14(10):R108. doi: 10.1186/gb-2013-14-10-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rath PC, Aggarwal BB. TNF-induced signaling in apoptosis. J Clin Immunol. 1999;19(6):350–364. doi: 10.1023/A:1020546615229. [DOI] [PubMed] [Google Scholar]
- 33.Brosius FC. Mouse models of diabetic nephropathy. J Am Soc Nephrol. 2009;20(12):2503–2512. doi: 10.1681/ASN.2009070721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubin A, Salzberg AC, Imamura Y, Grivitishvilli A, Tombran-Tink J. Identification of novel targets of diabetic nephropathy and PEDF peptide treatment using RNA-seq. BMC Genomics. 2016;17(1):936. doi: 10.1186/s12864-016-3199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, et al. Transcriptional profile of kidney from type 2 diabetic db/db mice. J Diabetes Res. 2017;2017:8391253. doi: 10.1155/2017/8391253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang HM, et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med. 2015;21(1):37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacob CO, McDevitt HO. Tumour necrosis factor-α in murine autoimmune ‘lupus’ nephritis. Nature. 1988;331(6154):356–358. doi: 10.1038/331356a0. [DOI] [PubMed] [Google Scholar]
- 38.Ryffel B, Eugster H, Haas C, Le Hir M. Failure to induce anti-glomerular basement membrane glomerulonephritis in TNF α/β deficient mice. Int J Exp Pathol. 1998;79(6):453–460. doi: 10.1046/j.1365-2613.1998.00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colhoun HM, Marcovecchio ML. Biomarkers of diabetic kidney disease. Diabetologia. 2018;61(5):996–1011. doi: 10.1007/s00125-018-4567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Awad AS, et al. Macrophage-derived tumor necrosis factor-αmediates diabetic renal injury. Kidney Int. 2015;88(4):722–733. doi: 10.1038/ki.2015.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lampropoulou IT, Stangou M, Papagianni A, Didangelos T, Iliadis F, Efstratiadis G. TNF-α and microalbuminuria in patients with type 2 diabetes mellitus. J Diabetes Res. 2014;2014:394206. doi: 10.1155/2014/394206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.