Abstract
African Americans develop end-stage renal disease at a higher rate compared with European Americans due to 2 polymorphisms (G1 and G2 risk variants) in the apolipoprotein L1 (APOL1) gene common in people of African ancestry. Although this compelling genetic evidence provides an exciting opportunity for personalized medicine in chronic kidney disease, drug discovery efforts have been greatly hindered by the fact that APOL1 expression is lacking in rodents. Here, we describe a potentially novel physiologically relevant genomic mouse model of APOL1-associated renal disease that expresses human APOL1 from the endogenous human promoter, resulting in expression in similar tissues and at similar relative levels as humans. While naive APOL1-transgenic mice did not exhibit a renal disease phenotype, administration of IFN-γ was sufficient to robustly induce proteinuria only in APOL1 G1 mice, despite inducing kidney APOL1 expression in both G0 and G1 mice, serving as a clinically relevant “second hit.” Treatment of APOL1 G1 mice with IONIS-APOL1Rx, an antisense oligonucleotide (ASO) targeting APOL1 mRNA, prior to IFN-γ challenge robustly and dose-dependently inhibited kidney and liver APOL1 expression and protected against IFN-γ–induced proteinuria, indicating that the disease-relevant cell types are sensitive to ASO treatment. Therefore, IONIS-APOL1Rx may be an effective therapeutic for APOL1 nephropathies and warrants further development.
Keywords: Nephrology, Therapeutics
Keywords: Drug therapy, Genetic diseases, Mouse models
An APOL1 antisense oligonucleotide therapeutic, IONIS-APOL1Rx, demonstrates efficacy in a physiologically-relevant APOL1 transgenic mouse model.
Introduction
In 2010, 2 mutually exclusive polymorphisms (G1 and G2) in the apolipoprotein L1 (APOL1) gene, commonly found in people of African ancestry, were identified to increase the risk of chronic kidney disease (CKD) in homozygous or compound heterozygote individuals by 7- to 30-fold, accounting for the higher prevalence of kidney failure in African Americans (1–4). These mutations originated in ancestral populations in West Africa and are now being described as associated with CKD risk in other descendant populations, highlighting West African ancestry as the ethnically linked population (5). The APOL1 risk alleles have been shown to strongly associate with various forms of nondiabetic nephropathy previously thought to be unrelated, including focal segmental glomerulosclerosis (FSGS), HIV-associated nephropathy (HIVAN), hypertensive-attributed kidney disease, lupus nephritis, and IFN therapy–related collapsing glomerulopathy (1, 3, 6, 7). Furthermore, APOL1 risk allele carriers with CKD demonstrate faster GFR decline and disease progression than CKD patients that harbor the G0 reference allele (3, 8). As roughly 13% of African Americans are homozygous for the APOL1 risk alleles (~6 million individuals at risk; ref. 9), it is estimated that there is a large population suffering from APOL1 nephropathies that may potentially benefit from an APOL1 therapeutic aimed at targeting the underlying molecular basis of their disease.
APOL1 is a newly evolved gene that is functional only in humans and select nonhuman primates (i.e., gorilla and baboon); it is highly expressed in the liver and placenta, with lower levels of expression in kidney, heart, and lung (10, 11). APOL1 protein is secreted into circulation predominantly from the liver and has been identified as the trypanolytic factor in human serum, forming pores in the lysosomal membrane of trypanosomes and triggering chloride influx (12–14). While the trypanosome strain Trypanosoma brucei brucei is sensitive to human serum, Trypanosoma brucei rhodesiense (T. b. rhodesiense) is resistant, as it has evolved to evade the lytic activity of APOL1, causing African sleeping sickness in humans. The G1 and G2 mutations of APOL1, however, overcome the serum resistance of T. b. rhodesiense, protecting individuals harboring at least one APOL1 G1/G2 allele from this pathogenic strain (1). As a result, APOL1 G1/G2 polymorphisms have been enriched in populations in sub-Saharan Africa where T. b. rhodesiense is endemic and, thus, in people of African descent, despite the propensity for CKD in homozygous carriers. While APOL1’s function as a plasma trypanolytic factor is well defined, the mechanism of increased risk for renal disease with the APOL1 risk variants remains unclear.
Discoveries surrounding APOL1’s role in renal disease and, as a result, drug discovery efforts, have been greatly hindered by the fact that rodents do not express APOL1. To date, 2 APOL1-transgenic mouse models have been reported, both driven by a podocyte-specific (Nephrin) promoter, resulting in high and supraphysiological APOL1 expression levels only in podocytes (15, 16). In one case, expression of the APOL1 risk alleles did not cause a renal phenotype, while in the second case, inducible podocyte-specific APOL1 expression resulted in an FSGS-like phenotype resembling human disease. Although this latter model has utility in studying the role of APOL1 G1/G2 proteins in podocytes, evaluating the role of APOL1 in other cell types and organs and assessing their contribution to renal disease is not possible. Moreover, in risk allele homozygous individuals, APOL1 nephropathy is thought to be induced by a second hit, and expression of APOL1 has been shown to be upregulated by various proinflammatory stimuli in vitro (6, 11, 17). Therefore, disease models that effectively recapitulate all aspects of APOL1 biology critical for understanding its role in renal disease are in great need.
Here, we sought to develop a physiologically relevant mouse model of APOL1 nephropathy that can be used to study APOL1 systemically and across cell types as well as achieve proof of concept for an antisense oligonucleotide (ASO) inhibitor of APOL1. Antisense technology is a powerful drug discovery approach for identifying oligonucleotide analogs that can specifically modify RNA expression through multiple mechanisms, including RNase H1–mediated degradation of RNA (18). Recently, the FDA approved mipomersen (Kynamro) as the first systemically administered ASO, and additional ASOs have since been approved for a variety of human diseases (19–22). We used a human APOL1-containing fosmid to generate APOL1 G0– and G1–transgenic mice that express APOL1 in similar tissues as that observed in humans and at similar relative expression levels. While naive APOL1-transgenic mice failed to show a renal phenotype, a single dose of IFN-γ induced robust proteinuria only in APOL1 G1-transgenic mice (APOL1 G1 mice), despite inducing APOL1 expression in both APOL1 G0-transgenic mice (APOL1 G0 mice) and APOL1 G1 mice. We also report on the discovery of the first APOL1 inhibitor, a generation 2.5 APOL1 ASO (IONIS-APOL1Rx) that displays an attractive activity and safety profile in vivo. Administration of IONIS-APOL1Rx to APOL1 G1 mice effectively reduced APOL1 expression in kidney and liver and completely prevented IFN-γ–induced proteinuria, demonstrating potent ASO activity in disease-relevant cell types.
Results
Generation and characterization of genomic APOL1-transgenic mice.
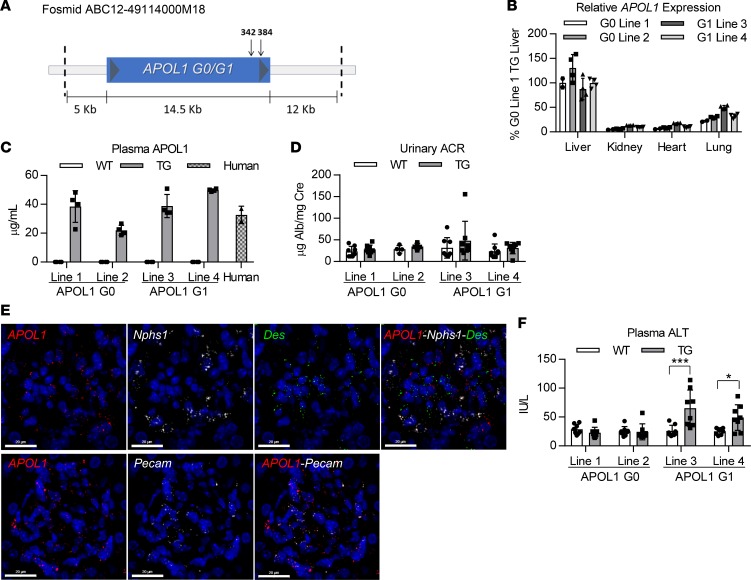
Human APOL1-transgenic mice were generated through pronuclear injection of C57BL/6 oocytes with an approximately 32-kb fosmid fragment containing the entire human APOL1 gene (either G0 or G1) as well as >-5 kb upstream and downstream regions to increase the likelihood of retaining the endogenous regulatory elements and promoter region of the human APOL1 gene (Figure 1A). This approach results in transgenic mice that express APOL1 in similar tissues as those identified in humans and at similar relative levels of expression, which was confirmed in offspring from 2 separate founders for each genotype. As observed in human tissues (11), APOL1 mRNA is highly expressed in livers in APOL1-transgenic mice, with approximately 10-fold less expression in kidney, heart, and lung (Figure 1B). Liver and kidney APOL1 protein expression was confirmed by Western blot (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.126124DS1), and APOL1 was shown to be effectively secreted to plasma, with levels resembling those of human plasma, by ELISA (Figure 1C).
Figure 1. Genomic APOL1-transgenic mice express APOL1 in similar tissues as those observed in humans and at similar relative physiological levels.
(A) Schematic of the human APOL1-containing fosmid fragment used to generate APOL1 G0– and G1–transgenic mice. Arrows mark the protein coding sequence location of the 2 point mutations that constitute the G1 genotype. (B) Relative APOL1 expression levels in livers, kidneys, hearts, and lungs of transgenic mice (n = 2–4) were measured by qRT-PCR and normalized to total RNA. Data from 2 different founder lines are shown for each genotype. (C) APOL1 plasma levels in transgenic mice (n = 4) and human pooled plasma (n = 2) were measured by ELISA. (D) Urine albumin levels of APOL1-transgenic and WT littermate mice (n = 4–8) were measured by albumin ELISA and normalized to urine creatinine levels. (E) FISH was used to evaluate APOL1 mRNA expression in podocytes (Nphs1), mesangial cells (Des), and endothelial cells (Pecam) in APOL1 G1 mouse kidneys (n = 4). Representative images shown (scale bar: 20 μm). (F) Plasma ALT levels of APOL1-transgenic and WT littermate mice (n = 8) were measured using a clinical chemistry analyzer. Two-way ANOVA with Bonferroni’s multiple comparisons test, *P < 0.05; ***P < 0.001. All data are presented as mean ± SD.
To determine whether expression of APOL1 G1 results in a differential renal phenotype in comparison to APOL1 G0, APOL1 G0– and G1–transgenic mice were characterized up to approximately 8 months of age, and age-matched littermates that lacked the APOL1 transgene were used as WT controls. Urinary albumin levels were measured in young and aged animals; however, proteinuria was not detected in either APOL1 G0 or G1 mice (Figure 1D and Supplemental Figure 2A). Similarly, kidney morphology was not perturbed in either APOL1 G0 or G1 mice, even at approximately 8 months of age (Supplemental Figure 2B). To ensure that the APOL1 transgene was being expressed in disease-relevant cell types in the kidney, FISH was performed to label mRNAs in kidney sections from APOL1 G1 mice. APOL1 mRNA colocalized with Nphs1 mRNA, Pecam mRNA, and Des mRNA in the glomerulus, demonstrating that APOL1 is expressed in podocytes, endothelial cells, and mesangial cells, respectively (Figure 1E), similar to that observed in human kidney (23). Interestingly, although APOL1 G1 mice lacked a discernable kidney phenotype, we did observe elevations in plasma alanine aminotransferase (ALT) in both founder lines of young APOL1 G1 mice (Figure 1F).
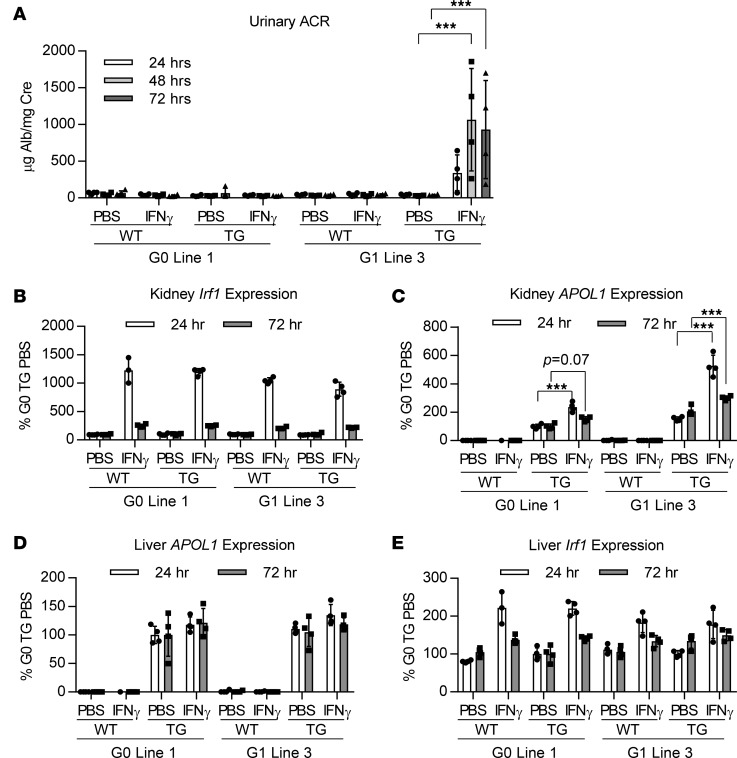
IFN-γchallenge induces proteinuria in APOL1 G1–transgenic mice. As these APOL1 genomic transgenic mice express APOL1 at low physiological levels in the kidney and only a subset of humans homozygous for APOL1 risk alleles develop kidney disease, we suspected that, similar to the disease etiology in humans, a second hit may be needed to cause a renal disease phenotype in APOL1 G1 mice. It has been shown that IFN is a potent inducer of APOL1 gene expression in podocytes and endothelial cells in vitro (6). Moreover, it has been reported that individuals homozygous for APOL1 risk variants undergoing IFN therapy are at increased risk for developing collapsing glomerulopathy that is reversed upon discontinuation of therapy (6, 24). Based on these observations, APOL1 G0–line 1 and G1–line 3 transgenic mice and their WT littermate controls were challenged with IFN-γ and evaluated for proteinuria after 24–72 hours. Indeed, a single administration of IFN-γ resulted in increases in urinary albumin levels after 24 hours in APOL1 G1–, but not APOL1 G0–, transgenic mice, which significantly increased after 48 hours (Figure 2A), establishing a differentiating kidney phenotype between APOL1 G0 and G1 mice. Evaluation of IFN regulatory factor 1 (Irf1) mRNA expression, an IFN-γ response gene shown to be involved in regulation of APOL1 expression in vitro (6), revealed robust kidney induction in all mice 24 hours after IFN-γ challenge, including WT littermates, demonstrating similar activation of the IFN-γ response pathway across genotypes (Figure 2B). Despite the robust induction of proteinuria in APOL1 G1 mice, however, histopathological analysis failed to reveal any morphological changes 24 or 72 hours after IFN-γ challenge (Supplemental Figure 3A). Similarly, plasma blood urea nitrogen (BUN) or creatinine levels and kidney expression of Wt1, a podocyte marker gene, were not significantly perturbed in APOL1 G1 mice compared with APOL1 G0 24 or 72 hours after IFN-γ challenge (Supplemental Figure 3, B–D). On the other hand, expression of Nphs1, encoding for the podocyte slit diaphragm protein Nephrin, was significantly reduced in APOL1 G1 mice 24 hours after IFN-γ challenge in comparison to APOL1 G0 mice and WT littermates (Supplemental Figure 3E), suggestive of podocyte injury. By 72 hours after IFN-γ challenge, however, Nphs1 levels returned to control levels. As proteinuria persists 72 hours after IFN-γ challenge (Figure 2A), there may be focal detachment of podocytes that is not reflected in total Nphs1 mRNA levels resulting in the robust proteinuria observed at this time point.
Figure 2. IFN-γ induces kidney APOL1 expression in APOL1 G0 and G1 mice, but proteinuria only in G1 mice.
Female APOL1 G0– and G1–transgenic and WT littermate mice (n = 3–4) were challenged with a single dose of IFN-γ (1.125 × 107 U/kg) or vehicle (PBS). (A) Urine was collected 24, 48, and 72 hours after IFN-γ challenge, and urinary albumin was measured by ELISA and normalized to urine creatinine. (B) Kidney Irf1 expression was measured by qRT-PCR 24 and 72 hours after IFN-γ challenge and normalized to Cyp expression. (C) Kidney and (D) liver APOL1 expression were measured by qRT-PCR 24 and 72 hours after IFN-γ challenge and normalized to Cyp expression. (E) Liver Irf1 expression was measured by qRT-PCR 24 and 72 hours after IFN-γ challenge and normalized to Cyp expression. All data are presented as mean ± SD. Two-way ANOVA with Tukey’s multiple comparisons test, ***P < 0.001.
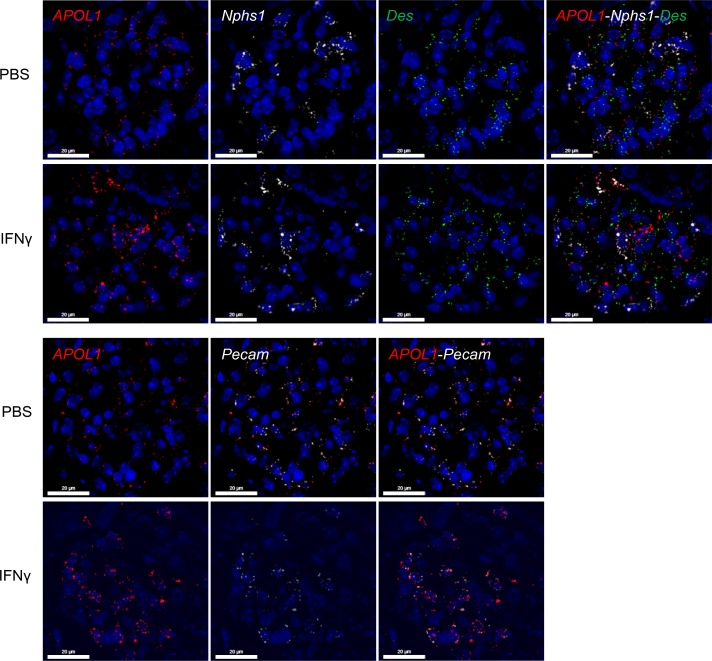
IFN-γinduces expression of kidney APOL1 in APOL1-transgenic mice. As these mice contain the endogenous human promoter, we next evaluated whether IFN-γ upregulates APOL1 expression in these mice. Indeed, 24 hours after IFN-γ challenge we observed robust induction of kidney APOL1 mRNA expression in both APOL1 G0 and G1 mice, with increased upregulation observed in APOL1 G1 mice (2.4-fold vs. 3.4-fold, respectively; Figure 2C). Increases in kidney APOL1 protein were confirmed by Western blot (Supplemental Figure 4, A and B). We also measured IFN-γ–induced APOL1 mRNA expression 72 hours after IFN-γ administration and observed a significant decline in kidney APOL1 mRNA levels in comparison to the 24-hour time point (Figure 2C). This decline in APOL1 induction correlated with reduced Irf1 mRNA levels observed 72 hours, in comparison to 24 hours, after IFN-γ administration (Figure 2B), indicating the transient regulation of APOL1 expression by IFN-γ. Liver APOL1 mRNA, on the other hand, was only modestly upregulated 24 hours after IFN-γ administration in APOL1 G0 and G1 mice (Figure 2D), in agreement with the approximately 5-fold lower induction of liver Irf1 mRNA (Figure 2E), in comparison to kidney Irf1 (Figure 2B), in mice of all genotypes. Consequently, plasma APOL1 levels were not increased following IFN-γ administration, consistent with the majority of plasma APOL1 being produced by the liver (Supplemental Figure 4C). FISH was used to determine in which kidney cell types of APOL1 G1 mice IFN-γ induces APOL1 mRNA expression, revealing strong upregulation of APOL1 in podocytes and endothelial cells, but not mesangial cells, 48 hours after IFN-γ challenge (Figure 3).
Figure 3. IFN-γ induces kidney APOL1 expression in podocytes and endothelial cells in the kidney.
Female APOL1 G1–transgenic mice (n = 4) were challenged with a single dose of IFN-γ (1.125 × 107 U/kg) or vehicle (PBS). FISH was used to evaluate APOL1 mRNA expression in podocytes (Nphs1), mesangial cells (Des), and endothelial cells (Pecam) in APOL1 G1 mouse kidneys 48 hours after IFN-γ challenge. Representative images shown (scale bars: 20 μm).
IFN-γ challenge resulted in stronger induction of kidney APOL1 expression to higher absolute levels in APOL1 G1–line 3 mice compared with APOL1 G0–line 1 mice, begging the question of whether absolute levels of APOL1 expression, regardless of genotype, are responsible for inducing proteinuria. To address this question, IFN-γ was administered to APOL1 G0–line 2 mice, which demonstrate higher basal levels of APOL1 mRNA expression compared with both APOL1 G0–line 1 and APOL1 G1–line 3 mice (Figure 1B), and proteinuria was evaluated 24 to 72 hours later. Induction of kidney APOL1 mRNA was higher in APOL1 G0–line 2 mice compared with APOL1 G1–line 3 mice 72 hours after IFN-γ challenge (Supplemental Figure 5A); yet, robust proteinuria was not observed in APOL1 G0–line 2 mice (Supplemental Figure 5B), indicating that APOL1 G1 expression is the main driver for this phenotype. As observed previously, kidney Irf1 mRNA levels were similarly induced by IFN-γ across mice of all genotypes (Supplemental Figure 5C).
Transcriptome analysis of IFN-γ–challenged APOL1-transgenic mice demonstrates clinical relevance of model. Transcriptome analysis was performed on kidney samples from APOL1 G0 and G1 mice challenged with either IFN-γ or PBS, uncovering 88 genes that were significantly differentially regulated in both APOL1 G0 and G1 mice upon IFN-γ administration compared with PBS controls (Supplemental Figure 6A and Supplemental Table 1). Gene ontology analysis revealed significant enrichment in genes involved in vesicle membranes and lysosome function (Supplemental Figure 6B) in agreement with previous reports on the function of APOL1 (14, 16, 25, 26). This transcriptome signature not only distinguished PBS- and IFN-γ–challenged animals, but also separated APOL1 G0 and G1 IFN-γ–challenged animals by principal components analysis (Supplemental Figure 6C). Importantly, comparing this APOL1-IFN-γ gene set against previously reported gene expression profiles from CKD patients (27) was sufficient to stratify patients by either their histopathology or APOL1 Risk status (Supplemental Figure 6D), suggesting relevance of the model to human APOL1 nephropathies.
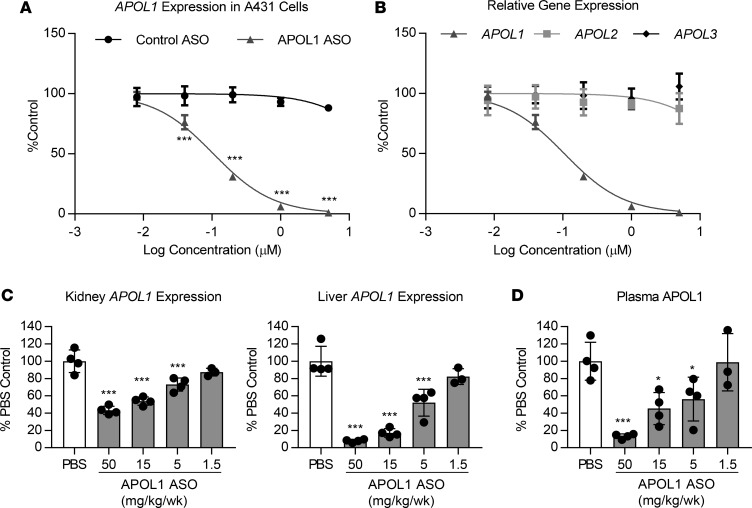
Treatment with IONIS-APOL1Rx, an APOL1 ASO, prevents IFN-γ–induced proteinuria. Over 4000 generation 2.5 ASOs, which bind various sites within the human APOL1 gene, were designed and evaluated for reduction of APOL1 mRNA in vitro. IONIS-APOL1Rx was selected as the APOL1 clinical candidate based on its consistent and potent activity in vitro (half-maximal inhibitory concentration, IC50 = 105 nM; Figure 4A) as well as in vivo in genomic APOL1-transgenic mice. Furthermore, chronic administration of IONIS-APOL1Rx at high doses was well tolerated in rodents and monkeys (data not shown). We also evaluated the specificity of IONIS-APOL1Rx and found that it is not cross-reactive toward other closely related APOL family members (Figure 4B). Subcutaneous administration of IONIS-APOL1Rx to APOL1 G0–transgenic mice for 4 weeks resulted in dose-dependent reductions in kidney and liver APOL1 mRNA, exhibiting a median effective dose (ED50) of 26.1 and 5.1 mg/kg/wk, respectively (Figure 4C). Maximum target reductions of 57% and 92% were achieved in the kidney and liver, respectively, with the highest dose in comparison to PBS controls. Moreover, APOL1 plasma protein levels were also dose-dependently reduced with an ED50 of 9.6 mg/kg/wk (Figure 4D) in agreement with liver APOL1 mRNA reduction.
Figure 4. IONIS-APOL1Rx is an APOL1 ASO that reduces APOL1 mRNA levels in vitro and in vivo.
(A) APOL1 expression in A431 cells in vitro (n = 4) was measured by qRT-PCR after 72 hours free uptake of IONIS-APOL1Rx or control ASO and normalized to GAPDH expression. IONIS-APOL1Rx significantly reduced APOL1 expression compared with control ASO. (B) A431 cells were treated with IONIS-APOL1Rx for 72 hours by free uptake (n = 4) and expression of APOL1, APOL2, and APOL3 were measured by qRT-PCR followed by normalization to GAPDH expression. (C and D) APOL1 G0–transgenic mice were treated with IONIS-APOL1Rx 1 time per week for 4 weeks and sacrificed 48 hours after the last dose (n = 3–4). (C) Liver and kidney APOL1 expression was measured by qRT-PCR and normalized to Cyp expression. IONIS-APOL1Rx significantly reduced APOL1 mRNA expression compared with PBS control. (D) Plasma APOL1 levels were measured by ELISA and are shown relative to PBS control levels. IONIS-APOL1Rx significantly reduced plasma APOL1 compared with PBS control. All data are presented as mean ± SD. Two-way ANOVA with Bonferroni’s multiple comparisons test for A and one-way ANOVA with Dunnett’s multiple comparison’s test for C and D, *P < 0.05; ***P < 0.001.
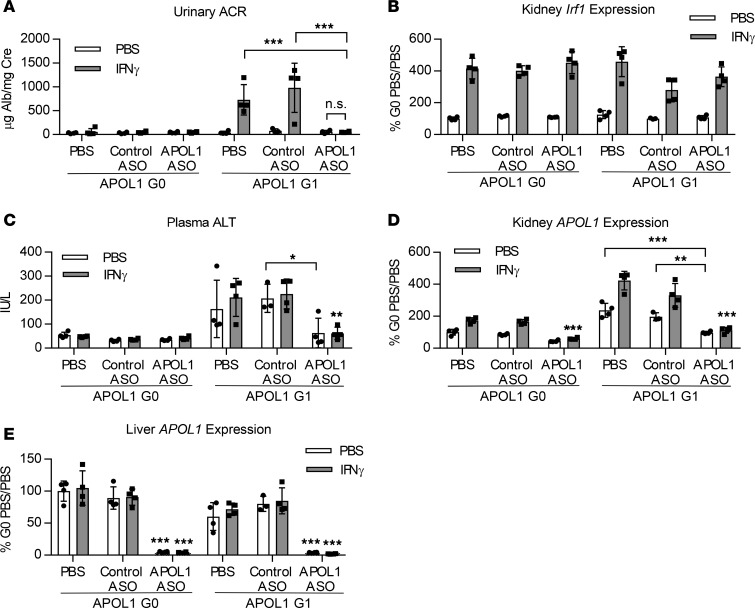
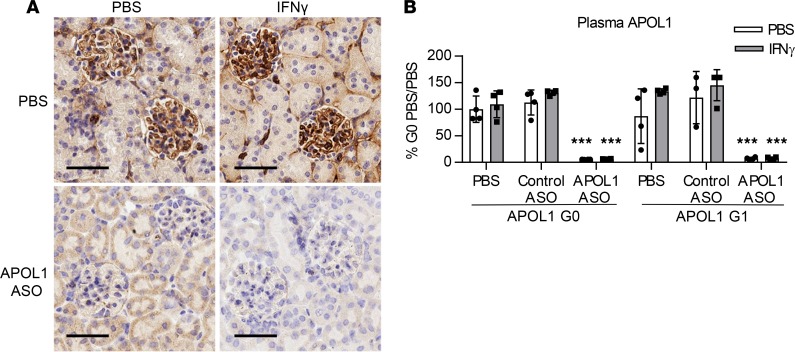
To demonstrate that an ASO targeting APOL1 would be effective in providing protection against IFN-γ–induced proteinuria, APOL1 G0– and G1–transgenic mice were treated with 50 mg/kg/wk IONIS-APOL1Rx or control ASO for 4 weeks prior to IFN-γ challenge. As observed previously, IFN-γ challenge resulted in significant increases in urinary albumin levels in PBS- or control ASO–treated APOL1 G1–transgenic mice 48 hours after IFN-γ administration but not in APOL1 G0 mice (Figure 5A). Treatment with IONIS-APOL1Rx, however, completely protected against IFN-γ–induced proteinuria without affecting Irf1 induction (Figure 5, A and B). Elevations in plasma ALT levels in APOL1 G1 mice were also significantly decreased with IONIS-APOL1Rx treatment (Figure 5C). As expected, IFN-γ administration to PBS- or control ASO–treated G0 or G1 mice resulted in robust inductions of kidney APOL1 mRNA (Figure 5D), while liver APOL1 was unaffected (Figure 5E). Treatment with IONIS-APOL1Rx, on the other hand, significantly prevented IFN-γ–induced APOL1 expression and resulted in an approximately 50%–60% reduction of kidney and 95% reduction of liver APOL1 mRNA levels. Reductions in APOL1 protein were confirmed by IHC and ELISA, respectively (Figure 6). Moreover, protection against IFN-γ–induced proteinuria with IONIS-APOL1Rx was potent and dose dependent (Supplemental Figure 7A), as treatment with doses as low as 6.25 mg/kg/wk IONIS-APOL1Rx that resulted in only 20%–30% reduction of kidney APOL1 mRNA in comparison to control ASO treatment provided significant protection against IFN-γ–induced proteinuria (Supplemental Figure 7, A–C).
Figure 5. Administration of IONIS-APOL1Rx prevents IFN-γ–induced proteinuria.
Female APOL1 G0– and G1–transgenic mice (n = 3–4) were treated with 50 mg/kg IONIS-APOL1Rx or control ASO 1 time per week for 4 weeks and challenged with a single dose of IFN-γ (1.125 × 107 U/kg) or vehicle (PBS). Study endpoints were evaluated 48 hours after IFN-γ challenge. (A) Urine was collected prior to sacrifice 48 hours after IFN-γ challenge, and urinary albumin was measured by ELISA and normalized to urine creatinine. (B) Kidney Irf1 expression was measured by qRT-PCR and normalized to Cyp expression. (C) Plasma ALT levels were measured using a clinical chemistry analyzer. (D) Kidney and (E) liver APOL1 expression were measured by qRT-PCR and normalized to Cyp expression. All data are presented as mean ± SD. Two-way ANOVA with Tukey’s multiple comparisons test, *P < 0.05; **P < 0.01; ***P < 0.001. Unless otherwise indicated, similar statistical significance was achieved when comparing APOL1 ASO–treated groups challenged with PBS or IFN-γ in comparison to respective PBS- or control ASO–treated groups.
Figure 6. Administration of IONIS-APOL1Rx reduces kidney and plasma APOL1 protein levels.
Female APOL1 G0– and G1–transgenic mice (n = 3–4) were treated with 50 mg/kg IONIS-APOL1Rx or control ASO 1 time per week for 4 weeks and challenged with a single dose of IFN-γ (1.125 × 107 U/kg) or vehicle (PBS). Study endpoints were evaluated 48 hours after IFN-γ challenge. (A) IHC analysis of APOL1 protein in kidney tissues from IONIS-APOL1Rx– or PBS-treated APOL1 G1–transgenic mice 48 hours after IFN-γ or PBS challenge. Representative images are shown (scale bar: 50 μm). (B) Plasma APOL1 levels were measured by ELISA. Data are presented as mean ± SD. Two-way ANOVA with Tukey’s multiple comparisons test, ***P < 0.001. Similar statistical significance was achieved when comparing APOL1 ASO–treated groups challenged with PBS or IFN-γ in comparison to respective PBS- or control ASO–treated groups.
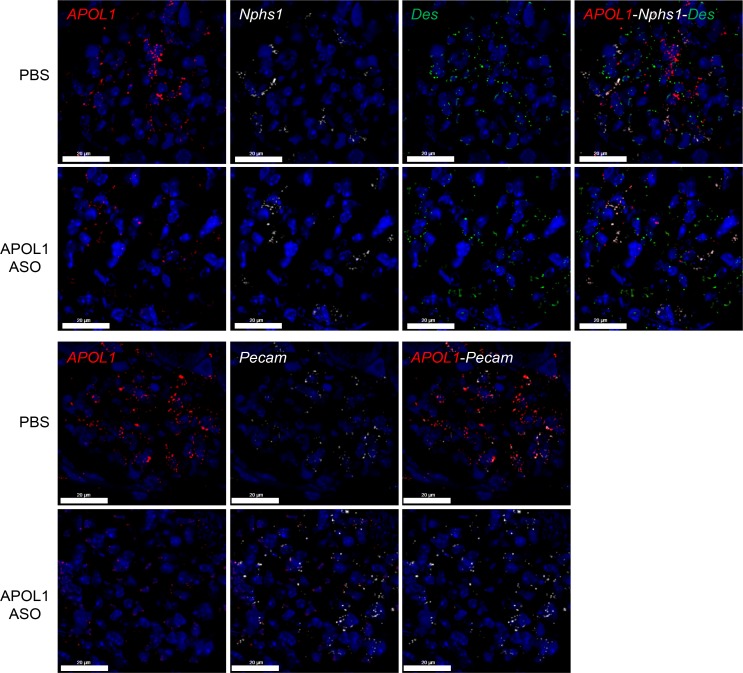
Generation 2.5 ASOs broadly distribute to peripheral organs upon systemic administration, yet sensitivity to ASO treatment varies for different cell types (28). As the kidney is a heterogenous organ, we used FISH to evaluate APOL1 reduction in a cell type–specific manner. We observed substantial reductions of APOL1 mRNA expression in all cell types evaluated, including podocytes, endothelial cells, and mesangial cells, demonstrating potent ASO activity in glomerular cells expressing APOL1 (Figure 7).
Figure 7. IONIS-APOL1Rx reduces APOL1 mRNA expression in kidney glomerular cells.
Female APOL1 G1–transgenic mice (n = 4) were treated with PBS or 50 mg/kg IONIS-APOL1Rx 1 time per week for 4 weeks and challenged with a single dose of IFN-γ (1.125 × 107 U/kg). FISH was used to evaluate APOL1 mRNA expression in podocytes (Nphs1), mesangial cells (Des), and endothelial cells (Pecam) in kidneys from IONIS-APOL1Rx– or PBS-treated APOL1 G1 mice 48 hours after IFN-γ challenge. Representative images are shown (scale bars: 20 μm).
To determine the effect of IONIS-APOL1Rx treatment on a global scale, we performed transcriptome analysis on both whole kidney and liver samples from IFN-γ–challenged APOL1 G1 and G0 mice that had been pretreated with PBS, control ASO, or IONIS-APOL1Rx and compared their transcriptome profiles using significantly (P < 0.05) differentially expressed genes between IFN-γ–challenged PBS-treated G0 mice (G0-PBS mice) and IFN-γ–challenged PBS-treated G1 mice (G1-PBS mice) for each tissue (Supplemental Tables 2 and 3). Principal components analysis demonstrated that IONIS-APOL1Rx treatment was able to shift the kidney transcriptional profiles of G1-PBS and G0-PBS mice closer together, suggesting that by reducing APOL1 levels the expression profiles of G1 mice were partially “normalized” toward those of G0 mice (Supplemental Figure 8A). The kidney transcriptional profiles of control ASO–treated G1 mice were also shifted in relation to the profiles of G1-PBS mice; however, in comparison to the alterations induced by IONIS-APOL1Rx treatment, the control ASO–induced shift did not result in as much overlap with the transcriptional profiles of G0-PBS mice and was likely a reflection of both the heterogeneity of whole kidney and variable response to IFN-γ. Principal components analysis of liver transcriptome profiles even more clearly demonstrates that the expression profiles of IONIS-APOL1Rx–treated G1 mice are partially “normalized” toward those of G0-PBS mice, while control ASO treatment had limited effects (Supplemental Figure 8B).
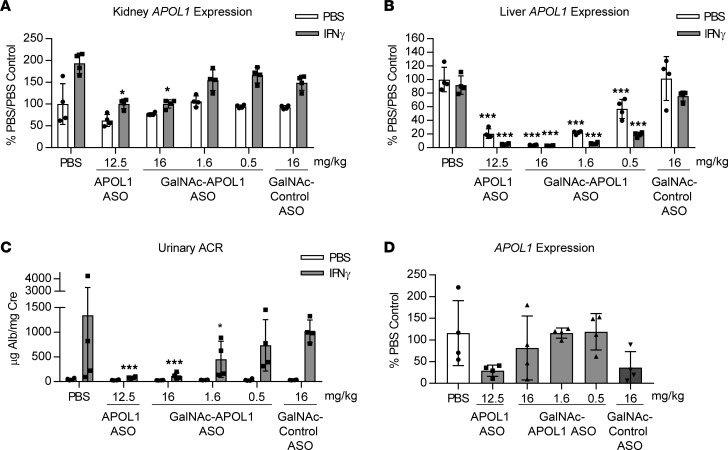
Kidney APOL1 is a significant contributor to IFN-γ–induced proteinuria in APOL1 G1–transgenic mice. Since our model exhibits expression of APOL1 in both liver and kidney and at relative physiological levels to those observed in humans, we used a hepatocyte-targeting version of IONIS-APOL1Rx to address whether liver- or kidney-derived APOL1 contributes to IFN-γ–induced proteinuria. By conjugating ASOs to a GalNAc moiety, which is a high-affinity ligand for the hepatocyte-specific asialoglycoprotein receptor, ASO distribution can be shifted almost entirely toward hepatocytes, resulting in an approximately 10-fold increase in ASO activity for hepatocyte targets in mice (29). GalNAc-IONIS-APOL1Rx and IONIS-APOL1Rx were administered at submaximal doses of 1.6 and 12.5 mg/kg/wk, respectively, for 4 weeks prior to IFN-γ challenge to achieve similar levels of APOL1 mRNA reduction in the liver but differing levels of APOL1 knockdown in kidney (Figure 8, A and B). Additional doses of GalNAc-IONIS-APOL1Rx were also administered to evaluate dose-dependent effects of the GalNAc-ASO on pharmacology. Assessment of urinary albumin levels 48 hours after IFN-γ administration demonstrated that despite reducing liver APOL1 to similar degrees, IONIS-APOL1Rx was more effective than GalNAc-IONIS-APOL1Rx at preventing IFN-γ–induced proteinuria (Figure 8C). Likewise, high-dose GalNAc-IONIS-APOL1Rx treatment (16 mg/kg/wk), which significantly reduced kidney APOL1 mRNA in addition to liver APOL1 mRNA (Figure 8, A and B), significantly prevented IFN-γ–induced proteinuria to a similar degree as IONIS-APOL1Rx (Figure 8C). Interestingly, despite having no effect on kidney APOL1 mRNA at 1.6 mg/kg/wk (Figure 8A), GalNAc-IONIS-APOL1Rx significantly, but incompletely, reduced IFN-γ–induced proteinuria in comparison to control groups (Figure 8C), suggesting that liver APOL1 may play a minor role in APOL1 nephropathies, albeit to a much lesser degree than kidney APOL1.
Figure 8. Hepatocyte-targeted IONIS-APOL1Rx provides incomplete protection against IFN-γ–induced proteinuria.
Female APOL1 G1–transgenic mice (n = 4) were treated with IONIS-APOL1Rx, hepatocyte-targeted IONIS-APOL1Rx, or control ASO 1 time per week for 4 weeks and challenged with a single dose of IFN-γ (1.125 × 107 U/kg) or vehicle (PBS). Study endpoints were evaluated 48 hours after IFN-γ challenge. (A) Kidney and (B) liver APOL1 expression were measured by qRT-PCR and normalized to Cyp expression. (C) Urine was collected prior to sacrifice 48 hours after IFN-γ challenge, and urinary albumin was measured by ELISA and normalized to urine creatinine. (D) Urinary shed cells were collected from PBS-challenged mice, and APOL1 expression was measured by qRT-PCR and normalized to 36B4/Rplp0 expression. All data are presented as mean ± SD. Statistics were performed (A–C) by comparing each APOL1 ASO–treated PBS- or IFN-γ–challenged group to the respective control ASO group. Two-way ANOVA with Tukey’s multiple comparisons test, *P< 0.05; ***P < 0.001.
APOL1 mRNA expression in urinary shed cells may be a novel kidney-specific target engagement biomarker.
Target engagement biomarkers are critical for clinical drug development, enabling measurement of drug activity in humans. While circulating APOL1 protein is an obvious biomarker candidate for liver-derived APOL1, biomarkers for kidney-derived APOL1 have not yet been identified. To establish a potentially direct assessment of APOL1 mRNA expression in kidney, we measured APOL1 mRNA in urinary shed cells collected from our APOL1-transgenic mice. Urinary shed cells are a mixture of various cell types composed of podocytes, proximal tubule epithelial cells, and kidney stem/progenitor cells (30). We explored whether expression of APOL1 mRNA in urinary shed cells could be used as a noninvasive method to measure APOL1 mRNA levels in kidney. Analysis of urinary shed cells collected from IONIS-APOL1Rx–treated APOL1 G0–transgenic mice demonstrated reductions in APOL1 mRNA levels that correlated with APOL1 mRNA reductions in whole kidney (Supplemental Figure 9, A and B). To validate that urinary shed cell APOL1 mRNA is a biomarker of kidney-derived APOL1, we measured APOL1 expression in urinary shed cells collected from APOL1 G1–transgenic mice that had been treated with IONIS-APOL1Rx or GalNAc-IONIS-APOL1Rx. Although not statistically significant due to variability observed in the PBS group, samples collected from IONIS-APOL1Rx–treated mice demonstrated a reduction in APOL1 mRNA in urinary shed cells, while those collected from mice treated with GalNAc-IONIS-APOL1Rx at doses that did not reduce kidney APOL1 (but did reduce liver APOL1) exhibited similar APOL1 levels to those of the PBS control group, correlating with whole kidney APOL1 mRNA levels (Figure 8, A and D). Importantly, evaluation of APOL1 mRNA expression in urinary shed cells isolated from healthy individuals revealed that it can also be robustly detected in humans (Supplemental Figure 9C).
Autophagy suppression in IFN-γ–challenged APOL1 G1–transgenic mice is reduced with IONIS-APOL1Rx treatment. APOL1 risk variants have been shown to alter autophagic flux in vitro and in vivo (16, 17, 31, 32), although the mechanism by which these polymorphisms affect autophagy has yet to be elucidated. To determine whether autophagy is perturbed in genomic APOL1 G1–transgenic mice challenged with IFN-γ, liver and whole kidney samples from APOL1 G0 and G1 mice challenged with and without IFN-γ were evaluated for autophagy activity by Western blot. Indeed, autophagy was significantly suppressed in APOL1 G1 livers 24 hours after IFN-γ challenge, as indicated by increases in the LC3BII/I ratio and accumulation of p62, while APOL1 G0 livers were unaffected (Supplemental Figure 10, A and B). To determine whether treatment with IONIS-APOL1Rx restores autophagy, we evaluated autophagic activity in liver samples from PBS, control ASO, and IONIS-APOL1Rx–treated APOL1 G0 and G1 mice that had been challenged with and without IFN-γ for 48 hours, the time point at which we see the greatest effect on proteinuria. While IFN-γ challenge caused greater p62 accumulation in APOL1 G1 mice in comparison to G0 mice, IONIS-APOL1Rx treatment reduced p62 levels in both APOL1 G0 and G1 mice, suggesting that ASO treatment was able to decrease IFN-γ–induced suppression of autophagy in liver of genomic APOL1-transgenic mice (Supplemental Figure 11, A and B). Similarly, in whole kidney tissue we observed suppressed autophagy 24 hours after IFN-γ challenge in APOL1 G1 mice, in comparison to APOL1 G0 mice, that was evident from LC3BII/I ratios, but less clear when evaluating accumulation of p62 levels (Supplemental Figure 10, C and D). We were unable to detect suppression of autophagy in whole kidney 48 hours after IFN-γ challenge, likely due to both the heterogenous nature of the kidney and the fact that APOL1 mRNA induction was on the decline 48 hours after IFN-γ administration in comparison to that at 24 hours (Supplemental Figure 11, C and D).
Discussion
The identification of the APOL1 risk alleles and their strong association with various forms of nondiabetic nephropathy, including HIVAN and FSGS, in CKD patients of African descent was a breakthrough discovery in CKD pathogenesis, accounting for the disproportionately high number of African Americans developing end-stage renal disease in comparison to other ethnic groups (1, 3). While the prevalence of HIVAN has decreased with widespread use of HAART therapy, FSGS remains a significant unmet medical need with limited therapeutic options (33). Currently, treatment options for FSGS patients are largely limited to nonspecific antiinflammatory agents, such as corticosteroids, and are often administered in combination with immunomodulatory drugs, such as calcineurin inhibitors. This aggressive treatment paradigm is accompanied by adverse side effects, poor remission, and high rates of relapse, as determined by proteinuria levels (34). Ultimately, many patients progress toward end-stage renal disease and dialysis, demonstrating a need for more effective therapies that target the underlying cause of the disease. As up to 70% of African Americans with FSGS harbor 2 APOL1 risk alleles (3), a therapeutic targeting APOL1 itself may be effective in reducing proteinuria and preventing disease progression in this patient population. Moreover, as APOL1 risk alleles have been associated with other forms of nondiabetic nephropathy, such as hypertensive-attributed end-stage renal disease, lupus nephritis, and sickle cell nephropathy, an APOL1 therapeutic may potentially prove to be beneficial in these diseases as well, collectively providing a rare opportunity for personalized medicine in CKD.
Whether inhibiting APOL1 therapeutically will be effective in these disease settings depends heavily on whether the G1/G2 polymorphisms are loss-of-function or toxic gain-of-function mutations. A case report of an APOL1-null individual, identified by Trypanosoma evansi (T. evansi) infection, a trypanosome strain normally sensitive to normal serum, was described as having no renal abnormalities and appeared otherwise healthy, despite lacking APOL1 protein (35–37). Moreover, the fact that APOL1 is not an evolutionarily conserved protein (10) also suggests that its function is not essential for kidney development or homeostasis. While these lines of reasoning support that APOL1 G1/G2 polymorphisms are not inhibiting APOL1 function and that reducing APOL1 therapeutically will not have any detrimental effects other than increasing susceptibility to trypanosomiasis in specific geographical regions, they do not illustrate that these mutations are toxic gain of function.
In doxycycline-inducible podocyte-specific APOL1-transgenic mice, expression of APOL1 G1/G2, but not APOL1 G0, was shown to cause proteinuria and global/segmental glomerulosclerosis, suggesting that APOL1 risk variants cause renal disease via a toxic gain of function, as mice lack the APOL1 gene (16). As APOL1 expression in these mice is driven exclusively by the Nephrin promoter, APOL1 levels are likely exceedingly high and supraphysiological. In contrast, in constitutive podocyte-specific APOL1-transgenic mice, expression of the APOL1 risk variants did not cause proteinuria or glomerulosclerosis (15), perhaps due to lower APOL1 expression. Here, we generated a genomic APOL1-transgenic mouse model driven by the endogenous human APOL1 promoter resulting in expression of APOL1 G0 or G1 in similar tissues as those identified in humans and at similar relative levels of expression. Furthermore, these mice contain the endogenous human APOL1 regulatory elements, enabling induction of APOL1 expression by physiologically relevant stimulants. As APOL1 is expressed at low levels in the kidneys of unchallenged mice, naive APOL1 G1–transgenic mice do not develop a renal phenotype. This observation agrees with the fact that not all homozygous APOL1 risk allele carriers develop kidney disease and that a second hit is required. A single administration of IFN-γ, on the other hand, which has been shown to upregulate APOL1 expression in vitro (6), is sufficient to induce robust proteinuria only in APOL1 G1 mice, despite upregulating kidney APOL1 expression in both G0 and G1 mice. This effect is specific to the G1 genotype, further supporting that the APOL1 risk alleles result in a toxic gain of function once expression surpasses a certain threshold and reconciling the differences in phenotype observed for the 2 podocyte-specific APOL1-transgenic mice. Moreover, these data suggest that reducing APOL1 may be an effective approach to treating APOL1 nephropathies.
IONIS-APOL1Rx is a first-in-class generation 2.5 APOL1 ASO therapeutic that is systemically administered to reduce APOL1 mRNA levels through RNase H1–mediated degradation. Administration of IONIS-APOL1Rx to genomic APOL1-transgenic mice significantly reduces APOL1 liver and kidney mRNA and protein and prevents IFN-γ–induced proteinuria in APOL1 G1 mice. As the mechanism of renal pathogenesis and the different cell types involved in APOL1 nephropathies remain unclear, it was important for us to develop a physiological model of APOL1-mediated renal disease, as described here, and demonstrate efficacy with an APOL1 ASO as proof of concept. The robust and dose-dependent prevention of IFN-γ–induced proteinuria with IONIS-APOL1Rx demonstrates that the disease-relevant cell types are indeed sensitive to ASO activity and that the efficacious doses are likely to be achievable in humans (38).
It is becoming increasingly clear that an environmental second hit, such as IFN, is required to induce APOL1 nephropathy in homozygous risk allele carriers (39), likely by upregulating APOL1 expression. In fact, many manifestations of nondiabetic nephropathy strongly associated with APOL1 risk alleles are subtypes that develop within the context of high IFN, including HIVAN, lupus nephritis, and IFN therapy–related glomerulopathy (40). Interestingly, these diseases are all characterized by collapsing glomerulopathy, which may be a pathological feature of the IFN second hit and the pattern of APOL1 induction. Whether additional second hits result in differing patterns of APOL1 induction and, consequently, other subtypes of APOL1 nephropathy, remains to be determined. As the genomic APOL1-transgenic mice described here contain the endogenous human regulatory elements of APOL1, they will be useful in addressing these questions and will be a focus of future studies.
In genomic APOL1-transgenic mice, IFN-γ causes robust induction of APOL1 in glomerular endothelial cells and lower levels of induction in podocytes. While expression of APOL1 G1/G2 in mouse podocytes has been shown to be sufficient to induce renal pathology resembling human disease (16), it is unclear what role, if any, APOL1 expression in glomerular endothelial cells is having in APOL1 nephropathies. APOL1 contains a signal peptide and is the only protein of a family of highly conserved APOL proteins that is secreted (10). One intriguing idea is that APOL1 G1/G2 may be driving renal disease both in an autocrine and paracrine manner in podocytes. This hypothesis is supported by previous work that demonstrated that human podocytes can uptake recombinant APOL1 in vitro and may take up APOL1 protein in a paracrine manner physiologically as well (23). An alternative hypothesis for the role of APOL1 in glomerular endothelial cells in APOL1 nephropathies is that APOL1 G1/G2 drives renal disease by acting on both glomerular endothelial cells and podocytes, as injury to glomerular endothelial cells has been shown to precede podocyte damage and proteinuria (41–43). Whether induced expression of APOL1 risk variants strictly in endothelial cells in mice is sufficient to recapitulate features of human APOL1 nephropathies, in particular arterionephrosclerosis, is an interesting question worth pursuing. Importantly, regardless of which cell type is critical for renal disease development, IONIS-APOL1Rx reduces APOL1 expression in all glomerular cells in genomic APOL1-transgenic mice, demonstrating robust activity in both endothelial cells and podocytes.
APOL1 is present at high concentrations in the serum, which is predominantly due to secretion from the liver (12, 44). Data from transplant studies revealed that kidneys from African American deceased donors homozygous for APOL1 risk alleles are associated with higher risk for allograft failure after transplantation (45, 46), while transplant outcome did not depend on recipient APOL1 genotype (47). These data suggest that endogenous kidney APOL1 is the critical factor in APOL1 nephropathy, but it remains to be determined whether there is any contribution from liver-derived APOL1 in circulation. We took advantage of the systemic and physiological expression of APOL1 in our model to directly address this question. The first clue that liver APOL1 is not the main driver in APOL1 nephropathy comes from the fact that liver APOL1 mRNA and, as a result, APOL1 protein in circulation are not induced by IFN-γ administration in genomic APOL1-transgenic mice. This is in agreement with reports showing that plasma APOL1 levels do not correlate with renal disease (48, 49). Additionally, comparison between IONIS-APOL1Rx and a GalNAc-conjugated version of IONIS-APOL1Rx, which results in preferential ASO uptake by hepatocytes (29), resulted in incomplete protection of IFN-γ–induced proteinuria by the GalNAc-conjugated APOL1 ASO. These data suggest that kidney APOL1 plays a critical role in APOL1 nephropathy, but we cannot rule out a minor contribution from liver-derived circulating APOL1. As IONIS-APOL1Rx is effective at significantly inhibiting both liver and kidney APOL1, both tissue sources of APOL1 will be affected resulting in maximal efficacy.
In search of a kidney-specific target engagement biomarker for IONIS-APOL1Rx, we evaluated APOL1 mRNA levels in urinary shed cells, which contain podocytes and therefore can enable noninvasive measurements of APOL1 target reduction in a disease-relevant cell type. Comparison of urinary cell APOL1 mRNA levels measured from APOL1-transgenic mice treated with GalNAc-conjugated or unconjugated IONIS-APOL1Rx demonstrated that urinary cell APOL1 correlates with kidney, and not liver, APOL1 mRNA levels. As we were able to readily detect APOL1 mRNA in human urinary cells, APOL1 mRNA in these cells may be utilized as a viable clinical biomarker for kidney APOL1 target reduction.
An interesting finding in naive genomic APOL1 G1–transgenic mice was the elevation in ALT in mice from 2 different APOL1 G1 founder lines. While APOL1 risk alleles have not been reported to be associated with liver injury, the elevations observed in these mice are modest and variable, such that they may be overlooked in humans as within normal range. As a result, a close analysis of ALT levels in APOL1 risk allele carriers in comparison to those that lack these alleles has likely not been performed and may be warranted. Interestingly, hydrodynamic gene delivery (HGD) of APOL1 G1 to mice resulted in widespread liver necrosis and macrophage infiltration, while delivery of APOL1 G0 did not drastically affect liver pathology (50). How APOL1 risk variants cause toxicity remains unclear; however, several different mechanisms have been proposed, including altered autophagic flux due to effects on endolysosomal trafficking (16, 17, 31, 32, 51). Our data support this hypothesis, as we observed enhanced suppression of autophagy following IFN-γ challenge in APOL1 G1 mice in comparison to APOL1 G0 mice. This effect is clearer in the liver than in the kidney, likely due to heterogeneity in the kidney. Future studies are needed to elucidate this further. Importantly, reduction of APOL1 with IONIS-APOL1Rx reduces plasma ALT elevations and suppression of autophagy upon IFN-γ challenge in liver, indicating that ALT elevations and enhanced suppression of autophagy in APOL1 G1 mice is APOL1 dependent. While other mechanisms of APOL1 risk allele–mediated toxicity, including inflammasome activation and altered electrolytic flux, have also been proposed, IONIS-APOL1Rx, by inhibiting APOL1 itself, will likely be able to suppress all modes of toxicity, including PKR activation by APOL1 G1/G2 RNA stem loops (52).
One limitation of our study is that this is an acute model of IFN-γ–induced proteinuria that lacks kidney pathology, and, as such, we could only evaluate IONIS-APOL1Rx in a prevention treatment paradigm. The transient regulation of APOL1 by IFN-γ, as shown by our data, likely necessitates chronic administration of IFN-γ to maintain APOL1 levels and increase the likelihood of developing kidney pathology. While this strategy will be the focus of future studies, there are compelling preclinical and clinical data suggesting that reduction of APOL1 expression may also be effective in a therapeutic setting. Removal of doxycycline after establishment of severe glomerulopathy in doxycycline-inducible podocyte-specific APOL1 G1/G2 mice was sufficient to reduce proteinuria (16). Furthermore, it has been shown that APOL1 risk allele carriers that develop IFN therapy–mediated collapsing glomerulopathy exhibit disease reversal upon cessation of IFN therapy (6, 24), likely by removal of the second hit that induces APOL1 expression. Together, these data support the hypothesis that, if APOL1 expression is reduced with IONIS-APOL1Rx treatment, disease progression will be halted and may even be reversed.
In summary, we developed a physiologically relevant genomic APOL1-transgenic mouse model that only exhibits a renal disease phenotype upon exposure to a clinically relevant second hit, such as IFN-γ, and exhibits upregulation of kidney APOL1 G1 expression in various cell types, including glomerular endothelial cells and podocytes. We also discovered what we believe to be the first APOL1 inhibitor, IONIS-APOL1Rx, which is a potent APOL1 antisense therapeutic inhibiting APOL1 expression systemically in both liver and kidney. IONIS-APOL1Rx treatment robustly and dose-dependently ameliorated IFN-γ–induced proteinuria in APOL1 G1 mice, suggesting that IONIS-APOL1Rx may be an effective therapeutic for CKD patients with APOL1 nephropathies and warrants further development.
Methods
Generation of transgenic mice.
Genomic APOL1 G0–transgenic mice were generated by pronuclear injection of C57BL/6NTac fertilized eggs (performed by University of California Irvine Transgenic Mouse Facility) with a Pac I–digested APOL1-containing fragment of fosmid ABC12-49114000M18. This approximately 32-kb fragment contained the APOL1 gene as well as approximately 5 kb upstream and approximately 12 kb downstream regions. Fosmid ABC12-49114000M18 was mutated to introduce the G1 S342G and I384M point mutations and was used in a similar manner to generate APOL1 G1–transgenic mice. Transgenic mice were identified by genomic PCR analysis using primers specific for human APOL1 (forward primer 5′-AGTCATGGAAGTGTGGCTGA-3′, reverse primer 5′-TATATGGGGTCTTCTGCCGG-3′). Complete transgene integration was measured by genomic PCR analysis using primers that spanned the fosmid fragment. Two founders from each genotype (APOL1 G0 and G1) were backcrossed to C57BL/6J mice, and the transgenic progeny were characterized in comparison to WT littermate controls. Breeding was performed at The Jackson Laboratory.
ASOs.
Generation 2.5 ASOs used in this study were 16 nucleotides in length and chemically modified with phosphorothioate in the backbone and 2′-O-methoxyethyl (MOE) and/or 2′-4′ constrained ethyl (cEt) in the wings. Synthesis of Gen 2.5 ASOs was previously described (53). For hepatocyte-targeted ASOs, ASOs were conjugated to triantennary N-acetylgalactosamine (GalNAc3) as previously described (54). ASOs were dissolved in PBS (without Mg2+ or Ca2+; Life Technologies) for in vivo administration.
Mouse experiments.
Mice were housed in microisolator cages on a constant 12-hour light/dark cycle, with controlled temperature and humidity and food and water provided ad libitum. For IFN-γ challenge studies, mice were administered a single intravenous dose (1.125 × 107 U/kg in PBS) of recombinant murine IFN-γ (BioLegend) and sacrificed at the time points indicated. For ASO studies, mice were administered ASO once weekly via subcutaneous administration for 4 weeks. WT control mice used consisted of WT littermates of APOL1-transgenic mice. The Ionis Pharmaceuticals vivarium has been accredited by Assessment and Accreditation of Laboratory Animal Care.
Cell culture experiments.
Human A431 cells (ATCC) were cultured in growth medium (DMEM + 10% FBS; Thermo Fisher) at 37°C and 5% CO2. Cells were trypsinized, counted, and diluted to 1.1 × 105 cells/mL in room temperature growth medium before adding 100 μL cell suspension to wells of collagen I–coated 96-well culture plates. Immediately after plating the cells, 11 μL 10× ASO in water was added to the appropriate wells. Four replicate 96-well plates were treated, and the cell culture plates were incubated at 37°C and 5% CO2. After 48 hours, cells were washed once with PBS before lysing for RNA isolation. Before purifying RNA, lysates from 2 replicate plates were combined to increase RNA yield. RNA was purified using a glass fiber filter plate (Pall) and chaotropic salts. mRNA expression levels were quantitated with quantitative reverse transcription PCR (qRT-PCR) on the 7900HT and QuantStudio 7 instruments (Applied Biosystems) using the following primer probe sets: human APOL1, forward primer 5′-GCTACTCCTGCTGACTGATAATG-3′, reverse primer 5′-AAGGTTGTCCAGAGCTTTACG-3′, probe 5′-FAM-TGCCCAGGAATGAGGCAGATGAG-IOWA-BLACK-3′; human APOL2, forward primer 5′-GATCCACACAGCTCAGAACA-3′, reverse primer 5′-TTCCTCTTCCCTCACTCTCA-3′, probe 5′-FAM-TTGGACAAGAGGACCCTGCCTTG-IOWA-BLACK-3′; human APOL3, forward primer 5′-CTGAATTGCCCAGGGATGA-3′, reverse primer 5′-CTCATCTTTCTGCTGCACATATTC-3′, probe 5′-FAM-AGAGCTTCGTAGAGAGCATCTGCC-IOWA-BLACK-3′; and human GAPDH, forward primer 5′-GAAGGTGAAGGTCGGAGTC-3′, reverse primer 5′-GAAGATGGTGATGGGATTTC-3′, probe 5′-VIC-CAAGCTTCCCGTTCTCAGCC-TAMRA-3′. Briefly, 5 μL qRT-PCR reactions containing 1 μL RNA were run using AgPath-ID One-Step RT-PCR Reagent (Life Technologies) following the manufacturer’s recommendations. Reactions were set up with the ECHO liquid handler (Labcyte). Relative gene expression was normalized to expression of GAPDH housekeeping gene. Data are presented as percentage untreated control. GraphPad Prism (v6.01) was used for curve fitting and IC50 calculations.
qRT-PCR analysis of mouse tissue samples.
Mouse tissues were homogenized in Guanidine Isothiocyanate solution (Life Technologies) containing 8% β-mercaptoethanol (MilliporeSigma), and total RNA was isolated using the RNeasy 96 Kits (Qiagen) according to the manufacturer’s recommendations. TaqMan qRT-PCR was performed on StepOnePlus Real-Time PCR Systems (Applied Biosystems) using One-Step SuperScript qRT-PCR kits (Life Technologies). TaqMan Gene Expression Assays were used to measure mouse Irf1 (Mm01288580_m1, Life Technologies), mouse Wt1 (Mm01337048_m1, Life Technologies), and mouse Nphs1 (Mm01176615_g1, Life Technologies) mRNA expression. The sequences of additional primers/probes used in this study are as follows: human APOL1, forward primer 5′-GGCAGCCTTGTACTCTTGGAA-3′, reverse primer 5′-GCTGGTAATCCCGGTCAAAG-3′, probe 5′-FAM-CTGGGATGGAGTTGGGAATCACAGCC-TAMRA-3′ and mouse CypA, forward primer 5′-TCGCCGCTTGCTGCA-3′, reverse primer 5′-ATCGGCCGTGATGTCGA-3′, probe 5′-FAM-CCATGGTCAACCCCACCGTGTTC-TAMRA-3′. When relative gene expression was compared across different tissue types, expression was normalized to total RNA, as measured by Quant-iT RiboGreen RNA assay (Molecular Probes); when relative gene expression was measured for a single tissue, then expression was normalized to the Cyp housekeeping gene.
Western blot analysis.
Mouse liver and kidney tissues were processed with T-PER Tissue Protein Extraction Reagent (Life Technologies) supplemented with 1× Halt Protease Inhibitor Cocktail, EDTA-Free (Life Technologies) and 1× Halt Phosphatase Inhibitor Cocktail (Life Technologies). Tissue samples were homogenized with Lysing Beads - Matrix D tubes (MP Biomedicals) using a FastPrep-24 5G Homogenizer (MP Biomedicals), and protein concentration was determined by Lowry DC protein assay (Bio-Rad). Samples were denatured in LDS (Life Technologies) supplemented with 4% β-mercaptoethanol (MilliporeSigma) at 75°C for 10 minutes. Equivalent protein amounts were loaded on Novex 4%–12% Bis-Tris SDS-PAGE gels (Life Technologies) and resolved. Proteins were transferred to 0.4-μm nitrocellulose or 0.2-μm PVDF membranes using standard methods, and membranes were blocked in StartingBlock T20 (TBS) Blocking Buffer (Life Technologies). Primary antibodies were diluted in blocking solution and were incubated with membranes at 4°C overnight. IFR fluorophore–conjugated secondary antibodies (LI-COR) were diluted in blocking solution and incubated with membranes at room temperature for 1 hour. Western blots were detected with the OdysseyCLx imager (LI-COR). Protein quantification was performed using Image Studio (LI-COR) analysis software, and band densities were normalized to α-tubulin (11224-1-AP; Proteintech Group). Additional antibodies used include the following: polyclonal anti-APOL1 (HPA018885-100UL, MilliporeSigma), polyclonal anti-LC3B (ab48394; Abcam), and polyclonal anti-p62/SQSTM1 (C-terminus; GP62-C, Progen). See complete unedited blots in the supplemental material.
Plasma chemistry analysis.
Mouse blood samples were collected via cardiac puncture and transferred to collection tubes containing EDTA. Samples were spun down to separate plasma, and plasma ALT, BUN, and creatinine were measured using the AU480 Clinical Chemistry Analyzer (Beckman Coulter). Plasma APOL1 levels were measured using an APOL1-specific ELISA (Proteintech) following manufacturer’s recommendations.
Urine chemistry analysis.
Mouse urine albumin levels were measured using an albumin-specific ELISA (Alpco). Mouse urine creatinine was measured using the AU480 Clinical Chemistry Analyzer (Beckman Coulter).
RNA in situ hybridization.
RNA in situ hybridization experiments were performed using RNAscope, a previously described RNA in situ hybridization technique (55). Briefly, mouse FFPE kidney samples were sectioned at 5 μm onto SuperFrost Plus slides. Each sample was evaluated for RNA integrity with a 3-plex positive control probe (catalog 320888 for mouse, Advanced Cell Diagnostics) and a 3-plex dapB negative control probe (catalog 320878, Advanced Cell Diagnostics). Using the positive and negative control probes, optimization was performed to establish the following pretreatment conditions: 15-minute epitope retrieval at 95°C and 15-minute Protease III (catalog 322340, Advanced Cell Diagnostics) at 40°C. The RNAscope Fluorescent Multiplex Reagent Kit (Advanced Cell Diagnostics) was used according to the manufacturer’s instructions. Paired double-Z oligonucleotide probes were designed against target RNA using custom software to specifically detect the genes of interest. The following probes (all from Advanced Cell Diagnostics) were used in this study: Mm-Nphs1-C3 (catalog 433578-C3), Mm-Des (catalog 407928), Mm-Pecam1-C3 (catalog 316728-C3), and Hs-APOL1-No-XMm-C2 (catalog 459798-C2). Probes were assigned to the following fluorophore configurations: channel 1 (C1) in fluorescein, channel 2 (C2) in Cy3, and channel 3 (C3) in Cy5. Tissues were counterstained with a DAPI nuclear stain prior to adding coverslips. Staining was performed at ACD Newark by the Pharma Assay Services group.
Immunohistochemistry.
Mouse FFPE kidney samples were sectioned at 4 μm, dried overnight, and baked at 60°C. Slides were deparaffinized, hydrated in ethyl alcohol, and washed in tap water using the HistoCore Spectra ST Autostainer (Leica Biosystems). Antigen retrieval was carried out in Tris-EDTA pH 9.0 in a 98°C incubator for 20 minutes using the PT Module (Thermo Fisher). The remaining IHC protocol was carried out using the Autostainer 720 (Thermo Fisher). Endogenous peroxidase activity was blocked in 3% H2O2 for 10 minutes, and endogenous biotin was blocked with Background Buster (Innovex Biosciences). Sections were blocked in 2% BSA + 5% normal goat serum in 1× TBS-Tween for 1 hour at room temperature. Slides were incubated at room temperature for 1 hour with anti-APOL1 primary antibody (11486-2-AP, Proteintech or HPA018885-100UL, MilliporeSigma; dilution 1:500), followed by goat anti-rabbit HRP secondary antibody (111-036-003, Jackson ImmunoResearch; dilution 1:200) for 30 minutes. Staining was visualized using DAB.
qRT-PCR analysis of urinary shed cells.
Human urine (200 mL) was collected from 5 healthy donors and centrifuged for 15 minutes at 4°C. The supernatant was discarded, and the pelleted urinary shed cells were lysed in 350 μl Buffer RLT (Qiagen). Total RNA was isolated with the RNeasy Micro Kit (Qiagen) using the QIAcube instrument (Qiagen), according to manufacturer’s instructions, including a DNase digestion step. RNA concentration was measured using a Nanodrop spectrophotometer (Saveen Werner). Reverse transcription was performed on a Gradient Thermal Cycler (MJ Research) using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) following manufacturer’s recommendations. APOL1 mRNA expression was measured by qPCR using TaqMan Gene Expression Assay (Hs01066280_m1, Applied Biosystems) and normalized using expression of the reference gene, 36B4/RPLP0 (MilliporeSigma) and calculated using 2-dCt. The sequences of primers/probe for human 36B4/RPLP0 are as follows: forward primer 5′-CCATTCTATCATCAACGGGTACAA-3′, reverse primer 5′-AGCAAGTGGGAAGGTGTAATCC-3′, probe (VIC-TAMRA) 5′-TCTCCACAGACAAGGCCAGGACTCGT-3′. Total RNA from urinary shed cells collected from APOL1-transgenic mice was extracted and subjected to reverse transcription as described above. Quantitative PCR was also performed as above, except the reference gene used was mouse 36B4/Rplp0. The sequence of primers/probe for mouse 36B4/Rplp0 are as follows: forward primer 5′-GAGGAATCAGATGAGGATATGGGA-3′, reverse primer 5′-AAGCAGGCTGACTTGGTTGC-3′, probe (VIC-TAMRA) 5′-TCGGTCTCTTCGACTAATCCCGCCAA-3′.
Transcriptional profiling and analysis.
Digital gene expression analysis was performed by sequencing 3′-enriched fragment libraries from purified total RNA. Briefly, first-strand synthesis was performed using custom, barcoded oligo-dT priming. Libraries were synthesized with a 3′ QuantSeq multiplexing kit (Lexogen) using random hexamers and barcoded amplification. Pair-barcoded samples were sequenced (60 base pair reads) on a NextSeq 500 (Illumina) to an average depth of 5.2 million uniquely mapped reads per sample. Demultiplexed reads were mapped using STAR alignment tool version 2.5.1b with the following run parameters: outFilterMultimapNmax 10, outFilterMatchNmin 23, seedSearchStartLmax 50. Aligned reads showed an average mapping rate of 75%. Transcript quantification was performed using Salmon version 0.7.1 using quasimapping-based mode with automated libtype detection (56, 57). Expression values are reported as transcripts per million (TPM) and were computed for each gene by aggregating transcript counts and normalizing by total number of mapped reads. For alignment and expression, gene model indexes were built upon Ensembl Mus musculus build-88 cDNA sequences. Differentially expressed genes, in comparison to gene expression levels in the reference cohorts, were identified by assuming a negative binomial distribution-derived model of gene-expression variance. Each comparison applied the variance model symmetrically, alternating both case and control groups as a reference for gene variation, followed by list aggregation. Gene-specific P values for each biological replicate were calculated and median aggregated. Genes having a minimum P value of less than or equal to 0.05 in all replicates within a group were considered significant and used in downstream analysis. All data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE121286.
Statistic.
All data are presented as mean ± SD unless otherwise indicated. Statistical analyses were performed by either 1-way or 2-way ANOVA with multiple comparison’s tests as indicated. Differences were considered statistically significant at P < 0.05 and not significant at P > 0.05.
Study approval.
All animal husbandry and procedures were approved by the Institutional Animal Care and Use Committee at Ionis Pharmaceuticals.
Author contributions
M. Aghajan was involved in study conception and design; acquisition, analysis, and interpretation of data; study supervision; and writing of the manuscript. SLB was involved in acquisition and analysis of data for in vivo studies. M. Althage was involved in conception and design of biomarker experiments, analysis and interpretation of data, study supervision, and review of the manuscript. CEH was involved in analysis and interpretation of transcriptome data and review of the manuscript. AE and CA were involved in design of in vivo studies, interpretation of data, and review of the manuscript. IM, JO, AML, SK, and DG were involved in acquisition and analysis of data. JH was involved in review of the manuscript. ATW was involved in design of in vitro experiments; acquisition, analysis, and interpretation of data; and study supervision. JAE was involved in histopathological analysis and review of the manuscript. BPM was involved in conception and supervision of the study and review of the manuscript. MCM was involved in conception and design of biomarker experiments, data interpretation, study supervision, and review of the manuscript. SG was involved in study conception and design, data interpretation, study supervision, and review of the manuscript.
Supplementary Material
Acknowledgments
We thank S. Freier (Ionis Pharmaceuticals) for design of human APOL1 ASOs; TaconicArtemis GmbH for generating the APOL1 G1 fosmid fragment; S. Murray (Ionis Pharmaceuticals) and the University of California Irvine Transgenic Mouse Facility for assistance with transgenic mice generation; Advanced Cell Diagnostics Pharma Assay Services for RNA in situ hybridization; B. DeBrosse-Serra and X. Xiao (Ionis Pharmaceuticals) for immunohistological procedures; and W. Sullivan (Ionis Pharmaceuticals) for graphics support. Funding for this study was provided by Ionis Pharmaceuticals and AstraZeneca.
Version 1. 06/20/2019
Electronic publication
Footnotes
MCM’s present address is: Janssen Research & Development, Boston, Massachusetts, USA.
Conflict of interest: MA, SLB, CEH, JO, SK, DG, ATW, JAE, BPM, and SG are employees and shareholders of Ionis Pharmaceuticals. MA, AE, IM, AML, JH, CA, and MCM are or were employees and shareholders of AstraZeneca.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(12):e126124. https://doi.org/10.1172/jci.insight.126124.
Contributor Information
Mariam Aghajan, Email: maghajan@ionisph.com.
Magnus Althage, Email: Magnus.Althage@astrazeneca.com.
Christopher E. Hart, Email: chart@ionisph.com.
Anette Ericsson, Email: Anette.E.Ericsson@astrazeneca.com.
Ingela Maxvall, Email: Ingela.Maxvall@astrazeneca.com.
Joseph Ochaba, Email: jochaba@ionisph.com.
Angela Menschik-Lundin, Email: Angela.Menschik-Lundin@astrazeneca.com.
Steven Kuntz, Email: skuntz@ionisph.com.
Danielle Gattis, Email: dgattis@isisph.com.
Jeffery A. Engelhardt, Email: jengelhardt@ionisph.com.
Brett P. Monia, Email: bmonia@isisph.com.
Maria Chiara Magnone, Email: magnonem2004@yahoo.it.
Shuling Guo, Email: sguo@ionisph.com.
References
- 1.Genovese G, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329(5993):841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzur S, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128(3):345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopp JB, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22(11):2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saran R, et al. US Renal Data System 2016 Annual Data Report: Epidemiology of Kidney Disease in the United States. Am J Kidney Dis. 2017;69(3 Suppl 1):A7–A8. doi: 10.1053/j.ajkd.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gribouval O, et al. APOL1 risk genotype in European steroid-resistant nephrotic syndrome and/or focal segmental glomerulosclerosis patients of different African ancestries. Nephrol Dial Transplant. doi: 10.1093/ndt/gfy176. [published online ahead of print July 9, 2018]. [DOI] [PubMed] [Google Scholar]
- 6.Nichols B, et al. Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int. 2015;87(2):332–342. doi: 10.1038/ki.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsen CP, Beggs ML, Saeed M, Walker PD. Apolipoprotein L1 risk variants associate with systemic lupus erythematosus-associated collapsing glomerulopathy. J Am Soc Nephrol. 2013;24(5):722–725. doi: 10.1681/ASN.2012121180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsa A, et al. APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med. 2013;369(23):2183–2196. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman DJ, Kozlitina J, Genovese G, Jog P, Pollak MR. Population-based risk assessment of APOL1 on renal disease. J Am Soc Nephrol. 2011;22(11):2098–2105. doi: 10.1681/ASN.2011050519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith EE, Malik HS. The apolipoprotein L family of programmed cell death and immunity genes rapidly evolved in primates at discrete sites of host-pathogen interactions. Genome Res. 2009;19(5):850–858. doi: 10.1101/gr.085647.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monajemi H, Fontijn RD, Pannekoek H, Horrevoets AJ. The apolipoprotein L gene cluster has emerged recently in evolution and is expressed in human vascular tissue. Genomics. 2002;79(4):539–546. doi: 10.1006/geno.2002.6729. [DOI] [PubMed] [Google Scholar]
- 12.Shukha K, et al. Most ApoL1 Is Secreted by the Liver. J Am Soc Nephrol. 2017;28(4):1079–1083. doi: 10.1681/ASN.2016040441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanhamme L, et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422(6927):83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- 14.Pérez-Morga D, et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science. 2005;309(5733):469–472. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- 15.Bruggeman LA, et al. APOL1-G0 or APOL1-G2 Transgenic Models Develop Preeclampsia but Not Kidney Disease. J Am Soc Nephrol. 2016;27(12):3600–3610. doi: 10.1681/ASN.2015111220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beckerman P, et al. Transgenic expression of human APOL1 risk variants in podocytes induces kidney disease in mice. Nat Med. 2017;23(4):429–438. doi: 10.1038/nm.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhaorigetu S, Wan G, Kaini R, Jiang Z, Hu CA. ApoL1, a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy. 2008;4(8):1079–1082. doi: 10.4161/auto.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crooke ST, Witztum JL, Bennett CF, Baker BF. RNA-Targeted Therapeutics. Cell Metab. 2019;29(2):501. doi: 10.1016/j.cmet.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Raal FJ, et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;375(9719):998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- 20.Finkel RS, et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016;388(10063):3017–3026. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]
- 21.Mercuri E, et al. Nusinersen versus Sham Control in Later-Onset Spinal Muscular Atrophy. N Engl J Med. 2018;378(7):625–635. doi: 10.1056/NEJMoa1710504. [DOI] [PubMed] [Google Scholar]
- 22.Ackermann EJ, et al. Suppressing transthyretin production in mice, monkeys and humans using 2nd-Generation antisense oligonucleotides. Amyloid. 2016;23(3):148–157. doi: 10.1080/13506129.2016.1191458. [DOI] [PubMed] [Google Scholar]
- 23.Ma L, et al. Localization of APOL1 protein and mRNA in the human kidney: nondiseased tissue, primary cells, and immortalized cell lines. J Am Soc Nephrol. 2015;26(2):339–348. doi: 10.1681/ASN.2013091017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markowitz GS, Nasr SH, Stokes MB, D’Agati VD. Treatment with IFN-{alpha}, -{beta}, or -{gamma} is associated with collapsing focal segmental glomerulosclerosis. Clin J Am Soc Nephrol. 2010;5(4):607–615. doi: 10.2215/CJN.07311009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan X, et al. APOL1 risk variants enhance podocyte necrosis through compromising lysosomal membrane permeability. Am J Physiol Renal Physiol. 2014;307(3):F326–F336. doi: 10.1152/ajprenal.00647.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomson R, Finkelstein A. Human trypanolytic factor APOL1 forms pH-gated cation-selective channels in planar lipid bilayers: relevance to trypanosome lysis. Proc Natl Acad Sci USA. 2015;112(9):2894–2899. doi: 10.1073/pnas.1421953112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sampson MG, et al. Integrative Genomics Identifies Novel Associations with APOL1 Risk Genotypes in Black NEPTUNE Subjects. J Am Soc Nephrol. 2016;27(3):814–823. doi: 10.1681/ASN.2014111131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hung G, et al. Characterization of target mRNA reduction through in situ RNA hybridization in multiple organ systems following systemic antisense treatment in animals. Nucleic Acid Ther. 2013;23(6):369–378. doi: 10.1089/nat.2013.0443. [DOI] [PubMed] [Google Scholar]
- 29.Prakash TP, et al. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 2014;42(13):8796–8807. doi: 10.1093/nar/gku531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliveira Arcolino F, et al. Human Urine as a Noninvasive Source of Kidney Cells. Stem Cells Int. 2015;2015:362562. doi: 10.1155/2015/362562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng D, et al. Biogenesis and cytotoxicity of APOL1 renal risk variant proteins in hepatocytes and hepatoma cells. J Lipid Res. 2015;56(8):1583–1593. doi: 10.1194/jlr.M059733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan G, Zhaorigetu S, Liu Z, Kaini R, Jiang Z, Hu CA. Apolipoprotein L1, a novel Bcl-2 homology domain 3-only lipid-binding protein, induces autophagic cell death. J Biol Chem. 2008;283(31):21540–21549. doi: 10.1074/jbc.M800214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg AZ, Kopp JB. Focal Segmental Glomerulosclerosis. Clin J Am Soc Nephrol. 2017;12(3):502–517. doi: 10.2215/CJN.05960616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korbet SM. Treatment of primary FSGS in adults. J Am Soc Nephrol. 2012;23(11):1769–1776. doi: 10.1681/ASN.2012040389. [DOI] [PubMed] [Google Scholar]
- 35.Johnstone DB, et al. APOL1 null alleles from a rural village in India do not correlate with glomerulosclerosis. PLoS ONE. 2012;7(12):e51546. doi: 10.1371/journal.pone.0051546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joshi PP, et al. Human trypanosomiasis caused by Trypanosoma evansi in India: the first case report. Am J Trop Med Hyg. 2005;73(3):491–495. doi: 10.4269/ajtmh.2005.73.491. [DOI] [PubMed] [Google Scholar]
- 37.Vanhollebeke B, et al. Human Trypanosoma evansi infection linked to a lack of apolipoprotein L-I. N Engl J Med. 2006;355(26):2752–2756. doi: 10.1056/NEJMoa063265. [DOI] [PubMed] [Google Scholar]
- 38.Hong D, et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci Transl Med. 2015;7(314):314ra185. doi: 10.1126/scitranslmed.aac5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langefeld CD, et al. Genome-wide association studies suggest that APOL1-environment interactions more likely trigger kidney disease in African Americans with nondiabetic nephropathy than strong APOL1-second gene interactions. Kidney Int. 2018;94(3):599–607. doi: 10.1016/j.kint.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedman DJ, Pollak MR. Apolipoprotein L1 and Kidney Disease in African Americans. Trends Endocrinol Metab. 2016;27(4):204–215. doi: 10.1016/j.tem.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daehn I, et al. Endothelial mitochondrial oxidative stress determines podocyte depletion in segmental glomerulosclerosis. J Clin Invest. 2014;124(4):1608–1621. doi: 10.1172/JCI71195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun YB, Qu X, Zhang X, Caruana G, Bertram JF, Li J. Glomerular endothelial cell injury and damage precedes that of podocytes in adriamycin-induced nephropathy. PLoS ONE. 2013;8(1):e55027. doi: 10.1371/journal.pone.0055027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dimke H, Maezawa Y, Quaggin SE. Crosstalk in glomerular injury and repair. Curr Opin Nephrol Hypertens. 2015;24(3):231–238. doi: 10.1097/MNH.0000000000000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duchateau PN, et al. Plasma apolipoprotein L concentrations correlate with plasma triglycerides and cholesterol levels in normolipidemic, hyperlipidemic, and diabetic subjects. J Lipid Res. 2000;41(8):1231–1236. [PubMed] [Google Scholar]
- 45.Freedman BI, et al. Apolipoprotein L1 gene variants in deceased organ donors are associated with renal allograft failure. Am J Transplant. 2015;15(6):1615–1622. doi: 10.1111/ajt.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reeves-Daniel AM, et al. The APOL1 gene and allograft survival after kidney transplantation. Am J Transplant. 2011;11(5):1025–1030. doi: 10.1111/j.1600-6143.2011.03513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee BT, et al. The APOL1 genotype of African American kidney transplant recipients does not impact 5-year allograft survival. Am J Transplant. 2012;12(7):1924–1928. doi: 10.1111/j.1600-6143.2012.04033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bruggeman LA, et al. Plasma apolipoprotein L1 levels do not correlate with CKD. J Am Soc Nephrol. 2014;25(3):634–644. doi: 10.1681/ASN.2013070700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kozlitina J, et al. Plasma Levels of Risk-Variant APOL1 Do Not Associate with Renal Disease in a Population-Based Cohort. J Am Soc Nephrol. 2016;27(10):3204–3219. doi: 10.1681/ASN.2015101121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomson R, et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci USA. 2014;111(20):E2130–E2139. doi: 10.1073/pnas.1400699111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kruzel-Davila E, et al. APOL1-Mediated Cell Injury Involves Disruption of Conserved Trafficking Processes. J Am Soc Nephrol. 2017;28(4):1117–1130. doi: 10.1681/ASN.2016050546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heymann J, Winkler CA, Hoek M, Susztak K, Kopp JB. Therapeutics for APOL1 nephropathies: putting out the fire in the podocyte. Nephrol Dial Transplant. 2017;32(suppl_1):i65–i70. doi: 10.1093/ndt/gfw402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seth PP, et al. Short antisense oligonucleotides with novel 2’-4’ conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J Med Chem. 2009;52(1):10–13. doi: 10.1021/jm801294h. [DOI] [PubMed] [Google Scholar]
- 54.Prakash TP, et al. Comprehensive Structure-Activity Relationship of Triantennary N-Acetylgalactosamine Conjugated Antisense Oligonucleotides for Targeted Delivery to Hepatocytes. J Med Chem. 2016;59(6):2718–2733. doi: 10.1021/acs.jmedchem.5b01948. [DOI] [PubMed] [Google Scholar]
- 55.Wang F, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14(1):22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.