Abstract
An incomplete understanding of the biology of the human kidney, including the relative abundances of and interactions between intrinsic and immune cells, has long constrained the development of therapies for kidney disease. The small amount of tissue obtained by renal biopsy has previously limited the ability to use patient samples for discovery purposes. Imaging mass cytometry (IMC) is an ideal technology for quantitative interrogation of scarce samples, permitting concurrent analysis of more than 40 markers on a single tissue section. Using a validated panel of metal-conjugated antibodies designed to confer unique signatures on the structural and infiltrating cells comprising the human kidney, we performed simultaneous multiplexed imaging with IMC in 23 channels on 16 histopathologically normal human samples. We devised a machine-learning pipeline (Kidney-MAPPS) to perform single-cell segmentation, phenotyping, and quantification, thus creating a spatially preserved quantitative atlas of the normal human kidney. These data define selected baseline renal cell types, respective numbers, organization, and variability. We demonstrate the utility of IMC coupled to Kidney-MAPPS to qualitatively and quantitatively distinguish individual cell types and reveal expected as well as potentially novel abnormalities in diseased versus normal tissue. Our studies define a critical baseline data set for future quantitative analysis of human kidney disease.
Keywords: Nephrology
Keywords: Cellular immune response, Chronic kidney disease
Imaging mass cytometry was used to develop a 2-dimensional atlas of the normal human kidney, yielding quantitative information about known and novel cell types.
Introduction
Despite considerable research in the field of acute kidney injury (AKI), no therapies exist for the most common intrinsic renal cause of AKI, acute tubular injury (ATI). Additionally, advances in understanding based on animal models have as yet yielded few therapies for chronic kidney disease, tubulointerstitial diseases, and many glomerular diseases (1). The lack of therapeutic options for many forms of kidney disease stems in part from our incomplete understanding of the gene expression and spatial organization of the healthy and injured human kidney (2–4). Renal biopsy is a useful tool that has for decades been used for descriptive analysis of human renal pathologies at the cellular level; however, there has not yet been a concerted effort to provide quantitative information about the tubular, stromal, and nonresident cell populations in disease. In fact, the basis for the classification of most forms of kidney disease remains these pathologic descriptions, rather than clinical presentation, pathophysiology, biomarkers, or response to therapies (5, 6). Unfortunately, the limited amount of tissue that is obtained by renal biopsy has restricted the types of analyses that can be performed on patient samples, thereby hampering discovery at the molecular and even cellular level.
Several emerging technologies, including single-cell and single-nucleus transcriptome sequencing, have catalyzed efforts towards more granular interrogation of kidney tissue. Such approaches have yielded quantitative data delineating the number of cell types present in the “normal” and diseased human kidney, including tubuloepithelial, stromal, and immune cells (7, 8). These data suggest at least 16 distinct cell types that comprise the adult human kidney, including a multitude of resident and infiltrating immune cells (7–9). Many of these cell populations are distinguished by specific differentially expressed genes (DEGs), which include transporters, secreted proteins, and transcription factors. Single-cell approaches have also identified insights into disease and identified novel cell populations in the murine kidney (10, 11). Such methods, however, rely on protease digestion of tissues, resulting in underrepresentation of specific cell types (9, 12). Furthermore, technique-specific biases may be exacerbated by injury or fibrosis, limiting the ability to perform comparative analysis between uninjured and injured tissue (12). Critically, while transcriptional analysis is powerful for characterizing the cell types comprising a scarce tissue sample, it does not offer spatial context. Yet, in perhaps no other solid organ — save for the brain — is function so closely tied to the 2- and 3-dimensional relationship between cells as it is in the kidney. We sought to utilize highly multiplexed imaging to faithfully and reproducibly determine the composition of the human kidney in the normal and diseased state, and to afford us with potentially novel information regarding the identity, location, and numbers of — as well as interactions between — resident and immune cells.
Imaging mass cytometry (IMC) is a technology that supports simultaneous detection of more than 40 protein markers on a single section of formalin-fixed, paraffin-embedded tissue, with the resultant reconstructions achieving 1-μm resolution (13, 14). In the kidney, endogenous fluorescence, particularly in tubules, can reduce signal-to-noise ratio and compromise image quantitation in fluorescence-based imaging, making a mass spectrometry–based analytic technique particularly appealing (15). IMC has been used to characterize the tumor microenvironment in solid malignancies, yielding information about the identity, number, and spatial relationship between immune and stromal cells (13, 16). Additionally, the immune milieu of the human pancreas has recently been characterized using IMC (17, 18). Studies in murine models, including IMC of murine dendritic cell populations, suggest that interactions between resident renal cells and cells of immune origin may be critical in both homeostasis and disease, though there is greater uncertainty in the human kidney (19–22). We postulated that IMC could be used to address this important question and sought to develop a panel of antibodies and establish the computer-based analysis pipeline needed to faithfully characterize and quantify the 2-dimensional cellular architecture of the human kidney. Herein we describe the successful application of IMC in the human kidney, using a combined imaging and analysis approach that we term Kidney-MAPPS (multiplexed antibody-based profiling with preservation of spatial context). For our sources of normal human kidney, we have chosen biopsies from living donors as well as nephrectomy tissue from areas remote from tumors. Using Kidney-MAPPS, we have successfully generated a 2-dimensional quantitative atlas of 22 distinct cell populations that comprise the human kidney.
Results
Antibody selection and validation for interrogation of the human kidney.
In order to define the spatial localization of the cell types that comprise the human kidney using a multiplexed immunostaining approach, we first sought to optimize a panel of commercially available antibodies directed against the protein products of cell-type-specific DEGs. Importantly, only commercial antibodies that are available in BSA-free and carrier-free formulations are suitable for conjugation to rare earth metals and use with IMC. Monoclonal antibodies were preferentially tested over polyclonal antibodies in order to minimize effects of batch variability. We first performed immunofluorescent staining with individual candidate antibodies using a single, common protocol on formalin-fixed, paraffin-embedded human kidney tissue obtained by nephrectomy. Each intrinsic renal marker was evaluated by a renal pathologist blinded to the antibody used, and who identified the morphological characteristics of the stained cells or regions. Antibodies in which positive staining was detected in expected cells with a maximal signal-to-noise ratio, and in which significant ectopic or background staining was not observed, were included in the final intrinsic renal antibody panel. The antibodies were conjugated to rare-earth metals for detection by IMC and tissue specificity was validated in groups of 2–5 antibodies to confirm that binding specificity had not changed through the conjugation process (Supplemental Figure 1 and data not shown; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.129477DS1). Antibody validation data generated as a part of this work are publicly available through the NIH (Re)Building a Kidney database — https://www.rebuildingakidney.org/chaise/recordset/#2/Antibody:Antibody_Tests@sort(RID) (23).
For immune markers, commercially available metal-conjugated antibodies were validated using IMC in human lymph node and abscess tissue, as well as by immunofluorescence in human kidneys known to harbor severe interstitial nephritis (Supplemental Figure 2 and data not shown). In lymph node tissue, CD20, a B cell marker, was detectable in areas corresponding morphologically to germinal centers, with CD3-positive T cells detectable in adjacent regions (Supplemental Figure 2A). CD4 and CD8 expression was found to be mutually exclusive, and each colocalized with CD3 (Supplemental Figure 2A). All lymphocyte markers had a characteristically annular appearance, though the ability to distinguish this is somewhat limited by the 1-μm resolution and the high cell density. CD68, a marker of macrophages, was detectable in morphologically distinct cells that did not overlap with lymphocyte markers (Supplemental Figure 2, A and B). CD66b, a marker of granulocytes, was found to be present in abscess tissue and was distinct from CD68 expression (Supplemental Figure 2B). Our final panel of IMC-validated antibodies is shown in Table 1.
Table 1. Antibodies, clones, dilutions, metals, and suppliers.
Defining the normal human kidney for analysis.
To define the 2-dimensional spatial relationship between distinct cell types in the normal human kidney, we next sought to obtain histopathologically normal kidney from available sources. Living kidney donors are carefully screened for evidence of chronic kidney disease as well as comorbidities such as diabetes or hypertension; therefore, living donors represent a useful potential source for normal kidney tissue (24). We prospectively accrued living donor samples, obtained by back-bench biopsy at the time of organ donation, and scored by a renal pathologist in a blinded manner. All samples were graded as “histopathologically normal” or “less than five percent interstitial fibrosis,” and were therefore included in our analysis (Supplemental Table 1).
Nephrectomy tissue taken from tumor-remote regions is another commonly used source of “normal” kidney tissue due to its ready availability (25, 26). In this study, nephrectomy tissue was obtained both prospectively and from banked samples. In total, we collected 61 nephrectomy samples prospectively. Of these, 35 underwent detailed analysis of tumor remote (>2 cm from tumor) regions by a clinical renal pathologist. The plurality of these samples was noted to feature arterio- or arteriolo-nephrosclerosis, interstitial fibrosis, tubular injury, or interstitial nephritis (data not shown). However, 7 of the 35 were reported to exhibit either “normal histology” or “minimal” histopathologic abnormalities. Of those 7, one was excluded because the remaining tissue was degraded and could not be imaged. The remainder of the normal samples were selected for imaging by IMC (Supplemental Table 1).
To image additional samples, we identified banked formalin-fixed, paraffin-embedded tissue collected from surgical nephrectomies performed at 2 campuses over a period of 9 years. Nine hundred eighty-eight pathology reports were queried for notes of “benign,” “tumor-remote,” or “uninvolved” tissue. This yielded 55 detailed pathology reports of tumor-remote tissue, which included only 5 samples in which pathology in tumor-remote regions was specifically noted to have “no histopathologic abnormalities.” In sum, we obtained 16 total reference kidney samples meeting criteria for “no” or “minimal” histopathological abnormalities (Supplemental Table 1), obtained 1 month to 9 years prior to our IMC analysis. Chart review was performed for each patient whose sample was included. As compared with patients undergoing nephrectomy, living donors trended to be younger with lower prevalence of hypertension and were more often female, although none of these trends reached statistical significance. There were no differences between the groups in serum creatinine levels or estimated glomerular filtration rate (Supplemental Table 1).
Qualitative assessment of highly multiplexed imaging of the human kidney with IMC.
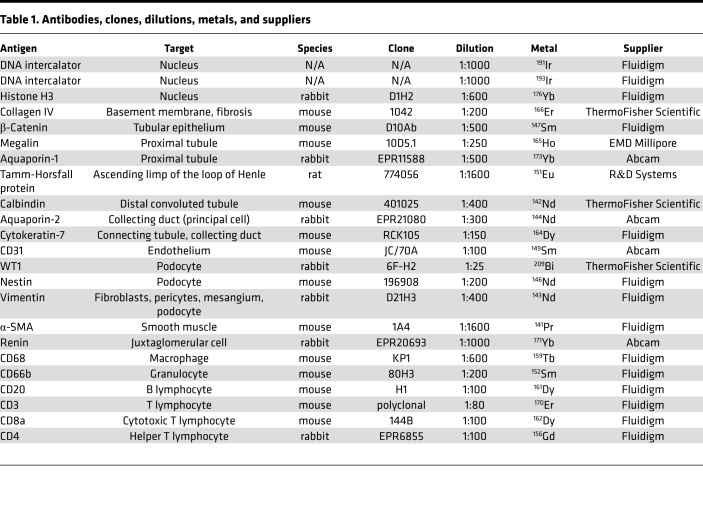
To define the spatial relationship between cell types in the human kidney, 5-μm sections of each of the 16 formalin-fixed, paraffin-embedded reference kidneys were labeled in parallel using a single cocktail of 23 markers (Table 1). IMC was then performed in randomized order on sequential days over a 6-week period. We imaged regions spanning from the cortex to the outer medulla with IMC (Figure 1, A–C). In order to avoid the immune-cell-rich subcapsular area, imaging of the cortex was initiated 600–800 μm from the capsule, which was detectable on all samples (boxed area in Figure 1, A and C).
Figure 1. IMC can be used to recapitulate human kidney morphology.
(A–C) H&E stain (A), trichrome stain (B), and light microscopic image of immunostained core biopsy from a living donor kidney (C). The inset in A and corresponding leftmost box in C show the region selected for IMC ablation (visualization of selected markers depicted in D and E). (C) Additional boxes show additional regions ablated. (D) Selected tubule and endothelial markers detected by IMC, pseudocolored as indicated, and overlaid on a pre-ablation image of the tissue. PT, proximal tubule; tALH, thick ascending loop of Henle; DCT, distal convoluted tubule; T, transitional area; MEG, megalin; THP, Tamm-Horsfall protein; CALB, calbindin; CK7, cytokeratin-7; AQP2, aquaporin-2. (E) Selected interstitial and immune markers detected by IMC, pseudocolored as indicated, and overlaid on a pre-ablation image of the tissue. aSMA, α-smooth muscle actin; COL IV, collagen IV; REN, renin; NES, nestin. (F) Inset from D. Glom, glomerulus; Ar, artery; Cy, cyst. (G) Inset from E. Arrowhead indicates renin-positive cells. Arrows indicate immune cells all designated using the same cyan color. Scale bars: 1 mm (A), 300 μm (D), and 100 μm (F).
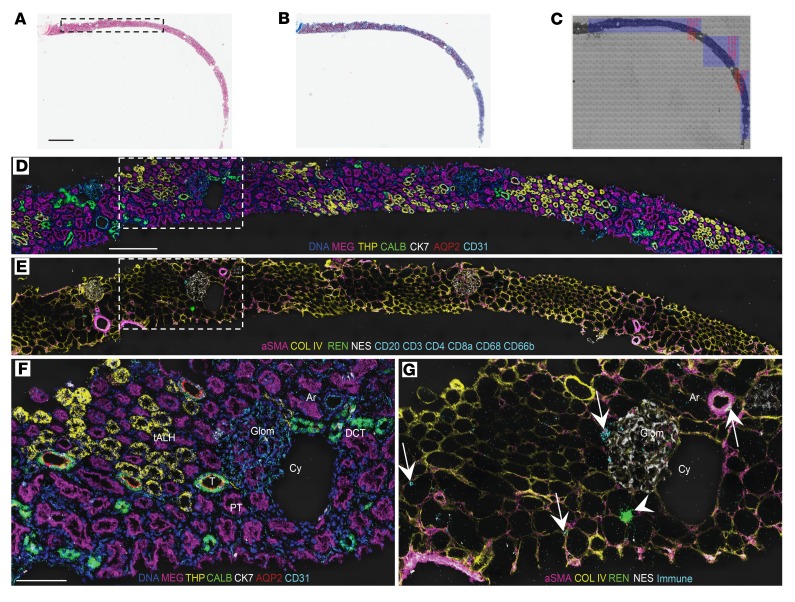
Qualitative analysis, performed using pseudocolored images of the IMC data from human cortex, revealed faithful labeling of canonical tubule segments consistent with expected histologic findings. We used 7 tubule-specific antibodies simultaneously to identify proximal tubule (PT; aquaporin-1–positive, megalin-positive, β-catenin–positive), thick ascending limb of the loop of Henle (tALH; Tamm-Horsfall protein–positive [THP-positive], β-catenin–positive), distal convoluted tubule (DCT; calbindin-positive, β-catenin–positive), connecting tubule/collecting duct (cytokeratin-7–positive, aquaporin-2–positive, β-catenin–positive), and transition elements between tubular segments (Figure 1D and Figure 2, A–H). In the cortex, there was an obvious preponderance of PT as compared with other cell types. In contrast to the cortex, the renal medulla had few detectable calbindin-positive DCT cells (Figure 2M), but revealed a population of megalin-low, aquaporin-1–positive, cytokeratin-7–positive, aquaporin-2–negative, β-catenin–negative tubules that most likely represent the thin descending limb of the loop of Henle (Figure 2, I–K, N, and P). Similarly, the THP-positive tubule segments in the medulla were composed of smaller cells than those in the cortex and had a smaller cuboidal profile, consistent with thin ascending limb of the loop of Henle (Figure 2, D and L).
Figure 2. Defining tubular heterogeneity and cellular identity with IMC.
(A–G) Individual tubule markers pseudocolored and depicted on a single region of cortex from living donor kidney. Arrow in E indicates distal convoluted tubule, as denoted by high calbindin staining. (H) Merged image for all 7 channels, overlaid on a pre-ablation light microscopic image of the tissue. Panels E, F, and H highlight a transitional area in which cytokeratin-7–positive, calbindin-negative cells (arrowheads) are interspersed in the same tubule. (I–O) Identical tubule markers from medulla on the same kidney, with images processed identically to those in A–G. (P) Merged image of all 7 channels, overlaid on a light image of the pre-ablation tissue. Arrows in J, N, and P denote an aquaporin-1–positive, megalin-low, cytokeratin-7–positive, β-catenin–negative tubule. Arrowheads in L, N, and P denote a THP-positive, aquaporin-1–negative, cytokeratin-7–negative tubule. β-CAT, β-catenin; AQP1, aquaporin-1; AQP2, aquaporin-2; MEG, megalin; THP, Tamm-Horsfall protein; CALB, calbindin; CK7, cytokeratin-7. Scale bar: 100 μm (A).
In addition to characterizing well-defined tubule segments, multiplexed imaging allowed us to characterize the cellular makeup of the less-well-characterized transitional tubular regions. In the superficial cortex, transition areas were identified in which calbindin-positive, cytokeratin-7–positive, and aquaporin-2–positive cells were detectable in the same tubular cross section, in tubules of qualitatively larger luminal diameter than the DCT (Figure 1, D and F, and Figure 2A). These areas most likely correspond to regions where the DCT is transitioning to connecting tubule (Supplemental Figure 3). More uniform cytokeratin-7–positive, aquaporin-2–positive tubules were found to be of higher abundance in the medulla, consistent with the presence of large numbers of collecting ducts in that region (Figure 2, N–P). Cytokeratin-7 in particular showed mosaic expression in transitional areas, as well as in connecting tubule and collecting duct.
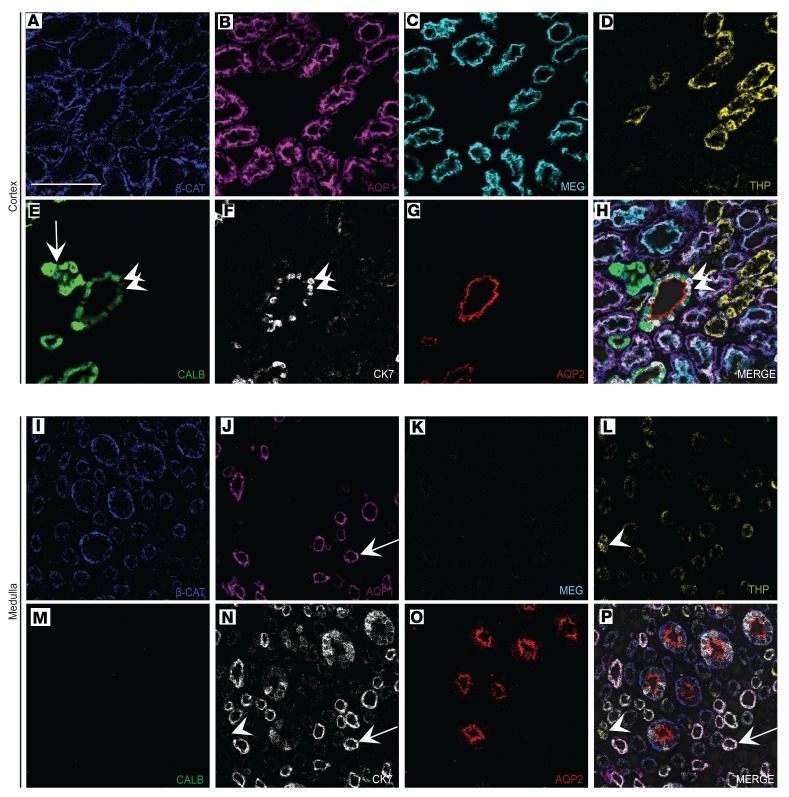
The renal interstitium includes the vascular and stromal cell populations that surround and support the tubular epithelium. The interstitium also includes the collagenous matrix surrounding the tubules, which is believed to undergo extensive changes during acute and chronic disease (27). Interstitial markers analyzed in this experiment include collagen IV, a marker of glomerular and tubular basement membrane, as well as α-smooth muscle actin (α-SMA) and vimentin, which mark stromal cells including fibroblasts, myofibroblasts, and pericytes (Figure 1, E and G, and Figure 3). In reference kidneys, a collagen mesh of basement membrane was visualized by IMC, with greater membrane thickness qualitatively apparent in the medulla than in cortex (Figure 3, B vs. H). The lacy network of cortical peritubular capillaries was made apparent by the large number of cells positive for CD31, an endothelial marker, between the collagen IV–positive basement membranes (Figure 3C). Vessels with smooth muscle investment, including arterioles and arteriola recta in clusters, were detected identified by closely associated α-SMA and CD31 staining (Figure 3, I, K, and L).
Figure 3. IMC elucidates stromal, vascular, and endothelial cell populations.
(A–E) Selected markers shown individually on an identical region of cortex from a living donor kidney and pseudocolored. (F) Merged image for the individual markers shown in A–E and overlaid on a pre-ablation image. (G–K) Selected markers shown individually on an identical region of medulla from the same living donor kidney, with images processed identically to those in A–E. (L) Merged image for the individual markers shown in G–K overlaid on a pre-ablation image of the tissue. Arrows in I–L indicate CD31-positive, vimentin-positive, α-SMA–positive arteriole recta. COL IV, collagen IV; VIM, vimentin; aSMA, α-smooth muscle actin. Scale bar: 100 μm (A).
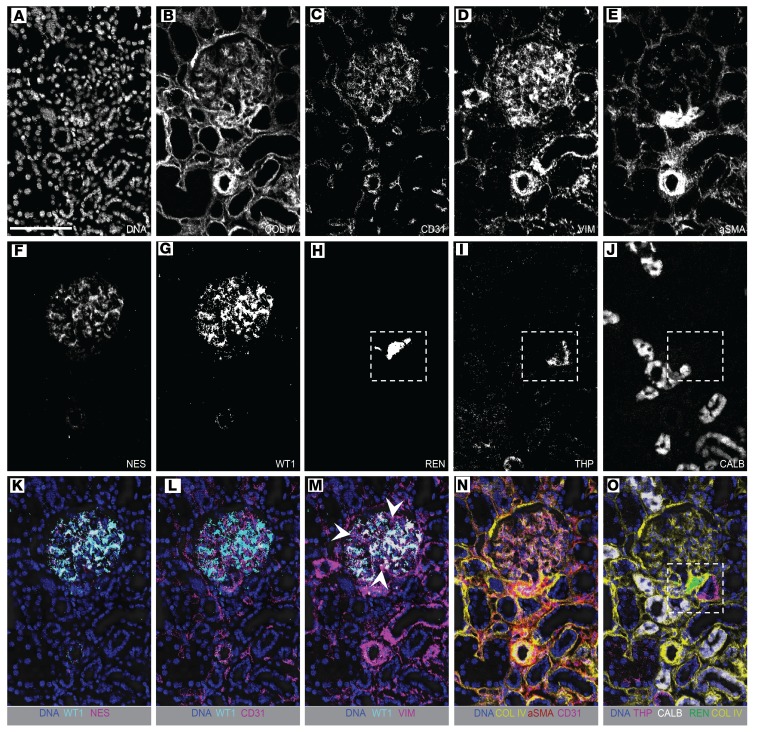
Within the glomerulus, collagen IV was found, as expected, to delineate the glomerular basement membrane, with highest staining intensity at the vascular pole and juxtaglomerular apparatus (JGA) (Figure 4, A and B). Both intraglomerular and extraglomerular endothelial cells were detectable by CD31 expression (Figure 4, C and N). Podocytes, an epithelial cell type positioned on the urinary side of glomerular capillaries, exhibit a complex 3-dimensional structure. In this study, podocytes were detected by 2 specific antibodies; anti-nestin (intermediate filament protein, Figure 4F) and anti–WT-1 (directed against the cytoplasmic form of the protein, Figure 4G). These were found to overlap to a high degree (Figure 4K), and podocytes were observed in direct apposition to the endothelium (Figure 4L).
Figure 4. Characterization of glomerular and juxtaglomerular cells with IMC.
(A–J) IMC images for individual markers as indicated, showing a glomerulus, vasculature, and surrounding tubules. (K–O) Merged, pseudocolored images selected from A–J, illustrating the associations between glomerular and juxtaglomerular cells. (K) Overlapping signal between 2 podocyte markers, WT-1 and nestin. (L) Close association between WT-1–positive podocytes and CD31-positive endothelial cells in capillary loops. (M) Vimentin is expressed in podocytes and mesangial cells, and overlaps with WT-1 expression in podocytes. Arrowheads indicate vimentin-positive, WT-1–negative cells, which represent intraglomerular mesangium. (N) Basement membrane and vascular markers show the vascular nature of the glomerulus. (O) Channels highlighting the morphology of the juxtaglomerular apparatus. Insets in H–J and O show that the juxtaglomerular apparatus is characterized by renin-positive cells separated by basement membrane from immediately adjacent THP-negative macula densa cells. The surrounding cells of the thick ascending limb are THP positive and transition to calbindin-positive tubular cells. COL IV, collagen IV; VIM, vimentin; aSMA, α-smooth muscle actin; NES, nestin; THP, Tamm-Horsfall protein; REN, renin; CALB, calbindin. Scale bar: 100 μm (A).
Mesangial cells are specialized contractile mesenchymal cells that maintain the structure of the glomerulus. Mesangial cells, like many podocytes, express low levels of α-SMA), as well as high levels of the intermediate filament protein vimentin (Figure 4, D and E). Mesangial cells could be visually distinguished from podocytes in this study by positivity for vimentin in the absence of expression of nestin and WT-1 (Figure 4M). Our antibody against α-SMA, which detected α-SMA on smooth muscle cells surrounding arteries and arterioles, was not useful for mesangial cell identification (Figure 4E)
The JGA is a specialized region of the nephron with critical roles in tubuloglomerular feedback and systemic blood pressure regulation. Recently, this region has gained increasing interest due to the putative role of renin-positive cells in mediating podocyte regeneration (28, 29). The components of the JGA include the aforementioned renin-positive cells (Figure 4H), as well as associated tubular segments that comprise the transition point from tALH to DCT (Figure 4, I and J). Within the thick ascending limb, THP-negative cells comprising the macula densa were found to be tightly opposed to — though separated by the collagen IV–positive basement membrane from — the specialized α-SMA–positive, vimentin-positive, renin-positive cells of the JGA (Figure 4, I and O). Remaining cells in this tubular segment were THP-positive (Figure 4I). Calbindin-positive DCT was always found in the vicinity of renin-positive cells, though not directly in contact with them (Figure 4, J and O).
Immune cells detectable in normal human kidneys using IMC included macrophages (CD68-positive), T cells (CD3-positive), helper T cells (CD3-positive, CD4-positive), cytotoxic T cells (CD3-positive, CD8a-positive), B cells (CD20-positive), and granulocytes (CD66b-positive). All were abundant in lymph node or skin abscess tissue (Supplemental Figure 2) and were detectable in low abundance in the interstitium of reference kidneys (Figure 1G and Supplemental Figure 4).
Quantitative assessment of the cellular anatomy of human kidney using Kidney-MAPPS.
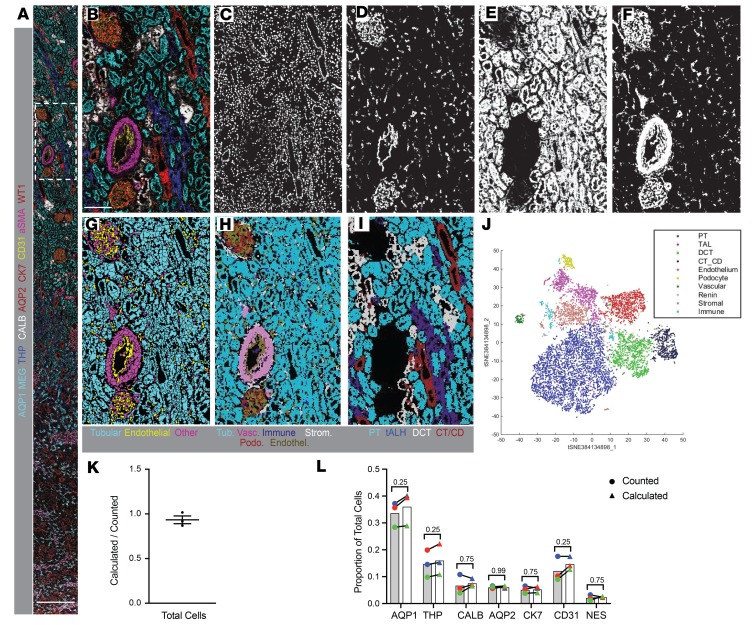
To quantitatively assess the cellular composition of normal human kidneys, we used an approach analogous to that which Bodenmiller and colleagues applied to cancer tissue, adapted for the unique and highly varied morphology of the kidney (13, 30). IMC data from relevant channels in 4 representative living donor and nephrectomy samples (Figure 5, A and B) were used to create distinct image stacks: one stack each representing nuclei, tubules, endothelial cells, and all remaining cell types (including stroma, podocytes, and immune cells). These image stacks were used to train a machine-learning algorithm to differentiate between signal and background in each data set and assign a probability to each pixel. After training on representative samples, this algorithm was extended to the entire cohort of samples to be analyzed by IMC. The nuclear probability image (Figure 5C) was used to define the existence of cells. The remaining probability images (Figure 5, D–F) were then iteratively used to categorize each nucleus as belonging to a particular cell type (tubular, endothelial, other) and to delineate the borders of each individual cell. These 3 resulting partial cell masks were merged into one unified mask defining every individual cell in the entirety of each ablated region (Figure 5G). Once cells were individually segmented, raw data from IMC was overlaid onto these segmentation masks using HistoCAT, a program designed for IMC analysis (30). Individual phenotypes were thereby ascribed to each cell in an unsupervised manner based on combinatorial expression of each of the 23 markers, allowing for classification and quantitation of each cell type within each region of interest (Figure 5, H–J). In cortex, 13 separate clusters of cells were detected, with phenotypes manually assigned based on the pattern of antigen expression (Figure 5J). Importantly, when phenotypes were assigned to individual cells based on the Kidney-MAPPS pipeline and overlaid on the segmentation masks (Figure 5, H and I), the resultant images closely recapitulated the patterns observed from raw data (Figure 5B). This consistent detection demonstrated that our cell segmentation and phenotyping algorithm faithfully represented the morphology of the kidney.
Figure 5. Single-cell segmentation, characterization, and quantification of a representative region imaged with IMC.
(A and B) Merged, pseudocolored images showing the entirety (A) and inset (B) of a region of a nephrectomy sample imaged by IMC, with selected markers depicted as indicated. (C–F) Probability images generated from a machine-learning algorithm applied to processed multiplexed images, in order to define pixels with the highest probability of belonging to nuclei (C), endothelial cells (D), tubular cells (E), and stromal cells, mesangium, and podocytes (F). These probability images were used as the basis to generate a single-cell segmentation mask. (G) Single-cell segmentation mask, with borders defined for each cell, and colors depicting tubular, endothelial, and all other cell types. (H and I) The segmentation mask in G was used as the basis for analysis of raw data from this IMC experiment. Based on raw data from each of the 23 input channels, each individual cell was clustered by expression by HistoCAT and manually assigned a phenotype. Resultant pseudocolored images are depicted. (H) Depicts cells assigned to tubular phenotypes, as well as vascular cells, immune cells, stromal cells, podocytes, and endothelial cells. (I) Shows specific tubular subtypes, as indicated. Colors were selected to approximate those used in the raw data in B. (J) Two-dimensional t-distributed stochastic neighbor embedding (t-SNE) projection showing the clusters and relative abundances of cells, their phenotypic relationship to each other, as well as their manually assigned phenotypic classes, with colors indicated on the associated legend. Four separate immune clusters were manually grouped together as one in this projection (cyan blue). (K) Quantification of the ratio of cells counted versus those calculated by the above machine-learning methodology for 3 selected regions. Bars indicate mean ± standard error of the mean. (L) Comparison of cells positive for the indicated markers detected either by manual counting (circles) or using the automated methodology (triangles). Mean values are shown by bars. n = 3. Statistical comparisons were made using Wilcoxon’s matched-pairs signed-ranked test, with P values indicated. Tub, tubular; Endothel., endothelial; Vasc, vascular; Strom., stromal; Podo., podocyte; PT, proximal tubule; tALH, thick ascending limb of the loop of Henle; DCT, distal convoluted tubule; CT/CD, connecting tubule and collecting duct; AQP1, aquaporin-1; AQP2, aquaporin-2; CALB, calbindin; CK7, cytokeratin-7; MEG, megalin; aSMA, α-smooth muscle actin; THP, Tamm-Horsfall protein; NES, nestin. Scale bars: 600 μm (A) and 150 μm (B).
Validation of the Kidney-MAPPS pipeline.
A series of experiments were performed to evaluate both the validity and reproducibility of data obtained by the Kidney-MAPPS data analysis pipeline. In representative regions from 3 kidneys, we manually counted and scored cells in a blinded manner, while simultaneously performing unsupervised quantitative analysis using the Kidney-MAPPS methodology (Figure 5K). Kidney-MAPPS was found to underestimate the total number of cells by an average of 6.6%, potentially reflecting the stringency with which we selected nuclei for inclusion in the machine-learning algorithm. To determine whether there was a bias toward dropout of specific cell types, cells positive for markers representing a wide array of cellular phenotypes were quantified both manually and with Kidney-MAPPS (Figure 5L). For 7 markers defining key structures of tubules and surrounding stroma, there were no significant differences in the proportions of positive cells detected by manual versus automated quantification, out of over 10,000 cells scored. This suggests that the slight underdetection of cells by the Kidney-MAPPS methodology is distributed proportionally among all cell types and confirms the strength of Kidney-MAPPS for assessment of cell phenotypes.
To address reproducibility, we selected a subset of tumor-remote nephrectomy samples with the most abundant tissue and for each sample performed a second distinct biopsy-sized ablation of the renal cortex (Supplemental Figure 5, A–C) (31). In comparing the originally imaged regions to the second sample from the same kidney section, we found no significant differences between the numbers of individual cortical cell types (Supplemental Figure 5D). We also performed staining with an identical antibody cocktail prepared 4 months after the initial data set was obtained. Using this validation cocktail, we stained adjacent sections from the same 4 kidneys, and selected areas corresponding closely to the original regions of interest for ablation and analysis. We found no significant differences in cell abundances when comparing the original staining and IMC ablation data to the validation data set (Supplemental Figure 5E).
Comparative analysis of nephrectomy and living donor tissue.
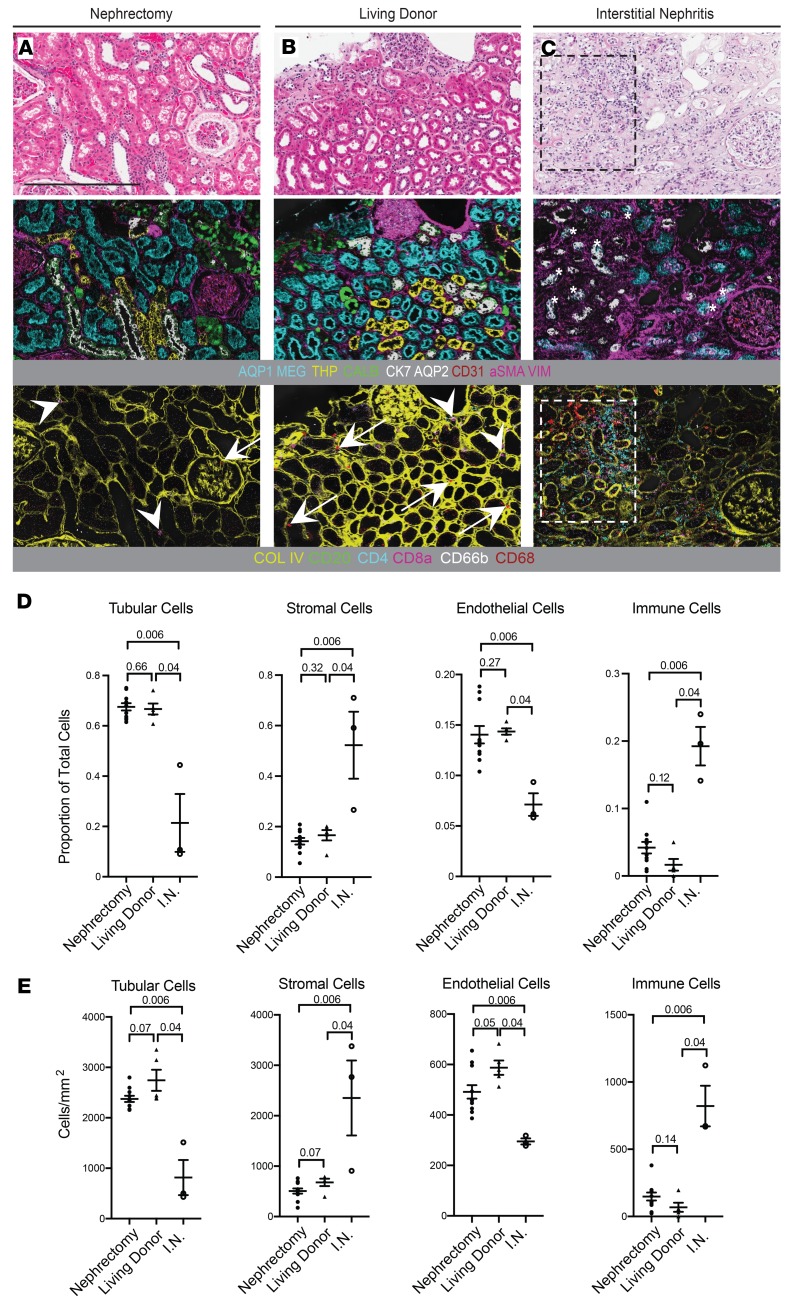
Because of the limited size and availability of histopathologically normal human biopsy samples, nephrectomy samples are frequently employed as a surrogate for normal kidney tissue. To determine whether pathologically normal nephrectomy samples are an appropriate surrogate for normal kidney tissue, we compared these to living kidney donor samples using Kidney-MAPPS. We analyzed an area from each of the 16 reference kidney samples that approximated the cross-sectional tissue area obtained in a single section of a typical needle kidney biopsy (31). The mean total tissue area for nephrectomy samples was 3.9 mm2 for cortex (n = 11) and 2.4 mm2 for medulla (n = 7). For living donors, the mean regions of interest were 2.0 mm2 for cortex (n = 5) and 1.7 mm2 for medulla (n = 2). Medulla was not available for 5 of the nephrectomy specimens and 3 of the living donor biopsies. By conventional analysis, including H&E, trichrome, and periodic acid–Schiff staining, no differences were readily apparent between nephrectomy and living donor tissue (Figure 6, A and B, and data not shown). Similarly, images generated by IMC showed no qualitative differences for tubular, stromal, or immune markers (Figure 6, A and B). Quantification of cell proportions with the Kidney-MAPPS pipeline revealed no significant differences between nephrectomy and living donor tissue in the proportion of tubular cells, stromal cells, or endothelial cells, normalized to either total number of cells or to total tissue area (Figure 6, D and E). There was a trend toward fewer endothelial cells in nephrectomy tissue than in living donor tissue, though this did not reach statistical significance (Figure 6, D and E).
Figure 6. Comparative analysis of nephrectomy tissue, living donor tissue, and interstitial nephritis with IMC reveals variability in stromal and immune makeup.
(A–C) Representative H&E and pseudocolored IMC images from adjacent sections of nephrectomy (A), living donor (B), and interstitial nephritis (C) tissue. The 2 IMC images for each sample represent the same region imaged at the same time, with a selection of tubular and stromal markers shown in the upper panels, and collagen IV and immune markers shown in the lower panels. Arrows mark macrophages and arrowheads mark CD8a-positive T cells in the nephrectomy and living donor tissue in A and B. Inset in C shows an area of significant interstitial nephritis, consisting of a mixed infiltrate including macrophages, CD4-positive, and CD8-positive T cells. Asterisks in C denote areas of de novo expression of cytokeratin-7 in megalin-positive tubules. (D and E) Quantification of tubular, stromal, endothelial, and immune cells, as a proportion of total cells (D) and per square millimeter (E), detected by automated analysis of IMC data. n = 11 for nephrectomy, n = 5 for living donor, and n = 3 for interstitial nephritis (I.N.). Statistical comparisons were made using the Mann-Whitney test, with P values between each group indicated. Bars show mean ± standard error of the mean. Scale bar: 800 μm (A). AQP1, aquaporin-1; AQP2, aquaporin-2; COL IV, collagen IV; VIM, vimentin; aSMA, α-smooth muscle actin; THP, Tamm-Horsfall protein; MEG, megalin; CALB, calbindin; CK7, cytokeratin-7.
To determine whether our Kidney-MAPPS algorithm could successfully identify differences in cell proportions and architecture between normal kidney and diseased kidney, we analyzed renal cortex from 3 tumor adjacent nephrectomy specimens histologically graded as having interstitial nephritis or immune infiltrate (mean area of 2.9 mm2). These kidneys had readily identifiable immune infiltrates visible by H&E staining (Figure 6C). IMC revealed persistence of tubule structures in areas of infiltrate, though with de novo expression of cytokeratin-7 in some megalin-positive PT cells (Figure 6C). The makeup of immune infiltrate could be readily determined with IMC and consisted largely of CD4-positive T cells, CD8-positive T cells, and CD68-positive macrophages (Figure 6C).
Quantitative analysis of cell numbers in the interstitial nephritis specimens using Kidney-MAPPS demonstrated significantly fewer tubule cells, fewer endothelial cells, and significantly more stromal and infiltrating immune cells as compared with both populations of reference kidneys (Figure 6, D and E). Taken as a whole, these results demonstrate that IMC coupled with Kidney-MAPPS can be used to qualitatively and quantitatively discern individual cell types and reveal expected as well as potentially novel abnormalities in injured tissue.
The normal human kidney.
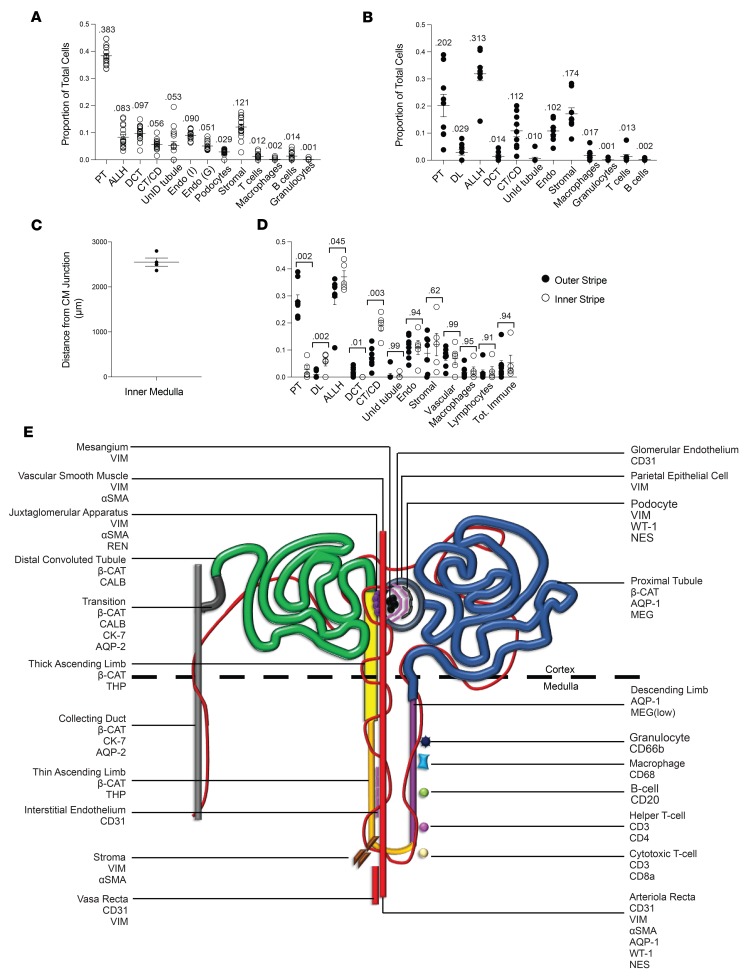
Given the functional equivalence between living donor and nephrectomy tissue in our analysis, these data sets were combined in order to generate an atlas of the human kidney (Figure 7). Cell types, their locations, and their relative abundances were defined for both cortex and medulla (Figure 7, A and B). In the medulla, we detected high variability across samples in terms of the proportion of PT observed (Figure 7B and Supplemental Figure 6A). When all medullary samples were averaged, the sum of the number of cells in the proximal and descending limb (PT + DL) was comparable to the ascending limb cells (ALLH), as would be expected (Figure 7B and Supplemental Figure 6A). However, within individual samples there was a skew towards either the descending or ascending segments, with the proportion of PT correlating inversely with the distance from the region analyzed to the corticomedullary junction (Supplemental Figure 6, A and B). This suggests that there is regional heterogeneity in tubule makeup that is not representative of the medulla as a whole, and that any single core biopsy can be expected to reflect this heterogeneity. We subdivided medulla into outer and inner stripe based on the depth at which the 95th percentile cutoff of PT was reached in each sample, with the outer/inner boundary found to be approximately 2.5 mm from the corticomedullary junction (Figure 7C). Indeed, we found that the proportions of all tubular segments, but not immune, vascular, or stromal cells, was highly dependent on which region of medulla — inner versus outer stripe — was imaged (Figure 7D).
Figure 7. The quantitative cellular makeup of the reference human renal cortex and medulla.
(A and B) Quantification of cells of each indicated phenotype, as a proportion of total cells in human renal cortex (A) and medulla (B); n = 16 for cortex and n = 9 for medulla. Numbers show total proportions. (C) Distance from the corticomedullary junction that defined the inner stripe of medulla, as ascertained by the 95th percentile proximal tubule cell depth in the medulla. n = 4. (D) Comparative quantification of cell types between outer and inner medullary stripe. Outer stripe n = 8. Inner stripe n = 5. Comparisons were made using the Mann-Whitney test with P values indicated. Bars show mean ± standard error of the mean. (E) Cartoon depiction of the nephron, with protein markers and cell types depicted. PT, proximal tubule; ALLH, ascending limb of the loop of Henle (includes thick and thin); DCT, distal convoluted tubule; CM, corticomedullary; CT/CD, connecting tubule and collecting duct; UnID, unidentified; Endo (I), interstitial endothelium; Endo (G), glomerular endothelium; DL, descending limb of the loop of Henle. AQP, aquaporin; COL IV, collagen IV; VIM, vimentin; THP, Tamm-Horsfall protein; MEG, megalin; CALB, calbindin; CK-7, cytokeratin-7; β-CAT, β-catenin; REN, renin; NES, nestin.
Based on the signatures conferred by the 23 markers utilized in this study as well as the spatial information conferred by IMC, 13 distinct cell types could be distinguished via clustering algorithm, and 22 distinct cellular populations recognized by overlaying clustering data onto spatial representations of the kidney (Figure 7E), with an additional 5% of tubular cells that remain unidentified using our current antibodies. We propose that the mean values obtained by applying IMC and Kidney-MAPPS to multiple regions of these 16 kidneys provides a close approximation of the expected proportions in the cortex and medulla of normal human kidneys.
Kidney-MAPPS reveals unexpected cellular heterogeneity, potentially novel cell populations, and ectopic expression of canonical markers.
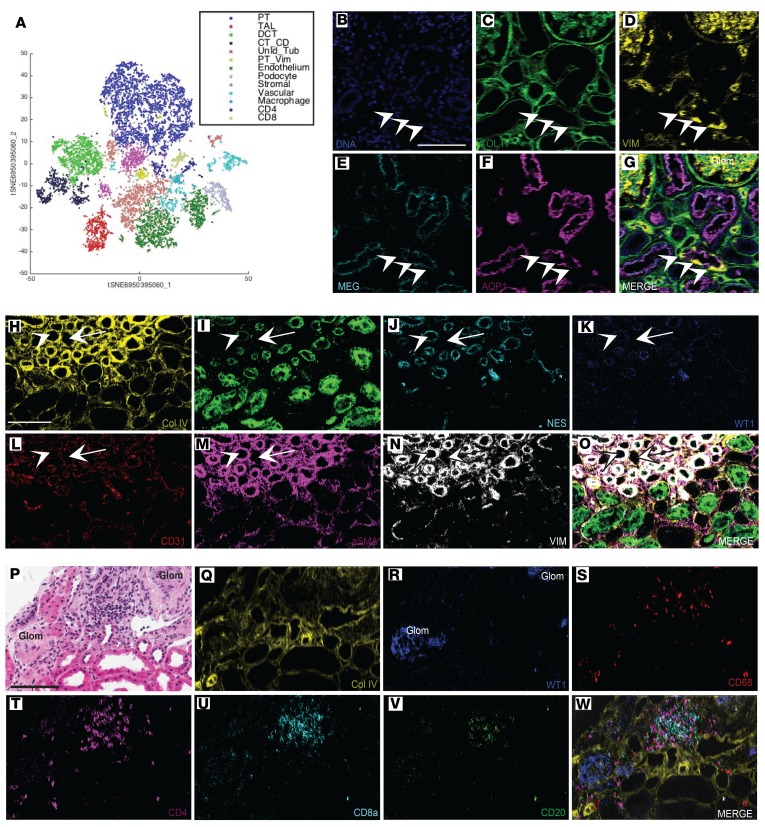
The ability to achieve single-cell identification with the Kidney-MAPPS methodology has more quantitatively defined the transition area between DCT and connecting tubule (Figure 1F, Figure 2H, and Supplemental Figure 3). Our automated methodology has also revealed a population of megalin-positive, aquaporin-1–positive PT cells that is also positive for the intermediate filament vimentin. This rare population was found in low abundance in 7 out of the 16 total reference kidney samples (Figure 8A) and could be clearly identified as single cells within the tubular basement membrane (Figure 8, B–G); these cells may represent an injured, fibrotic, or even regenerative tubule cell type (32).
Figure 8. Kidney-MAPPS reveals potentially novel cell populations, unexpected marker expression, and potentially novel cellular relationships.
(A) t-Distributed stochastic neighbor embedding (t-SNE) from a living donor cortex sample revealing 13 distinct cell populations, including a subset of proximal tubule cells that also expressed vimentin (yellow). (B–G) Pseudocolored IMC reconstructions of a selected region from this kidney showing several vimentin-positive proximal tubule cells. Arrowheads indicate positive cells, which are located within the tubular basement membrane. (H–O) IMC reconstructions showing vasa (arrow) and arteriolae (arrowhead) recta. Arteriolae are characterized by a thicker α-SMA–positive wall, which is also positive for nestin and WT-1. (P) H&E stain of a living donor kidney showing 2 sclerotic glomeruli (Glom). (Q–W) Adjacent section of the same kidney processed by IMC, showing immune infiltrate consisting of CD68-positive cells, CD4-positive cells, CD8a-positive cells, and CD20-positive cells in the region between the 2 sclerotic glomeruli. Scale bars: 100 μm. AQP1, aquaporin-1; Col IV, collagen IV; VIM, vimentin; MEG, megalin; NES, nestin; aSMA, α-smooth muscle actin.
The arteriolae and vasa recta comprise a series of vascular structures that run in parallel in the medulla. We were able to identify these structures in this study based on morphology and coexpression of the vascular markers vimentin, α-SMA, and CD31 (Figure 3L). Unexpectedly, these regions were also found to express the canonical podocyte markers WT-1 and nestin, which have previously been described as being coexpressed in putative vasculogenic niches (Figure 8, H–O) (33).
Immune cells were found in low overall numbers in the reference kidney and were found to consist largely of T cells and macrophages (Supplemental Figure 4, Figure 7, A and B, and Figure 8, P–W). In particular, T cells were associated with sclerotic glomeruli (Figure 8, P–W). In this study, granulocytes were rarely detected, and most granulocytes found in the kidney were also positive for the marker CD68, which can be coexpressed in human neutrophils (Supplemental Figure 3) (34).
Discussion
It is increasingly understood that the development of viable therapies for many forms of kidney disease will require refinements to our understanding of the biology of the normal human kidney, including the interactions between immune and intrinsic renal cells (3, 35). We describe herein the development of a robust technique for multiplexed imaging as well as an associated pipeline for automated data analysis, which can be used for rapid, quantitative, 2-dimensional analysis of the cell types comprising the human kidney.
The heterogeneity and complex spatial arrangement of tubular and interstitial cell types comprising the mammalian kidney have long been appreciated. The various segments of the nephron were first distinguished from each other by painstaking morphological analysis, and more recently have been distinguished by function, consensus protein marker expression, and transcriptome (7, 9). While emerging technologies, including single-cell and single-nucleus RNA sequencing have been important in furthering efforts towards this molecular characterization of the human kidney, dissociation protocols may introduce bias towards specific populations, particularly in fibrotic or injured kidneys (12). Our data will serve as a useful reference for integration with dissociation-based interrogation of the normal kidney. Subsequent work will serve to provide complementary spatial and quantitative data of the injured state, which will further inform our knowledge of human kidney disease.
There are several emerging spatially preserved means of interrogating human kidney tissue that we view as complementary to IMC. Tissue cytometry has been shown to yield quantitative 3-dimensional information about immune cell subsets in human samples simultaneously stained for 6 markers and analyzed by fluorescence (36). Antibody removal–based fluorescent multiplexing can allow for dozens of antibodies to be analyzed on a single tissue section with subcellular resolution, with imaging occurring in an iterative manner (14, 37). Mass spectrometry imaging (MSI) has been used to yield quantitative information about metabolites in human kidney with a resolution ranging from 10 μm to 200 μm (38). An IMC-based approach can complement data obtained with these additional technologies by offering a high degree of multiplexing, the absence of background fluorescence, the ability to perform imaging in a single acquisition, reproducibility both in tissue and longitudinally, and the resultant ability to quantitatively analyze all channels in a single data set in an unsupervised fashion and without image subtraction, adjustment, or normalization. Additionally, IMC capabilities have recently been expanded to also include simultaneous localization of protein and mRNA targets (39).
Weaknesses of current IMC technology include the limitations on resolution (1 μm) owing to the width of the ablating laser pulse. Fluorescence-based imaging, by offering higher resolution and 3-dimensional capabilities, will complement findings by IMC and MSI, particularly in subcellular localization and in characterizing cells in the tightly interwoven structure of the glomerulus. Notably, the related mass cytometry platform multiplexed ion beam imaging (MIBI) supports multiplexed imaging of metal-conjugated antibodies at a resolution approaching 200 nm (40). Future studies employing the antibody reagents and analysis pipeline defined here may take advantage of that increased resolution.
Human AKI is rarely biopsied, unless glomerular disease or acute interstitial nephritis is suspected, and the ability to analyze human AKI specimens will initially rely on archival patient tissue, while larger-scale efforts are underway to prospectively bank human AKI specimens (3, 35). A critical advantage of our approach lies in the fact that Kidney-MAPPS can be used on both prospectively acquired and archived kidney biopsies. In fact, we have demonstrated the use of this technique on samples banked over 8 years prior to analysis and found no evidence of signal attenuation. Because preserved samples provide excellent labeling, imaging and analysis of control and injured samples can occur in parallel, minimizing potential technical confounders. We also believe that our strategy for cell segmentation could be readily adapted for many other solid organs, including the liver, which are composed of a complex network of epithelial, stromal, and endothelial cells.
As the field of nephrology moves toward comparative interrogation of the reference and diseased human kidney, an outstanding question remains: what source of tissue should be used as the reference standard? Absent biopsy tissue from healthy normal donors, several sources have been proposed. These include biopsies from living donors as well as nephrectomy tissue in areas remote from tumors, biopsies from patients with minimal change disease, and deceased donor tissue (24, 41). In this study, we use Kidney-MAPPS to demonstrate that careful selection of rare nephrectomy specimens that have been scored as morphologically normal can serve as appropriate surrogates for reference kidneys in future analyses.
An ongoing concern in the interpretation of human kidney biopsies is the degree to which each biopsy specimen is subject to sampling bias, and how the cellular makeup varies across randomly sampled regions of a kidney (31). Dropout of particular cell types, such as endothelium and specific tubule subsets, has been postulated to play a role in AKI and chronic kidney disease (42, 43). But a robust analysis of this requires a baseline understanding of how much those cell types vary in the normal kidney, and how that variability is influenced by kidney location and sampling bias. This study has allowed us to quantify the degree of variability in biopsy-sized regions of normal kidney samples, as well as between remote regions and serial sections of the same kidney. In the medulla, we have demonstrated that the tubular cells are organized in complex macroscopic arrays that can greatly influence the cellular makeup of a biopsy-sized region. For example, the relative proportion of PT and ALLH varies according to the distance from the corticomedullary junction that is being sampled, implying that human kidney biopsies exhibit a normal range of region-dependent sample bias for certain cell types. By providing a quantitative assessment of each cell type in each region of the kidney, these data will allow researchers, pathologists, and clinicians to directly compare cell proportions in each biopsy to the expected variability of the normal kidney, thus rapidly identifying the critical cell compartments that are impacted by specific pathologic insults. We demonstrate an application of this approach in our analysis of kidneys containing tumor-associated interstitial infiltrate, in which endothelial and tubular dropout were quantified and the stromal and immune compartments found to be vastly expanded. Indeed, we believe that the degree of endothelial dropout noted in areas of chronic tumor-associated infiltrate is a novel finding that bears further exploration in other forms of chronic kidney disease.
Additional potentially novel findings in this study included unexpected heterogeneity in tubular regions and ectopic expression of canonical markers. Cells of the mammalian DCT are characterized by expression of the intracellular calcium-binding protein calbindin (8, 44). However, marked heterogeneity of calbindin expression was observed, with 1–3 cells per tubular cross section clearly not expressing this protein (Figure 1D and Figure 2E). Although small numbers of intercalated cells are believed to populate distal regions of the DCT, the calbindin-negative cells were found throughout the DCT and appear larger and more cuboidal than a typical intercalated cell (45). Thus, these may represent a novel cell type relevant to future investigations. The transitional region between DCT and connecting tubule was characterized by cells with mosaic expression of calbindin, cytokeratin-7, and aquaporin-2, in which cytokeratin-7 and aquaporin-2 were coexpressed in cells that were mutually exclusive from those that expressed calbindin; these cytokeratin-7–positive, aquaporin-2–positive cells likely represent principal cells. While we did not seek to further explore heterogeneity in the collecting duct or to further delineate the ratios between principal and intercalated cells in this study, this could be an interesting area for future application of this technique, particularly given emerging evidence about the importance of intercalated cell dropout in human disease (10). Additional cell types relevant to human disease that could be better characterized with this methodology include CD133-positive cells, which have been proposed as tubule progenitor cells in the adult human kidney (46, 47).
Using β-catenin, aquaporin-1, and megalin, we were able to identify the vast majority of the PT, though we were not able to distinguish between the S1, S2, and S3 segments via unsupervised analyses, based on the markers selected. Furthermore, a population of unidentified tubular cells, comprising approximately 5% of total cells, was found to consist predominantly of PT cells with artifactually low levels of staining. As with the case of distinguishing between principal and intercalated cells, we predict that an expanded antibody panel will limit the number of unidentified cells and allow us to better distinguish the major cell constituents of the PT. Our unbiased analysis technique did identify a subset of PT cells, found in 7 of 16 human samples, which coexpressed the intermediate filament vimentin. Such cells have been postulated to represent regenerating tubuloepithelium (32). In several samples, these cells were found to associate closely with CD4+ T cells (data not shown). Additional work using this technique, including costaining for markers of proliferation and injury such as Kim-1, Ki-67, and MCP1 will be useful in determining whether these cells are in fact regenerating tubular cells, and whether there are important associations between immune and intrinsic renal cells that influence injury, regeneration, and repair (48, 49).
In summary, we have developed an IMC-based protocol and analysis pipeline that was used to generate a spatially preserved single-cell-resolution atlas of the normal human kidney, with greater than 20 markers analyzed simultaneously. We have used this technique to identify potentially novel and unexpected cell types and to contrast the normal and abnormal kidney. We believe that expansion of our antibody panel to include specific subtypes of kidney cell injury, greater immune cell coverage, and novel cell types identified by single-cell and single-nucleus RNA sequencing holds the potential to greatly expand our understanding of the pathogenesis of human kidney disease.
Methods
Antibody selection.
Antigens were selected from canonical DEGs in previously published single-cell and single-nucleus RNA sequencing from human kidney (7, 9). Commercial antibodies against intrinsic renal cell types were selected based on citations in the literature where available, validation data presented on the manufacturer’s website, and validation data made publicly available through the (Re)Building a Kidney consortium (23). BSA-free, glycerol-free formulations of antibodies were used for validation steps as delineated below. Immune markers were selected from validated commercially available metal-conjugated antibodies (Fluidigm Corporation) and were subjected to the additional validation steps defined below.
Antibody validation and conjugation.
Antibodies were validated by performing immunostaining using a single common protocol on human kidney and lymph node. For intrinsic renal markers, staining was analyzed morphologically by a renal pathologist blinded to the antibody used. Further validation was performed for selected antibodies by counterstaining for additional markers of the same cell type as indicated. Antibody validation data have been deposited on the (Re)Building a Kidney website https://www.rebuildingakidney.org/chaise/recordset/#2/Antibody:Antibody_Tests@sort(RID) (23). Immune markers were validated on human lymph node tissue, with staining confirmed by IMC. Intrinsic renal antibodies that were validated by immunofluorescence were conjugated using the Maxpar conjugation kit (Fluidigm Corporation) following the manufacturer’s protocol, scaled for 100 μg of carrier-free antibody. Metals were selected based on expected signal strength and to minimize overlap due to isotopic impurity. After conjugation, validation was performed by IMC (Supplemental Figure 1); for approximately 70% of antibodies, conjugation did not affect the observed binding pattern, and these antibodies were therefore included in the final antibody panel (Table 1).
IMC.
Immunostaining protocols are available on the (Re)Building a Kidney website (23). Except for the serial section validation cohort, immunostaining was performed on all 16 formalin-fixed, paraffin-embedded samples on a single day using the same antibody cocktail, and IMC was performed over a 6-week span, alternating between nephrectomy and living donor samples. Of injured kidneys, one was stained in parallel with all normal samples, and the other 2 were stained from a separate antibody cocktail of identical composition. For nephrectomy tissue, regions of interest were selected to encompass contiguous areas from cortex to medulla in areas free of large discontinuities in the tissue. For donor tissue, regions of interest were selected to span the entire length of the available tissue from cortex to medulla. After selection of regions of interest, IMC was performed as described using the Hyperion Imaging System (Fluidigm Corporation). Tissue of 5-μm thickness was ablated with a Nd:YAG 213 nm laser and analyzed in a Fluidigm CyTOF mass cytometer (13). Images were analyzed using MCD Viewer (Fluidigm, version 1.0). For all images shown, thresholding was performed in MCD Viewer with gamma set to 1, minimum color range set to zero, and maximum color range set to 100. Thresholding was performed to remove background signal, as assessed morphologically. For all comparative images, signal thresholds were set identically for each marker between samples to allow for unbiased comparison. Analyses of cortex and medulla were performed separately; medulla was defined morphologically.
Pixel classification.
For machine learning purposes, upper and lower intensity thresholds were set manually for each channel in MCD Viewer to accentuate signal and reduce noise. Four separate tiff image stacks were created for each sample by overlaying relevant channels: nuclei (histone H3, DNA intercalator 1, DNA intercalator 2); tubular membranes (β-catenin, aquaporin-1, megalin, THP, calbindin, aquaporin-2, cytokeratin-7); endothelial membranes (CD31); and interstitial/glomerular membranes (all other membrane channels). For each sample, individual channels were excluded for segmentation purposes if poor signal or high background staining was likely to hinder cell border determination. Each of these 4 image stacks was uploaded into Ilastik version 1.3.0 (https://www.ilastik.org/), an open-source software for pixel classification. In Ilastik, pixels were identified as belonging to nuclei, 1 of the 3 membrane types above, or background in an initial training set consisting of 4 samples (2 nephrectomy and 2 living donor). This classification was then applied to all remaining images using a machine-learning approach, and probability images were generated.
Cell segmentation.
Probability images for each sample were ultimately exported as tiff files into Cell Profiler version 3.1.8 for cell segmentation and mask generation (50). First, nuclei were identified by size and shape as primary objects based on the nuclear probability image. Tubular cells were defined by creating secondary objects based on the tubular membrane image and filtering for objects containing high tubular probability; nuclei assigned to tubular cells were then removed. From the remaining non-tubular nuclei, secondary objects were created using the endothelial membrane image and filtered to select objects containing a higher integrated probability intensity of endothelial membranes than other interstitial/glomerular membranes. Endothelial nuclei were then removed. Finally, all remaining nuclei were assigned to secondary objects based on the interstitial/glomerular membrane probability image. These 3 filtered cellular “sub-masks” (tubular, endothelial, and other) were subtracted from one another to remove pixel overlap and then merged to create a single segmentation mask for that entire image.
Multiplexed data analysis.
Raw images for each channel were exported from MCD Viewer without any adjustments or thresholding. Samples and their corresponding masks were cropped where necessary to allow for separate analysis of cortex and medulla. Each sample and mask was imported into HistoCAT version 1.75, an open source software specifically developed for single-cell analysis of multiplexed cytometry data (30). Unbiased clustering was performed using the PhenoGraph algorithm, a nearest-neighbor–based clustering approach, and each cluster was assigned a phenotype based on its expression profile (51). Similar clusters were merged to obtain a total cell number for each phenotype.
Data validation.
To validate our method for cell segmentation and quantification, results obtained by Kidney-MAPPS on selected representative sections were compared to results obtained by manual quantification of the same sections. Manual quantification was performed by a blinded observer using direct counting of cells of each phenotype on thresholded tiff images using Microsoft Paint. Results for selected markers were directly compared to results obtained through the above data processing methods. To further validate our approach, we imaged multiple areas from 4 large nephrectomy samples and used the Kidney-MAPPS data analysis pipeline to quantify intrasample variability. Areas selected for validation were greater than 1 cm from previously imaged regions, and qualitatively comprised the same ratio of cortex and medulla. In an additional validation step, a new antibody cocktail was utilized in order to stain adjacent sections of the same 4 kidneys, and regions corresponding to the original regions of interest were selected for ablation and analysis.
Medullary depth.
Medulla and cortex were distinguished morphologically based on distinguishing characteristics including the exclusion of glomeruli in the medulla and were analyzed separately after cropping images that spanned both regions. After clustering and phenotyping, each medullary sample that was noted morphologically to span both the outer and inner stripes was exported from HistoCAT as a csv file, which includes both the cluster assignment and the coordinates of each cell. Of the cells contained within clusters identified as PT, the 95th percentile x or y coordinate was calculated in Microsoft Excel, depending on the tissue orientation. These values were utilized to calculate the depth at which the 95th percentile medullary PT cell was located from the corticomedullary junction.
Statistics.
Wilcoxon’s matched-pairs signed-rank test was used to compare total cell number, as well as the number of cells expressing specific markers, as ascertained by manual counting versus automated methodologies. This test was further used to compare total cell proportions from original versus validation regions of interest from the same kidneys. The Mann-Whitney test was used to compare proportions of total cells between nephrectomy, living donor, and interstitial nephritis tissue, as well as to compare cellular proportions in outer versus inner medullary stripe. The Student’s 2-tailed t test was used to compare the ages as well as serum creatinine levels between nephrectomy and living donor patients. Fisher’s exact test was used to compare the proportion of female, hypertensive, and diabetic patients, as well as the proportion of patients with estimated glomerular filtration rate ≥60 mL/min/1.73 m2. All dot plots for quantitative data are presented to show mean ± standard error of the mean.
Study approval.
Human tissue was obtained in accordance with the policies of Yale University’s Human Investigations Committee (HIC). Written informed consent for research use of patient samples was obtained from each patient prior to study inclusion at the time of sample collection by the performing clinician. Tumor remote samples from nephrectomies were collected prospectively from 2016 until 2018. Living donor samples were collected prospectively from 2018 onward. All prospectively accrued samples were deposited fresh into 10% formalin, fixed overnight at room temperature, and dehydrated in 70% ethanol prior to paraffin embedding and sectioning to 5-μm thickness. Additional banked nephrectomy samples obtained between 2009 and 2017 were acquired retrospectively after chart review. Patient charts were reviewed in accordance with guidelines of Yale’s Joint Data Analytics Team, to ensure that informed consent had not been withdrawn in the intervening time after sample collection. For nephrectomy patients and living donors, the following information, when available, was obtained from the electronic medical record, from the time of sample collection: patient age, sex, race, serum creatinine level, estimated glomerular filtration rate, history of hypertension, history of diabetes, and tissue diagnosis (specifically for patients undergoing nephrectomy). All data were deidentified and maintained on a secure database. For nephrectomy specimens, formal pathology reports were used. For living donor samples, samples were read by a renal pathologist in a blinded manner.
Author contributions
NS and LGC conceived of and designed all experiments. NS, ZMA, and JAK performed all experiments. JAK and ZMA developed the data analysis pipeline and ZMA performed data analysis. NS, ZMA, BMS, and LGC designed human investigative committee protocols. NS, ZMA, and BMS obtained human samples. NS and ZMA performed patient chart review. GWM is the pathologist who performed antibody validation and pathological scoring. RRM provided conceptual information and reagent guidance. NS compiled all figures and tables and wrote the manuscript. All authors reviewed and contributed to the final manuscript.
Supplementary Material
Acknowledgments
We thank Shelly Ren and the Yale CyTOF facility for technical assistance and Daniel Carvajal-Hausdorf, David Rimm, Kurt Schalper, and Franz Villarroel for helpful discussion and technical guidance. We thank Marta Boeke, Lisa Gras, and Yalai Bai for assistance with collecting human specimens. We thank Madeleine Hamilton for contributions to antibody optimization. We thank Ricarda Tomlin and Sanjay Kulkarni for assistance with living donor sample acquisition. We thank our funding sources, including the National Institute of Diabetic and Digestive and Kidney Diseases via (Re)Building a Kidney and DiaComp grants (to LGC). Additional support came from the Robert E. Leet and Clara Guthrie Patterson Trust Mentored Research Award (to NS), NIH T32 (to NS and ZMA), and the TRENAL program (to JAK).
Version 1. 06/20/2019
Electronic publication
Footnotes
JAK’s present address is: Charité Universitätsmedizin, Berlin, Germany.
BMS’s present address is: Department of Urology, Ronald Reagan UCLA Medical Center, Los Angeles, California, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(12):e129477. https://doi.org/10.1172/jci.insight.129477.
Contributor Information
Nikhil Singh, Email: nikhil.singh@yale.edu.
Zachary M. Avigan, Email: zachary.avigan@yale.edu.
Judith A. Kliegel, Email: judith.kliegel@charite.de.
Brian M. Shuch, Email: bshuch@mednet.ucla.edu.
Ruth R. Montgomery, Email: ruth.montgomery@yale.edu.
Gilbert W. Moeckel, Email: gilbert.moeckel@yale.edu.
Lloyd G. Cantley, Email: lloyd.cantley@yale.edu.
References
- 1.Norton JM, Ketchum CJ, Narva AS, Star RA, Rodgers GP. Complementary initiatives from the NIDDK to advance kidney health. Clin J Am Soc Nephrol. 2017;12(9):1544–1547. doi: 10.2215/CJN.02120217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonventre JV, et al. AKI: a path forward. Clin J Am Soc Nephrol. 2013;8(9):1606–1608. doi: 10.2215/CJN.06040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonventre JV, et al. The kidney research national dialogue: gearing up to move forward. Clin J Am Soc Nephrol. 2014;9(10):1806–1811. doi: 10.2215/CJN.07310714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gluck C, Ko YA, Susztak K. Precision medicine approaches to diabetic kidney disease: tissue as an issue. Curr Diab Rep. 2017;17(5):30. doi: 10.1007/s11892-017-0854-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang WR, Parikh CR. Biomarkers of acute and chronic kidney disease. Annu Rev Physiol. 2019;81:309–333. doi: 10.1146/annurev-physiol-020518-114605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kretzler M, et al. Repuncturing the renal biopsy: strategies for molecular diagnosis in nephrology. J Am Soc Nephrol. 2002;13(7):1961–1972. doi: 10.1097/01.ASN.0000020390.29418.70. [DOI] [PubMed] [Google Scholar]
- 7.Wu H, et al. Single-cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J Am Soc Nephrol. 2018;29(8):2069–2080. doi: 10.1681/ASN.2018020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu H, Uchimura K, Donnelly EL, Kirita Y, Morris SA, Humphreys BD. Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell Stem Cell. 2018;23(6):869–881.e8. doi: 10.1016/j.stem.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu H, Kirita Y, Donnelly EL, Humphreys BD. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol. 2019;30(1):23–32. doi: 10.1681/ASN.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park J, et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360(6390):758–763. doi: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindström NO, De Sena Brandine G, Ransick A, McMahon AP. Single-cell RNA sequencing of the adult mouse kidney: from molecular cataloging of cell types to disease-associated predictions. Am J Kidney Dis. 2019;73(1):140–142. doi: 10.1053/j.ajkd.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Sullivan ED, Mylonas KJ, Hughes J, Ferenbach DA. Complementary roles for single-nucleus and single-cell RNA sequencing in kidney disease research. J Am Soc Nephrol. 2019;30(4):712–713. doi: 10.1681/ASN.2019020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giesen C, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11(4):417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 14.Bodenmiller B. Multiplexed epitope-based tissue imaging for discovery and healthcare applications. Cell Syst. 2016;2(4):225–238. doi: 10.1016/j.cels.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, et al. Spectral characteristics of autofluorescence in renal tissue and methods for reducing fluorescence background in confocal laser scanning microscopy. J Fluoresc. 2018;28(2):561–572. doi: 10.1007/s10895-018-2217-4. [DOI] [PubMed] [Google Scholar]
- 16.Carvajal-Hausdorf DE, et al. Multiplexed (18-plex) measurement of signaling targets and cytotoxic T cells in trastuzumab-treated patients using imaging mass cytometry. Clin Cancer Res. 2019;25(10):3054–3062. doi: 10.1158/1078-0432.CCR-18-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang YJ, et al. Multiplexed in situ imaging mass cytometry analysis of the human endocrine pancreas and immune system in type 1 diabetes. Cell Metab. 2019;29(3):769–783.e4. doi: 10.1016/j.cmet.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Damond N, et al. A map of human type 1 diabetes progression by imaging mass cytometry. Cell Metab. 2019;29(3):755–768.e5. doi: 10.1016/j.cmet.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huen SC, Cantley LG. Macrophages in renal injury and repair. Annu Rev Physiol. 2017;79:449–469. doi: 10.1146/annurev-physiol-022516-034219. [DOI] [PubMed] [Google Scholar]
- 20.Huen SC, Huynh L, Marlier A, Lee Y, Moeckel GW, Cantley LG. GM-CSF promotes macrophage alternative activation after renal ischemia/reperfusion injury. J Am Soc Nephrol. 2015;26(6):1334–1345. doi: 10.1681/ASN.2014060612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berry MR, et al. Renal sodium gradient orchestrates a dynamic antibacterial defense zone. Cell. 2017;170(5):860–874.e19. doi: 10.1016/j.cell.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 22.Brähler S, et al. Opposing roles of dendritic cell subsets in experimental GN. J Am Soc Nephrol. 2018;29(1):138–154. doi: 10.1681/ASN.2017030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oxburgh L, et al. (Re)Building a Kidney. J Am Soc Nephrol. 2017;28(5):1370–1378. doi: 10.1681/ASN.2016101077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muruve DA, et al. The biobank for the molecular classification of kidney disease: research translation and precision medicine in nephrology. BMC Nephrol. 2017;18(1):252. doi: 10.1186/s12882-017-0669-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko YA, et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol. 2013;14(10):R108. doi: 10.1186/gb-2013-14-10-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woroniecka KI, Park AS, Mohtat D, Thomas DB, Pullman JM, Susztak K. Transcriptome analysis of human diabetic kidney disease. Diabetes. 2011;60(9):2354–2369. doi: 10.2337/db10-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montgomery TA, et al. Breast regression protein-39/chitinase 3-like 1 promotes renal fibrosis after kidney injury. J Am Soc Nephrol. 2017;28(11):3218–3226. doi: 10.1681/ASN.2017010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eng DG, et al. Detection of renin lineage cell transdifferentiation to podocytes in the kidney glomerulus with dual lineage tracing. Kidney Int. 2018;93(5):1240–1246. doi: 10.1016/j.kint.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomez RA. Fate of renin cells during development and disease. Hypertension. 2017;69(3):387–395. doi: 10.1161/HYPERTENSIONAHA.116.08316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schapiro D, et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Methods. 2017;14(9):873–876. doi: 10.1038/nmeth.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Visconti L, et al. Renal biopsy: Still a landmark for the nephrologist. World J Nephrol. 2016;5(4):321–327. doi: 10.5527/wjn.v5.i4.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smeets B, et al. Proximal tubular cells contain a phenotypically distinct, scattered cell population involved in tubular regeneration. J Pathol. 2013;229(5):645–659. doi: 10.1002/path.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasuri F, et al. Nestin and WT1 expression in small-sized vasa vasorum from human normal arteries. Histol Histopathol. 2012;27(9):1195–1202. doi: 10.14670/HH-27.1195. [DOI] [PubMed] [Google Scholar]
- 34.Amanzada A, et al. Identification of CD68(+) neutrophil granulocytes in in vitro model of acute inflammation and inflammatory bowel disease. Int J Clin Exp Pathol. 2013;6(4):561–570. [PMC free article] [PubMed] [Google Scholar]
- 35.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372(9):793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winfree S, et al. Quantitative three-dimensional tissue cytometry to study kidney tissue and resident immune cells. J Am Soc Nephrol. 2017;28(7):2108–2118. doi: 10.1681/ASN.2016091027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolognesi MM, et al. Multiplex staining by sequential immunostaining and antibody removal on routine tissue sections. J Histochem Cytochem. 2017;65(8):431–444. doi: 10.1369/0022155417719419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grove KJ, et al. Imaging mass spectrometry reveals direct albumin fragmentation within the diabetic kidney. Kidney Int. 2018;94(2):292–302. doi: 10.1016/j.kint.2018.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schulz D, et al. Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. Cell Syst. 2018;6(4):531. doi: 10.1016/j.cels.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Angelo M, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20(4):436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer MB, Vichot AA, Cantley LG, Moeckel GW. Quantification and localization of M2 macrophages in human kidneys with acute tubular injury. Int J Nephrol Renovasc Dis. 2014;7:415–419. doi: 10.2147/IJNRD.S66936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basile DP, Anderson MD, Sutton TA. Pathophysiology of acute kidney injury. Compr Physiol. 2012;2(2):1303–1353. doi: 10.1002/cphy.c110041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kramann R, Wongboonsin J, Chang-Panesso M, Machado FG, Humphreys BD. Gli1. J Am Soc Nephrol. 2017;28(3):776–784. doi: 10.1681/ASN.2016030297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee CT, Ng HY, Lee YT, Lai LW, Lien YH. The role of calbindin-D28k on renal calcium and magnesium handling during treatment with loop and thiazide diuretics. Am J Physiol Renal Physiol. 2016;310(3):F230–F236. doi: 10.1152/ajprenal.00057.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reilly RF, Ellison DH. Mammalian distal tubule: physiology, pathophysiology, and molecular anatomy. Physiol Rev. 2000;80(1):277–313. doi: 10.1152/physrev.2000.80.1.277. [DOI] [PubMed] [Google Scholar]
- 46.Sagrinati C, et al. Isolation and characterization of multipotent progenitor cells from the Bowman’s capsule of adult human kidneys. J Am Soc Nephrol. 2006;17(9):2443–2456. doi: 10.1681/ASN.2006010089. [DOI] [PubMed] [Google Scholar]
- 47.Romagnani P, Remuzzi G. CD133+ renal stem cells always co-express CD24 in adult human kidney tissue. Stem Cell Res. 2014;12(3):828–829. doi: 10.1016/j.scr.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 48.Han WK, Bailly V, Abichandani R, Thadhani R, Bonventre JV. Kidney injury molecule-1 (KIM-1): a novel biomarker for human renal proximal tubule injury. Kidney Int. 2002;62(1):237–244. doi: 10.1046/j.1523-1755.2002.00433.x. [DOI] [PubMed] [Google Scholar]
- 49.Moledina DG, et al. Plasma monocyte chemotactic protein-1 is associated with acute kidney injury and death after cardiac operations. Ann Thorac Surg. 2017;104(2):613–620. doi: 10.1016/j.athoracsur.2016.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamentsky L, et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011;27(8):1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levine JH, et al. Data-Driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162(1):184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.