Abstract
Inhibition of Bruton tyrosine kinase (BTK) is a breakthrough therapy for certain B cell lymphomas and B cell chronic lymphatic leukemia. Covalent BTK inhibitors (e.g., ibrutinib) bind to cysteine C481, and mutations of this residue confer clinical resistance. This has led to the development of noncovalent BTK inhibitors that do not require binding to cysteine C481. These new compounds are now entering clinical trials. In a systematic BTK mutagenesis screen, we identify residues that are critical for the activity of noncovalent inhibitors. These include a gatekeeper residue (T474) and mutations in the kinase domain. Strikingly, co-occurrence of gatekeeper and kinase domain lesions (L512M, E513G, F517L, L547P) in cis results in a 10- to 15-fold gain of BTK kinase activity and de novo transforming potential in vitro and in vivo. Computational BTK structure analyses reveal how these lesions disrupt an intramolecular mechanism that attenuates BTK activation. Our findings anticipate clinical resistance mechanisms to a new class of noncovalent BTK inhibitors and reveal intramolecular mechanisms that constrain BTK’s transforming potential.
Keywords: Hematology, Oncology
Keywords: Lymphomas, Protein kinases
A mutagenesis screen identifies mutations in Bruton Tyrosine Kinase (BTK) that contribute to resistance to second generation BTK inhibitors and unleash acquired transforming activity.
Introduction
Pharmacological inhibitors of Bruton tyrosine kinase (BTK) show striking clinical activity against B cell chronic lymphatic leukemia and some forms of B cell lymphoma (1–6). However, the emergence of clinical resistance remains a critical concern. For example, the covalent BTK inhibitor (BTKi) ibrutinib acts by binding to BTK cysteine (C481), and mutations of this residue are associated with clinical ibrutinib resistance (7–10). Second-generation, noncovalent BTKis, such as RN486, GDC-0853, and SNS-062, do not require binding to C481 and are designed to retain activity in C481-mutant cancers (11–19). These new compounds are now entering clinical testing, and potential causes of resistance have not been reported (14, 17).
BTK mediates the signal transduction from the B cell receptor (BCR) to downstream NF-κB and PI3K/AKT molecules (20, 21). BTK is required for normal B cell development and function, and inherited BTK loss-of-function mutations can result in X-linked agammaglobulinemia (22–24). Most components of the BCR signaling pathway, including BTK, are targets of recurrent somatic mutations in lymphoma (25–32). However, neither wild-type BTK nor the mutant BTK alleles observed in treatment-naive and ibrutinib-treated patients are considered oncogenic (22, 33). Here, we report results of a BTK mutagenesis screen that uncovers BTK residues that are required for noncovalent inhibitor interactions and that constrain BTK’s transforming activity.
Results
Probing BTK interactions with covalent and noncovalent inhibitors.
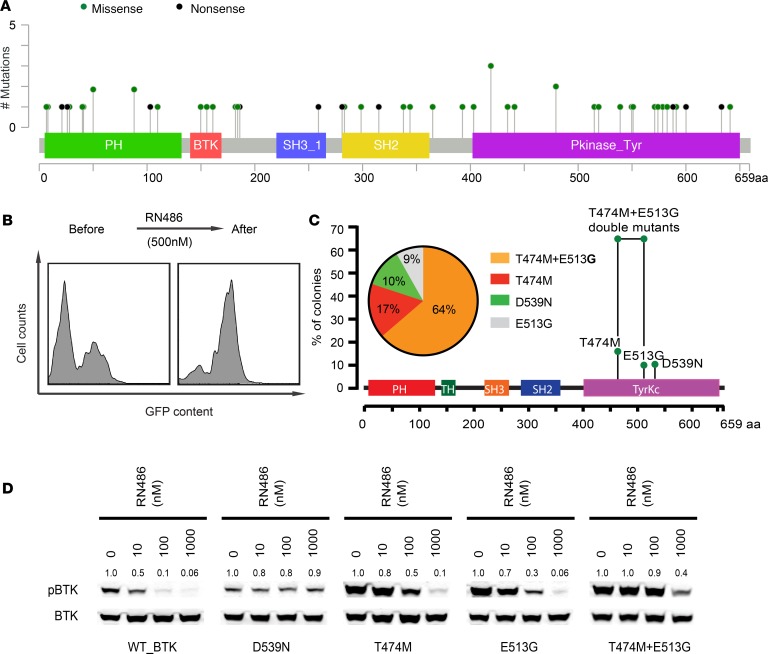
In ibrutinib-naive patients, BTK mutations occur with a frequency of approximately 3%, and no apparent mutation hotspot has been reported (refs. 26–29) (Figure 1A, Supplemental Figure 1A, and Supplemental Tables 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.127566DS1). We comprehensively mutagenized the BTK coding sequence (CDS) using repair-defective E. coli, reaching an overall nucleotide variant frequency of 90%, and screened the resulting library of mutant BTK CDSs (expressed along the GFP reporter) for escape from ibrutinib and the new, noncovalent BTKis in murine BCL1 lymphoma cells (Supplemental Figure 1, B–D; Supplemental Figure 2, A and B; and Supplemental Tables 3 and 4). Ibrutinib covalently binds to BTK at cysteine C481, and mutation of this site confers resistance (7–9, 34, 35). We readily observed outgrowth of library and GFP-expressing BCL1 cells under ibrutinib (50 nM, 4 weeks), confirming the performance of our assay (Supplemental Figure 1, B and C). In detail, 74% of 350 clones showed BTK C481 mutations (C481Y, C481R); other mutations were new to our knowledge, including L528W (25%, 88/350) and T474M (1%; 5/350; Supplemental Figure 1D). The L528W mutation has been seen in ibrutinib-naive lymphoma patients at low frequency and in 1 chronic lymphatic leukemia (CLL) patient upon ibrutinib relapse but had not been linked to ibruitinib resistance before (refs. 27, 28, 36; Figure 1A, Supplemental Figure 1A, and Supplemental Tables 1 and 2). These data indicate the accuracy and ability of our screening system to identify relevant BTKi resistance mutations.
Figure 1. BTK mutations affecting noncovalent BTKi.
(A) BTK mutations identified in 50 ibrutinib-naive lymphoma specimens from 1575 sequenced samples in this study. Detailed amino acid change and patient data are included in Supplemental Table 1. (B) FACS analysis showing GFP and mutant BTK enrichment under RN486 treatment. (C) Summary of BTK sequencing results from 281 enriched clones; notably BTK (T474M and E513G) occurs as a double mutant in cis. (D) Immunoblot analysis of BTK Y223 autophosphorylation in HEK293T cells that express the indicated BTK alleles and lack endogenous BTK, treated with RN486. Total BTK was used as a control and quantification was done with ImageJ (NIH).
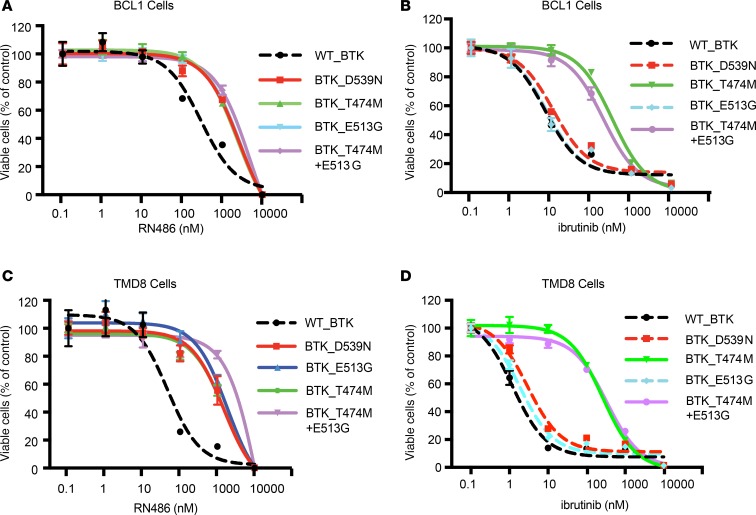
Noncovalent BTKis do not require binding to C481 to block BTK activity; RN486 represents this class of drugs (11, 13–16). We screened our mutant BTK CDS library to identify residues that are critical for RN486 activity (500 nM, 4 weeks; Figure 1B). As expected, the spectrum of enriched mutations was completely different, and surprisingly, the majority (64% of clones; 179/281) contained a BTK CDS with 2 mutations (T474M and E513G; Figure 1C). The T474M and E513G mutations were individually detected at lower frequencies (T474M: 17%; E513G: 9%), and another, D539N, was enriched in up to 10% of refractory clones. We confirmed the effect of BTK mutations on BTK Y223 autophosphorylation in HEK293T cells (that do not express endogenous BTK; refs. 7, 8; Figure 1D and Supplemental Figure 1E). We also confirmed protection from cell proliferation inhibition by ibrutinib and RN486 in BCL1 and TMD8 lymphoma cells expressing the BTK mutants (Figure 2 and Supplemental Figure 3). Although the C481 mutations did not affect RN486, both the T474M and the double-mutant BTK (T474M and E513G) impaired the effects of ibrutinib and RN486 (Figure 2 and Supplemental Figure 3). We confirmed these effects with another covalent BTKi, acalabrutinib, and noncovalent BTKi, GDC-0853, whereas SNS-062 showed insufficient BTK inhibition in our hands (Supplemental Figure 4). The BTK D539N mutation has been observed in ibrutinib-naive lymphoma patients at low frequency (Figure 1A and Supplemental Table 1). Clinical data on resistance mutations to the noncovalent inhibitors are not yet available. Our data anticipate potential mechanisms of resistance to noncovalent BTKis, including a mutation in a gatekeeper residue (T474 → M747) that require 2 nucleotide changes (ACT → ATG).
Figure 2. Validation of screening results with different BTKis.
(A and B) Cell proliferation assay of BCL1 lymphoma cells expressing the BTK wild-type or mutant alleles treated with RN486 (A) or ibrutinib (B) at the indicated dose range for 72 hours. Viable cells were determined by CellTiter-Glo and normalized to DMSO treatment. (C and D) Analogous results from TMD8 cells treated with RN486 (C) or ibrutinib (D).
Modeling the structural effects of L528W and T474M mutations.
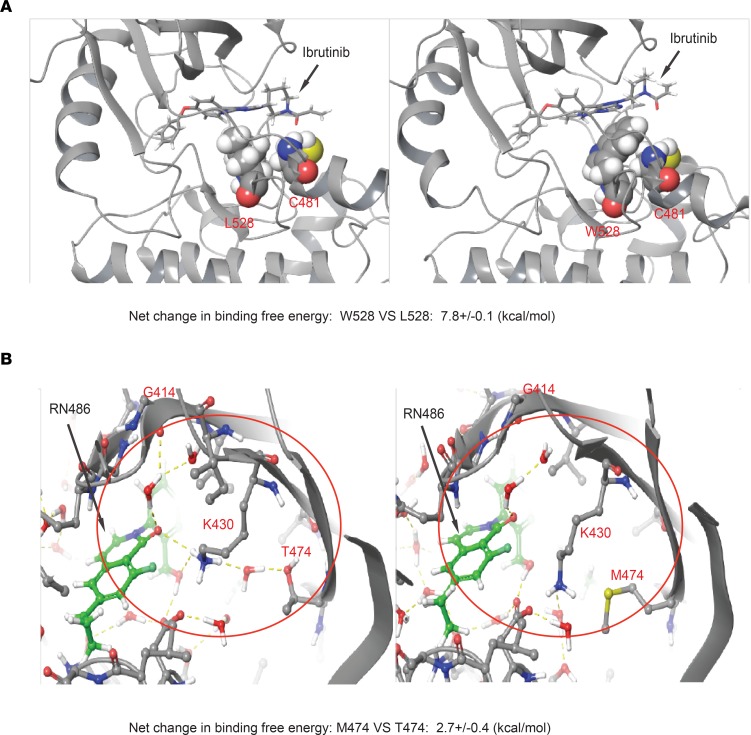
The BTK mutations L528W and T474M impair BTKi action, and we performed computational structure modeling to understand how they interact with the inhibitors. The L528W mutation causes steric hindrance such that ibrutinib is pushed away from the C481 attachment site, causing a net calculated increase in free energy for ibrutinib binding to C481 by 7.8 kcal/mol (Figure 3A). The T474M mutation affects both covalent and noncovalent BTKis. Briefly, wild-type T474 residue engages with an H-bonded network that spans from K430, to T474, to G414 and coordinates inhibitor (RN486) binding (Figure 3B, highlighted in yellow). In the T474M-mutant protein, this hydrogen network is disrupted, which results in an increase in free energy for RN486 binding to BTK of 2.7 kcal/mol (Figure 3B). This pattern is very much reminiscent of the T315I gatekeeper mutation in the fused BCR-ABL (37). Current free energy models are not able to precisely model the effects of double mutants such as T474M and E513G. Hence, the T474 residue is a gatekeeper that controls BTK binding to covalent and noncovalent inhibitors.
Figure 3. Structural modeling of the binding to BTK L528W and T474M mutants.
(A) The structural context of ibrutinib binding in BTK W528 compared with L528. The BTK protein is shown in cartoon representation. Ibrutinib and its covalent attachment point C481, as well as L528/W528, are highlighted in all-atom CPK representation. The mutation of L528 to W528 sterically destabilizes interactions of ibrutinib with C481. (B) Structural modeling of wild-type BTK (T474) and BTK T474M mutation (M474) binding to RN486. The H-bonded network involving K430, T474, and G414 backbone is shown in yellow dotted lines and highlighted within the red circle. This water network is disrupted by the mutation of T474 to M474.
A doubly mutated BTK allele acquires in vivo transforming activity.
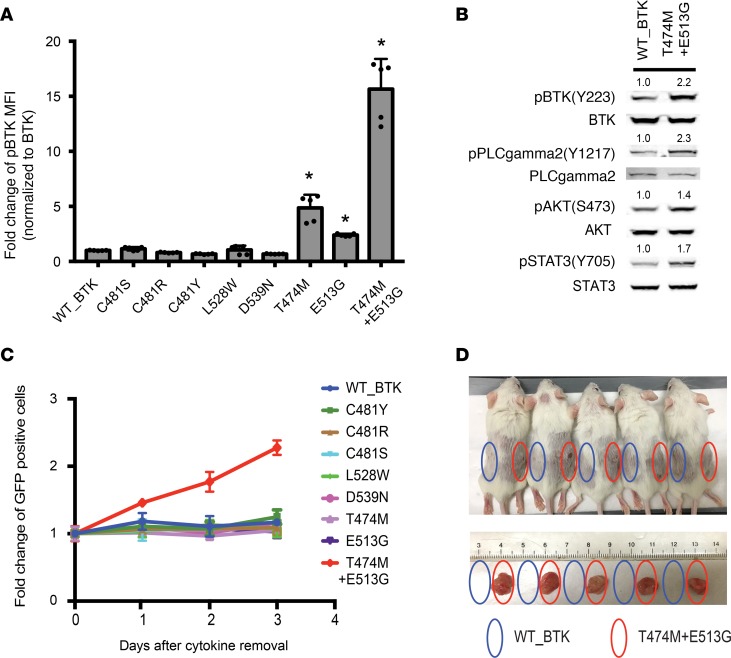
To examine how different BTK mutations affect BTK function, we measured BTK Y223 autophosphorylation by FACS in HEK293T cells expressing the wild-type and mutant alleles. Y223 provides the most direct readout of BTK activity, whereas other phosphorylation sites are also substrates of other kinases (7, 38, 39). The C481S mutation showed no change; C481R and C481Y reduced autophosphorylation; T474M and E513G showed 4.8-fold and 2.4-fold increases, respectively (Figure 4A and Supplemental Figure 5A). Strikingly, the double-mutant T474M and E513G protein showed a 15-fold increase in Y223 phosphorylation over wild-type BTK (Figure 4A). By comparison, the known (E41K) gain-of-function BTK mutation caused only a 1.7-fold increase in the same assay (refs. 40, 41; Supplemental Figure 5B). Overexpressing the double mutant (T474M and E513G) in human B cell lymphoma cell line TMD8 (which is already driven by an activated BTK and therefore is an established model of ibrutinib sensitivity) led to a further increase in BTK Y223 autophosphorylation and additional although more variable activation of BTK downstream signaling (ref. 25 and Supplemental Figure 5, C–D). Similarly, the double-mutant BTK (T474M and E513G) in Ba/F3 murine pro-B cells caused increased phosphorylation of known BTK downstream molecules (Figure 4B). Interleukin-3–independent (IL-3–independent) growth in Ba/F3 cells is an established surrogate of in vitro transformation (42). Notably, the BTK double mutant (T474M and E513G), but not wild-type or any other BTK allele, was sufficient to confer IL-3–independent growth in Ba/F3 cells (Figure 4C). Furthermore, subcutaneous implantation of Ba/F3 cells expressing the double-mutant BTK (1 × 107 cells into NOD/SCID-γ [NSG] mice) demonstrated in vivo tumorigenesis with 100% penetrance; in contrast, expression of wild-type BTK was completely unable to cause tumors in vivo (Figure 4D). Hence, a double-mutant BTK allele is insensitive to pharmacological inhibition and shows in vitro and in vivo transforming and tumorigenic activity.
Figure 4. Functional analysis of drug-resistant BTK mutants.
(A) FACS analysis of BTK (Y223) autophosphorylation comparing wild-type BTK and the indicated mutant BTK alleles. Data are represented as mean ± SD from 2 independent experiments. *P < 0.05 vs. WT_BTK determined by Student’s t test. (B) Immunoblot of Ba/F3 lysates expressing wild-type and mutant BTK probed for indicated BTK downstream molecules. Total protein was used as a control and quantification was done with ImageJ. (C) Proliferation of Ba/F3 cells expressing mutant BTK and GFP in the absence of IL-3. (D) In vivo tumorigenicity of 1 × 107 Ba/F3 cells expressing wild-type or mutant BTK (T474M and E513G) injected into the flanks of NSG mice; below, tumors harvested after 4 weeks.
The T474M gatekeeper mutation cooperates with several kinase domain mutations.
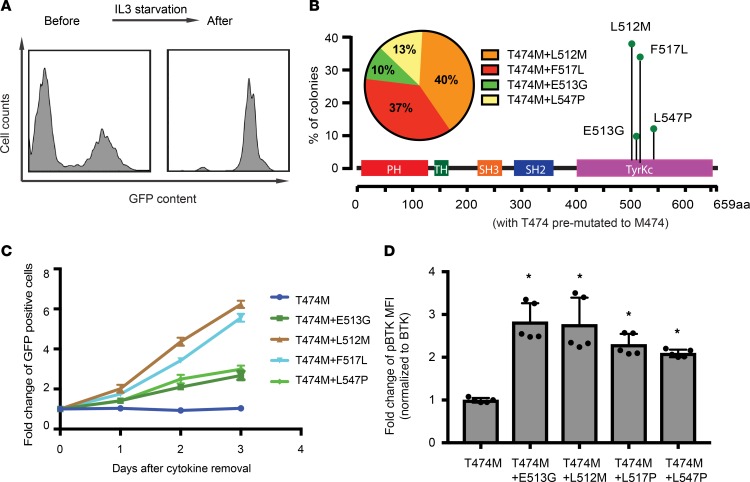
We wondered whether other BTK lesions would similarly cooperate with the T474 gatekeeper. Briefly, we used the T474M gatekeeper mutation as a baseline CDS and generated random mutations in this CDS using the same approach as described above. We screened this new library (T474M plus X) for the ability to confer IL-3 independence in Ba/F3 cells as a surrogate for transformation. After 2 weeks of selection in IL-3–depleted medium, cells attained an enrichment to more than 95%, indicating outgrowth of IL-3–independent cells (Figure 5A). Sequence analysis revealed several cooperating mutations that were all located in the BTK kinase domain: L512M, E513G, F517L, and L547P (Figure 5B). We confirmed IL-3–independent growth (Figure 5C) and found increased BTK autophosphorylation at Y223 for all double-mutant BTK alleles compared with the BTK T474M mutant (Figure 5D). Hence, the gatekeeper T474M lesion cooperates with several kinase domain mutations to activate BTK’s transforming potential.
Figure 5. Sensitized screen for transforming BTK mutations in the context of the BTKT474M gatekeeper allele.
(A) FACS analysis of Ba/F3 cells shows enrichment of GFP (coexpressed with the mutant BTKT474M library) after IL-3 starvation. (B) Sequence analysis of 156 colonies from Ba/F3 cells indicates frequency and location of secondary mutations in the context of the T474M mutation. (C) Confirmation of IL-3–independent growth for the indicated BTK mutants coexpressed with GFP and measured relative to nontransduced parental cells (indicated as percentage of GFP-positive cells). (D) FACS analysis of BTK autophosphorylation (Y223) in HEK293T cells expressing the indicated BTK alleles. Data are represented as mean ± SD from 2 independent experiments. *P < 0.05 vs. BTK_T474M determined by Student’s t test.
Modeling and testing the cooperative effects of the BTK double mutein.
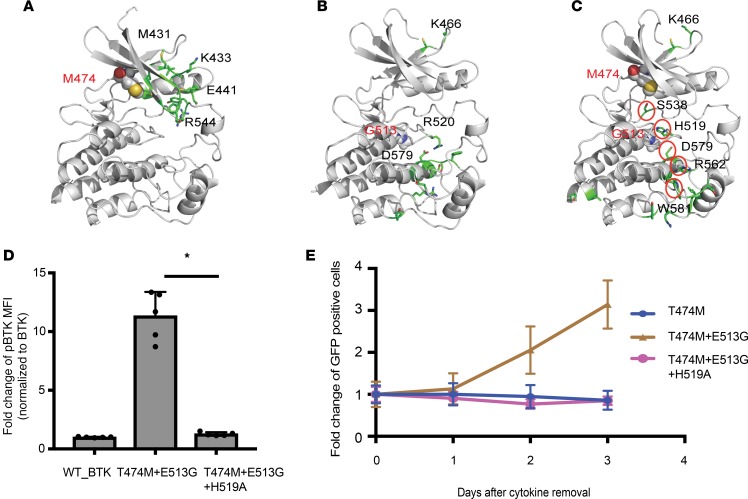
The cooperation between kinase domain mutations and the distant T474 residue is very surprising and suggests an intramolecular mechanism that constrains BTK activity. We performed molecular dynamics (MD) simulations of wild-type, single-mutant (T474M or E513G), and double-mutant (T474M and E513G) BTK proteins (Figure 6, A–C, and Supplemental Figure 6). The gatekeeper and kinase domain lesions localize to the N-lobe and C-lobe of BTK, respectively, and they are distant from BTK’s activation loop and previously identified critical residues implicated in activation (D579, H519, and F540; refs. 43, 44). MD simulations compared the frequency of contacts between all pairs of residues in wild-type and mutant BTK (Figure 6, A–C, and Supplemental Figure 6). Residues with changed contact patterns between wild-type BTK and the single and double BTK muteins are highlighted in stick representation in the protein model (Figure 6, A–C). For example, several residues in the N-lobe showed a differential contact pattern for T474M (Figure 6A), and weaker signals propagated to the C-lobe (Supplemental Figure 6D). For the E513G mutation, differential contact patterns were found to propagate to other residues in the C-lobe, including D579 (Figure 6B and Supplemental Figure 6D). The double mutant (T474M and E513G) showed a striking pattern of differential contact dynamics for a small set of residues connecting the 2 mutations to residues in the C-lobe, including D579 and H519, implicated in BTK activation (Figure 6C). This simulation of the double mutant predicts that its ability to activate BTK involves critical activation loop residues, such as H519. We directly tested this predicted mechanism by mutating the H519 residue to alanine (H519A). This change completely abrogated BTK activation as measured by BTK Y223 autophosphorylation (Figure 6D). It also relinquished the ability of the BTK double mutein to support IL-3–independent growth of Ba/F3 cells (Figure 6E). Together, these results identify an intramolecular mechanism that constrains BTK’s kinase and transforming activity.
Figure 6. Modeling and testing the cooperative effects of the BTK double mutein.
(A–C) All-atom MD simulations comparing T474M (A), E513G (B), and the double mutants (T474M and E513G) (C) with wild-type BTK. The residues computed to have differential frequency of contacts in the mutant compared with the wild-type BTK are highlighted in stick representation in the protein model. Mutated residues are highlighted in all-atom CPK representation. A structural pattern of residues with differential contacts emerges for the double mutant, connecting the mutations to distant residues implicated in activation, viz. D579 and H519. (D) H519A mutation reverts the increased BTK Y223 autophosphorylation of cells expressing BTK_T474M+E513G to wild-type BTK levels as measured by FACS in HEK393T cells. Data are represented as mean ± SD from 2 independent experiments. *P < 0.05 determined by Student’s t test. (E) H519A further abrogates IL-3–independent growth in Ba/F3 cells expressing BTK_T474M+E513G.
Discussion
Our study provides new insight into the function of BTK and its interaction with covalent and noncovalent inhibitors. Specifically, we identify a potentially new gatekeeper mutation (T474M) that attenuates inhibitor access to BTK. Achieving the T → M mutation requires 2 nucleotide changes, which may explain the relative rarity of this lesion. Importantly, changes at this gatekeeper residue showed a surprising cooperativity with kinase domain mutations, resulting in an approximately 15-fold gain of kinase activity, in vitro transforming, and in vivo tumorigenic activity. Mechanistically, the T474 gatekeeper functions in a manner similar to the homologous T315 residue in the related BCR-ABL kinase (37). The cooperative activation of the kinase by T474 and kinase domain lesions reveals a potentially new auto-inhibitory mechanism that attenuates BTK’s transforming activity. Wild-type BTK has no oncogenic activity and the reported BTK mutations (including kinase domain and T474 mutations) are individually insufficient to activate BTK’s oncogenic activity. This unique requirement for 2 mutations may explain why BTK — a major kinase in the oncogenic BCR pathway — is not a target of activating, somatic mutations while both up- and downstream molecules are known drivers of lymphomagenesis. Our results anticipate clinically relevant mechanisms of resistance to noncovalent BTKis. Noncovalent inhibitors have been designed to overcome the cysteine C481 mutations that block ibrutinib activity in patients. These new drugs are now entering clinical trials, and response data or relapse samples are not yet available (14). We noticed that both T474 and kinase domain mutations have been reported in patients with CLL or lymphoma (27, 28, 36). Our findings indicate these mutations may be more frequently seen in patients exposed to noncovalent inhibitors. Co-occurrence of T474 and kinase domain mutations is expected to remain a rare event because each individual mutation causes only a modest advantage in the absence of the inhibitor and may not facilitate acquisition of the second mutation. In summary, our findings provide new insight into the intramolecular mechanism that constrains BTK activity and anticipate mutational mechanisms that may incur clinical resistance to noncovalent BTKis.
Methods
Cell culture.
The following cell lines were purchased from American Type Culture Collection (ATCC) and cultured according to the instructions from ATCC: BCL1 (TIB197), Phoenix-GP (CRL-3215), and HEK293T (CRL-3216). The TMD8 cell line was a gift of Ari M. Melnick (Division of Hematology/Oncology, Department of Medicine, Weill Cornell Medicine, Cornell University, New York, NY, USA). TMD8 cells were maintained in RPMI 1640 with 10% FBS, supplemented with l-glutamine, HEPES, and penicillin/streptomycin. The Ba/F3 cell line was provided by Craig B. Thompson (Cancer Biology and Genetics Program, Memorial Sloan-Kettering Cancer Center, New York, NY, USA) and maintained in RPMI 1640 with 10% FBS, 10% WEHI-3B cell–conditioned (DSMZ, catalog ACC26) medium, and 1 ng/mL mouse IL-3, supplemented with l-glutamine and penicillin/streptomycin. Cell lines were authenticated by short tandem repeat analysis.
Molecular cloning.
Wild-type BTK (WT_BTK) open reading frame was PCR amplified from WT_BTK/pGFPN1 (a gift of C.I. Edward Smith [Department of Laboratory Medicine, Karolinska Institute Clinical Research Center, Huddinge, Sweden]) and subcloned into pMIG (a gift from William C. Hahn, Addgene plasmid 9044) retroviral vector with internal ribosome entry site–GFP to generate WT_BTK/pMIG CDS plasmid. BTK_E41K/pMIG plasmid was generated by PCR amplification of BTK_E41K CDS from BTK_E41K/pGFPN1 (a gift of C.I. Edward Smith) and then subcloned into pMIG (a gift from William Hahn, Addgene plasmid 9044). BTK_C481S/pMIG and BTK_T474M+E513G+H519A plasmids were generated by site-directed mutagenesis using QuikChange II XL Site-Directed Mutagenesis Kit from Agilent following manufacturer’s instructions using WT_BTK/pMIG and BTK_T474M+E513G as templates, respectively. Other drug-resistant, BTK-mutant plasmids were generated by PCR amplifying these mutant CDSs from PGEMT colonies and then subcloned into pMIG. All mutations were confirmed by Sanger sequencing. All primers are listed in Supplemental Table 5.
Immunoblot assays and flow cytometry.
HEK293T cells transiently transfected with wild-type and mutant BTK plasmids were treated with DMSO, ibrutinib, or RN486 for 1 hour. RIPA buffer (Boston Bioproducts) was used to prepare whole-cell lysates from Ba/F3 or TMD8 cells with or without anti-IgM stimulation. TMD8 cells were stimulated for 5 minutes with 10 ug/mL goat anti–human IgM (Jackson ImmunoResearch, 109-006-129) immediately followed by adding 3.3 mM H2O2. Immunoblot assays were performed according to standard procedures using antibodies from Cell Signaling Technology: p-BTK (Y223) (no. 5082), total BTK (no. 8547), p-PLCG2 (Y1217) (no. 3871), total PLCG2 (no. 3872). p-AKT (S473) (no. 4058), total AKT (no. 2938), p-STAT3 (Y705) (no. 9131), total STAT3 (no. 12640), p-P65 (Ser536) (no. 3033), and total P65 (no. 8242). Total protein was used as a control for each corresponding phospho-protein, and quantification was done with ImageJ (NIH). For BTK intracellular flow cytometry assay, HEK293T cells transiently transfected with wild-type or mutated BTK plasmids were fixed with paraformaldehyde, permeabilized, and stained for phospho-flow as described (7). Data were acquired with a BD Fortessa cell analyzer and analyzed with FlowJo software (Tree Star). BV421 mouse anti–Btk p-Y223 (clone N35-86) and Alexa Fluor 647 mouse anti-Btk (clone 53/BTK) antibodies from BD Biosciences were used for flow cytometry. For mutagenesis screen, data were acquired with a Guava easyCyte Flow Cytometer (MilliporeSigma).
Random mutagenesis screen.
WT_BTK/pMIG plasmid was used as a template to generate randomly mutated BTK CDS libraries using XL1-Red cells (Agilent) similar to a previously described method (37). Two BTK mutation libraries were generated independently. The complexity of both libraries was confirmed by Illumina HiSeq with paired-end run, 2 × 75 bp. FASTQ files generated from HiSeq were aligned with GRCh37/hg19 to identify the nucleotide variance. VSVG-pseudotyped retroviruses were generated from these libraries in Phoenix-GP cells. Then these viruses were used to infect the BTKi-sensitive BCL1 lymphoma cells with low multiplicity of infection by spin infection as described (37). To select for drug-resistant, BTK-mutant clones, the infected cells were treated with either 50 nM ibrutinib or 500 nM RN486 (both from Chemietek). Old medium was replaced every 3 to 5 days with fresh medium containing the inhibitor after washing cells 3 times with PBS. The treatment was stopped when GFP-positive cells were enriched close to 100%. Genomic DNA was isolated from these drug-resistant cells. The integrated BTK CDSs were recovered by PCR followed by TA cloning and Sanger sequencing of individual clones. To identify BTK mutations, alignments of sequencing results and human BTK mRNA (NM_000061.2) were performed with the SeqMan Pro program. The numbering of amino acid residues in this paper is according to the BTK isoform NP_000052.1. Presented results are combined data of screens from 2 mutation libraries. The sensitized screen was performed similarly except we used BTK_T474M/pMIG plasmid as a mutation template to make the library and IL-3 starvation for screening in Ba/F3 cells.
Cell proliferation assay.
Ba/F3 cells partially transduced with indicated BTK alleles were washed with PBS and then were cultured in RPMI 1640 containing 10% FBS but without conditioned medium of WEHI-3B cells or mouse IL-3 (day 0). The percentages of GFP-positive cells were determined by Guava every day from day 0 to day 3. The fold change of GFP percentage in days 1 to 3 compared with day 0 was used to show the growth advantage of transduced cells over nontransduced cells under IL-3 starvation. For cell proliferation assay under drug treatment, BCL1 or TMD8 lymphoma cells expressing the indicated BTK wild-type or drug-resistant mutant alleles were treated with BTKi at the indicated dose range for 72 hours. Viable cells were determined by CellTiter-Glo and normalized to DMSO treatment. Data were analyzed in Microsoft Excel and GraphPad Prism. Ibrutinib and RN486 were from Chemietek, Acalabrutinib was from Selleckchem, and GDC-0853 and SNS-062 were from Medchemexpress.
In vivo transformation assay.
NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, Jackson Laboratory number 005557) that were 6–8 weeks old were purchased and kept under standard laboratory conditions according to the guidelines of the National Cancer Institute (NCI). The pure population of Ba/F3 cells infected with WT_BTK or the double mutant (T474M and E513G) were washed with PBS and suspended in a 1:1 mixture of PBS and Matrigel (BD, 354234). The cell suspensions were then subcutaneously injected into both left (WT_BTK) and right (T474M and E513G) flanks of each animal. We injected 1 × 107 cells per flank. Tumors were monitored twice weekly. After 4 weeks, mice were euthanized, and tumors were dissected and imaged. All experiments were approved by the Memorial Sloan-Kettering Cancer Center (MSKCC) IACUC (protocol 07-01-002).
Free energy calculations.
The reported free energy calculations were initiated from protein models based on the x-ray structure Protein Data Bank (PDB) ID 3GEN for ibrutinib (45) and PDB ID 4OTR for RN486 (11). The structure 3GEN was chosen for ibrutinib because of the high shape similarity between the co-crystallized ligand and ibrutinib. These crystal structures were separately imported into the Maestro program, where all crystallographic waters were deleted, hydrogens were added to the structures to assume a pH 7 environment, and the position of the heavy atoms was locally optimized using the Protein Prep Wizard utility. This procedure optimized the protein-protein and protein-ligand hydrogen bonding and relieved unfavorable steric clashes without introducing any significant changes to the crystallographically determined positions of the heavy atoms. Once the prepared protein structures of the 2 complexes were obtained, the initial binding mode for ibrutinib was constructed through a standard docking protocol using Glide. Once these prepared complex structures for ibrutinib were obtained, free energy calculations of protein residue mutations were performed with the FEP+ program (46, 47) using default settings. The version of the FEP+ program used in this work employs the OPLS3 force field, replica exchange with solute scaling 2 enhanced sampling, and the Desmond 3.9 graphics processing unit MD engine as described (46). Each λ window of the free energy calculation was simulated for a 100-ps equilibration followed by a 5-ns production simulation. The default λ schedule was applied throughout this work, and 12 λ windows were used to perform each of the reported free energy calculations. To compute a meaningful binding free energy difference of the inhibitor binding to the wild-type protein versus the mutant form, the protein residue in question was alchemically mutated in the presence of the inhibitor and then again alchemically mutated in the absence of the inhibitor, with the difference in these results yielding the relative binding free energy of the inhibitor to the mutant form of the protein versus the wild-type protein.
MD simulations.
The MD simulations for the wild-type and mutants BTK_T474M, BTK_E513G, and BTK_T474M+E513G were performed starting from the PDB ID 4OTR, without any ligand bound. The initial protein models were prepared with the same protocol as for the FEP+ calculations described above, and each simulation was run for 400 ns. The last 200 ns of each of the MD trajectories was analyzed to compare the dynamic residue-residue contacts in the protein. A contact matrix was constructed from the MD simulations for all the constructs. A contact between 2 residues was defined as the percentage of time any heavy atom from 2 residues was within 4.5 Å of each other. Thus, residues in contact for the entire duration of the simulation would be given a value of 1, and those that were never in contact with each other would be assigned a value of 0. To understand the impact of a mutation on the protein dynamics, a contact difference map was created for each mutant, subtracting the contact matrix for the wild-type simulation from that of the mutant MD simulations. These contact difference maps have a scale that ranges from 1 to –1, where a value of 1 indicates a new contact was formed in the mutant simulations that did not exist in the wild-type MD simulations and –1 indicates the converse. This allowed us to identify any region within the mutant that differed from the wild-type BTK in terms of dynamic contacts. We mapped the top 20 peaks to their location on the structure of the protein, to identify corresponding residues.
Additional methods are available in supplemental methods.
Statistics.
All statistical tests were 2-tailed Student’s t tests, type 1. Data are represented as mean ± SD. P < 0.05 was considered significant.
Study approval.
Animal experiments were approved by the MSKCC IACUC (protocol 07-01-002).
Author contributions
SW designed and performed experiments, performed data analysis, and cowrote the manuscript. CZ and MB assisted with experiments. SM, PG, and RA performed the structural modeling and helped with writing related to the modeling. CLB, AD, VES, MRG, and AY provided the clinical data. HGW designed the study and cowrote the manuscript.
Supplementary Material
Acknowledgments
We thank C.B. Thompson and A.M. Melnick for cell lines, C.I.E. Smith and W.C. Hahn for plasmids, J. Chodera and M. Balbas (MSKCC) for helpful discussion about the screen, N.D. Socci (MSKCC) for bioinformatics analysis, J.H. Schatz (University of Miami Miller School of Medicine, Miami, Florida, USA) for cell line authentication, and all members in the Wendel group for the thoughtful discussions related to this study. We thank A. Viale, members of the MSKCC Integrated Genomics Operation core, E. de Stanchina, and members of the MSKCC Antitumor Assessment core and MSKCC Flow Cytometry core. SW is supported by a postdoctoral fellowship from the Swedish Research Council (VR) and by the 2015 American Association for Cancer Research–Millennium Fellowship in Lymphoma Research, grant number 15-40-38-WANG. CLB is supported by the ASCO Young Investigators Award, ASH Clinical Scholars Award, Lymphoma Research Foundation Clinical Research Mentoring Program, Solomon Genomics Program, Steven Greenberg Lymphoma Research Foundation, and Memorial Sloan-Kettering Lymphoma SPORE P50 CA192937-01A1. AD, VES, and AY are supported by NCI SPORE 1 P50 CA192937. MRG is supported by NIH R01CA201380, the MD Anderson Cancer Center NCI CORE grant P30 CA016672, and the Schweitzer Family Fund. AY is also supported by Lymphoma Society Specialized Center for Research grant 7014-17. HGW is supported by the National Institutes of Health grants 5 R01CA190384‑03, 1 P50 CA192937-01A1, 1 R01 CA207217-01, 5 R01 CA183876-03, P50 CA217694, Leukemia and Lymphoma Society Fund (RSG-1318-15 and GC227931), Lymphoma Research Foundation (GC227729), American Cancer Society grant (RSG-13-048-01-LIB), Starr Cancer Consortium grant (I10-0064), Steven A. Greenberg Startup Grant (GC238719), Memorial Sloan Kettering Cancer Center Core grant (P30 CA008748) and Technology Development Fund (GC230724), Clinical and Translational Science Collaborative Fund EPAR5927 and New York State Stem Cell Science II IRP grant (C028131). HGW is a scholar of the Leukemia and Lymphoma Society.
Version 1. 06/20/2019
Electronic publication
Footnotes
CZ’s present address is: Functional Genomics, Target Sciences and Technologies, Pfizer Inc., Pearl River, New York, New York, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(12):e127566. https://doi.org/10.1172/jci.insight.127566.
Contributor Information
Shenqiu Wang, Email: wangs1@mskcc.org.
Sayan Mondal, Email: sayan.mondal@schrodinger.com.
Chunying Zhao, Email: chunying2011@gmail.com.
Marjan Berishaj, Email: berisham@mskcc.org.
Phani Ghanakota, Email: phani.ghanakota@schrodinger.com.
Ahmet Dogan, Email: dogana@mskcc.org.
Venkatraman E. Seshan, Email: seshanv@mskcc.org.
Robert Abel, Email: robert.abel@schrodinger.com.
Anas Younes, Email: younesa@mskcc.org.
Hans-Guido Wendel, Email: wendelh@mskcc.org.
References
- 1.Wilson WH, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21(8):922–926. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Treon SP, et al. Ibrutinib in previously treated Waldenstrom’s macroglobulinemia. N Engl J Med. 2015;372(15):1430–1440. doi: 10.1056/NEJMoa1501548. [DOI] [PubMed] [Google Scholar]
- 3.Wang ML, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2013;369(6):507–516. doi: 10.1056/NEJMoa1306220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrd JC, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369(1):32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Advani RH, et al. Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765) has significant activity in patients with relapsed/refractory B-cell malignancies. J Clin Oncol. 2013;31(1):88–94. doi: 10.1200/JCO.2012.42.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noy A, et al. Targeting Bruton tyrosine kinase with ibrutinib in relapsed/refractory marginal zone lymphoma. Blood. 2017;129(16):2224–2232. doi: 10.1182/blood-2016-10-747345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woyach JA, et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370(24):2286–2294. doi: 10.1056/NEJMoa1400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furman RR, et al. Ibrutinib resistance in chronic lymphocytic leukemia. N Engl J Med. 2014;370(24):2352–2354. doi: 10.1056/NEJMc1402716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu L, et al. Acquired mutations associated with ibrutinib resistance in Waldenström macroglobulinemia. Blood. 2017;129(18):2519–2525. doi: 10.1182/blood-2017-01-761726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiron D, et al. Cell-cycle reprogramming for PI3K inhibition overrides a relapse-specific C481S BTK mutation revealed by longitudinal functional genomics in mantle cell lymphoma. Cancer Discov. 2014;4(9):1022–1035. doi: 10.1158/2159-8290.CD-14-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lou Y, et al. Structure-based drug design of RN486, a potent and selective Bruton’s tyrosine kinase (BTK) inhibitor, for the treatment of rheumatoid arthritis. J Med Chem. 2015;58(1):512–516. doi: 10.1021/jm500305p. [DOI] [PubMed] [Google Scholar]
- 12.Young WB, et al. Potent and selective Bruton’s tyrosine kinase inhibitors: discovery of GDC-0834. Bioorg Med Chem Lett. 2015;25(6):1333–1337. doi: 10.1016/j.bmcl.2015.01.032. [DOI] [PubMed] [Google Scholar]
- 13.Di Paolo JA, et al. Specific Btk inhibition suppresses B cell- and myeloid cell-mediated arthritis. Nat Chem Biol. 2011;7(1):41–50. doi: 10.1038/nchembio.481. [DOI] [PubMed] [Google Scholar]
- 14.Byrd JC, et al. First-in-human phase 1 study of the BTK inhibitor GDC-0853 in relapsed or refractory B-cell NHL and CLL. Oncotarget. 2018;9(16):13023–13035. doi: 10.18632/oncotarget.24310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lou Y, Owens TD, Kuglstatter A, Kondru RK, Goldstein DM. Bruton’s tyrosine kinase inhibitors: approaches to potent and selective inhibition, preclinical and clinical evaluation for inflammatory diseases and B cell malignancies. J Med Chem. 2012;55(10):4539–4550. doi: 10.1021/jm300035p. [DOI] [PubMed] [Google Scholar]
- 16.Thompson PA, Burger JA. Bruton’s tyrosine kinase inhibitors: first and second generation agents for patients with Chronic Lymphocytic Leukemia (CLL) Expert Opin Investig Drugs. 2018;27(1):31–42. doi: 10.1080/13543784.2018.1404027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiff SD, et al. Noncovalent inhibition of C481S Bruton tyrosine kinase by GDC-0853: a new treatment strategy for ibrutinib-resistant CLL. Blood. 2018;132(10):1039–1049. doi: 10.1182/blood-2017-10-809020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crawford JJ, et al. Discovery of GDC-0853: a potent, selective, and noncovalent Bruton’s tyrosine kinase inhibitor in early clinical development. J Med Chem. 2018;61(6):2227–2245. doi: 10.1021/acs.jmedchem.7b01712. [DOI] [PubMed] [Google Scholar]
- 19.Johnson AR, et al. Battling Btk mutants with noncovalent inhibitors that overcome Cys481 and Thr474 mutations. ACS Chem Biol. 2016;11(10):2897–2907. doi: 10.1021/acschembio.6b00480. [DOI] [PubMed] [Google Scholar]
- 20.Corneth OBJ, Klein Wolterink RGJ, Hendriks RW. BTK signaling in B cell differentiation and autoimmunity. Curr Top Microbiol Immunol. 2016;393:67–105. doi: 10.1007/82_2015_478. [DOI] [PubMed] [Google Scholar]
- 21.Satterthwaite AB, Witte ON. The role of Bruton’s tyrosine kinase in B-cell development and function: a genetic perspective. Immunol Rev. 2000;175:120–127. doi: 10.1111/j.1600-065X.2000.imr017504.x. [DOI] [PubMed] [Google Scholar]
- 22.Smith CI. From identification of the BTK kinase to effective management of leukemia. Oncogene. 2017;36(15):2045–2053. doi: 10.1038/onc.2016.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vetrie D, et al. The gene involved in X-linked agammaglobulinaemia is a member of the src family of protein-tyrosine kinases. Nature. 1993;361(6409):226–233. doi: 10.1038/361226a0. [DOI] [PubMed] [Google Scholar]
- 24.Tsukada S, et al. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. Cell. 1993;72(2):279–290. doi: 10.1016/0092-8674(93)90667-F. [DOI] [PubMed] [Google Scholar]
- 25.Davis RE, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109(10):3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krysiak K, et al. Recurrent somatic mutations affecting B-cell receptor signaling pathway genes in follicular lymphoma. Blood. 2017;129(4):473–483. doi: 10.1182/blood-2016-07-729954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddy A, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171(2):481–494.e15. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morin RD, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122(7):1256–1265. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapuy B, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24(5):679–690. doi: 10.1038/s41591-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitz R, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378(15):1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okosun J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46(2):176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pal Singh S, Dammeijer F, Hendriks RW. Role of Bruton’s tyrosine kinase in B cells and malignancies. Mol Cancer. 2018;17(1):57. doi: 10.1186/s12943-018-0779-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamasy A, et al. Substitution scanning identifies a novel, catalytically active ibrutinib-resistant BTK cysteine 481 to threonine (C481T) variant. Leukemia. 2017;31(1):177–185. doi: 10.1038/leu.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng S, Guo A, Lu P, Ma J, Coleman M, Wang YL. Functional characterization of BTK(C481S) mutation that confers ibrutinib resistance: exploration of alternative kinase inhibitors. Leukemia. 2015;29(4):895–900. doi: 10.1038/leu.2014.263. [DOI] [PubMed] [Google Scholar]
- 36.Maddocks KJ, et al. Etiology of Ibrutinib therapy discontinuation and outcomes in patients with chronic lymphocytic leukemia. JAMA Oncol. 2015;1(1):80–87. doi: 10.1001/jamaoncol.2014.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burgess MR, Skaggs BJ, Shah NP, Lee FY, Sawyers CL. Comparative analysis of two clinically active BCR-ABL kinase inhibitors reveals the role of conformation-specific binding in resistance. Proc Natl Acad Sci U S A. 2005;102(9):3395–3400. doi: 10.1073/pnas.0409770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honigberg LA, et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci U S A. 2010;107(29):13075–13080. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohammad DK, Nore BF, Hussain A, Gustafsson MO, Mohamed AJ, Smith CI. Dual phosphorylation of Btk by Akt/protein kinase b provides docking for 14-3-3ζ, regulates shuttling, and attenuates both tonic and induced signaling in B cells. Mol Cell Biol. 2013;33(16):3214–3226. doi: 10.1128/MCB.00247-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li T, et al. Activation of Bruton’s tyrosine kinase (BTK) by a point mutation in its pleckstrin homology (PH) domain. Immunity. 1995;2(5):451–460. doi: 10.1016/1074-7613(95)90026-8. [DOI] [PubMed] [Google Scholar]
- 41.Mohamed AJ, et al. Bruton’s tyrosine kinase (Btk): function, regulation, and transformation with special emphasis on the PH domain. Immunol Rev. 2009;228(1):58–73. doi: 10.1111/j.1600-065X.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 42.Azam M, Seeliger MA, Gray NS, Kuriyan J, Daley GQ. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat Struct Mol Biol. 2008;15(10):1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao C, Zhou M, Uckun FM. Crystal structure of Bruton’s tyrosine kinase domain suggests a novel pathway for activation and provides insights into the molecular basis of X-linked agammaglobulinemia. J Biol Chem. 2001;276(44):41435–41443. doi: 10.1074/jbc.M104828200. [DOI] [PubMed] [Google Scholar]
- 44.Joseph RE, Xie Q, Andreotti AH. Identification of an allosteric signaling network within Tec family kinases. J Mol Biol. 2010;403(2):231–242. doi: 10.1016/j.jmb.2010.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marcotte DJ, et al. Structures of human Bruton’s tyrosine kinase in active and inactive conformations suggest a mechanism of activation for TEC family kinases. Protein Sci. 2010;19(3):429–439. doi: 10.1002/pro.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abel R, et al. Accelerating drug discovery through tight integration of expert molecular design and predictive scoring. Curr Opin Struct Biol. 2017;43:38–44. doi: 10.1016/j.sbi.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 47.Steinbrecher T, Abel R, Clark A, Friesner R. Free energy perturbation calculations of the thermodynamics of protein side-chain mutations. J Mol Biol. 2017;429(7):923–929. doi: 10.1016/j.jmb.2017.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.