Abstract
BACKGROUND
Little is known about the genomic differences between metastatic lower tract urothelial carcinoma (LTUC) and upper tract urothelial carcinoma (UTUC). We compare genomic features of primary and metastatic UTUC and LTUC tumors in a cohort of patients with end-stage disease.
METHODS
We performed whole-exome sequencing on matched primary and metastatic tumor samples (n = 37) collected via rapid autopsy of 7 patients with metastatic urothelial carcinoma. Inter- and intrapatient mutational burden, mutational signatures, predicted deleterious mutations, and somatic copy number variations (sCNVs) were analyzed.
RESULTS
We investigated 3 patients with UTUC (3 primary samples, 13 metastases) and 4 patients with LTUC (4 primary samples, 17 metastases). We found that somatic single-nucleotide variant (sSNV) burden was higher in metastatic LTUC compared with UTUC. Moreover, the apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC), mutational signature was pervasive in metastatic LTUC and less so in UTUC. Despite a lower overall sSNV burden, UTUC displayed greater inter- and intra-individual genomic distances at the copy number level between primary and metastatic tumors than LTUC. Our data also indicate that metastatic UTUC lesions can arise from small clonal populations present in the primary cancer. Importantly, putative druggable mutations were found across patients with the majority shared across all metastases within a patient.
CONCLUSIONS
UTUC demonstrated a lower overall mutational burden but greater structural variability compared with LTUC. Our findings suggest that metastatic UTUC displays a greater spectrum of copy number divergence from LTUC. Importantly, we identified druggable lesions shared across metastatic samples, which demonstrate a level of targetable homogeneity within individual patients.
FUNDING
NIH, Seattle Translation Tumor Research Program in Bladder Cancer, Howard J. Cohen Bladder Cancer Foundation, Johns Hopkins Greenberg Bladder Cancer Institute, Department of Defense Prostate Cancer Research Program, American Association for Cancer Research, Burroughs Wellcome Fund, David Matthews, and the Stinchcomb Memorial Funds.
Keywords: Oncology
Keywords: Cancer, Urology
Metastatic upper tract urothelial carcinoma demonstrates a lower overall mutational burden but greater structural variability compared with lower tract urothelial carcinoma.
Introduction
Urothelial carcinoma (UC) can occur anywhere along the urinary system. The vast majority of UC cases arise from the lower tract (called LTUC) involving the bladder, where up to 30% present with muscle invasion. Standard-of-care locoregional treatment options include either radical cystectomy and pelvic lymph node dissection or chemoradiation therapy (1–3). Neoadjuvant cisplatin-based chemotherapy achieves a complete response in up to 40% of patients (4–8), yet the overall 5-year survival benefit is only 5% to 10% (9). Once patients progress, metastatic bladder cancer has a median survival time of 14 to 15 months (10). For these patients, treatment options may include chemotherapy and immune-modulating approaches (3). Although most patients may achieve initial response, metastatic bladder cancer remains a lethal condition with significant morbidity and mortality worldwide.
Upper tract urothelial carcinoma (UTUC) is a less common entity, accounting for 5% to 10% of all urothelial tumors, and involves the ureter or renal pelvis (11). Patients with UTUC often present at an older age, with higher stage disease, which can be associated with worse survival (12–14). Standard-of-care therapy includes radical nephroureterectomy with consideration for neoadjuvant or adjuvant cisplatin-based chemotherapy in select patients. Platinum-based chemotherapy has shown efficacy in a proportion of patients (15, 16), but the overall survival of metastatic UTUC is poor (17, 18).
Given the disease burden and morbidity of both UTUC and LTUC, a better understanding of the pathophysiology of disease progression is required to identify therapeutic targets and predictive biomarkers to improve outcomes. Genomic sequencing of localized LTUC has led to new insights into the genetic determinants of bladder cancer and potential therapeutic opportunities (19–26). However, less is known about the genomic landscape of UTUC (27–31). For example, recent work has shown that both cancer types share similar mutations; however, UTUC exhibits more alterations in fibroblast growth factor receptors (FGFRs), and higher incidence of microsatellite instability, whereas LTUC demonstrates more frequent mutations in tumor protein 53 (TP53) (27–30). Clinical studies also suggest that UTUC and LTUC exhibit differential responses to specific systemic treatment regimens, raising the question of discordant biology (32–34).
Although prior studies of localized urothelial cancers have improved the understanding of disease physiology with potential treatment implications, exceedingly little is known about the genomic architecture of metastatic UC. Only a singular report conducted genomic analyses across multiple metastatic specimens from the same patients and described early branching evolution in LTUC (35). However, no studies to date to our knowledge have used contemporary genomic sequencing and analysis to compare primary and metastatic LTUC and UTUC tumors. Furthermore, the extent to which intrapatient heterogeneity exists in individuals with disseminated UTUC is unknown. In this study, we have evaluated a unique set of patient tumors acquired through the University of Washington Urothelial Cancer Rapid Autopsy Program between 2015 and 2017. In particular, we performed whole-exome sequencing on 37 primary and metastatic tumors from 7 UTUC and LTUC patients. A comparative analysis based on mutational burden, mutational signatures, somatic single-nucleotide variants (sSNVs), and somatic copy number variations (sCNVs) revealed unique distinctions between these topologically divergent disease entities across and within patients. In addition, we delineated the clinical implications of putative druggable lesions across multiple metastases. Taken together, the genomic landscape of metastatic UTUC and LTUC reveals new differences between the 2 disease phenotypes and raises the promise of therapeutic opportunities.
Results
The mutational burden differs between localized and metastatic UTUC and LTUC.
We conducted whole-exome sequencing across multiple tumor specimens in 7 patients with metastatic LTUC and UTUC (Figure 1A and Supplemental Tables 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.128728DS1). Three patients had UTUC originating from the renal pelvis (n = 2) and ureter (n = 1), and the remainder had LTUC of the bladder (n = 4). Mean age at diagnosis was 67.3 (±11.6 years). Most patients were White and men (n = 6 and 5, respectively). Three of the 7 patients had a history of smoking, and 2 endorsed a history of chemical exposures. A family history of bladder cancer was present in 2 patients while nearly all patients (6 of 7) had a family history of other cancers. All patients had predominant urothelial histology, with 3 patients having small areas of squamous differentiation (Supplemental Table 1).
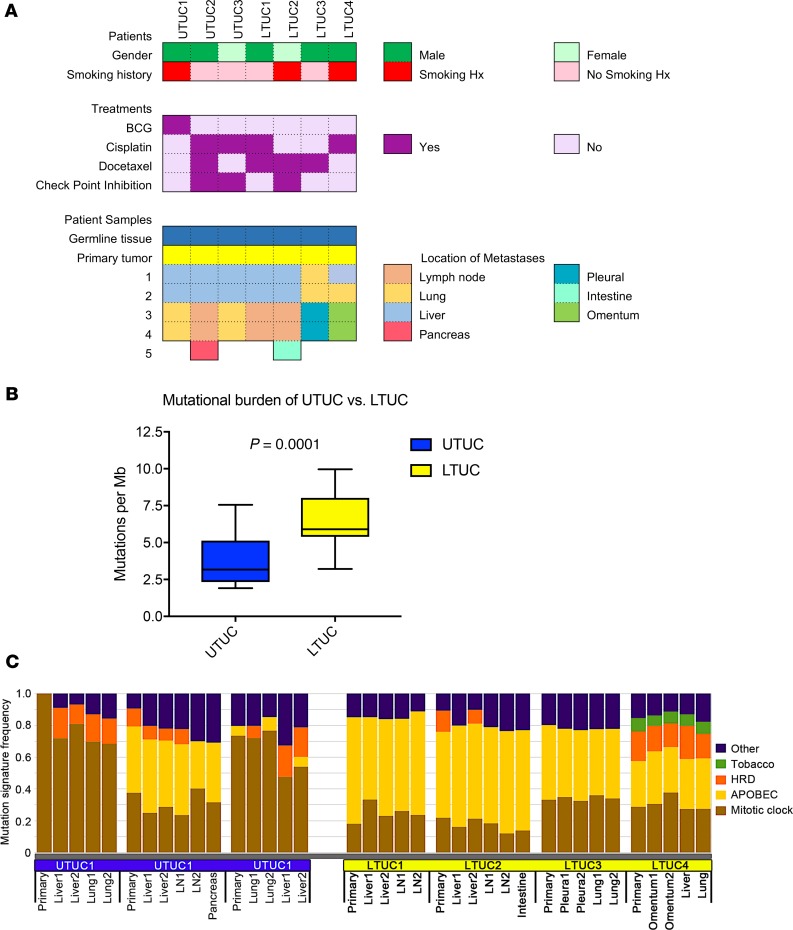
Figure 1. Whole-exome sequencing of metastatic UTUC and LTUC reveals mutational heterogeneity and distinct mutational signatures.
(A) Clinical features of rapid autopsy patients (n = 7), including sex, smoking history, treatment history, and location of metastatic lesions. Of note, the “intestine metastasis” (LTUC2) refers to an implant to the small bowel serosa from carcinomatosis. (B) Overall mutational burden of each sequenced tumor stratified by UTUC (n = 16) and LTUC (n = 21) (box-and-whisker plot representing median and IQR; P values calculated using Mann-Whitney U test). (C) Mutational signature analysis across each primary and metastatic UTUC and LTUC tumor specimen represented as a proportion of all mutations within a given tumor. All mutation analyses were performed following baseline read filtering (see Methods). HRD, homologous recombination deficiency; APOBEC, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like.
To delineate differences between localized and metastatic UTUC and LTUC, we analyzed the exomes of 13 metastatic tumor specimens from patients with UTUC and 17 metastatic tumor specimens from patients with LTUC (Figure 1A). Primary cancer specimens and germline tissues were also analyzed. In the 2 patients with a known family history of bladder cancer (UTUC1, UTUC2), no germline pathogenic SNVs were identified. Interestingly, several patients with LTUC without clear family histories possessed germline loss-of-function (LOF) mutations, including LTUC1 (ALDH1B1; NM00692.5:c.346C>T; g.chr9:38396091-38396091; p.Arg116*), LTUC2 (ATM; NM_000051.3:c.7630-2A>C; g.chr11:108202604-108202604; p.Leu2544Splice), and LTUC4 (ALDH16A1; NM_001145396.1:c.47C>A; g.chr19:49956635-49956635; p.Ser16*). We observed that the average LTUC mutational burden was significantly higher compared with UTUC (average mutations/megabase per specimen, 6.50 Mut/Mb, vs. 3.71 Mut/Mb; P = 0.0001; Mann-Whitney U test) (Figure 1B and Supplemental Table 3). When stratified by primary or metastatic samples, UTUC versus LTUC tumors were also significantly different (6.08 Mut/Mb vs. 2.85 Mut/Mb in primaries; 6.57 Mut/Mb vs. 3.90 in metastases; P = 0.002; Mann-Whitney U test) (Supplemental Figure 1). These findings are slightly lower than the mutational burden of primary LTUC observed in The Cancer Genome Atlas (TCGA) for bladder cancer (8.2 Mut/Mb) (20). Consistent with a recent report in primary UTUC (28), our results demonstrated a lower mutational burden in UTUC than LTUC in both primary and metastatic tumors.
Mutational signatures remain consistent from primary to metastatic disease but differ between UTUC and LTUC.
Next, we sought to determine whether our tumor samples comprised specific mutational signatures using the Catalogue Of Somatic Mutations In Cancer (COSMIC, SBS V2) signatures of mutational processes in human cancer. Mutational signatures were largely conserved between primary and metastatic tumors in both UTUC and LTUC (Figure 1C). Overall, we observed mutational signatures for tobacco use, HRD, and mitotic clock. Interestingly, the HRD signature was not associated with any pathogenic SNVs to HRD-associated genes or biallelic loss of BRCA1/2 (Supplemental Tables 4 and 5, and Supplemental Figure 2). However, monoallelic losses of HRD-associated genes have also been described to contribute to defects in homologous recombination (36–38). Interestingly, our patients with the HRD mutational signature (UTUC1–3 and LTUC2 and LTUC4) all possessed multiple monoallelic losses of genes implicated in homologous DNA recombination (Supplemental Tables 5 and 6).
In addition to the HRD signature, a major fraction of LTUC mutations were enriched for the APOBEC signature, which is consistent with previous reports in lower tract disease (Figure 1C; refs. 20, 35). The APOBEC family of enzymes function as cytosine deaminases and can edit RNA or ssDNA (39). In tumor cells they are thought to be responsible for hypermutation at cytosine bases in exposed ssDNA and can promote cancer phenotypes (40). We observed that these signatures were maintained between primary tumors and metastatic LTUC tumors and did not appear to change with subsequent therapies (Figure 1C). Together, these findings suggest that APOBEC mutagenesis is likely a common event in the development of LTUC that is preserved from primary to metastatic disease.
Although the APOBEC signature was detected in LTUC tumors, it was observed at high levels in only 1 of the UTUC patients. Patient UTUC2 was more similar to our LTUC patients with a predominant APOBEC signature (Figure 1C) while UTUC1 and UTUC3 had mutations enriched for the mitotic clock signature commonly seen in cancer. Notably, UTUC1 and UTUC3 had the lowest somatic mutation burden out of all of our patients as well (mean 2.6 with range 2.3–3.0 Mut/Mb vs. 6.4 with range 5.4–8.4 Mut/Mb in the remaining patients; P < 0.001; 2-tailed t test). Interestingly, the APOBEC mutations accounted for nearly all the differences in mutation rate between UTUC2 and LTUC1–4 and UTUC1 and UTUC3 (Supplemental Figure 3). Together, these findings suggest that APOBEC-mediated mechanisms may account for the higher mutational load of UTUC2 and LTUC1–4 because the nucleotide-editing activity of APOBEC is known to drive somatic DNA mutagenesis in cancer (40). Furthermore, the consistency of APOBEC predominance shared across all tumor samples within individual patients suggests this may represent a driving force in progression.
UTUC primary tumors share fewer deleterious mutations with metastatic counterparts compared with LTUC.
To determine how sSNVs affect gene function, we adopted an algorithm for calling predicted deleterious/functional driver mutations (Supplemental Figure 4). We found that UTUC tumors had fewer predicted deleterious mutations compared with LTUC (Supplemental Figure 2 and Supplemental Table 4). Mean and median burdens of predicted deleterious mutations were significantly higher in LTUC compared with UTUC (mean genes mutated in LTUC 59 ± 15 vs. UTUC 33 ± 14; median in LTUC 55 [IQR 49–68] vs. UTUC 28 [IQR 24–38]; 2-tailed t test, P < 0.001) (Supplemental Table 7). These findings are consistent with the overall higher mutational burden (Mut/Mb) of LTUC patients compared with UTUC patients (Figure 1B). In addition, we sought to determine the extent to which different patients with LTUC and UTUC share functional mutations. We observed in our small cohort that there were very few shared mutations between patients (highlighted by the blue areas, Figure 2A). However, when we compared mutations from different tumors of the same patient, we observed a greater amount of homogeneity (Figure 2A). When the mutated genes were further stratified into shared (present within every patient tumor), semishared (present in 2 or more tumors), or private (found in only 1 tumor per patient) within individuals, we found that patients with LTUC had a greater proportion of shared mutations across primary and metastatic samples compared with patients with UTUC (27% vs. 13%, respectively; P < 0.001; χ2 test) (Figure 2B). Reciprocally, UTUC tumors had a significantly higher proportion of private mutations across tumors (45% vs. 27%, respectively; P < 0.001; χ2 test). This relationship was maintained when the primary specimens were excluded from the analysis (48% vs. 22%; P < 0.001; χ2 test) (Figure 2B). Thus, among these driver mutations, LTUC patients appeared to possess relatively more shared deleterious mutations compared with UTUC patients. These findings suggest increased clonal genomic diversity between metastases within UTUC patients compared with LTUC patients. Interestingly, a recent publication analyzing LTUC and UTUC in patients with a history of both tumors identified clonal relatedness but different mutational patterns, indicating the importance of intra-individual mutagenesis (30).
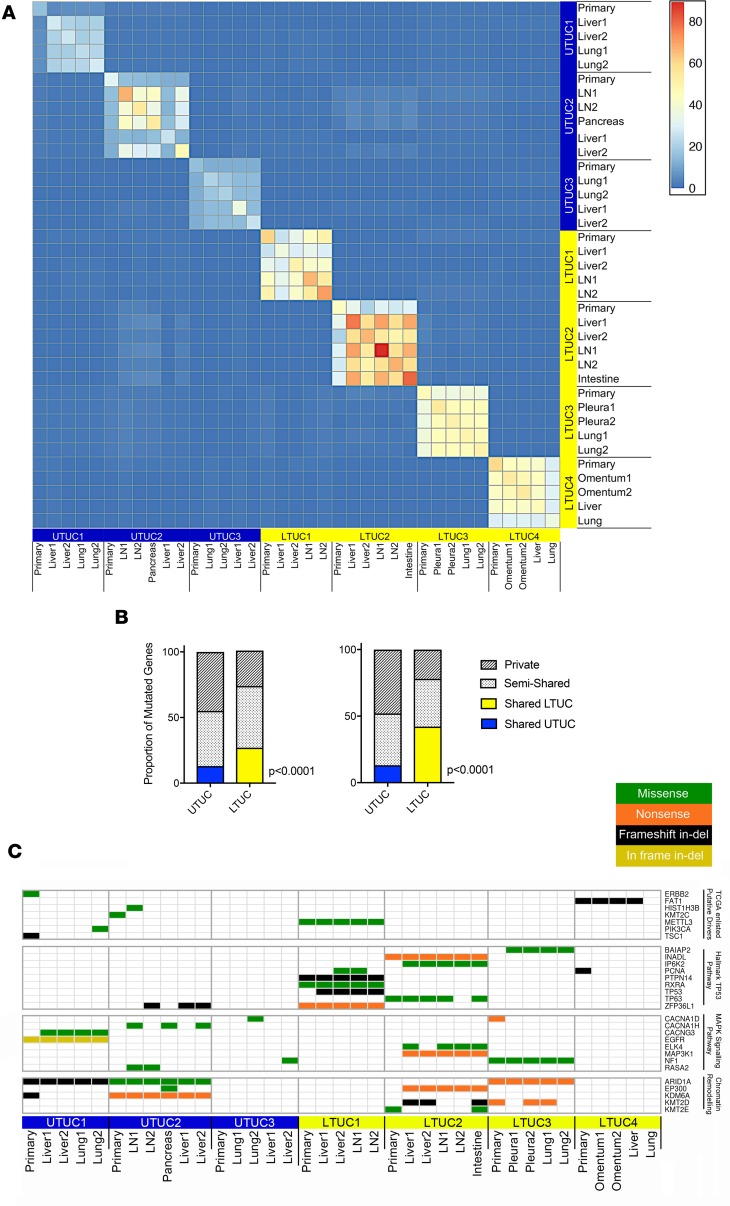
Figure 2. Metastatic UTUC tumors exhibit more deleterious private mutations compared with metastatic LTUC tumors.
(A) Heatmap representing the absolute number of predicted deleterious genes mutated that are shared between all pairs of tumors, revealing limited interpatient homogeneity across both UTUC (n = 16) and LTUC (n = 21). Here, we constructed a per-gene binary indicator vector for every tumor and calculated the number of affected genes that are shared between all pairs of tumors, which are plotted in the matrix. The numbers of shared mutations are indicated by the color scale. LN, lymph node. (B) Intrapatient predicted deleterious mutations across UTUC (blue) and LTUC (yellow) tumors represented as a percentage of all shared (colored), semishared (present in >1 sample), and private (mutated only in single samples) mutations. Left, both primary and metastatic specimens; right, metastases only (P values calculated using the χ2 test). (C) Representative bladder cancer TCGA mutations identified in our patient cohort as well as genes identified by gene set enrichment analysis (tumor-suppressive p53 pathway FDR = 6.09 × 10–4, and oncogenic MAPK signaling pathway FDR = 8.98 × 10–6).
UC metastases can arise from nondominant primary tumor populations.
Next, we sought to determine how specific deleterious mutations differ within and among patients with metastatic LTUC and UTUC. We did not observe any significant enrichment of specific mutations unique to UTUC or LTUC based on an unsupervised analysis of our small patient cohort despite having adequate power to detect shared mutations across individual patients (Figure 2C and Supplemental Figures 2 and 5). However, we performed focused analysis and identified mutations reported in the UC TCGA, including ERBB2 (S310F), KMT2C (novel D348N), PIK3CA (E545K), and TSC1 (novel G147fs) (Figure 2C and Supplemental Figure 2). We further conducted gene set enrichment analysis of our functional mutations identified in both LTUC and UTUC and found that a series of cancer-associated pathways were altered. These included the tumor-suppressive p53 pathway (FDR = 6.09 × 10–4) and the oncogenic MAPK signaling pathway (FDR = 8.98 × 10–6), both of which are deregulated in bladder cancer (Figure 2C and ref. 20). In addition, we were powered to observe that both UTUC and LTUC patients harbored a number of LOF mutations to genes associated with chromatin remodeling, including ARID1A, EP300, KDM6A, KMT2D, and KMT2E (Figure 2C and Supplemental Figure 2).
UTUC intrapatient tumor analysis uncovered mutations present in all metastases but not present at high levels in the primary tumor. An example of this can be seen with the SWI/SNF (switch/sucrose non-fermentable) complex member AT-rich interaction domain 1A (ARID1A), which is commonly deregulated in UC (20, 41–43). ARID1A encodes a component of the SWI/SNF protein complex that regulates transcription by altering chromatin structure. In patient UTUC1, we found that the primary tumor harbored a LOF ARID1A frameshift mutation (2-bp deletion) at position chr1:27107094. However, this was not found in any of the metastatic specimens despite more than 20× coverage at that position. Instead, all the metastatic sites shared another ARID1A LOF mutation at position chr1:27087900 (17-bp deletion). Interestingly, upon further analysis of chr1:27087900 in the primary specimen, we found that the 17-bp deletion was indeed present but at very low read depth (Supplemental Figure 6). Interestingly, this finding was not isolated to ARID1A, and a similar phenomenon was observed after baseline filtering for ADAM29, SCNN1A, CFHR4, and MAGI1 in patient UTUC1 (Supplemental Table 8). Importantly, the decrease in the mutational load of these genes in the primary specimen was not secondary to a lower tumor cellularity (Supplemental Table 8). Together, these findings raise the important point that UTUC metastases can arise from nondominant clonal populations. Because this observation was made within a single patient, it requires validation in a larger cohort, and it remains to be determined whether this also occurs in LTUC.
Intra- and interpatient copy number heterogeneity is prevalent in UTUC.
To determine intra- and interpatient variability in gene structure, we conducted genome-wide copy number analysis. Metastatic tumors exhibited more similarities to other metastases within the same patient than to the primary tumor (Figure 3A). We found no consistent large copy number amplifications or losses that were uniform across both LTUC and UTUC. However, we observed 8p heterozygous loss across all LTUC tumors, which has been described in UC (44). Next, we estimated per nucleotide scale Euclidean distances between tumor samples to delineate how similar or different each tumor was within a patient and among patients at the copy number level. In Figure 3B, each patient specimen is represented as a single point, and the distance between each point underscores its relationship to all other tumors. Our analysis revealed that UTUC tumors exhibit substantial differences at the copy number level within patients compared with LTUC tumors. Moreover, as a group, LTUC patients were more similar to one another at the copy number level compared with UTUC patients (Figure 3B). These findings suggest that UTUC tumors undergo a wider variety of copy number alterations compared with LTUC both within and among patients. This is further supported by hierarchical clustering of gene-restricted copy number profiles, which revealed a great genomic difference between primary tumors and metastases in UTUC that was less pronounced in LTUC (Figure 3B, inset).
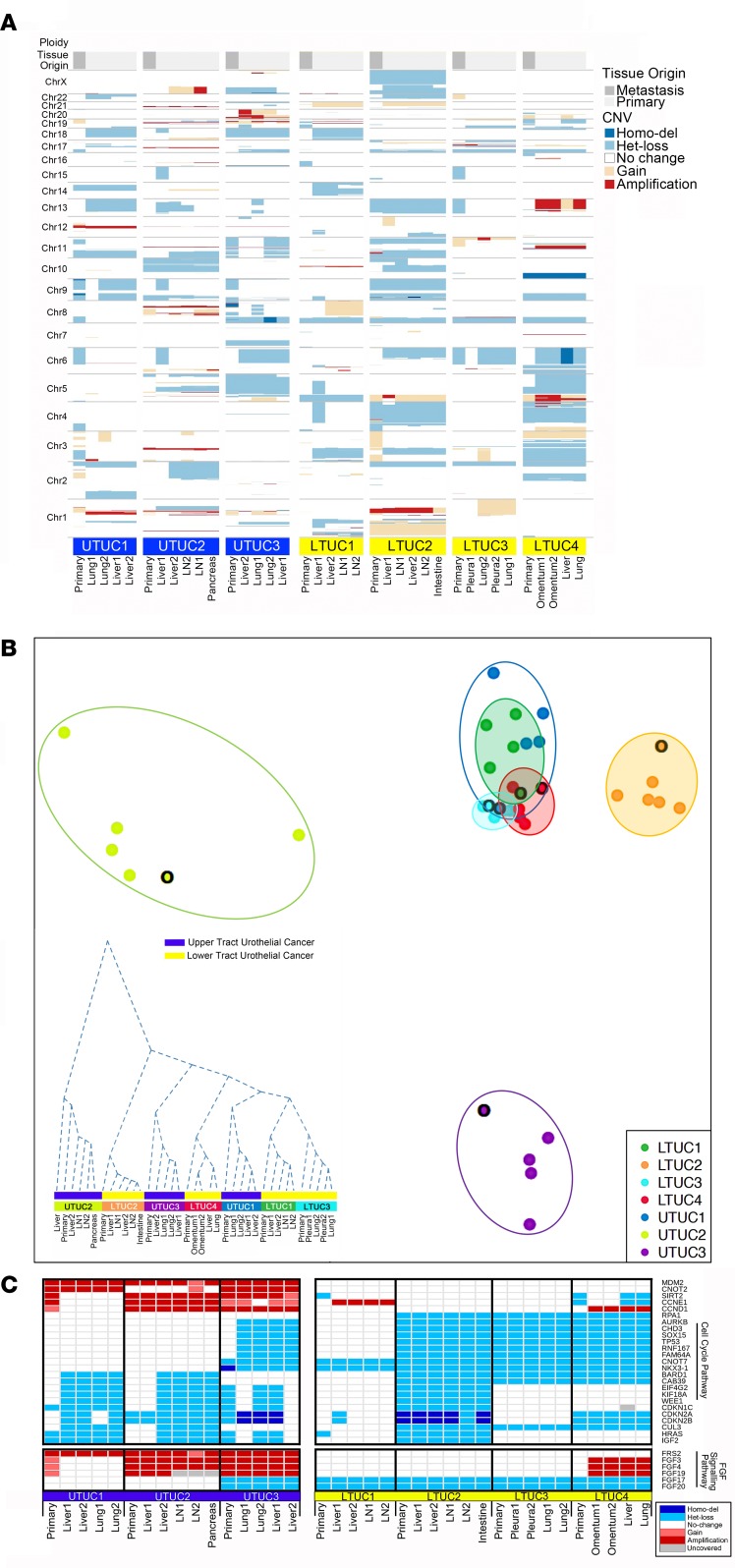
Figure 3. Metastatic UTUC exhibits significant intra- and interpatient copy number heterogeneity compared with metastatic LTUC.
(A) Copy number analysis at the chromosome level normalized for baseline ploidy across each primary and metastatic UTUC (n = 16) and LTUC tumor (n = 21). (B) Multidimensional (2D) scaling (MDS) analysis using estimated genome-wide absolute copy number (UTUC: unshaded circles; LTUC: shaded circles). Results are stratified by primary (black border) versus metastatic samples per patient. Axes represent multidimensional intersample distances projected to 2 dimensions using classical MDS. Inset: hierarchical clustering dendrogram using genome-wide, gene-restricted copy number profiles. The height of the dendrogram delineates the cumulative copy number differences among tumor specimens. (C) Representative gene-specific copy number analysis reveals altered cell cycle genes (delineated by gene set enrichment analysis, FDR = 5.78 × 10–13) as well as a group of FGFR family genes. Het loss, heterozygous loss; homo del, homozygous deletion.
Next, we analyzed how copy number alterations affected specific genes and oncogenic pathways (Figure 3C). We observed that a number of cell cycle genes were significantly altered in both UTUC and LTUC patients through gene set enrichment analysis (FDR = 5.78 × 10–13). These included amplification of cyclin D1 and deletions of the cell cycle inhibitors CDKN2A and CDKN2B (Figure 3C). MDM2 was amplified exclusively in patients with UTUC across every tumor, whereas we observed no amplification in patients with LTUC (Figure 3C) in our patient population. Interestingly, although we did not observe sSNV mutations to the FGFRs (Supplemental Figure 2), their ligands, including FGF3, FGF4, FGF19, and the FGFR adapter FRS2, were amplified in our UTUC cohort but not in our LTUC cohort. These gene-specific findings suggest that copy number alterations may drive oncogenic processes through different mechanisms in LTUC and UTUC.
Analysis of putative druggable genomic alterations reveals a remarkable level of similarity between metastases.
Given the multiplicity of tumor specimens we had for each patient, we next asked how different tumors within the same patient would be predicted to respond to targeted therapeutics. To do so, we evaluated mutations and structural alterations identified in each tumor for putative druggable targets using the Drug Gene Interaction Database (DGIdb), OncoKB, and literature review. Potential therapeutic vulnerabilities included LOF mutations of ARID1A, an activating PIK3CA E545K mutation, and an LOF TSC1 mutation. We also observed changes at the gene structure level that may predict for drug sensitivity, including amplification of cyclin D1, homozygous deletion of the cyclin-dependent kinase inhibitor CDKN2A, and genomic gains of FGFR1, FGFR3, and PIK3CA (Figure 4). Interestingly, when considering only metastatic tissues, approximately 70% (15 of 22) of mutations were present in all samples from the same patient. This finding demonstrates that putative druggable lesions can be homogenous within a metastatic urothelial cancer patient and highlights the potential utility of using a biopsy from a single metastatic site to guide treatment decision-making.
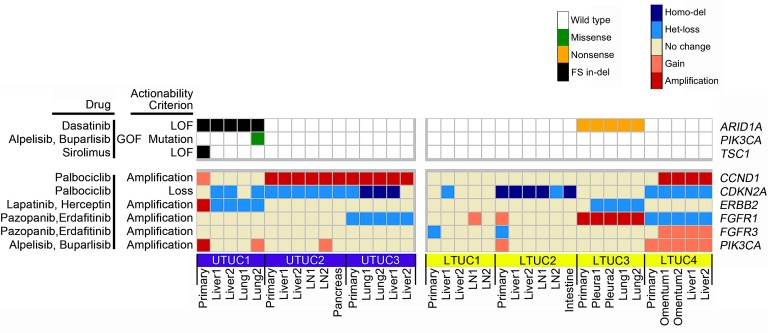
Figure 4. Marked homogeneity of druggable genomic alterations is present in UC patients.
All deleterious sSNVs and copy number alterations were screened for potential druggability using DGIdb, OncoKB, and published literature. This analysis reveals that out of 22 mutation targets observed within metastatic specimens with therapeutic potential, 15 (~70%) were present in all metastatic tumor specimens within a given individual. However, out of 20 mutations present in primary tumors, only 11 (55%) were also present in all metastatic tumors. LOF, loss-of-function mutation; GOF, gain-of-function mutation; Homo-del, homozygous deletion; Het-loss, heterozygous loss.
Discussion
We report the first genomic analysis to our knowledge of primary and metastatic UTUC and LTUC tumors across multiple sites within the same patient and observe several differences between UTUC and LTUC. First, we observed that both primary and metastatic UTUC tumors had on average lower Mut/Mb than LTUC tumors, consistent with previous reports (20, 28). However, a recent study demonstrated that microsatellite instability (MSI) and Lynch syndrome in patients with UTUC is associated with higher mutational burden (30). Interestingly in our study, patients with UTUC had a lower mutational burden than patients with LTUC. None of our patients with UTUC had evidence of perturbations to Lynch syndrome–associated genes, such as MSH2, MSH6, and MLH1, or MSI (Supplemental Tables 4 and 9, Supplemental Figure 7), which may explain the lower mutation rate among the patients with UTUC whose tumors we analyzed.
A major question with regard to the intrinsic differences between UTUC and LTUC has been, what are the genomic idiosyncrasies of the two urothelial cancer phenotypes? UTUC more frequently harbors pathological mutations to FGFR3, HRAS, CDKN2B, KMT2D, KDM6A, and NOTCH, while LTUC enriches for TP53, RB1, ATM, and ERBB2 mutations (27–30). In addition to these findings, our landscape view of copy number alterations across metastatic specimens from the same patients revealed that UTUC tumors exhibited more substantial differences at the copy number level compared with LTUC. This was evident both within and among patients and suggests that the fidelity of genome architecture in UTUC may be more disparately affected during tumor initiation and progression than LTUC. However, at the gene-specific level, our population revealed distinct oncogenic pathways consistently mutated in UTUC. For example, we observed the amplification of MDM2 in all of our primary and metastatic UTUC patient samples (Figure 3C). This finding suggests that there may be a selective pressure to preserve MDM2 amplification from primary disease to metastatic disease. This is consistent with MDM2’s role as an E3 ubiquitin ligase involved in proteasomal degradation of the tumor suppressor p53. Recently, it has been shown that MDM2 amplification is associated with increased stage and risk of distant recurrence in UTUC (45). Notably, MDM2 amplification was not observed in our LTUC cohort yet is described to be present in 6% of LTUCs (20), which may underlie differences in our small, heavily treated patient population.
Our study also highlights the potential for putative druggable targets across tumor sites within an individual patient. For example, we found that out of 22 mutations observed within metastatic specimens with therapeutic potential, 15 (~70%) were present in all metastatic tumor specimens in a given individual (Figure 4). This suggests that specific mutations may be clonally represented in the majority of tumors within a patient. Thus, a biopsy of a single metastatic site may be used in the setting of advanced-stage bladder cancer to help guide treatment decisions. This phenomenon has also been observed in an integrative genomic analysis of metastatic prostate cancer patient specimens through rapid autopsy. Indeed, Kumar et al. found limited genetic diversity within patients compared with substantial differences among patients (46). However, these findings are tempered by the fact that approximately 30% of mutation targets in our cohort were present only in a subset of metastases. For example, patient UTUC1 had only 1 PIK3CA mutation in a singular lung metastasis. Three other distinct metastases from UTUC1 did not exhibit the PIK3CA E545K mutations despite more than 33× coverage of the genomic region, more than 80% power to detect the mutation, and similar tumor cellularity (Figure 4, Supplemental Figures 2 and 5). Furthermore, we observed that patient UTUC1 had a TSC1 LOF mutation that was present only in a primary tumor but not present in any metastatic tumors (Figure 4 and ref. 47). The emerging technology of circulating tumor DNA (ctDNA) sequencing may represent a potential solution to complement tumor tissue sampling. For example, ctDNA testing of patients with advanced LTUC can detect various germline and somatic genomic alterations, including MDM2 amplification, so this testing has the potential to guide therapies and monitor treatment response in a serial manner (42, 48). Future experiments comparing tumor DNA from multiple LTUC and UTUC metastases within a patient and matched ctDNA are needed to further assess its clinical utility.
We observed that metastatic UTUC may not arise from the dominant clonal population in a primary tumor. In particular, patient UTUC1 had an ARID1A frameshift mutation at chr1:27107094 that was not present in any metastatic specimens. Instead, the metastatic lesions shared an ARID1A frameshift mutation at chr1:27087900 (Supplemental Figure 6). This observation points to the potential importance of ARID1A loss in bladder cancer pathogenesis, which is supported by large genomic studies in bladder cancer and in vivo studies in other cancer models (49, 50). Importantly, we found that the primary tumor specimen did in fact contain the mutation found in the metastatic tissues, but the mutation was much less than our read count cutoffs. This was observed for a series of other genes as well, including ADAM29, SCNN1A, CFHR4, and MAGI1, after baseline filtering and adjusting for tumor cellularity (Supplemental Table 8). As such, it is possible that the metastases in UTUC1 were seeded by a nondominant clone present in the patient’s primary tumor. This may be due to sampling bias; however, our primary tumor DNA specimens were derived from shavings spanning a region or multiple punches across a tumor block. Moreover, this type of observation has been made in other cancers. For example, it has been shown in prostate cancer that a small tumor clone characterized by PTEN, SPOP, and TP53 mutations, which was present in a patient’s primary tumor, seeded all the eventual metastases (51). These findings demonstrate the importance of obtaining genomic data dynamically from patients’ tumors because archival tissues may not adequately represent the diversity or even the clonal makeup of metastatic lesions.
Our study has some inherent limitations. The number of patients analyzed is limited, and although we evaluated multiple tumors within patients, they received varying numbers of systemic treatments, which may influence the observed differences. All patients in this cohort came from a single center and agreed to participate in a rapid autopsy program and thus may not broadly represent all patients with UTUC and LTUC. Moreover, our analysis was focused on DNA sequencing, whereas RNA- and protein-based analyses may also identify important differences between UTUC and LTUC biology. Despite the limitations, this clinical work provides a much-needed genomic landscape view of metastatic LTUC and UTUC, which will lay the groundwork for future larger studies.
Conclusions.
In this rapid autopsy series, we demonstrate that this population of patients with metastatic UTUC exhibited lower mutational burden but higher private deleterious mutations within patients. At the DNA copy number level, we observed greater intra- and inter-individual divergence in patients with UTUC. Taken together, these findings may explain differences in clinical behavior and treatment response between UTUC and LTUC. Importantly, despite overall mutational differences, common druggable targets were identified across metastases, underlining opportunities for therapeutic targeting in metastatic UC.
Methods
Clinical cohort.
Metastases were obtained within 4 hours of death as part of the rapid autopsy program described previously (52). For the logistical framework of the rapid autopsy program, see Supplemental Figure 8. Specimens were obtained from 7 patients including histopathologically normal tissue (n = 7, fresh-frozen), primary tumor samples (n = 7; 5 FFPE, 2 fresh-frozen), and 4–5 metastases per patient (n = 30, fresh-frozen) (Supplemental Tables 1 and 2). All patients within this cohort had metastatic UC, 3 derived from UTUC and 4 derived from LTUC (see Supplemental Table 1 for clinical details); all patients had visceral metastases, while all but 1 had liver metastases. Primary or metastatic specimens selected for sequencing had more than 80% tumor cellularity by genitourinary pathology review.
Whole-exome sequencing.
FFPE and frozen total DNA were extracted using the Puregene DNA isolation kit (Qiagen) (see Supplemental Table 2 for listing of samples). Sequencing libraries were prepared from 1 μg of intact genomic DNA quantified on an Invitrogen Qubit 2.0 Fluorometer (Life Technologies, Invitrogen) and Trinean DropSense96 spectrophotometer (Caliper Life Sciences). DNA was fragmented using a Covaris LE220 Focused-ultrasonicator using factory settings for an average size of 200 bp. Sequencing libraries were prepared using the KAPA Hyper Prep DNA library preparation kit (Kapa Biosystems, Roche) following end repair and A-tailing in a single-tube protocol. Library size distributions were validated using an Agilent 2200 TapeStation (Agilent Technologies) and quantified using the Qubit 2.0 Fluorometer. Individually indexed KAPA Hyper libraries were hybrid captured to NimbleGen SeqCap EZ Exome V3+UTR probes (Roche) according to the manufacturer’s protocol on a Sciclone NGSx Workstation (PerkinElmer) using a 1-capture-per-library strategy. Library size distributions after capture were validated using Agilent 2200 TapeStation and quantified using a Trinean DropSense96 spectrophotometer. Additional quality control during blending of pooled indexed libraries and cluster optimization were performed using the Qubit 2.0 Fluorometer. Libraries from frozen tissues were clustered at 4-plex and FFPE libraries at 3-plex per lane on an Illumina v4 flow cell using an Illumina cBot. Sequencing was performed using an Illumina HiSeq 2500 in high-output 100-bp paired-end mode using v4 reagents (PE100). Image analysis and base calling were performed using Illumina’s Real Time Analysis v1.18.66.3 software, followed by demultiplexing of indexed reads and generation of FASTQ files, using Illumina’s bcl2fastq Conversion Software v1.8.4 (https://support.illumina.com). The raw sequencing data have been submitted to the Database of Genotypes and Phenotypes (accession phs001797.v1.p1).
Sequencing data analysis.
All sequencing reads were aligned to human genome reference Hg19 using Bowtie (53). GATK Best Practices were followed for preprocessing of all bam files (54). Average read depth was 51×, with an average of 93% of nucleotides with more than 10× coverage (Supplemental Table 10) (55). The intersect tool from bedtools (56) was used to calculate the on-target aligned read percentage. MuTect (57) and Strelka (58) were used to call somatic mutations and short indels. We considered loci for variant calls covered by at least 14 normal reads with variant allele loci with a minimum of 7 alternate alleles and higher than q20 phred score. Tumor variants were restricted to at least 10% variant allelic frequency to exclude machine errors, sample preservation, and Covaris fragmentation technique–associated low-frequency false-positive calls (59). Oncotator (60) and Annovar (61) were subsequently used for annotation of all called variants. Presumed deleterious, nonsynonymous sSNVs were predicted through a consensus call of the majority of 11 mutation functional impact assessor tools (>6 of 11 or 55%; Supplemental Table 11; refs. 60, 61). Additionally, all frameshift and nonframeshift insertions or deletions, splicing, and stop-gain mutations were included in the analysis (Supplemental Figure 2, Supplemental Table 4). As a quality control measure, we evaluated the sSNVs from a single tumor, which was bisected. Half was FFPE and half was fresh-frozen. This revealed 80% concordance at the sSNV level, demonstrating only a limited impact of the different preservation techniques on our analysis (Supplemental Figure 9A).
sSNV power analysis.
To assess whether each mutant locus had sufficient coverage to reliably determine its mutation status, we calculated the statistical power to call an sSNV given the tumor read depth (N), tumor cellularity (α), and tumor ploidy (ϕ). Let p be the expected variant allelic fraction for observing a heterozygous clonal sSNV, where p = α/(αϕ + 2[1 – α]). For every locus, we computed the theoretical power using the binomial test for observing 3 or more variant reads: p (X ≥ 3) = 1 – [Bin(0, N, p) + Bin(1, N, p) + Bin(2, N, p)]. For each sSNV, we extracted the reference and variant read counts and total read depth using the R function pileLettersAt from R/Bioconductor package GenomicAlignments (v 1.18.1). Indels were excluded from these power calculations. Tumor cellularity and tumor ploidy were estimated by Sequenza. For our analyses, we determined loci to have sufficient coverage for calling somatic mutations if the power estimate was greater than 80%. Overall, we showed more than 80% power to detect mutations across individual patients for approximately 90% of mutations identified (Supplemental Figure 5).
Mutational signature analysis.
Mut/Mb stats included all called variants restricted to defined 104.8-Mb target capture regions (Supplemental Table 3). Mutational signatures were evaluated using the DeconstructSigs (62), which used trinucleotide somatic mutation frequencies normalized to the exome capture region, followed by weighted reconstruction of the predominant signatures of each sample. The deconvoluted mutation signature frequency was derived using 30 predefined COSMIC SBS V2 signature definitions. Final groups were then clustered based on biological phenotypes (e.g., mitotic clock, APOBEC, HRD, smoking, etc.)
Copy number analysis.
Sequenza was used to perform absolute copy number calling and estimation of tumor cellularity and ploidy (63). Exome sequences were aligned using the BWA aligner (64). A Q15 base quality cutoff was used to avoid DNA alignment biases. Furthermore, only high-quality (Q40) aligned reads were considered for all downstream copy number estimation analyses. To avoid bias related to genome-wide GC content depth, a normalization step was performed using Hg19 GC content data from the University of California Santa Cruz genome browser (63, 65). Initial segmentation calls were made using the circular binary segmentation algorithm from the Bioconductor DNAcopy R package, followed by Sequenza’s probabilistic model-based copy number estimation. The copy number estimation uses the average depth ratio (tumor vs. normal) and B allele frequency (the lesser of the 2 allelic fractions as measured at germline heterozygous positions) and performs the estimation considering the derived overall tumor ploidy/cellularity, genomic segment–specific copy number, and minor allele copy number (Supplemental Table 5). As a quality control measure, we evaluated the sCNV differences from a single tumor, half of which was FFPE and half of which was fresh-frozen. This revealed high concordance in terms of somatic copy number aberration, tumor ploidy, and percentage of genome loss estimation, suggesting the differential mode of tissue preservation had no significant effect on estimated sCNV profiles (Supplemental Figure 9B). Using the genomic segment–specific absolute copy number values, the genomic distance between every tumor pair was calculated as a sum of the per nucleotide Euclidean distances to derive intratumor and intertumor genomic distances. Mean centering of copy number variation was used to derive an estimate of percentage of genome loss and amplification for each tumor tissue. Gene-restricted copy number was derived using Gencode v19 annotation references. The recurrently copy-number-altered genes (altered in >68% of tumors in both LTUC and UTUC) were then checked for pathway enrichment using the GOrilla Gene Ontology term enrichment tool (http://cbl-gorilla.cs.technion.ac.il/).
MSI evaluation.
MSIsensor was used to detect replication slippage variants at microsatellite regions for each tumor/normal pair and MSI scores were reported (Supplemental Table 9 and Supplemental Figure 7; ref. 66).
Actionable target analysis.
Actionable mutation prediction was based on called variants only. Both sSNVs and sCNVs were queried using established drug genotype databases, including DGIdb (67) and OncoKB (68), to identify potentially actionable variants. The final list was filtered based on previously published preclinical data and clinical trials in cancer.
Statistics.
Statistical analyses pertaining to each figure are included within the appropriate legends. For direct comparison of continuous variables, 2-tailed t test and/or Mann-Whitney U tests were used, whereas χ2 was used for categorical variables as appropriate.
Study approval.
All samples were obtained from patients with signed informed consent documents under the aegis of the Cancer Donor Program at the University of Washington (University of Washington IRB 2341).
Author contributions
ACH, JLW, HML, and RBM conceived of and designed the project. BRW, NDS, SJ, SA, HB, LM, GH, and ACH acquired, analyzed, and interpreted the data. FVL conducted all pathological analyses. HHC, MTS, EYY, PG, JKL, and ACH obtained patients’ consent for this study. LK provided support for the rapid autopsy program. SKH provided statistical support. BRW, NDS, and ACH wrote the manuscript. BRW, NDS, SA, HB, SJ, FVL, HHC, MTS, EYY, PG, JKL, LK, SKH, LM, GH, PSN, RBM, JLW, HML, and ACH reviewed and approved the manuscript.
Supplementary Material
Acknowledgments
We are grateful to the patients and their families who participated in this study. We thank members of the Hsieh laboratory for helpful advice and criticism. We are thankful for the support of the University of Washington Genitourinary Cancer Research Laboratory and the rapid autopsy team. We thank A. Wyatt for critical discussion and review of this work. We thank J.M. Shen for editing the manuscript. This work was supported by the Seattle Translation Tumor Research Program in Bladder Cancer, the Howard J. Cohen Bladder Cancer Foundation, seed funding from David Matthews, and the Stinchcomb Memorial Funds. BRW is a recipient of a Young Investigator Award from the Johns Hopkins Greenberg Bladder Cancer Institute fund. NDS is a recipient of a Department of Defense (DOD) Prostate Cancer Research Program (PCRP) Early Investigator Research Award (W81XWH-17-1-0380). SJ is a recipient of the Fred Hutch Interdisciplinary Training Grant and a DOD PRCRP Horizon Award. HML is a recipient of a New Investigator Award from the Fred Hutch/University of Washington Cancer Consortium (P30CA015704). ACH is a V Foundation Scholar and is funded by a NextGen Grant for Transformative Cancer Research from the American Association for Cancer Research, a Burroughs Wellcome Fund Career Award for Medical Scientists, and NIH grants K08CA175154 and R37CA230617. This research was also supported by the Genomics Shared Resource of the Fred Hutch/University of Washington Cancer Consortium (P30CA015704).
Version 1. 05/30/2019
In-Press Preview
Version 2. 07/11/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(13):e128728. https://doi.org/10.1172/jci.insight.128728.
Contributor Information
Brian R. Winters, Email: wintersb@uw.edu.
Navonil De Sarkar, Email: ndesarka@fredhutch.org.
Sonali Arora, Email: sarora@fredhutch.org.
Hamid Bolouri, Email: hbolouri@fredhutch.org.
Sujata Jana, Email: sjana@fredhutch.org.
Funda Vakar-Lopez, Email: fvakar@uw.edu.
Heather H. Cheng, Email: hhcheng@uw.edu.
Michael T. Schweizer, Email: schweize@uw.edu.
Evan Y. Yu, Email: evanyu@u.washington.edu.
Petros Grivas, Email: pgrivas@u.washington.edu.
John K. Lee, Email: jklee5@fredhutch.org.
Lori Kollath, Email: lkollath@uw.edu.
Sarah K. Holt, Email: sholt@uw.edu.
Lisa McFerrin, Email: anomolisa@gmail.com.
Gavin Ha, Email: gha@fredhutch.org.
Peter S. Nelson, Email: pnelson@fredhutch.org.
Robert B. Montgomery, Email: rbmontgo@u.washington.edu.
Jonathan L. Wright, Email: jlwright@u.washington.edu.
Hung-Ming Lam, Email: minglam@uw.edu.
Andrew C. Hsieh, Email: ahsieh@fredhutch.org.
References
- 1.Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009;374(9685):239–249. doi: 10.1016/S0140-6736(09)60491-8. [DOI] [PubMed] [Google Scholar]
- 2.Spiess PE, et al. Bladder Cancer, version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2017;15(10):1240–1267. doi: 10.6004/jnccn.2017.0156. [DOI] [PubMed] [Google Scholar]
- 3.Flaig TW, et al. NCCN Guidelines insights: Bladder Cancer, version 5.2018. J Natl Compr Canc Netw. 2018;16(9):1041–1053. doi: 10.6004/jnccn.2018.0072. [DOI] [PubMed] [Google Scholar]
- 4.Dash A, et al. A role for neoadjuvant gemcitabine plus cisplatin in muscle-invasive urothelial carcinoma of the bladder: a retrospective experience. Cancer. 2008;113(9):2471–2477. doi: 10.1002/cncr.23848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeshchina O, et al. Relative efficacy of perioperative gemcitabine and cisplatin versus methotrexate, vinblastine, adriamycin, and cisplatin in the management of locally advanced urothelial carcinoma of the bladder. Urology. 2012;79(2):384–390. doi: 10.1016/j.urology.2011.10.050. [DOI] [PubMed] [Google Scholar]
- 6.Plimack ER, et al. Accelerated methotrexate, vinblastine, doxorubicin, and cisplatin is safe, effective, and efficient neoadjuvant treatment for muscle-invasive bladder cancer: results of a multicenter phase II study with molecular correlates of response and toxicity. J Clin Oncol. 2014;32(18):1895–1901. doi: 10.1200/JCO.2013.53.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee FC, et al. Pathologic response rates of gemcitabine/cisplatin versus methotrexate/vinblastine/adriamycin/cisplatin neoadjuvant chemotherapy for muscle invasive urothelial bladder cancer. Adv Urol. 2013;2013:317190. doi: 10.1155/2013/317190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zargar H, et al. Multicenter assessment of neoadjuvant chemotherapy for muscle-invasive bladder cancer. Eur Urol. 2015;67(2):241–249. doi: 10.1016/j.eururo.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grossman HB, et al. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. N Engl J Med. 2003;349(9):859–866. doi: 10.1056/NEJMoa022148. [DOI] [PubMed] [Google Scholar]
- 10. Guzzo TJ, Vaughn DJ. Management of invasive and metastatic bladder cancer. In: McDougal W, et al., eds. Campbell-Walsh Urology. Philadelphia, Pennsylvania, USA; 2016:2223–2241.e7. [Google Scholar]
- 11.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 12.Emamekhoo H, et al. Prognostic factors and risk stratification in invasive upper tract urothelial carcinoma. Clin Genitourin Cancer. 2018;16(4):e751–e760. doi: 10.1016/j.clgc.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 13.David KA, Mallin K, Milowsky MI, Ritchey J, Carroll PR, Nanus DM. Surveillance of urothelial carcinoma: stage and grade migration, 1993-2005 and survival trends, 1993–2000. Cancer. 2009;115(7):1435–1447. doi: 10.1002/cncr.24147. [DOI] [PubMed] [Google Scholar]
- 14.Melamed MR, Reuter VE. Pathology and staging of urothelial tumors of the kidney and ureter. Urol Clin North Am. 1993;20(2):333–347. [PubMed] [Google Scholar]
- 15.Almassi N, et al. Impact of neoadjuvant chemotherapy on pathologic response in patients with upper tract urothelial carcinoma undergoing extirpative surgery. Clin Genitourin Cancer. 2018;16(6):e1237–e1242. doi: 10.1016/j.clgc.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Birtle AJ, et al. Results of POUT: A phase III randomised trial of perioperative chemotherapy versus surveillance in upper tract urothelial cancer (UTUC) J Clin Oncol. 2018;36(suppl_6):407 [Google Scholar]
- 17. Smith AK, Matin SF, Jarrett, TW. Urothelial tumors of the upper urinary tract and ureter. In: McDougal W, et al, eds. Campbell-Walsh Urology. Philadelphia, Pennsylvania, USA; 2016:1365-1402.e8. [Google Scholar]
- 18.Sternberg CN, et al. Methotrexate, vinblastine, doxorubicin, and cisplatin for advanced transitional cell carcinoma of the urothelium. Efficacy and patterns of response and relapse. Cancer. 1989;64(12):2448–2458. doi: 10.1002/1097-0142(19891215)64:12<2448::AID-CNCR2820641209>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson AG, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2018;174(4):1033. doi: 10.1016/j.cell.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damrauer JS, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A. 2014;111(8):3110–3115. doi: 10.1073/pnas.1318376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi W, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25(2):152–165. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sjödahl G, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res. 2012;18(12):3377–3386. doi: 10.1158/1078-0432.CCR-12-0077-T. [DOI] [PubMed] [Google Scholar]
- 24.Seiler R, et al. Impact of molecular subtypes in muscle-invasive bladder cancer on predicting response and survival after neoadjuvant chemotherapy. Eur Urol. 2017;72(4):544–554. doi: 10.1016/j.eururo.2017.03.030. [DOI] [PubMed] [Google Scholar]
- 25.Nickerson ML, et al. Molecular analysis of urothelial cancer cell lines for modeling tumor biology and drug response. Oncogene. 2017;36(1):35–46. doi: 10.1038/onc.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Allen EM, et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 2014;4(10):1140–1153. doi: 10.1158/2159-8290.CD-14-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sfakianos JP, et al. Genomic characterization of upper tract urothelial carcinoma. Eur Urol. 2015;68(6):970–977. doi: 10.1016/j.eururo.2015.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moss TJ, et al. Comprehensive genomic characterization of upper tract urothelial carcinoma. Eur Urol. 2017;72(4):641–649. doi: 10.1016/j.eururo.2017.05.048. [DOI] [PubMed] [Google Scholar]
- 29.Lee JY, et al. Molecular characterization of urothelial carcinoma of the bladder and upper urinary tract. Transl Oncol. 2018;11(1):37–42. doi: 10.1016/j.tranon.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Audenet F, et al. Clonal relatedness and mutational differences between upper tract and bladder urothelial carcinoma. Clin Cancer Res. 2019;25(3):967–976. doi: 10.1158/1078-0432.CCR-18-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullane SA, Bellmunt J. Re: John P. Sfakianos, Eugene K. Cha, Gopa Iyer, et al. Genomic characterization of upper tract urothelial carcinoma. Eur Urol 2015;68:970-7. Eur Urol. 2016;70(3):e71. doi: 10.1016/j.eururo.2016.01.036. [DOI] [PubMed] [Google Scholar]
- 32.Bellmunt J, et al. Randomized phase III study comparing paclitaxel/cisplatin/gemcitabine and gemcitabine/cisplatin in patients with locally advanced or metastatic urothelial cancer without prior systemic therapy: EORTC Intergroup Study 30987. J Clin Oncol. 2012;30(10):1107–1113. doi: 10.1200/JCO.2011.38.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrylak DP, et al. Ramucirumab plus docetaxel versus placebo plus docetaxel in patients with locally advanced or metastatic urothelial carcinoma after platinum-based therapy (RANGE): a randomised, double-blind, phase 3 trial. Lancet. 2017;390(10109):2266–2277. doi: 10.1016/S0140-6736(17)32365-6. [DOI] [PubMed] [Google Scholar]
- 34.Balar AV, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet. 2017;389(10064):67–76. doi: 10.1016/S0140-6736(16)32455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faltas BM, et al. Clonal evolution of chemotherapy-resistant urothelial carcinoma. Nat Genet. 2016;48(12):1490–1499. doi: 10.1038/ng.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikkilä J, et al. Heterozygous mutations in PALB2 cause DNA replication and damage response defects. Nat Commun. 2013;4:2578. doi: 10.1038/ncomms3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pathania S, et al. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat Commun. 2014;5:5496. doi: 10.1038/ncomms6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan SLW, et al. A class of environmental and endogenous toxins induces BRCA2 haploinsufficiency and genome instability. Cell. 2017;169(6):1105–1118.e15. doi: 10.1016/j.cell.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salter JD, Bennett RP, Smith HC. The APOBEC protein family: united by structure, divergent in function. Trends Biochem Sci. 2016;41(7):578–594. doi: 10.1016/j.tibs.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burns MB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494(7437):366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu JG, Shi DY, Liu X, Zheng XX, Wang L, Li Q. Chromatin-regulatory genes served as potential therapeutic targets for patients with urothelial bladder carcinoma. J Cell Physiol. 2019;234(5):6976–6982. doi: 10.1002/jcp.27440. [DOI] [PubMed] [Google Scholar]
- 42.Agarwal N, et al. Characterization of metastatic urothelial carcinoma via comprehensive genomic profiling of circulating tumor DNA. Cancer. 2018;124(10):2115–2124. doi: 10.1002/cncr.31314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beothe T, Zubakov D, Kovacs G. Homozygous losses detected by array comparative genomic hybridization in multiplex urothelial carcinomas of the bladder. Cancer Genet. 2015;208(9):434–440. doi: 10.1016/j.cancergen.2015.05.029. [DOI] [PubMed] [Google Scholar]
- 45.Bagrodia A, et al. Genomic biomarkers for the prediction of stage and prognosis of upper tract urothelial carcinoma. J Urol. 2016;195(6):1684–1689. doi: 10.1016/j.juro.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar A, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22(4):369–378. doi: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iyer G, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338(6104):221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vandekerkhove G, et al. Circulating tumor DNA reveals clinically actionable somatic genome of metastatic bladder cancer. Clin Cancer Res. 2017;23(21):6487–6497. doi: 10.1158/1078-0432.CCR-17-1140. [DOI] [PubMed] [Google Scholar]
- 49.Mathur R, et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet. 2017;49(2):296–302. doi: 10.1038/ng.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun X, et al. Arid1a has context-dependent oncogenic and tumor suppressor functions in liver cancer. Cancer Cell. 2017;32(5):574–589.e6. doi: 10.1016/j.ccell.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haffner MC, et al. Tracking the clonal origin of lethal prostate cancer. J Clin Invest. 2013;123(11):4918–4922. doi: 10.1172/JCI70354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roudier MP, et al. Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone. Hum Pathol. 2003;34(7):646–653. doi: 10.1016/S0046-8177(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 53.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.1033. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28(14):1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 59.Costello M, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013;41(6):e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramos AH, et al. Oncotator: cancer variant annotation tool. Hum Mutat. 2015;36(4):E2423–E2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Favero F, et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol. 2015;26(1):64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Janevski A, Varadan V, Kamalakaran S, Banerjee N, Dimitrova N. Effective normalization for copy number variation detection from whole genome sequencing. BMC Genomics. 2012;13(suppl 6):S16. doi: 10.1186/1471-2164-13-S6-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niu B, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30(7):1015–1016. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cotto KC, et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46(D1):D1068–D1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chakravarty D, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017:2017. doi:10.1200/PO.17.00011. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.