Abstract
Diabetic β cell failure is associated with β cell dedifferentiation. To identify effector genes of dedifferentiation, we integrated analyses of histone methylation as a surrogate of gene activation status and RNA expression in β cells sorted from mice with multiparity-induced diabetes. Interestingly, only a narrow subset of genes demonstrated concordant changes to histone methylation and RNA levels in dedifferentiating β cells. Notable among them was the α cell signature gene Gc, encoding a vitamin D–binding protein. Although diabetes was associated with Gc induction, Gc-deficient islets did not induce β cell dedifferentiation markers and maintained normal ex vivo insulin secretion in the face of metabolic challenge. Moreover, Gc-deficient mice exhibited a more robust insulin secretory response than normal controls during hyperglycemic clamp studies. The data are consistent with a functional role of Gc activation in β cell dysfunction and indicate that multiparity-induced diabetes is associated with altered β cell fate.
Keywords: Endocrinology, Metabolism
Keywords: Beta cells, Diabetes, Epigenetics
Integration of histone and RNA profiles in β cell from multiparity-induced diabetic mice revealed functional roles of α cell-restricted protein Gc in β cell dedifferentiation.
Introduction
Type 2 diabetes affects nearly 300,000,000 people worldwide (1) and is characterized by insulin resistance and pancreatic β cell failure (2). The latter can be attributed to reduced β cell function and mass and is in part genetically determined (3). Because treatments that restore β cell function have limited effect or durability (4), various therapeutic approaches have been proposed to increase or restore β cell mass (5). In either case, developing a mechanism-based intervention to redress β cell failure requires a detailed understanding of the relevant pathophysiology.
We and others have proposed that β cell failure arises from β cell dedifferentiation, i.e., loss of mature β cell features associated with regression to a progenitor-like stage (6–10). This process accounts for the partial clinical reversibility of β cell dysfunction (8, 11–16) and has been found to variable extents (17) in autopsy surveys of diabetic patients (18, 19). In addition, it may explain the observation of mixed-features α/β cells in humans (20) and lineage conversion in rodents (7), which dovetail with the increased glucagon “tone” in diabetes (21).
Recently, new information has emerged from mapping histone modifications and chromatin-remodeling events associated with β cell development, fate commitment, proliferation, regeneration (22–25), as well as abnormalities linked to type 2 diabetes (26–28). However, how these modifications relate to β cell dedifferentiation remains unknown.
To fill this gap in knowledge, we integrated an assessment of genome-wide histone modifications with gene expression data to identify effectors of β cell dedifferentiation. Specifically, we developed a model of multiparity-induced diabetes and isolated β cells to perform ChIP-Seq and survey histone H3 trimethylation at lysine 4 and lysine 27, 2 modifications that herald transcriptional activation and repression, respectively. Integration of these data with transcriptome data led us to the identification of a restricted subset of candidate mediators of β cell failure. We selected a gene emerging from these analyses for functional interrogation. Vitamin D–binding protein, Gc, displayed elevated mRNA as well as histone 3 lysine 4 trimethylation levels in dedifferentiated β cells. Compared with WT, Gc-ablated mice maintained normal insulin secretion when fed a high-fat diet and showed an increased insulin secretory response during hyperglycemic clamps. Thus, the vitamin D–binding protein, Gc, appears to contribute to β cell dysfunction and to be a marker of ongoing β cell dedifferentiation.
Results
Experimental approach.
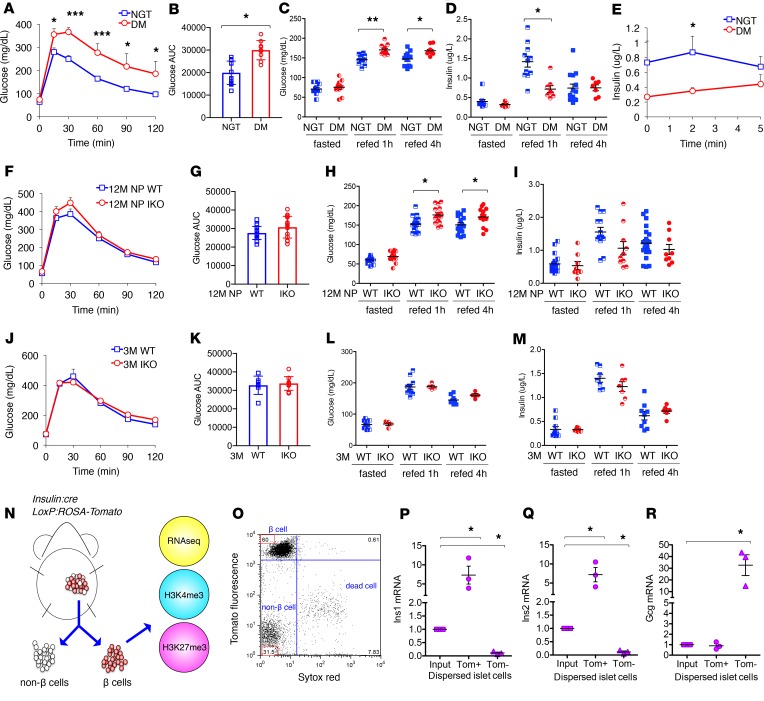
FoxO1 ablation results in age- or multiparity-dependent diabetes and β cell dedifferentiation (6). Therefore, we built our experimental approach on mice that developed diabetes following multiple pregnancies, as a result of the FoxO1 knockout. Consistent with prior data, 12-month-old, multiparous, β cell–specific FoxO1-knockout mice (I-KO) had impaired glucose tolerance, hyperglycemia, and reduced glucose-stimulated insulin secretion (Figure 1, A–E, and ref. 6), and are henceforth referred to as DM, while WT are indicated as NGT. As a control for changes induced by parity, we compared β cells isolated from age-matched (12-month-old) nulliparous (NP) animals (12M NP WT vs. 12M NP I-KO; Figure 1, F–I), whereas as a control for changes induced by aging, we used 3-month-old animals (3M WT vs. 3M I-KO; Figure 1, J–M). As a model to integrate chromatin modifications and gene expression changes occurring during β cell dedifferentiation, we compared β cells sorted by FACS from WT and I-KO mice (ref. 6 and Figure 1N). We labeled β cells by introducing an insulin-Cre–driven Rosa26 Tomato allele. The marked increase of fluorescence (Figure 1O) and the 7-fold enrichment of Insulin1 (Ins1) and Insulin2 (Ins2) in Tomato-positive cells (Figure 1, P and Q) indicated that they are indeed β cells. Tomato-negative cells showed a 10-fold depletion of Ins1 and Ins2 and a 30-fold enrichment of Glucagon (Gcg) compared with the input (Figure 1, P–R).
Figure 1. FACS isolates a highly pure β cell population from WT and I-KO mice.
(A) Oral glucose tolerance test in normal glucose tolerance (NGT) (n = 11) and diabetes mellitus (DM) (n = 9) mice. (B) Area under the curve (AUC) in A. (C) Glucose levels of fasted and refed NGT (n ≥ 10) and DM (n ≥ 7) mice. (D) Insulin levels of fasted and refed NGT (n ≥ 9) and DM (n ≥ 6) mice. (E) Acute insulin secretion following glucose stimulation in NGT (n = 5) and DM (n = 6) mice. (F) Oral glucose tolerance test in 12M NP WT (n = 17) and I-KO (n = 14) mice. (G) AUC in F. (H) Glucose levels of fasted and refed 12M NP WT (n ≥ 10) and I-KO (n ≥ 10) mice. (I) Insulin levels of fasted and refed 12M NP WT (n ≥ 10) and I-KO (n ≥ 8) mice. (J) Oral glucose tolerance test in 3M WT (n = 6) and I-KO (n = 9) mice. (K) AUC in J. (L) Glucose levels of fasted and refed 3M WT (n ≥ 6) and I-KO (n ≥ 3) mice. (M) Insulin levels of fasted and refed 3M WT (n ≥ 7) and I-KO (n ≥ 6) mice. (N) Schematic of β cell collection and experimental design. (O) A representative flow cytometric plot pregated on Tomato fluorescence–positive cells, where SYTOX Red (x axis) distinguishes live from dead cells and PE-TR (y axis) indicates the level of Tomato fluorescence. FACS was performed more than 10 times. Gene expression by reverse transcription quantitative PCR for (P) Ins1, (Q) Ins2, and (R) Gcg in dispersed pancreatic islet cells as input and Tomato fluorescence–positive and –negative cell populations (n = 3). Error bars represent ± SEM, *P < 0.05, **P < 0.01, and ***P < 0.005 by Student’s t test and ANOVA.
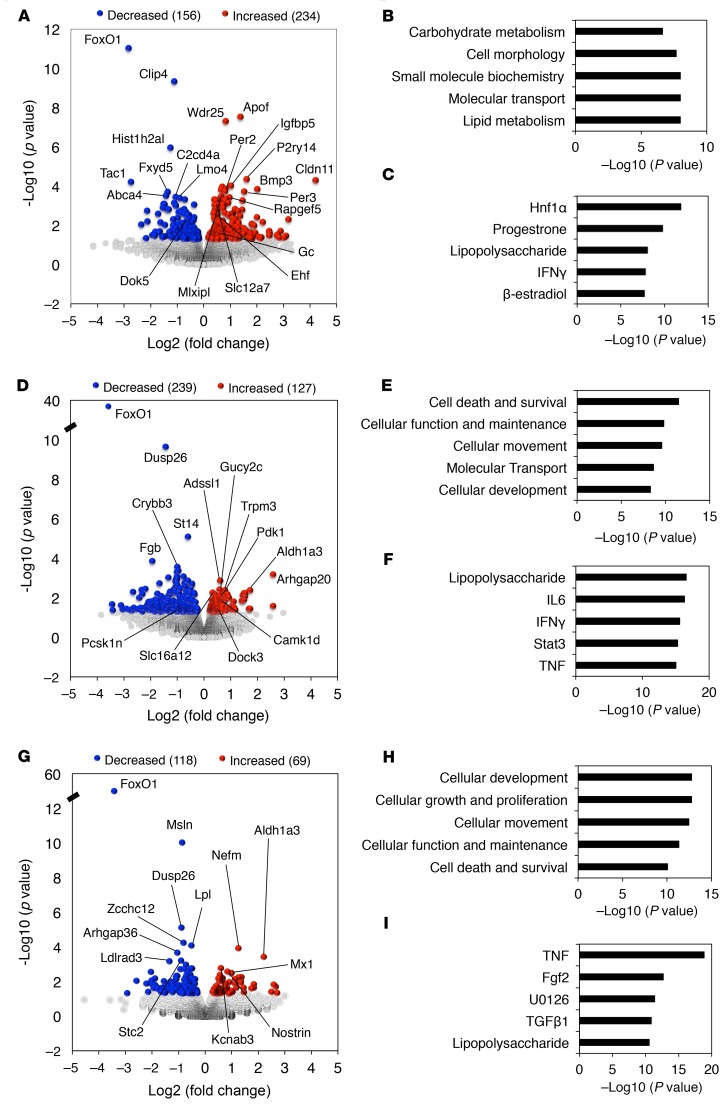
Transcriptome analysis inβcells. To investigate the mechanism of β cell dedifferentiation, we subjected sorted β cells from 3M WT and I-KO, 12M NP WT and I-KO, and NGT as well as DM mice to RNA-Seq. When we compared DM versus NGT, we found 390 genes with altered expression (156 upregulated and 234 downregulated; Figure 2A and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.128351DS1). Ingenuity pathway analysis categorized the enriched genes to carbohydrate metabolism, cell morphology, small molecule biochemistry, molecular transport, and lipid metabolism (Figure 2B), while top upstream regulators included Hnf1α, progesterone, lipopolysaccharide, IFN-γ, and β-estradiol (Figure 2C). In contrast with the multiparous group, key determinants of genotypic differences in age-matched (12M) nondiabetic NP WT versus I-KO did not include carbohydrate metabolism or Hnf1α. Rather, they included cell death and survival, cellular function, maintenance, movement, and development (Figure 2, D–F, and Supplemental Table 2). Although these mice were not diabetic, there were incipient signs of dedifferentiation in FoxO1-deficient β cells. They included increased expression of Aldh1a3 (29) and β cell “disallowed” genes Ldha, monocarboxylate transporter family members Slc16a3/MCT4 and Slc16a12/MCT12 (30, 31), and striking decreases in mRNAs encoding Sst, Ppy, and Pyy (Supplemental Table 2). In contrast, 3M NP WT and I-KO showed the fewest gene differences and no notable functional candidates among them (Figure 2, G–I, and Supplemental Table 3). Based on gene ontology results, absence of FoxO1 affected carbohydrate and lipid metabolism in β cells in the multiparity context, likely contributing to diabetes onset. Top upstream regulators pinpointed Hnf1α signaling as a key contributor to the process (32).
Figure 2. Genome-wide gene expression analysis in dedifferentiated β cells.
Volcano plots show 2-way comparison between (A) NGT versus DM, (D) 12M NP WT versus I-KO, and (G) 3M WT versus I-KO mice for RNA-Seq (n = 3 for each group), where y axis shows negative log10 (P value), and x axis represents log2 function of I-KO/WT. Ingenuity pathway analysis revealed top 5 molecular and cellular functions categorized by altered genes in (B) NGT versus DM, (E) 12M NP WT versus I-KO, and (H) 3M WT versus I-KO mice. Top 5 upstream regulators computed by ingenuity pathway analysis in (C) NGT versus DM, (F) 12M NP WT versus I-KO, and (I) 3M WT versus I-KO mice.
Integration of H3 trimethylation patterns with transcriptomes.
Next, we surveyed histone methylation using genome-wide ChIP-Seq with H3K27me3 as a mark of transcriptional repression and H3K4me3 as a mark of transcriptional activation. In this analysis, owing to costs, we focused on 12M animals (Figure 3) because the 3M control group showed minimal changes in RNA profile. As shown in Figure 2, A and D, 12M multiparae and nulliparae, respectively, showed similar numbers of altered transcripts but widely divergent numbers of altered histone marks (Supplemental Tables 4–7). Interestingly, we observed greater changes in nondiabetic (NP) β cells, irrespective of genotype (Figure 3).
Figure 3. Chromatin dynamics in dedifferentiated β cells.
Genome-wide survey of trimethylation at histone 3 lysine 4 (H3K4me3) and lysine 27 (H3K27me3) in NGT versus DM and 12M NP WT versus I-KO mice (n = 3 for each genotype).
Several genes linked to diabetes showed altered histone trimethylation. In NGT versus DM, Hhex (33), Adcy5 (34), and Lmo4 had decreased H3K4me3, while Rapgef5, Ehf, and Gc had increased H3K4me3 (Figure 3 and Supplemental Table 4). In 12M NP WT versus I-KO, Rfx6, Isl1, Neurod1, and Slc30a8 had decreased H3K4me3, while Camk2b, Camk2g, and Jag2 had increased H3K4me3 (Figure 3 and Supplemental Table 5). It is again interesting to note that genes associated with β cell dedifferentiation appeared poised to activate in nondiabetic but FoxO1-deficient islets.
To narrow down candidate genes for β cell dysfunction, we overlaid RNA-Seq and histone 3 trimethylation data and identified genes with increased expression levels and active H3K4me3 marks or decreased expression levels and increased repressive H3K27me3 marks. Among the genes that met these criteria, we focused on the vitamin D–binding protein, Gc, because its induction was specific to the diabetic state and because it is an α cell signature gene (35). Thus, its activation is consistent with the notion that diabetes is associated with cell fate conversion in β cells (6). Furthermore, Gc is an Hnf1α target (36, 37), consistent with altered Hnf1α signaling in DM β cells.
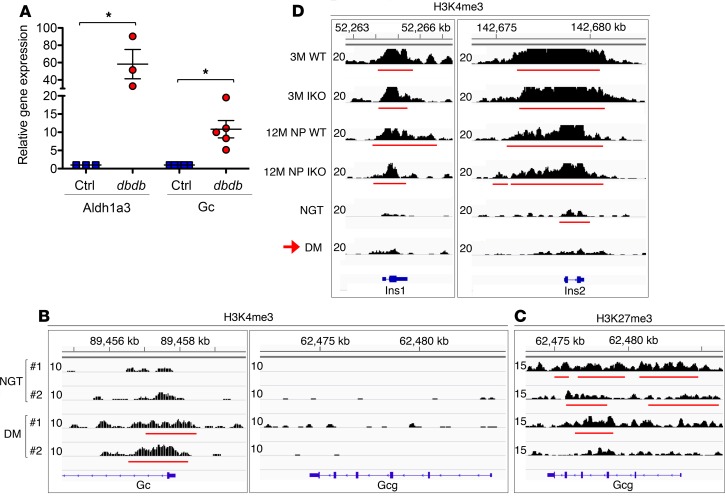
Activation of theαcell signature gene Gc. To ascertain the robustness of these observations, we analyzed Gc levels in 4 additional models of β cell dysfunction: Gc increased in triple–FoxO-KO β cells (32), in β cells with elevated levels of Aldh1a3 — a biomarker of failing β cells (29), and in sorted β cells of db/db mice (Figure 4A). Conversely, it decreased upon FoxO1 gain of function in Ins1 cells (38). In DM β cells, we detected an activation peak at the transcription start site of Gc that was absent in NGT (Figure 4B).
Figure 4. Induced Gc expression and altered histone profiles at Ins1 and Ins2 in dedifferentiated β cells.
(A) Gene expression analysis of Aldh1a3 and Gc in pancreatic β cells sorted by FACS in female control (n ≥ 3) and db/db (n ≥ 3) mice. Error bars represent ± SEM; *P < 0.05 by Student’s t test. (B) H3K4me3 profile at Gc and Gcg locus in NGT and DM β cells. (C) H3K27me3 profile at Gcg locus in NGT and DM β cells. (D) Histone 3 modifications in 3M and 12M NP WT and I-KO, NGT, and DM, where H3K4me3 profiles for Ins1 and Ins2 genes in β cells sorted by FACS are shown (n = 3 for each genotype). The red horizontal lines indicate the called peaks.
We sought further evidence supporting β cell fate conversion by analyzing H3 trimethylation marks at signature genes: Gcg, Ins1, and Ins2. In DM β cells, we didn’t detect activation peaks in Gcg (Figure 4B) but found a widespread loss of repressive marks, suggesting that the gene is poised for activation (Figure 4C and Supplemental Tables 6 and 7). Strikingly, Ins1 and Ins2 lost H3K4me3 in their promoters (Figure 4D), consistent with impaired gene activation. The breadth of the H3K4me3 regions, a marker of transcriptional consistency (39, 40), decreased at Ins1 and Ins2 of NGT and even further in DM β cells (Figure 4D). These histone profiles are consistent with β cell dedifferentiation and acquisition of α cell features.
Gc ablation improves islet function in mice fed a high-fat diet.
To understand the function of Gc, we characterized islets from Gc-deficient (Gc-KO) mice. Gc is largely derived from the liver; thus, systemic changes in insulin production or sensitivity in these mice cannot be interpreted as directly implicating the β cell. Therefore, we limited our analysis to isolated islets. In basal conditions, we did not find changes in expression of key islet genes (Supplemental Figure 1). However, because Gc induction is limited to states of β cell stress or dedifferentiation, we placed 3M male mice on a diet consisting of 60% fat for 12 weeks, then assessed expression of Aldh1a3 as a marker of β cell dedifferentiation (18, 29). Consistent with prior data (29), we observed an induction of Aldh1a3 expression in islets of WT mice fed a high-fat diet (HFD). In contrast, Gc-KO islets did not induce Aldh1a3 (Figure 5A). Islet immunohistochemistry with insulin and Aldh1a3 confirmed that Aldh1a3 expression arose in β cells of WT but not Gc-KO mice (Figure 5, B–G). In contrast, chromogranin A expression did not change in Gc-KO compared to WT (Supplemental Figure 2), indicating that there was no developmental defect in Gc-KO islets.
Figure 5. Gc ablation improves islet function in HFD-fed mice.
(A) Gene expression of Aldh1a3 in the islets of HFD-fed WT and Gc-KO mice. (B–G) Immunostaining for insulin (Ins) and Aldh1a3 in (B–D) WT and (E–G) Gc-KO islets. Scale bars: 50 μm. (H) Glucose-suppressed glucagon secretion in islets of HFD-fed WT and Gc-KO mice. (I) Pancreatic glucagon content in HFD-fed WT and Gc-KO mice. (J) Glucose-stimulated insulin secretion in islets of chow- or HFD-fed WT and Gc-KO mice. (K) Insulin secretion stimulated by glucose and arginine in islets of chow- or HFD-fed WT and Gc-KO mice. (L) Gene expression analysis of key genes in different islet cell types of HFD-fed WT and Gc-KO mice. n ≥ 9 mice per genotype, error bars represent ± SEM, and *P < 0.05 by Student’s t test.
Because Gc is expressed in α cells, we performed glucose-suppressed glucagon secretion in isolated islets of chow- or HFD-fed WT and Gc-KO mice. Glucagon secretion was decreased to a similar degree in Gc-KO and WT mice, suggesting that Gc ablation does not affect α cell function (Figure 5H). Furthermore, glucagon content was not altered in Gc-KO mice (Figure 5I). However, HFD impaired glucose-stimulated insulin secretion in WT islets, whereas Gc ablation significantly augmented insulin secretion stimulated by glucose and arginine from islets of HFD-fed mice (Figure 5, J and K). Moreover, islet gene expression analysis revealed increased levels of mRNA encoding key islet genes, including Pdx1, Nkx2-2, Ins1, Sur1, Gcg, and Sst in Gc-KO mice (Figure 5L). These data are consistent with the possibility that Gc contributes to β cell dysfunction in response to HFD.
Gc ablation preserves insulin secretory capacity in mice fed an HFD.
To investigate whether the improved insulin secretory capacity was due to increased β cell mass, we performed immunohistochemistry to assess the insulin-positive area in the pancreas of HFD-fed mice (Supplemental Figure 3, A–D). We did not detect a significant difference in β cell area or pancreas weight in Gc-KO compared to WT (Supplemental Figure 3, E and F).
Next, we performed hyperglycemic clamps in chow- or HFD-fed WT and Gc-KO mice to assess insulin secretory capacity in vivo and to eliminate confounders from the liver by clamping the glycemic levels to approximately 300 mg/dL (Figure 6, A–C). Consistent with ex vivo islet glucose-stimulated insulin secretion, HFD-fed Gc-KO mice showed a sizably increased rate of glucose infusion necessary to maintain hyperglycemia compared with WT (Figure 6D). This effect was not present in chow-fed mice (Figure 6, E and F). The increased glucose infusion rate in HFD-fed Gc-KO mice was due to increased insulin secretion (Figure 6, G–I). These data indicate that Gc ablation preserved islet function in response to HFD and suggest that activation of Gc in diabetic islets contributes to the functional deficit of β cells.
Figure 6. Hyperglycemic clamp studies in chow- and HFD-fed WT and Gc-KO mice assess in vivo insulin secretory capacity.
(A–C) Glucose levels during hyperglycemic clamp in (A) HFD-fed or (B) chow-fed WT and Gc-KO mice, where the quantification of AUC is shown in C. (D–F) Glucose infusion rates (GIRs) during hyperglycemic clamp in (D) HFD-fed or (E) chow-fed WT and Gc-KO mice, where AUC is graphed in F. (G–I) Plasma insulin levels during hyperglycemic clamp in (G) HFD-fed or (H) chow-fed WT and Gc-KO mice, where AUC is plotted in I. n ≥ 5 mice per genotype. Data represent mean ± SEM. *P < 0.05, and **P < 0.01 by Student’s t test and ANOVA.
Discussion
In this work, we integrated gene expression and histone modification analyses of β cells purified from a model of multiparity-induced diabetes to identify effectors of β cell dysfunction. Diabetic β cells activate expression and increase active histone modifications at the α cell marker, Gc. Consistent with a role of Gc induction in β cell dysfunction, Gc-deficient islets showed preserved insulin response to a hyperglycemic challenge in animals fed an HFD. Gc-deficient β cells also failed to induce Aldh1a3, a marker of β cell dedifferentiation, consistent with a healthier state of β cells. Gc is normally expressed in α cells, but its ablation did not affect glucagon secretion or glucagon content. Although our data indicate that Gc is not simply a marker of transition from β to α-like cells in response to metabolic challenges but plays a role in β cell dysfunction, the mechanism whereby it does so remains to be determined.
The present approach confirms our view that FoxO1 ablation accelerates β cell dysfunction by mimicking the natural course of this process. In this regard, it is interesting to compare the effect of the FoxO1 KO in NP versus multiparous mice. In NP mice, the FoxO1 KO appears to predispose to the loss of β cell features, as indicated by the activation of β cell dedifferentiation marker Aldh1a3 (29), the “disallowed” genes (Ldha, Slc16a family members; ref. 30, 31), and the loss of abundant mRNAs, such as Sst, Ppy, and Pyy. Interestingly, 2 pregnancy-induced mRNAs, growth hormone receptor (Ghr) and prolactin receptor (Prlr), also spontaneously increased. These changes were muted in the comparison of WT versus I-KO multiparous mice, consistent with the view that, as β cell function deteriorates, it comes to resemble the state induced by the FoxO1 KO. Our histone methylation data are also consistent with this view because the largest variation was observed in NP mice, as opposed to multiparous mice.
Gc transports the lipid-soluble vitamin D to target tissues. Given the recent report showing that injection of a synthetic vitamin D receptor ligand (calcipotriol) can preserve β cell function in db/db mice (41), and considering that vitamin D analogs represent only a fraction (~5%) of total Gc binding capacity and that only a small, unbound fraction of circulating vitamin D enters the cell (42), cargos other than vitamin D are likely responsible for the effect of Gc on β cells. Indeed, Gc also transports fatty acids and acts as a neutrophil cochemotactic agent for complement C5a (42). Furthermore, inflammation-primed lymphocytes can convert Gc to the potent macrophage-activating factor (43). Thus, it is possible that Gc secreted from β cells acts in an autocrine manner to initiate an inflammatory response, damaging β cell function. These hypotheses will require further work to be examined.
Our analysis of histone methylation indicated that both Ins1 and Ins2 lost histone activation marks in their promoters after multiple pregnancies and that diabetes resulted in a further loss of Ins2 activation. These data suggest that the loss of histone activation heralds reduced expression of insulin genes in β cells. In addition, we observed a loss of suppressive markers at the Gcg gene. It is likely that the dedifferentiation process in the multiparity model is not advanced enough to score Gcg activation. Additional potential candidate genes participating in the progression of β cell dysfunction include LIM domain only 4 (Lmo4) and docking protein 5 (Dok5). Lmo4 is a 165–amino acid transcription cofactor (44) required for Snail2-mediated epithelial-mesenchymal transition in neural crest cells (45) that regulates calcium-induced calcium release in the hippocampus (46). Dok5, also known as insulin receptor substrate 6 (IRS-6), is a susceptibility gene for obesity and type 2 diabetes in North Indians (47). It is phosphorylated in response to insulin and IGF-1 (48), although with lower kinetics compared with IRS-1 (~40 minutes vs. ~5 minutes; ref. 49). Unlike IRS-1, Dok5 does not contain YMXM and YXXM motifs to engage the Src homology 2 (SH2) domains of p85 (the catalytic domain of PI3K; ref. 48), nor does it interact with SH2 domain–containing proteins downstream of MAPK signaling, such RasGAP and Crk. Its expression is decreased in diabetic β cells, but its role in the β cell is unclear.
We found altered Hnf1α signaling in dedifferentiating β cells, consistent with a previous study of β cell–specific triple–FoxO-KOs (FoxO1, -3a, and -4) (32). Gc is a transcriptional target of Hnf1α in islets (37) but lacks a FoxO1-binding site (Kuo and Accili, unpublished observation). Based on ingenuity pathway analysis, Hnf1α signaling was activated in the absence of FoxO1. Moreover, Gc transcript levels were decreased in human embryonic stem cell–derived β-like cells with Hnf1α ablation (Supplemental Figure 4). These data suggest that FoxO1 antagonizes Hnf1α-induced Gc.
In conclusion, our approach demonstrates the unique power of integrating chromatin with expression analysis to help identify candidate genes predisposing to β cell failure. A systematic interrogation of additional interesting candidate genes identified in our analysis is currently underway to provide a detailed road map of β cell failure in diabetes.
Methods
Mice care and use.
We generated mice lacking FoxO1 in pancreatic β cells using RIP-Cre transgenics to excise the floxed FoxO1 gene. Gt(Rosa)26Sortm9(Cag−tdTomato)Hze mice (The Jackson Laboratory) were crossed with mice lacking FoxO1 in β cells to lineage trace β cells. Littermate control mice (WT) retained at least 1 WT FoxO1 allele. Mice were maintained on a mixed 129J-C57BL/6 background. For FACS, Cre recombinase is required to activate the Tomato allele; therefore, both WT and KO mice possessed the Cre allele. For Gc studies, we purchased the sperms from UCD Knockout Mouse Project Repository (Gctm1.1(KOMP)Vlcg) to generate Gc–total KO mice. All mice were fed chow diet unless otherwise noted and maintained on a 12-hour light/12-hour dark cycle (lights on at 7 am). Experiments were performed in female mice for the epigenetic studies and in male mice for Gc-KO validations. Littermate control mice for Gc-KO retained at least 1 WT Gc allele.
FACS.
Collagenase-purified pancreatic islets were isolated as described and allowed to recover in RPMI supplemented with 15% FBS overnight (50). The next day, islets were washed with PBS twice and treated with 1 mL of 0.05% trypsin for dissociation. FBS (200 μL) was added to quench trypsinization. Thereafter, the dispersed islets were spun down at 100 g for 3 minutes, washed with PBS, and incubated with SYTOX Red (Invitrogen, S34859) to identify live cells and DNAse I (Sigma-Aldrich, D4513) on ice until sorting. Before sorting, cells were filtered through a 35-μm cell strainer (Corning 352235) to remove clusters. Tomato-positive and -negative cells differed by approximately 2 orders of magnitude in Tomato fluorescence with excitation of 554 nm and emission of 581 nm by Influx cell sorter (BD Biosciences).
RNA-Seq and data analysis.
To obtain enough RNA, β cells sorted by FACS from multiple mice of the same condition were pooled as the following: 3M WT (n = 3) with a total of 10 mice, 3M I-KO (n = 3) with 14 mice, 12M NP WT (n = 3) with 20 mice, 12M NP I-KO (n = 3) with 15 mice, NGT (n = 3) with 9 mice, and DM (n = 3) with 12 mice. RNA was isolated with Nucleospin RNA kit (Macherey-Nagel) with DNAse I treatment. Directional polyA RNA-Seq libraries were prepared and sequenced as PE42 (42-bp paired-end reads) on Illumina NextSeq 500. For the analysis, 34,000,000 to 73,000,000 read pairs per sample were used. The reads were mapped to the mouse genome mm10 using STAR (v2.5.2b; ref. 51) algorithm with default settings. FeatureCounts from the Subread (v1.5.2; ref. 52) package was used to assign concordantly aligned read pairs to RefSeq genes. A 25-bp minimum as overlapping bases in a fragment was specified for read assignment. Raw read counts were used as input for DESeq2 (v1.14.1; ref. 53), which was further used to filter out genes with low counts, normalize the counts using library sizes, and perform statistical tests to find significant differential genes. For statistical analysis, a standard cutoff of P value less than 0.05 was applied. Differential calling results for RNA-Seq with NGT versus DM, 12M NP WT versus I-KO, and 3M WT versus I-KO are presented in Supplemental Tables 1, 2, and 3, respectively. Gene ontology was performed using Ingenuity Pathway Analysis software (QIAGEN Inc.; www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/).
RNA isolation and quantitative PCR.
We isolated total RNA from mouse pancreatic islets or β cells sorted with FACS using Nucleospin RNA kit (Macherey-Nagel) and followed previously described protocol for reverse transcription (54). We used the resulting cDNA to perform quantitative PCR with GoTaq master mix (Promega) and analyzed data with the standard ΔΔCt method. Hprt gene expression was used for internal normalization. Primer sequences are listed in Supplemental Table 9.
ChIP.
To obtain enough chromatin, β cells sorted by FACS from multiple mice of the same condition were pooled as the following for each histone modification: NGT (n = 3) with a total of 14 mice, DM (n = 3) with 12 mice, 12M NP WT (n = 3) with 17 mice, and 12M NP I-KO (n = 3) with 15 mice. Then, β cells were fixed with 2 mM disuccinimidyl glutarate for 20 minutes, followed by 2% formaldehyde for 10 minutes at room temperature. The reaction was quenched with 0.125 M glycine (final concentration). Chromatin was isolated with lysis buffer and disrupted with a Dounce homogenizer. The lysate samples were fragmented with a Covaris S220 sonicator to shear DNA into 300 to 500 bp in length, followed by preclearing with protein A (H3K4me3 and H3K27me3 samples) agarose beads (Santa Cruz Biotechnology). ChIP was performed with the following antibodies: anti-H3K4me3 (Active Motif, 39159, Research Resource Identification: AB_2615077) and anti-H3K27me3 (MilliporeSigma, 07-449, Research Resource Identification: AB_310624). The appropriate antibody was added to the samples and incubated overnight. The next day, magnetic beads (Active Motif) were added to samples, which were rotated for 2 hours at 4°C, followed by washes with RIPA buffer and protease inhibitor with NaCl or LiCl salt. Then, 10 mM Tris buffer supplemented with proteinase K, RNase, and 0.7% SDS was added to each sample for reverse cross-linking overnight at 65°C. Finally, ChIP DNA was purified with phenol/chloroform extraction and ethanol precipitation.
ChIP-Seq and data analysis.
ChIP DNA libraries were prepared from ChIP DNA with the following steps: end repair, dA addition, adaptor ligation, and PCR amplification, as previously described (55). The resulting DNA libraries were quantified with Bioanalyzer (Agilent), and sequenced on Illumina NextSeq 500 with 75-nt reads and single end. Reads were aligned to mouse genome mm10 using the Burrows-Wheeler Aligner algorithm with default settings (56). These reads passed Illumina’s purity filter and aligned with no more than 2 mismatches. Duplicate reads were removed, and only uniquely mapped reads with mapping quality greater than or equal to 25 were subjected to further analysis. Reads were extended to 200 bp at their 3′ ends, and bigwig files were created for visualization using Active Motif software (Active Motif Inc.). For H3K4me3, peak locations were determined using the MACS algorithm (v1.4.2) with a cutoff of P value = 1 × 10–7 (57). H3K27me3-enriched regions were identified using the SICER algorithm with FDR of 1 × 10–10, gap size of 200 bp (58). Peak coordinates from all 3 replicates were merged from each condition, and merged regions (referred to as “active regions”) were used for further analysis. DESeq2 (53) and its normalization method were applied for differential calling. Differential calling results for ChIP-Seq histone H3K4me3 in NGT versus DM, H3K4me3 in 12M NP WT versus I-KO, H3K27me3 in NGT versus DM, and H3K27me3 in 12M NP WT versus I-KO, are presented in Supplemental Tables 4–7, respectively. Statistics from total number of reads to filtered peaks are listed in Supplemental Table 8. Raw and processed sequencing data were deposited into the MINSEQE-compliant National Center for Biotechnology Information Gene Expression Omnibus database (GSE130998).
Immunofluorescence, immunohistochemical, and morphometric analyses.
Pancreata were removed from WT and Gc-KO mice on an HFD for 12 weeks, were weighed, and (a) were fixed for 4 hours in 4% paraformaldehyde-PBS at 4°C, followed by 20 hours in 30% sucrose-PBS, and embedded in optimal cutting temperature (OCT) compound (Tissue-TEK) or (b) were fixed for 16 hours in 10% formalin at 4°C, followed by 20 hours in 70% ethanol, and embedded in paraffin. For OCT-frozen samples, 5-μm-thick sections were stained for β cells using guinea pig anti-insulin antibody (DAKO, A0564), for endocrine cells using mouse anti–chromogranin A antibody (Santa Cruz Biotechnology, sc-393941), and for dedifferentiating β cells using rabbit anti-Aldh1a3 antibody (Novus Biologicals, NBP2-15339), followed by Alexa Fluor–conjugated goat antibodies as secondary (Thermo Fisher Scientific; goat anti–guinea pig, A21450; goat anti-mouse, A11031; goat anti-rabbit, A11034) antibodies. Images were taken using a Zeiss LSM 710 confocal laser-scanning microscope. For paraffin samples, several sections 200 μm apart were covered systematically by accumulating images from nonoverlapping fields with a Leica SCN 400 slide scanner at ×40 magnification, and we measured the stained area using Leica Biosystems Tissue image analysis tool. Results were expressed as a percentage of the total surveyed pancreatic area (pixel-converted mm2) occupied by β cells (pixel-converted mm2).
Metabolic parameters.
We performed oral glucose tolerance tests by gavaging glucose (2 g/kg) after a 16-hour fast. We measured insulin levels with an ELISA kit (Mercodia) and glucagon levels with a radioimmunoassay kit (MilliporeSigma). For hyperglycemic clamps, we placed an indwelling intravenous catheter in the right jugular vein at least 1 week before the clamp. Briefly, we inserted a polyurethane catheter (PU 10) filled with saline solution containing heparin (10 U/mL) into the target vessel. The catheter was tunneled through the back of the neck and placed under the back skin. A silk suture was tied to the catheter, and a small opening was made at the back of the neck. This partially exposed suture was removed on the day of the clamp. We raised plasma glucose levels to approximately 300 mg/dL by continuous glucose infusion and maintained them by adjusting the glucose infusion rate. Throughout the 120-minute period, we recorded glucose infusion rates and plasma glucose levels and collected plasma for insulin measurement.
Differentiation of embryonic stem cells into pancreaticβ-like cells. Human embryonic stem cells were grown to 80% to 90% confluence, followed by dissociation and suspension in mTeSR medium (STEMCELL Technology, 05850) with 10 μM Rho-associated kinase inhibitor Y27632 (Selleckchem, S1049), and then were plated in a 1:1 ratio into Matrigel-coated (Thermo Fisher Scientific, 354277) wells for differentiation. Differentiation was performed as previously described (59), and all differentiations were done for 25 days.
Statistics.
Pearson’s correlation coefficient was implemented with the R statistical package (http://www.r-project.org). Two-tailed Student’s t test and ANOVA were performed with Prism 8 (GraphPad) for quantitative PCR experiments. A P value less than 0.05 was considered significant.
Study approval.
The Columbia University Institutional Animal Care and Utilization Committee approved all experiments. Animals were maintained according to Columbia University animal welfare guidelines.
Author contributions
TK conceived the study, bred mice, designed and conducted experiments, collected and analyzed the data, acquired funding, and wrote the manuscript. MD analyzed the data and wrote the manuscript. BJG and DE contributed RNA-Seq data for the human embryonic stem cell–derived endocrine cells. MAL interpreted the data and wrote the manuscript. DA conceived the study, designed experiments, interpreted the data, acquired funding, and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank members of the Accili laboratory for discussions and Thomas Kolar, Ana Flete-Castro, Qiong Xu, Jun Feranil, and Lumei Xu (Columbia University) for technical support. This work was supported by NIH grants T32DK07328 and K01DK114372 (to TK), DK64819 (to DA), and DK63608 (to Columbia University Diabetes Research Center).
Version 1. 05/23/2019
In-Press Preview
Version 2. 07/11/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(13):e128351. https://doi.org/10.1172/jci.insight.128351.
Contributor Information
Taiyi Kuo, Email: tk2592@cumc.columbia.edu.
Manashree Damle, Email: damle@molbio.mgh.harvard.edu.
Bryan J. González, Email: bg2450@cumc.columbia.edu.
Mitchell A. Lazar, Email: lazar@pennmedicine.upenn.edu.
Domenico Accili, Email: da230@columbia.edu.
References
- 1.American Diabetes Association. Economic costs of diabetes in the U.S. in 2012. Diabetes Care. 2013;36(4):1033–1046. doi: 10.2337/dc12-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Accili D. Insulin action research and the future of diabetes treatment: The 2017 Banting Medal for Scientific Achievement Lecture. Diabetes. 2018;67(9):1701–1709. doi: 10.2337/dbi18-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaulton KJ, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015;47(12):1415–1425. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrannini E. The stunned β cell: a brief history. Cell Metab. 2010;11(5):349–352. doi: 10.1016/j.cmet.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Q, Melton DA. Pancreas regeneration. Nature. 2018;557(7705):351–358. doi: 10.1038/s41586-018-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150(6):1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chera S, et al. Diabetes recovery by age-dependent conversion of pancreatic δ-cells into insulin producers. Nature. 2014;514(7523):503–507. doi: 10.1038/nature13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, York NW, Nichols CG, Remedi MS. Pancreatic β cell dedifferentiation in diabetes and redifferentiation following insulin therapy. Cell Metab. 2014;19(5):872–882. doi: 10.1016/j.cmet.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puri S, Akiyama H, Hebrok M. VHL-mediated disruption of Sox9 activity compromises β-cell identity and results in diabetes mellitus. Genes Dev. 2013;27(23):2563–2575. doi: 10.1101/gad.227785.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor BL, Liu FF, Sander M. Nkx6.1 is essential for maintaining the functional state of pancreatic β cells. Cell Rep. 2013;4(6):1262–1275. doi: 10.1016/j.celrep.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Accili D, et al. When β-cells fail: lessons from dedifferentiation. Diabetes Obes Metab. 2016;18(suppl 1):117–122. doi: 10.1111/dom.12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blum B, et al. Reversal of β cell de-differentiation by a small molecule inhibitor of the TGFβ pathway. Elife. 2014;3:e02809. doi: 10.7554/eLife.02809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenz A, Toren-Haritan G, Efrat S. Redifferentiation of adult human β cells expanded in vitro by inhibition of the WNT pathway. PLoS One. 2014;9(11):e112914. doi: 10.1371/journal.pone.0112914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toren-Haritan G, Efrat S. TGFβ pathway inhibition redifferentiates human pancreatic islet β cells expanded in vitro. PLoS One. 2015;10(9):e0139168. doi: 10.1371/journal.pone.0139168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman-Mazursky O, Elkon R, Efrat S. Redifferentiation of expanded human islet β cells by inhibition of ARX. Sci Rep. 2016;6:20698. doi: 10.1038/srep20698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lean ME, et al. Primary care-led weight management for remission of type 2 diabetes (DiRECT): an open-label, cluster-randomised trial. Lancet. 2018;391(10120):541–551. doi: 10.1016/S0140-6736(17)33102-1. [DOI] [PubMed] [Google Scholar]
- 17.Butler AE, et al. β-Cell deficit in obese type 2 diabetes, a minor role of β-cell dedifferentiation and degranulation. J Clin Endocrinol Metab. 2016;101(2):523–532. doi: 10.1210/jc.2015-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cinti F, et al. Evidence of β-cell dedifferentiation in human type 2 diabetes. J Clin Endocrinol Metab. 2016;101(3):1044–1054. doi: 10.1210/jc.2015-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun J, et al. β-Cell dedifferentiation in patients with T2D with adequate glucose control and nondiabetic chronic pancreatitis. J Clin Endocrinol Metab. 2019;104(1):83–94. doi: 10.1210/jc.2018-00968. [DOI] [PubMed] [Google Scholar]
- 20.Spijker HS, et al. Conversion of mature human β-cells into glucagon-producing α-cells. Diabetes. 2013;62(7):2471–2480. doi: 10.2337/db12-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller WA, Faloona GR, Aguilar-Parada E, Unger RH. Abnormal α-cell function in diabetes. Response to carbohydrate and protein ingestion. N Engl J Med. 1970;283(3):109–115. doi: 10.1056/NEJM197007162830301. [DOI] [PubMed] [Google Scholar]
- 22.Xie R, et al. Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell. 2013;12(2):224–237. doi: 10.1016/j.stem.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Arensbergen J, et al. Derepression of Polycomb targets during pancreatic organogenesis allows insulin-producing β-cells to adopt a neural gene activity program. Genome Res. 2010;20(6):722–732. doi: 10.1101/gr.101709.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tennant BR, et al. Identification and analysis of murine pancreatic islet enhancers. Diabetologia. 2013;56(3):542–552. doi: 10.1007/s00125-012-2797-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avrahami D, Kaestner KH. Epigenetic regulation of pancreas development and function. Semin Cell Dev Biol. 2012;23(6):693–700. doi: 10.1016/j.semcdb.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhandare R, et al. Genome-wide analysis of histone modifications in human pancreatic islets. Genome Res. 2010;20(4):428–433. doi: 10.1101/gr.102038.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stitzel ML, et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010;12(5):443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasquali L, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46(2):136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim-Muller JY, et al. Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic β cells in diabetic mice. Nat Commun. 2016;7:12631. doi: 10.1038/ncomms12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemaire K, Thorrez L, Schuit F. Disallowed and allowed gene expression: two faces of mature islet β cells. Annu Rev Nutr. 2016;36:45–71. doi: 10.1146/annurev-nutr-071715-050808. [DOI] [PubMed] [Google Scholar]
- 31.Rutter GA, Pullen TJ, Hodson DJ, Martinez-Sanchez A. Pancreatic β-cell identity, glucose sensing and the control of insulin secretion. Biochem J. 2015;466(2):203–218. doi: 10.1042/BJ20141384. [DOI] [PubMed] [Google Scholar]
- 32.Kim-Muller JY, et al. Metabolic inflexibility impairs insulin secretion and results in MODY-like diabetes in triple FoxO-deficient mice. Cell Metab. 2014;20(4):593–602. doi: 10.1016/j.cmet.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, McKenna LB, Bogue CW, Kaestner KH. The diabetes gene Hhex maintains δ-cell differentiation and islet function. Genes Dev. 2014;28(8):829–834. doi: 10.1101/gad.235499.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van de Bunt M, et al. Transcript expression data from human islets links regulatory signals from genome-wide association studies for type 2 diabetes and glycemic traits to their downstream effectors. PLoS Genet. 2015;11(12):e1005694. doi: 10.1371/journal.pgen.1005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ackermann AM, Wang Z, Schug J, Naji A, Kaestner KH. Integration of ATAC-seq and RNA-seq identifies human α cell and β cell signature genes. Mol Metab. 2016;5(3):233–244. doi: 10.1016/j.molmet.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song YH, Ray K, Liebhaber SA, Cooke NE. Vitamin D-binding protein gene transcription is regulated by the relative abundance of hepatocyte nuclear factors 1α and 1β. J Biol Chem. 1998;273(43):28408–28418. doi: 10.1074/jbc.273.43.28408. [DOI] [PubMed] [Google Scholar]
- 37.Servitja JM, et al. Hnf1alpha (MODY3) controls tissue-specific transcriptional programs and exerts opposed effects on cell growth in pancreatic islets and liver. Mol Cell Biol. 2009;29(11):2945–2959. doi: 10.1128/MCB.01389-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buteau J, Shlien A, Foisy S, Accili D. Metabolic diapause in pancreatic β-cells expressing a gain-of-function mutant of the forkhead protein Foxo1. J Biol Chem. 2007;282(1):287–293. doi: 10.1074/jbc.M606118200. [DOI] [PubMed] [Google Scholar]
- 39.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benayoun BA, et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158(3):673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei Z, et al. Vitamin D switches BAF complexes to protect β cells. Cell. 2018;173(5):1135–1149.e15. doi: 10.1016/j.cell.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Speeckaert MM, Speeckaert R, van Geel N, Delanghe JR. Vitamin D binding protein: a multifunctional protein of clinical importance. Adv Clin Chem. 2014;63:1–57. doi: 10.1016/B978-0-12-800094-6.00001-7. [DOI] [PubMed] [Google Scholar]
- 43.Nagasawa H, et al. Gc protein (vitamin D-binding protein): Gc genotyping and GcMAF precursor activity. Anticancer Res. 2005;25(6A):3689–3695. [PubMed] [Google Scholar]
- 44.Manetopoulos C, Hansson A, Karlsson J, Jönsson JI, Axelson H. The LIM-only protein LMO4 modulates the transcriptional activity of HEN1. Biochem Biophys Res Commun. 2003;307(4):891–899. doi: 10.1016/S0006-291X(03)01298-1. [DOI] [PubMed] [Google Scholar]
- 45.Ferronha T, Rabadán MA, Gil-Guiñon E, Le Dréau G, de Torres C, Martí E. LMO4 is an essential cofactor in the Snail2-mediated epithelial-to-mesenchymal transition of neuroblastoma and neural crest cells. J Neurosci. 2013;33(7):2773–2783. doi: 10.1523/JNEUROSCI.4511-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qin Z, et al. LIM domain only 4 (LMO4) regulates calcium-induced calcium release and synaptic plasticity in the hippocampus. J Neurosci. 2012;32(12):4271–4283. doi: 10.1523/JNEUROSCI.6271-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tabassum R, et al. Evaluation of DOK5 as a susceptibility gene for type 2 diabetes and obesity in North Indian population. BMC Med Genet. 2010;11:35. doi: 10.1186/1471-2350-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai D, Dhe-Paganon S, Melendez PA, Lee J, Shoelson SE. Two new substrates in insulin signaling, IRS5/DOK4 and IRS6/DOK5. J Biol Chem. 2003;278(28):25323–25330. doi: 10.1074/jbc.M212430200. [DOI] [PubMed] [Google Scholar]
- 49.Weigert C, et al. Interplay and effects of temporal changes in the phosphorylation state of serine-302, -307, and -318 of insulin receptor substrate-1 on insulin action in skeletal muscle cells. Mol Endocrinol. 2008;22(12):2729–2740. doi: 10.1210/me.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuo T, Kim-Muller JY, McGraw TE, Accili D. Altered plasma profile of antioxidant proteins as an early correlate of pancreatic β cell dysfunction. J Biol Chem. 2016;291(18):9648–9656. doi: 10.1074/jbc.M115.702183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 53.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuo T, et al. Repression of glucocorticoid-stimulated angiopoietin-like 4 gene transcription by insulin. J Lipid Res. 2014;55(5):919–928. doi: 10.1194/jlr.M047860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuo T, Lew MJ, Mayba O, Harris CA, Speed TP, Wang JC. Genome-wide analysis of glucocorticoid receptor-binding sites in myotubes identifies gene networks modulating insulin signaling. Proc Natl Acad Sci U S A. 2012;109(28):11160–11165. doi: 10.1073/pnas.1111334109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25(15):1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sui L, et al. β-Cell replacement in mice using human type 1 diabetes nuclear transfer embryonic stem cells. Diabetes. 2018;67(1):26–35. doi: 10.2337/db17-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.