Abstract
Monosomy 7 and deletion of 7q, known as del(7q), are common clonal cytogenetic abnormalities associated with high-grade myelodysplastic syndrome (MDS) arising in inherited and acquired bone marrow failure. Current nontransplant approaches to treat marrow failure may be complicated by stimulation of clonal outgrowth. To study the biological consequences of del(7q) within the context of a failing marrow, we generated induced pluripotent stem cells (iPSCs) derived from patients with Shwachman-Diamond syndrome (SDS), a bone marrow failure disorder with MDS predisposition, and genomically engineered a 7q deletion. The TGF-β pathway was the top differentially regulated pathway in transcriptomic analysis of SDS versus SDSdel(7q) iPSCs. SMAD2 phosphorylation was increased in SDS relative to wild-type cells, consistent with hyperactivation of the TGF-β pathway in SDS. Phospho-SMAD2 levels were reduced following 7q deletion in SDS cells and increased upon restoration of 7q diploidy. Inhibition of the TGF-β pathway rescued hematopoiesis in SDS iPSCs and in bone marrow hematopoietic cells from SDS patients while it had no impact on the SDSdel(7q) cells. These results identified a potential targetable vulnerability to improve hematopoiesis in an MDS predisposition syndrome and highlighted the importance of the germline context of somatic alterations to inform precision medicine approaches to therapy.
Keywords: Hematology, Therapeutics
Keywords: Bone marrow, Drug therapy, Human stem cells
Induced pluripotent stem cell model of bone marrow failure and myelodysplastic syndrome predisposition identifies the TGFβ pathway as a potential therapeutic target.
Introduction
Monosomy 7 or deletion of 7q [del(7q)] frequently arises in the context of inherited and acquired bone marrow failure (1, 2). The appearance of this cytogenetic abnormality is associated with high-grade myelodysplastic syndrome (MDS) and leukemic transformation with poor prognosis. The current treatment of choice for bone marrow failure is hematopoietic stem cell transplantation, but outcomes are limited by regimen-related toxicities and donor availability. The development of nontransplant approaches to treat bone marrow failure without promoting outgrowth of malignant clones is limited by the paucity of disease models. Modeling in mice is challenging because the syntenic regions of human chromosome 7q map to several murine chromosomes. It is currently unknown whether the surrounding failing marrow provides a contextual relative fitness advantage for the monosomy 7 or del(7q) clone or whether the propensity to develop this cytogenetic abnormality in bone marrow failure results from a cell-intrinsic process.
Here we developed a human induced pluripotent stem cell (iPSC) model of del(7q) in the context of bone marrow failure. We derived iPSCs from patients with Shwachman-Diamond syndrome (SDS), a bone marrow failure syndrome characterized by pancreatic dysfunction, skeletal abnormalities, and a propensity for developing MDS and acute myeloid leukemia (3, 4). A recent genomic analysis of somatic mutations in MDS revealed that a significant subset (4%) of young MDS patients had SDS, suggesting that SDS is likely more prevalent than currently recognized (5). Biallelic mutations in the Shwachman-Bodian-Diamond syndrome (SBDS) gene are the most common genetic cause of SDS (6). Monosomy 7 or del(7q) frequently arises in SDS (4). SDS-derived iPSCs have been previously shown to phenocopy bone marrow failure (7). Therefore, we engineered a deletion of 7q in SDS iPSCs and studied the molecular and biological consequences of del(7q) with the goal of identifying a potential therapeutic strategy to improve bone marrow failure in an MDS predisposition syndrome.
Results
Generation of SDS iPSCs and SDSdel(7q) iPSCs.
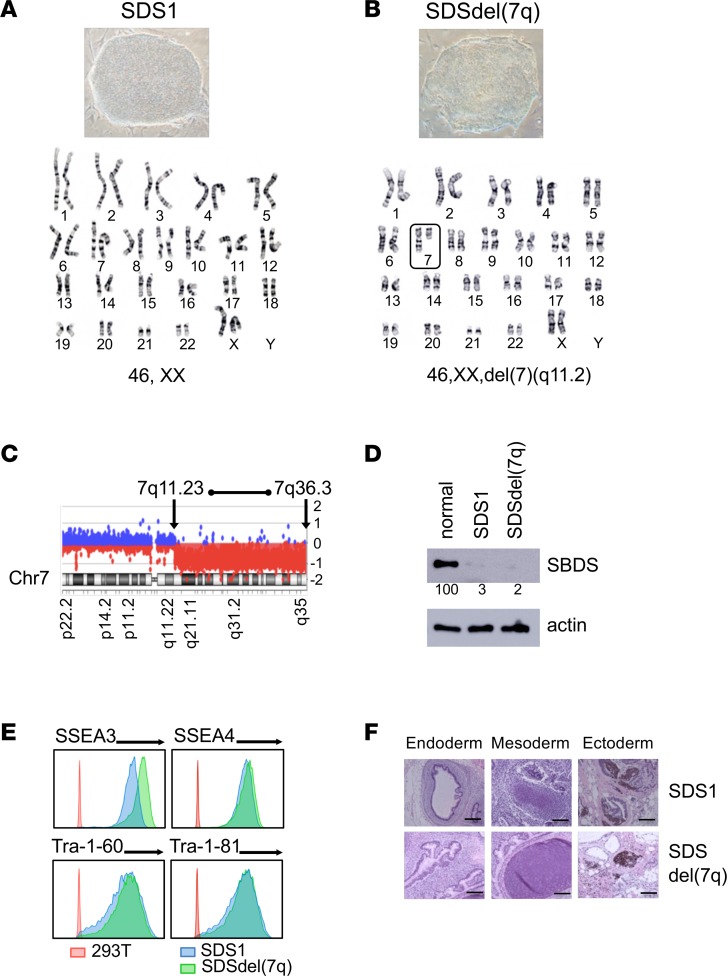
We generated iPSCs from bone marrow mononuclear cells of 2 patients with SDS (SDS1 and SDS2). Both patients carried homozygous c.258+2T>C SBDS mutations, the most common SBDS mutation found on at least 1 SBDS allele as noted in the North American SDS Registry (ref. 8, Figure 1A, and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.125157DS1). To model progression of SDS to MDS, we engineered del(7q). The long arm of chromosome 7 was deleted by targeted insertion of 2 inverted loxP sites into the long arm of chromosome 7 followed by transient expression of Cre-recombinase as previously published (ref. 9 and Supplemental Figure 2, A–C). Multiple clones were screened for the deletion of 7q by quantitative PCR and FISH (Supplemental Figure 2, D and E). The deletion of 7q was verified by karyotype analysis and mapped by array-based comparative genomic hybridization (aCGH) to span region 7q11.23 to 7q36.3, which encompasses the MDS-associated common deleted region (refs. 10–12 and Figure 1, B and C). All SDS iPSC lines were verified to retain the endogenous SBDS mutations (Supplemental Figure 1B and Supplemental Figure 2F) and express scant levels of SBDS protein, similar to the reduced levels found in patients (Figure 1D). All iPSC lines were confirmed to be pluripotent as determined by expression of markers of pluripotency (SSEA3, SSEA4, Tra-1-60, Tra-1-81) and by formation of teratomas in mice containing all 3 embryonic germ layers (Figure 1, E and F).
Figure 1. Generation of SDS iPSCs and SDSdel(7q) iPSCs.
(A and B) Representative iPSC colony morphology and karyotype for SDS patient–derived iPSCs (SDS1.5) before (A) and after (B) deletion of the long arm of chromosome 7 (box) (SDS1.5D5Cre4). Karyotype analysis was performed for all iPSC lines. (C) aCGH analysis showing deletion of the chromosome 7 region between bands q11.23 and q36.3 in 1 allele. (D) Western blot analysis of SBDS protein expression in SDS1 (SDS1.5) iPSCs and SDSdel(7q) (SDS1.5D5Cre4.9#9) iPSCs compared with normal (niPS) iPSC. Actin is shown as a loading control. Numbers below the bands indicate average densitometry quantitation of the SBDS band normalized to normal control sample value. (E) Flow cytometry of pluripotency surface markers SSEA3, SSEA4, Tra-1-60, and Tra-1-81 in SDS1 iPSCs (shown in blue, SDS1.2), SDSdel(7q) iPSCs (green, SDS1.5D5Cre4.9#2), and nonpluripotent cell line (red, HEK293T). (F) The indicated iPSCs were injected into immunodeficient mice. Histology of representative teratomas derived from SDS1 (SDS1.5) iPSCs and SDSdel(7q) (SDS1.5D5Cre4.9) iPSCs show differentiation into all 3 embryonic germ layers: endoderm (left), mesoderm (middle), and ectoderm (right). Scale bar: 100 μm.
Hematopoiesis from SDS and SDSdel(7q) iPSCs.
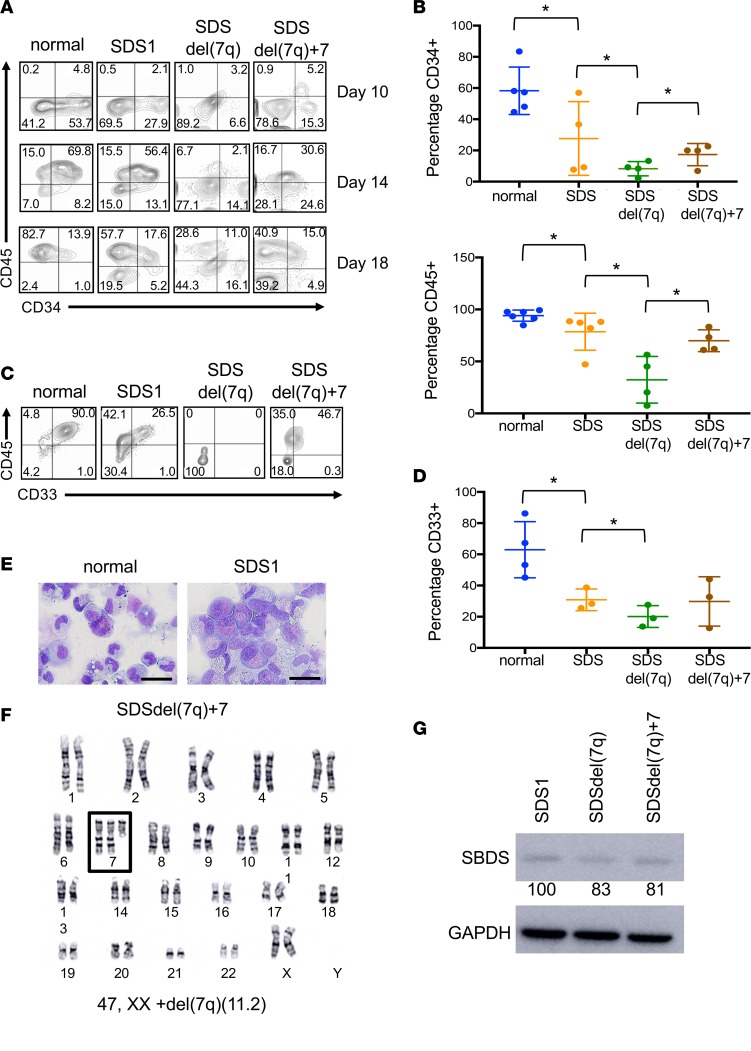
We investigated the hematopoietic differentiation potential of the SDS and SDSdel(7q) iPSCs. All clones tested from SDS1 (SDS1.2, SDS1.3, and SDS1.5) and SDS2 (SDS2.2 and SDS2.5) iPSCs demonstrated impaired hematopoiesis with decreased generation of CD34+ cells (Figure 2, A and B) and reduced differentiation to CD45+ cells (Figure 2, A and B) compared with normal iPSCs. The SDS iPSCs also demonstrated impaired differentiation to the CD33+ myeloid population compared with normal iPSCs (Figure 2). Deletion of 7q further reduced the production of CD34+ cells. The CD34+ cells with del(7q) showed markedly impaired differentiation to CD45+ cells and myeloid CD33+ cells (Figure 2, A–D). The cell growth and cell cycle profiles were not significantly different between the SDS and SDSdel(7q) cells for all clones tested (Supplemental Figure 3).
Figure 2. Effect of del(7q) on hematopoiesis of SDS iPSCs.
(A) iPSC-derived CD34+ and CD45+ cells at days 10, 14, and 18 of hematopoietic differentiation of normal, SDS1, SDSdel(7q), and SDSdel(7q)+7 iPSCs. (B) Graph summary of CD34 expression at day 10 of hematopoietic differentiation (top) and CD45 expression at day 18 of hematopoietic differentiation (bottom). Normal (N2.12 D1-1, 1157, n = 5), SDS (SDS1.5, SDS2.5, n = 4), SDSdel(7q) (SDS1.5D5Cre4.9#4, SDS1.5D5Cre4.9#9, n = 4), and SDSdel(7q)+7 (SDS1.5D5Cre4.9+7#1, SDS1.5D5Cre4.9+7#3, SDS1.5D5Cre4.9+7#4, n = 4). (C) iPSC-derived CD45+ and CD33+ cells at day 14 of myeloid differentiation for normal, SDS1, SDSdel(7q), and SDS del(7q)+7 iPSCs. (D) Comparative graph of percentage of CD33+ cells at day 14 of myeloid differentiation. Normal (N2.12 D1-1, 1157, n = 4), SDS (SDS1.5, SDS2.5, n = 3), SDSdel(7q) (SDS1.5D5Cre4.9#4, SDS1.5D5Cre4.9#9, n = 3), and SDSdel(7q)+7 (SDS1.5D5Cre4.9+7#3, SDS1.5D5Cre4.9+7#4, n = 3). For B and D, all results are represented as means ± SD. (E) Representative Wright-Giemsa stain of myeloid cells on day 7 of differentiation from normal (N2.12 D1-1, left) and SDS iPSCs (SDS1.5, right). Scale bar: 20 μm. (F) Representative karyotype of SDSdel(7q) iPSCs after spontaneous acquisition of additional chromosome 7 (box). (G) Western blot analysis of SBDS protein in SDS1 (SDS1.5), SDSdel(7q) (SDS1.5D5Cre4.9#2), and SDSdel(7q)+7 (SDS1.5D5Cre4.9+7#4) iPSCs. GAPDH is shown as a loading control. Numbers below the bands indicate average densitometry quantitation normalized to normal control sample values. *P < 0.05 by unpaired, 2-tailed Student’s t test.
As previously reported in del(7q) iPSCs in a normal (non-SDS) background (9), we observed spontaneous acquisition of an extra chromosome 7 in SDSdel(7q) iPSCs (Figure 2F and Supplemental Table 1). Upon spontaneous duplication of chromosome 7, hematopoietic differentiation and myeloid differentiation improved in 3 independent lines tested (SDS1.5Cre4.9+7#1, SDS1.5Cre4.9+7#3, and SDS1.5Cre4.9+7#4) (Figure 2, A–D), when compared with the parental SDSdel(7q) cells. Specifically, a 2-fold increase in CD34+ cells [8% ± 2% for SDSdel(7q) vs. 17% ± 4% for SDSdel(7q)+7], a 2.2-fold increase in CD45+ cells [32% ±11% for SDSdel(7q) vs. 70% ± 5% for SDSdel(7q)+7], and a 1.2-fold increase in CD33+ cells [20% ± 4% for SDSdel(7q) vs. 30% ± 9% for SDSdel(7q)+7] were observed upon spontaneous duplication of chromosome 7. The SDSdel(7q)+7 iPSCs expressed low SBDS protein levels similar to those in SDS and SDSdel(7q) cells (Figure 2G). Thus 7q haploinsufficiency severely impaired hematopoiesis in the context of a failing marrow.
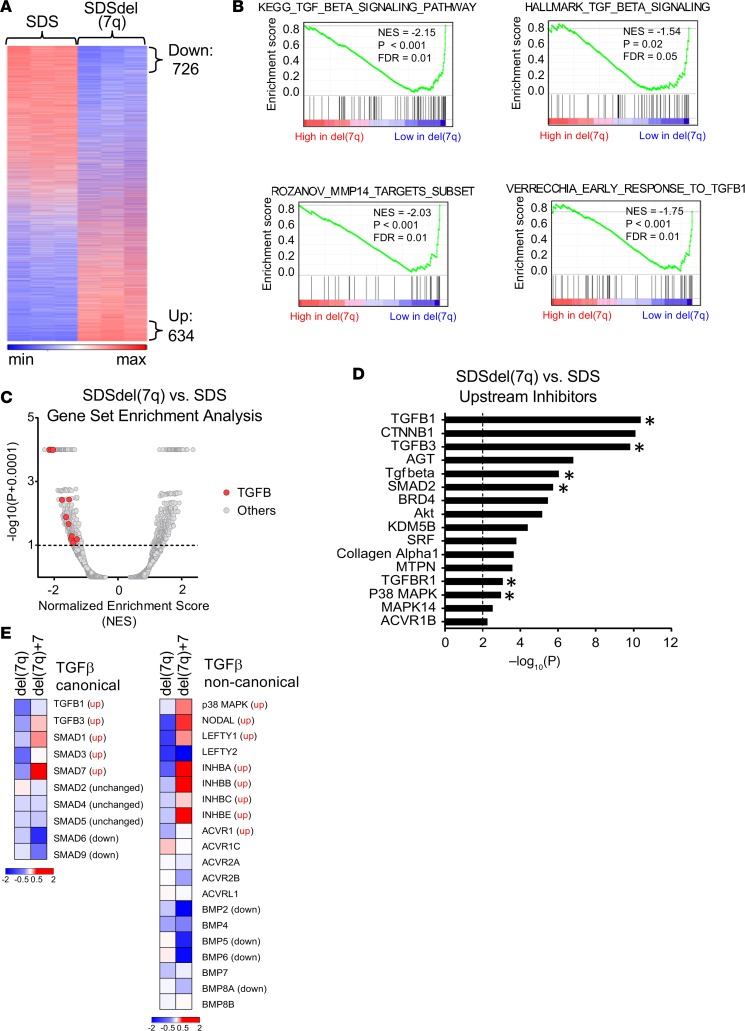
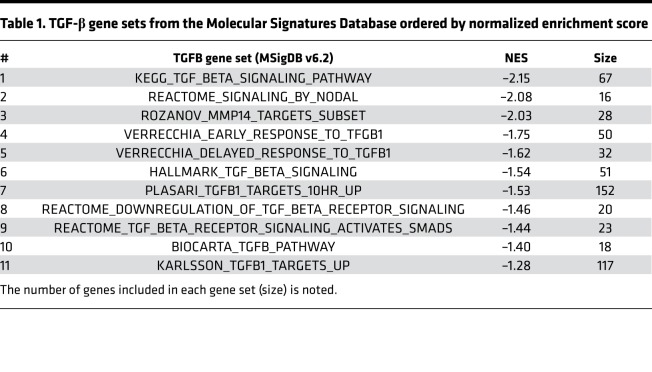
Differential activation of the TGF-βpathway in SDS versus SDSdel(7q). To explore the biological consequences of del(7q) in SDS, we conducted RNA-Seq analysis of SDS1 (SDS1.5Cre6), SDSdel(7q) (SDS1.5D5Cre4.9), and SDSdel(7q)+7 (SDS1.5D5Cre4.9+7#1) iPSCs (Supplemental Table 2 and Supplemental Figure 4). Following deletion of 7q in SDS iPSCs, 726 genes were downregulated and 634 genes upregulated (Figure 3A). Gene set enrichment analysis (GSEA) identified differential expression of inflammatory pathways, including decreased expression of the TGF-β pathway, in SDSdel(7q) relative to SDS (Figure 3, B and C, and Table 1). Six of the top 16 upstream regulators identified by Ingenuity Pathway Analysis (IPA) to be downregulated in SDSdel(7q) relative to SDS iPSCs belonged to the TGF-β family (Figure 3D and Supplemental Table 3). As predicted from pathway analysis, downstream TGF-β targets quantified by quantitative reverse transcription PCR (qRT-PCR) analysis demonstrated decreased expression in SDSdel(7q) iPSCs relative to the SDS iPSCs (Supplemental Figure 4C). These findings were consistent for TGF-β targets located on the long arm of chromosome 7 as well as those located elsewhere in the genome (Supplemental Table 3). Expression of TGF-β targets was increased in the SDS iPSCs relative to that in normal iPSCs (Supplemental Figure 4C), demonstrating hyperactivation of the TGF-β pathway in SDS. Restoration of 7q diploidy in the SDSdel(7q) cells reactivated the TGF-β pathway (Figure 3E and Supplemental Figure 4C). In contrast, analysis of non-SDS del(7q) iPSC transcriptomes previously published (9) showed downregulation of DNA repair and splicing pathways and increase in the TGF-β pathway compared with normal donor iPSCs (Supplemental Figure 4D). Thus, the del(7q)-associated, relative modulation of the TGF-β pathway was dependent on the germline genetic context of the deletion.
Figure 3. Differential activation of the TGF-β pathway in SDS iPSCs versus SDSdel(7q) iPSCs.
(A) Heatmap of RNA-Seq transcriptomic analysis of 12,993 genes with log2 fragments per kilobase of transcript per million mapped reads (FPKM) expression greater than 1 from SDS (SDS1.5) versus SDSdel(7q) (SDS1.5D5Cre4.9) iPSCs. Genes ranked based on log2 fold change FPKM at least 1.5 between SDSdel(7q) and SDS iPSCs with P value 0.05 or less in EdgeR; 634 genes were noted to have increased expression, and 726 genes had decreased expression in SDSdel(7q) iPSCs relative to SDS iPSCs. (B) Subset of the genome-wide GSEA canonical pathways and experimental gene sets with decreased expression in SDSdel(7q) iPSCs compared with SDS iPSCs. (C) Volcano plot of GSEA canonical pathways and experimental gene sets. Pathways associated with TGF-β are noted with red dots. (D) Top 16 upstream regulators identified by IPA to be inhibited in SDSdel(7q) compared with SDS iPSCs with activation score –3.0 or less. Upstream regulators of TGF-β pathways are highlighted (*). (E) Heatmap of TGF-β upstream regulators in SDSdel(7q) and SDSdel(7q)+7 iPSCs relative to SDS iPSCs for the canonical (left) and noncanonical (right) pathways.
Table 1. TGF-β gene sets from the Molecular Signatures Database ordered by normalized enrichment score.
The TGF-β pathway is an important regulator of hematopoiesis. Either hyperactivation or inhibition of the TGF-β pathway can impair hematopoiesis (13–15). Upregulation of the TGF-β pathway inhibits hematopoiesis in Fanconi anemia (16). We therefore hypothesized that hyperactivation of the TGF-β pathway in SDS cells may contribute to bone marrow failure and that inhibition of the TGF-β pathway might present a targetable vulnerability to exploit for the treatment of bone marrow failure in MDS predisposition syndromes.
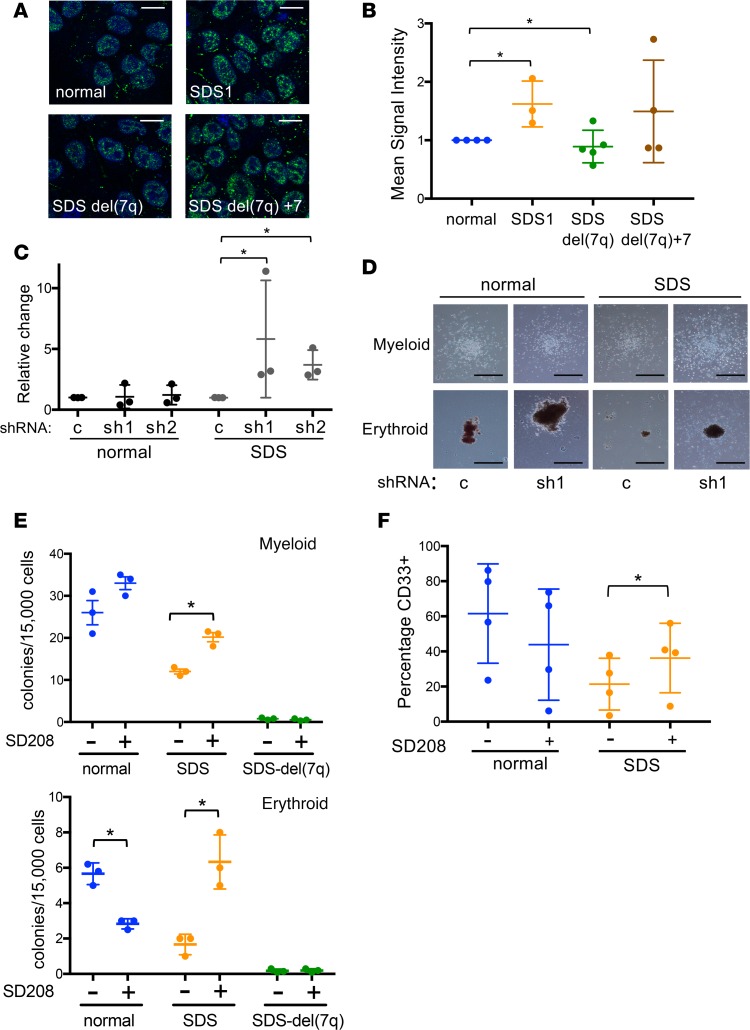
TGF-βpathway inhibition rescues hematopoiesis in SDS but not SDSdel(7q). To query the activation status of the canonical TGF-β pathway, we measured phosphorylated nuclear SMAD2 by immunofluorescence in the normal, SDS1, SDSdel(7q), and SDSdel(7q)+7 iPSCs. A significant increase in nuclear phospho-SMAD2 was observed in the SDS1 iPSCs compared with normal iPSCs (Figure 4, A and B). Deletion of 7q reduced phospho-SMAD2 levels, and restoration of 7q diploidy upregulated phosphorylation of SMAD2 (Figure 4, A and B). The increased phospho-SMAD2 signaling together with the transcriptome data were consistent with signaling through the canonical SMAD2/3 TGF-β pathway. To test whether the canonical TGF-β pathway plays a role in the hematopoietic impairment of SDS, we investigated the effect of knocking down SMAD3 expression. Two shRNA constructs targeting different regions of SMAD3 were verified to reduce SMAD3 mRNA and SMAD3 protein levels (ref. 16 and Supplemental Figure 5). The number and size of hematopoietic colonies from primary bone marrow mononuclear cells from 3 SDS patients were improved following knockdown of SMAD3 expression (Figure 4, C and D). No consistent effect of SMAD3 knockdown was observed in control bone marrow cells from healthy donors (Figure 4, C and D).
Figure 4. The TGF-β pathway is upregulated in SDS iPSCs and modulated following 7q deletion.
(A) Representative images of phospho-SMAD2 immunofluorescence (green) in normal, SDS1, SDSdel(7q), and SDSdel(7q)+7 iPSCs. Nuclei are stained with DAPI (blue). Scale bar: 10 μm. (B) Mean signal intensity for nuclear phospho-SMAD2 signal was quantified in individual cells (n = 100 cells/line/experiment) of normal (1157), SDS1 (SDS1.5), SDSdel(7q) (SDS1.5D5Cre4.9#9), and SDSdel(7q)+7 (SDS1.5D5Cre4.9+7#2) iPSCs. (C) Relative methylcellulose hematopoietic colony formation per 10,000 cells plated for normal or SDS primary marrow mononuclear cells following lentiviral transduction of the indicated shRNA: scrambled (c), SMAD3 shRNA 1 (sh1), and SMAD3 shRNA2 (sh2). Cells were plated in triplicate for each of 3 independent experiments (n = 3). (D) Representative images of myeloid (top) and erythroid (bottom) colonies from normal or SDS primary marrow with scrambled (c) versus SMAD3 shRNA (sh1). Scale bar: 1 mm. (E) Hematopoietic colony formation per 15,000 iPSC-derived cells plated in methylcellulose for normal (blue, N2.12 D1-1, n = 3), SDS (orange, SDS1.5, n = 3), and SDSdel(7q) (green, SDS1.5D5Cre4.9#9, n = 3) iPSCs in the presence or absence of 1 μM SD208. Mean ± SD are shown for myeloid (top) and erythroid (bottom) colonies. (F) CD33 expression at day 7 of myeloid differentiation showing means ± SD for normal (blue, N2.12 D1-1, n = 4) and SDS (orange, SDS1.5, n = 4) iPSCs. *P < 0.05 by unpaired, 2-tailed Student’s t test (B, C, E, and F).
We next used the small-molecule inhibitor SD208, an ATP mimetic that blocks the kinase activity of TGF-β receptor I and thereby inhibits downstream signaling (ref. 17 and Supplemental Figure 6A). Hematopoietic colony formation was decreased in SDS iPSCs compared with normal iPSCs, consistent with the bone marrow failure phenotype of SDS (Figure 4E). Both erythroid and myeloid colony number were improved in the SDS iPSCs with SD208 treatment. In contrast, no significant improvement in hematopoietic colony number was observed following TGF-β inhibition of normal or SDSdel(7q) cells (Figure 4E). Increased colony size of SDS iPSC–derived erythroid and myeloid colonies was also readily visible following TGF-β inhibition (Supplemental Figure 6, B and C). Addition of SD208 to the iPSC-derived CD34+ cells rescued myeloid differentiation of the SDS iPSCs without improving myelopoiesis of the normal or SDSdel(7q) cells (Figure 4F). Improved hematopoiesis upon treatment with either SD208 or AVID200, a ligand trap specific to TGF-βI and TGF-βIII, was also evident in primary marrow mononuclear cells from SDS patients (Shimamura Lab, unpublished observations). Thus, inhibition of the TGF-β pathway improves hematopoiesis selectively in the SDS cells wherein the TGF-β pathway is hyperactivated, but not in the SDSdel(7q) or normal non-SDS cells where the TGF-β pathway activity is relatively reduced (Figure 5).
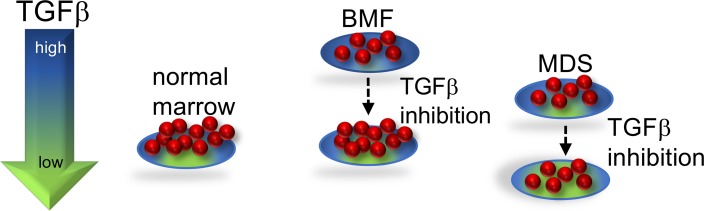
Figure 5. Proposed model of TGF-β pathway activity in bone marrow failure and MDS.
Either excessive or insufficient TGF-β signaling is deleterious. In SDS, the TGF-β pathway is hyperactivated, which inhibits hematopoiesis. TGF-β inhibitors relieve the hyperactivated state of TGF-β signaling in SDS, resulting in improved hematopoiesis. Because the TGF-β pathway activity is relatively lower in SDSdel(7q), reduction of TGF-β signaling results in insufficient TGF-β pathway activity and inhibits outgrowth of these cells.
Discussion
The molecular mechanisms leading to bone marrow failure in SDS remain poorly understood. SBDS-deficient cells exhibit impaired ribosome biogenesis (18–22), abnormal mitotic spindle dynamics (23), and increased responses to ER stress and DNA damage (24). Impaired hematopoiesis may result from either excessive activation or inhibition of the TGF-β pathway. Although hyperactivation of the TGF-β pathway inhibits hematopoiesis and affects stem cell quiescence (13, 14, 25), the TGF-β pathway plays an important role in hematopoiesis, and knock out of the TGF-β pathway is also deleterious (14). Suppression of hematopoiesis by hyperactivation of the TGF-β pathway has also been reported in Fanconi anemia, a bone marrow failure syndrome with impaired DNA repair (16), and iPSC models of Diamond-Blackfan anemia, which is caused by mutations affecting ribosome homeostasis and protein translation (26). Taken together, these data suggest that TGF-β likely exerts general effects on hematopoiesis in addition to affecting possible disease-specific pathways, such as homologous recombination versus nonhomologous end-joining in Fanconi anemia (16).
The selective pressures resulting in the frequent acquisition of monosomy 7 and del(7q) clones in bone marrow failure disorders remain unclear. Previous studies demonstrated that deletion of 7q failed to confer a proliferative advantage in iPSC MDS models (9). Our study extended these findings to show that deletion of 7q failed to confer a relative growth advantage even in the context of a failing marrow in this iPSC model. Indeed, other MDS-associated driver mutations have also failed to confer a proliferative advantage in murine models (27, 28). Monosomy 7 can be transient (28–31), suggesting that additional events may be required in addition to chromosome 7 deletion for MDS development.
Here we demonstrate a hematopoietic cell–intrinsic inhibitory effect of TGF-β on blood cell production; however, TGF-β effects are also context dependent, and the additional contribution of potential interactions between the bone marrow niche and hematopoietic cells remain to be explored. TGF-β regulation by the bone marrow niche affects hematopoiesis (32). Deletion of SBDS in the bone marrow niche promotes genotoxic stress in hematopoietic cells through activation of inflammatory signaling in SDS mouse models (33). Increased levels of inflammatory cytokines activating the TLR and TGF-β pathways have been implicated in MDS pathogenesis (34–36).
Taken together, our studies with iPSC models identified a differentially regulated pathway that could be therapeutically targeted in a bone marrow failure and MDS predisposition disorder with the goal of improving hematopoiesis. In this proposed model (Figure 5), TGF-β inhibition improves hematopoiesis of the SDS marrow, wherein the TGF-β pathway is hyperactivated, but further reduction of the already low TGF-β signaling in the del(7q) cells did not improve hematopoiesis of the del(7q) cells. Further studies are needed to understand the potential effects of TGF-β inhibition on the del(7q) clone. Activin receptor ligand traps to inhibit downstream TGF-β pathway signaling have shown promise in MDS models and in clinical trials of MDS (37). Additional studies to elucidate the mechanism(s) whereby the TGF-β pathway is activated in SDS and how this pathway impairs hematopoiesis will further inform treatment strategies. The distinct effects of del(7q) in SDS versus non-SDS stem cells highlights the importance of the germline context of somatic alterations to inform precision medicine approaches to therapy.
Methods
iPSC generation.
Cryopreserved bone marrow mononuclear cells were cultured in StemPro-34 serum-free medium (SFM, Gibco) with 1% nonessential amino acids (Gibco), 1 mM l-glutamine (Gibco), and 0.1 mM β-mercaptoethanol (Gibco) and supplemented with 100 ng/ml stem cell factor (SCF, R&D Systems), 100 ng/ml Flt-3 ligand (Flt3L, R&D Systems), 100 ng/ml thrombopoietin (TPO, R&D Systems), and 20 ng/ml IL-3 (R&D Systems) for 24 to 48 hours. Viral transduction with the excisable lentiviral vector CMV-fSV2A expressing OCT4, KLF4, c-Myc, and SOX2; reprogramming; selection of colonies by human induced pluripotent stem cell morphology (iPSC); and Cre-mediated vector excision were done as previously described (38). Characterization of pluripotency by flow cytometry and teratoma formation assay was conducted as previously described (9). Patients with SDS harbored biallelic SBDS mutations.
Cell culture.
Culture of human iPSCs on mouse embryonic fibroblasts (mitomycin C-treated, Applied Stem Cell, Inc.) or in feeder-free conditions using Matrigel (Gibco) was done as previously described (9). Normal iPSC (niPS) (female, gift of I. Bernstein, Fred Hutchinson Cancer Research Center, Seattle, WA USA) and niPS 1157 (male, gift of G. Daley, Boston Children’s Hospital, Boston, MA, USA) and HEK293T cells (gift of D.A. Williams, Boston Children’s Hospital, Boston, MA, USA) were cultured in Dulbecco’s modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (FBS). A549 cells (ATCC) were grown in F-12K (ATCC) media with 10% FBS (MilliporeSigma) and 1% penicillin/streptomycin (Gibco). Cells were treated with 1 μM SD208 (MilliporeSigma) or 0.6 μg/ml AVID200 (Formation Biologics) for 2 to 4 hours at 37°C/5% CO2 in the tissue culture incubator before harvest.
AAV-mediated gene targeting and selection of clones with 7q deletions.
The AAV targeting vector was previously described (9). Puromycin-resistant colonies were selected and Southern blot verification was conducted as previously described (9). Transduction with a Cre-expressing, integrase-deficient lentiviral vector was done as previously described (9). Ganciclovir (MilliporeSigma) selection was performed at a concentration of 150 μM for 14 days.
Karyotyping.
iPSCs were plated on Matrigel as single cells at a density of 400,000 cells in a T-25 flask. After 24 to 48 hours, cells were karyotyped by Cell Line Genetics.
aCGH.
aCGH was performed on SurePrint G3 Human Genome CGH+SNP Microarray Kit, a high-resolution array including SNPs, with coverage averaging every 25 kb with increased coverage in ISCA regions (5 kb), by Cell Line Technologies.
Western blot analysis.
iPSCs were harvested as single cells with Accutase (Stem Cell Technologies) and lysed in RIPA buffer (MilliporeSigma) supplemented with protease inhibitors (Inhibitab, Roche) and phosphatase inhibitor cocktails 2 and 3 (MilliporeSigma). Protein concentrations were determined by colorimetric assay (BCA Protein, Thermo Fisher Scientific), and 40 μg of protein was loaded on 12% SDS-PAGE gels and blotted on a PVDF membrane (MilliporeSigma). The membranes were blocked with 5% nonfat dry milk (VWR) diluted in Tris-buffered saline (MilliporeSigma) with 1% Tween-20 (VWR). Primary antibodies SBDS (23), GAPDH (clone 14C10, Cell Signaling Technology), total Smad2 (ab40855, Abcam), and phospho-Smad2 (ab3849-I, MilliporeSigma) were incubated overnight at 4°C. After washing, membranes were incubated with HRP-conjugated secondary antibodies (ECL anti-rabbit IgG NA934V and ECL anti-mouse, NA931V; GE Healthcare) and developed using SuperSignal West Pico Chemiluminescent substrate (Thermo Fisher Scientific). Detection of bands was conducted in the Amersham Imager 600 (GE Healthcare).
Flow cytometry.
For flow cytometry, the following antibodies were used: Alexa Fluor 647 SSEA-3 (clone MC-631, BioLegend), Alexa Fluor 647 SSEA-4 (clone MC-813-70, BioLegend), Alexa Fluor 647 Tra-1-60 (clone Tra-1-80-R, BioLegend), Alexa Fluor 647 Tra-1-81 (clone Tra-1-81, BioLegend), PECy7-CD34 (clone 8G12, BD Pharmingen), APC-CD45 (clone 2D1, BD Pharmingen), PE-CD33 (clone WM33, BD Pharmingen), PECy7-CD11b (clone ICRF44, BD Pharmingen), DAPI (MilliporeSigma), and propidium iodide (BD Pharmingen). Flow cytometry was conducted on an LSR Fortessa machine (BD) at the Boston Children’s Hospital and Harvard Stem Cell Institute Flow Cytometry Research Facility and data analyzed using FlowJo (FlowJo LLC).
Hematopoietic differentiation of iPSCs.
hPSC colonies were collected and grown in ultra-low attachment dishes (Corning) and subjected to cytokine media changes for 18 days as previously described (9). At the end of embryoid body (EB) differentiation culture (days 10, 14, 18), cells were dissociated with Accutase (Stem Cell Technologies) into single cells and analyzed by flow cytometry. For methylcellulose colony formation assays, the cells were dissociated at day 12, and 1.5 × 104 cells were resuspended in StemPro-34 SFM and added to 3 ml of methylcellulose (H4434, Stem Cell Technologies). Then 1 ml was plated in triplicate wells of 6-well Smartdishes (Stem Cell Technologies). After 14 days of growth at 37°C/5% CO2, colonies were imaged and scored using STEMVision (Stem Cell Technologies). The score was averaged for triplicate wells. For myeloid differentiation, the EBs were dissociated at day 18 and grown in media containing 100 ng/ml SCF (R&D Systems), 50 ng/ml granulocyte-colony stimulating factor (R&D Systems), 50 μg/ml ascorbic acid (MilliporeSigma), 40 ng/ml Flt3L (R&D Systems), 40 ng/ml IL-3, 20 ng/ml TPO (R&D Systems), and 20 ng/ml IL-6 (R&D Systems), with media changes every 3 to 4 days. We harvested 50,000 cells by cytospin at 300 rpm for 4 minutes (Shandon Cytospin 3) onto glass microscope slides (Thermo Fisher Scientific). Cells were allowed to dry before staining per manufacturer’s instructions (NovaUltra Hema-Diff Stain kit, IHC World).
Gene expression analysis by qRT-PCR.
RNA was isolated following manufacturer’s instructions for RNeasy Plus Mini Kit (QIAGEN Inc.). RNA was eluted in 30 μl of water. We used 200 ng to 5 μg of RNA for reverse transcription with Superscript III First Strand Synthesis using oligo-dT primer (Invitrogen). Quantitative PCR was performed with iTaq Universal SYBR Green Supermix (Bio-Rad) using GAPDH as an internal control with primers shown in Supplemental Table 4. Reactions were carried out in triplicate in a 7500 Fast Real-Time PCR System (Applied Biosystems) and analyzed using the ΔΔCT method.
RNA-Seq.
RNA quality was verified on an Agilent 2200 TapeStation for an RNA integrity number greater than 8. Library prep, quality control, and sequencing with the Illumina HiSeq 2500 platform were performed at the Fred Hutchinson Cancer Research Center Genomic Core. Reads were aligned using TopHat v2.13 against the hg19 assembly of the human genome. Counts for each gene were generated using htseq-count v0.6.1pl; genes with less than 1 count/million in at least 3 samples were removed. Gene expression was quantified as log2(1 + FPKM). Data were restricted to the genes with log2(1 + FPKM) expression greater than 1 in at least 1 sample: 12,993 genes for SDS del(7q) and 11,576 genes for SDS del(7q)+7. EdgeR v3.12.1 (39) and DEseq2 v1.20.0 (40) were used to normalize data, conduct significance testing, and pair samples from the same patient, with significance cutoffs as follows: absolute fold change for log2(1 + FPKM) expression ≥ 1.5; P ≤ 0.05; and Benjamini-Hochberg false discovery rate ≤ 0.05.
Genome-wide GSEA (v3.0, refs. 41, 42) of the different samples were compared to gene sets included in the MSigDB (v6.2, refs. 41, 43), while disregarding the chr7 genes. IPA v01-07 (QIAGEN Inc.; www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/) was used for upstream regulator analysis. The RNA-Seq data for this study have been deposited into the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) repository (44) and are accessible through the GEO series accession number GSE118378.
Immunofluorescence.
Cells were grown on Matrigel-covered coverslips at a density of 5 × 105 cells/coverslip for 24 hours. Cells were washed with PBS (Gibco), fixed with 4% (w/v) paraformaldehyde (MilliporeSigma) for 10 minutes at room temperature, washed 3 times with PBS, permeabilized with 0.3% Triton X-100 (VWR) for 10 minutes at room temperature, washed 3 times with PBS, and blocked for 1 hour at room temperature in solution with 1% FBS (MilliporeSigma) before overnight incubation at 4°C with primary antibody (phospho-SMAD2, 44-244G, Thermo Fisher Scientific). The coverslips were washed 3 times with PBS before incubation with secondary antibody (Alexa Fluor 488 goat anti–rabbit IgG, Life Technologies). Cells were mounted with DAPI (Vector Laboratories) for nuclear counterstaining and were imaged with a ×63 oil immersion objective of a confocal Leica SP5 microscope (Dana-Farber Cancer Institute core facility). Image analysis and quantification were done by Fiji (ImageJ, www.Fiji.sc).
FISH.
Cells were incubated 2 times in fixative (3:1 methanol/acetic acid) for 15 minutes each. Cells were attached to a glass coverslip (Thermo Fisher Scientific) by cytospin at 2000 rpm for 2 minutes (Shandon Cytospin 3). Cells were rehydrated in 2× SSC (MilliporeSigma) at 37°C for 2 minutes followed by dehydration in a series of ethanol solutions (70%, 80%, and 95%) for 2 minutes each. Fluorescent probes (Cytocell) were added to coverslips and incubated at 75°C for 2 minutes and then overnight at 37°C. Cells were washed for 2 minutes at 72°C in 0.4× SSC followed by 2× SSC/0.5% Tween-20 (MilliporeSigma) at room temperature for 30 seconds. Cells were mounted and imaged as described above. The fluorescent probes were chromosome 7 centromeric (Cytocell, D7Z1, 7p11.1–7q11.1, aqua) and chromosome 7 subtelomeric (Cytocell, LPT 07QR/G-A, Texas red).
Lentivirus production and transduction.
Lentivirus was produced in HEK293T cells seeded at approximately 50% confluence 24 hours before transfection. Transfection was performed with polyethylenimine (Life Technologies). Virus was harvested 24 hours after transfection, filtered through a 0.45-μm membrane (MilliporeSigma), and concentrated by ultracentrifugation at 48,490 g for 2 hours at 4°C. An MOI of 20 was used. Primary bone marrow–derived mononuclear cells were grown for 24 hours in StemSpan SFEM II (Stem Cell Technologies) supplemented with 100 ng/ml of SCF, TPO, and Flt3L and 20 ng/ml of IL-3 (PeproTech). Cells were resuspended at 1 × 106 cells/ml with 8 μg/ml polybrene (MilliporeSigma), and 200 μl of cell suspension was used per reaction. After addition of virus, cells were centrifuged at 887 g for 30 minutes at room temperature. Puromycin (1 μg/ml, Mirus) selection was conducted for 72 hours before cells were harvested for methylcellulose assay as described above. Scrambled shRNA (CAACAAGATGAAGAGCACCAA) (45), SMAD3 shRNA1 (CTGTGTGAGTTCGCCTTCAAT) (16), and SMAD3 shRNA2 (CCCAGCACATAATAACTTGGA) (16) were synthesized by Addgene.
Statistics.
Statistical analyses were conducted with Prism 7 (GraphPad Software) using 2-tailed, unpaired Student’s t test. P values less than 0.05 were considered significant.
Study approval.
Patient samples were obtained with patients’ informed consent under protocols approved by the Institutional Review Boards at Fred Hutchinson Cancer Research Center (Seattle, Washington, USA), Seattle Children’s Hospital (Seattle, Washington, USA), and Boston Children’s Hospital (Boston, Massachusetts, USA).
Author contributions
MRG, OVB, EPP, KS, and AS designed experiments. DWR and AGK designed and produced the AAV targeting vectors. KM and AS collected clinically phenotyped patient samples. MRG, OVB, and KB performed experiments. MRG, GA, and CEJ performed computational analyses. MRG, OVB, and AS analyzed data. MRG and AS wrote the manuscript, and MRG, OVB, GA, AGK, KB, CEJ, DWR, KS, KM, CDN, EPP, and AS provided critical reviews of the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH/National Institute of Diabetes and Digestive and Kidney Diseases R24 DK099808 (to AS), NIH/National Heart, Lung, and Blood Institute (NHLBI) R01 HL121570 (to EPP), NIH/National Cancer Institute T-32 CA009351-34 (to MRG), NHLBI K–12 (to MRG), National Cancer Institute R35 CA210030 (to KS), Department of Defense W81XWH-16-1-0054 (to AS), the Leukemia and Lymphoma Society (to AS and KS), the Aplastic Anemia and MDS Foundation Edward P. Evans Fellowship (to MRG), and the Butterfly Guild of Seattle Children’s Hospital (to AS). We would like to thank the lab of I. Bernstein (Fred Hutchinson Cancer Research Center, Seattle, WA USA) for assistance with teratoma assays and the lab of M. Fleming (Boston Children’s Hospital, Boston, MA USA) for processing the teratoma samples. We thank the Boston Children’s Hospital and Harvard Stem Cell Institute flow cytometry research facility. We thank M. Bonilla and K. Bisht for excellent technical assistance. We thank the SDS Registry for patient samples.
Version 1. 04/30/2019
In-Press Preview
Version 2. 06/20/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(12):e125157. https://doi.org/10.1172/jci.insight.125157.
Contributor Information
Melisa Ruiz-Gutierrez, Email: melisa.ruiz-gutierrez@childrens.harvard.edu.
Gabriela Alexe, Email: Gabriela_Alexe@DFCI.HARVARD.EDU.
Kaitlyn Ballotti, Email: kaitlyn.ballotti@childrens.harvard.edu.
Cailin E. Joyce, Email: cailin.joyce@gmail.com.
David W. Russell, Email: drussell@u.washington.edu.
Kimberly Stegmaier, Email: kimberly_stegmaier@dfci.harvard.edu.
Kasiani Myers, Email: kasiani.myers@cchmc.org.
Carl D. Novina, Email: carl_novina@dfci.harvard.edu.
Eirini P. Papapetrou, Email: eirini.papapetrou@mssm.edu.
Akiko Shimamura, Email: Akiko.Shimamura@childrens.harvard.edu.
References
- 1.Maciejewski JP, Selleri C. Evolution of clonal cytogenetic abnormalities in aplastic anemia. Leuk Lymphoma. 2004;45(3):433–440. doi: 10.1080/10428190310001602363. [DOI] [PubMed] [Google Scholar]
- 2.Glaubach T, Robinson LJ, Corey SJ. Pediatric myelodysplastic syndromes: they do exist! J Pediatr Hematol Oncol. 2014;36(1):1–7. doi: 10.1097/MPH.0000000000000046. [DOI] [PubMed] [Google Scholar]
- 3.Shimamura A, Alter BP. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010;24(3):101–122. doi: 10.1016/j.blre.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myers KC, Davies SM, Shimamura A. Clinical and molecular pathophysiology of Shwachman-Diamond syndrome: an update. Hematol Oncol Clin North Am. 2013;27(1):117–ix. doi: 10.1016/j.hoc.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindsley RC, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376(6):536–547. doi: 10.1056/NEJMoa1611604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boocock GR, et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33(1):97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 7.Tulpule A, et al. Pluripotent stem cell models of Shwachman-Diamond syndrome reveal a common mechanism for pancreatic and hematopoietic dysfunction. Cell Stem Cell. 2013;12(6):727–736. doi: 10.1016/j.stem.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myers KC, et al. Variable clinical presentation of Shwachman-Diamond syndrome: update from the North American Shwachman-Diamond Syndrome Registry. J Pediatr. 2014;164(4):866–870. doi: 10.1016/j.jpeds.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotini AG, et al. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat Biotechnol. 2015;33(6):646–655. doi: 10.1038/nbt.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Beau MM, Espinosa R, Davis EM, Eisenbart JD, Larson RA, Green ED. Cytogenetic and molecular delineation of a region of chromosome 7 commonly deleted in malignant myeloid diseases. Blood. 1996;88(6):1930–1935. [PubMed] [Google Scholar]
- 11.Döhner K, et al. Molecular cytogenetic characterization of a critical region in bands 7q35-q36 commonly deleted in malignant myeloid disorders. Blood. 1998;92(11):4031–4035. [PubMed] [Google Scholar]
- 12.Jerez A, et al. Loss of heterozygosity in 7q myeloid disorders: clinical associations and genomic pathogenesis. Blood. 2012;119(25):6109–6117. doi: 10.1182/blood-2011-12-397620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenet F, Kermani P, Spektor R, Rafii S, Scandura JM. TGFβ restores hematopoietic homeostasis after myelosuppressive chemotherapy. J Exp Med. 2013;210(3):623–639. doi: 10.1084/jem.20121610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blank U, Karlsson S. TGF-β signaling in the control of hematopoietic stem cells. Blood. 2015;125(23):3542–3550. doi: 10.1182/blood-2014-12-618090. [DOI] [PubMed] [Google Scholar]
- 15.Naka K, Hirao A. Regulation of hematopoiesis and hematological disease by TGF-β family signaling molecules. Cold Spring Harb Perspect Biol. 2017;9(9):ea027987. doi: 10.1101/cshperspect.a027987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, et al. TGF-β inhibition rescues hematopoietic stem cell defects and bone marrow failure in Fanconi anemia. Cell Stem Cell. 2016;18(5):668–681. doi: 10.1016/j.stem.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akhurst RJ, Hata A. Targeting the TGFβ signalling pathway in disease. Nat Rev Drug Discov. 2012;11(10):790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finch AJ, et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev. 2011;25(9):917–929. doi: 10.1101/gad.623011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong CC, Traynor D, Basse N, Kay RR, Warren AJ. Defective ribosome assembly in Shwachman-Diamond syndrome. Blood. 2011;118(16):4305–4312. doi: 10.1182/blood-2011-06-353938. [DOI] [PubMed] [Google Scholar]
- 20.Weis F, et al. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat Struct Mol Biol. 2015;22(11):914–919. doi: 10.1038/nsmb.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganapathi KA, et al. The human Shwachman-Diamond syndrome protein, SBDS, associates with ribosomal RNA. Blood. 2007;110(5):1458–1465. doi: 10.1182/blood-2007-02-075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burwick N, Coats SA, Nakamura T, Shimamura A. Impaired ribosomal subunit association in Shwachman-Diamond syndrome. Blood. 2012;120(26):5143–5152. doi: 10.1182/blood-2012-04-420166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Austin KM, et al. Mitotic spindle destabilization and genomic instability in Shwachman-Diamond syndrome. J Clin Invest. 2008;118(4):1511–1518. doi: 10.1172/JCI33764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ball HL, et al. Shwachman-Bodian Diamond syndrome is a multi-functional protein implicated in cellular stress responses. Hum Mol Genet. 2009;18(19):3684–3695. doi: 10.1093/hmg/ddp316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao X, et al. TGF-β inhibitors stimulate red blood cell production by enhancing self-renewal of BFU-E erythroid progenitors. Blood. 2016;128(23):2637–2641. doi: 10.1182/blood-2016-05-718320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge J, et al. Dysregulation of the transforming growth factor β pathway in induced pluripotent stem cells generated from patients with Diamond Blackfan anemia. PLoS One. 2015;10(8):e0134878. doi: 10.1371/journal.pone.0134878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu JJ, Smeets MF, Tan SY, Wall M, Purton LE, Walkley CR. Modeling human RNA spliceosome mutations in the mouse: not all mice were created equal. Exp Hematol. 2019;70:10–23. doi: 10.1016/j.exphem.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Wlodarski MW, Sahoo SS, Niemeyer CM. Monosomy 7 in pediatric myelodysplastic syndromes. Hematol Oncol Clin North Am. 2018;32(4):729–743. doi: 10.1016/j.hoc.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Wong JC, et al. Germline SAMD9 and SAMD9L mutations are associated with extensive genetic evolution and diverse hematologic outcomes. JCI Insight. 2018;3(14):e121086. doi: 10.1172/jci.insight.121086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pastor VB, et al. Constitutional SAMD9L mutations cause familial myelodysplastic syndrome and transient monosomy 7. Haematologica. 2018;103(3):427–437. doi: 10.3324/haematol.2017.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tesi B, et al. Gain-of-function SAMD9L mutations cause a syndrome of cytopenia, immunodeficiency, MDS, and neurological symptoms. Blood. 2017;129(16):2266–2279. doi: 10.1182/blood-2016-10-743302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamazaki S, et al. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell. 2011;147(5):1146–1158. doi: 10.1016/j.cell.2011.09.053. [DOI] [PubMed] [Google Scholar]
- 33.Zambetti NA, et al. Mesenchymal inflammation drives genotoxic stress in hematopoietic stem cells and predicts disease evolution in human pre-leukemia. Cell Stem Cell. 2016;19(5):613–627. doi: 10.1016/j.stem.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 34.Fang J, et al. Ubiquitination of hnRNPA1 by TRAF6 links chronic innate immune signaling with myelodysplasia. Nat Immunol. 2017;18(2):236–245. doi: 10.1038/ni.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider RK, et al. Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat Med. 2016;22(3):288–297. doi: 10.1038/nm.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mies A, Platzbecker U. Increasing the effectiveness of hematopoiesis in myelodysplastic syndromes: erythropoiesis-stimulating agents and transforming growth factor-β superfamily inhibitors. Semin Hematol. 2017;54(3):141–146. doi: 10.1053/j.seminhematol.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 37.Platzbecker U, et al. Luspatercept for the treatment of anaemia in patients with lower-risk myelodysplastic syndromes (PACE-MDS): a multicentre, open-label phase 2 dose-finding study with long-term extension study. Lancet Oncol. 2017;18(10):1338–1347. doi: 10.1016/S1470-2045(17)30615-0. [DOI] [PubMed] [Google Scholar]
- 38.Papapetrou EP, Sadelain M. Generation of transgene-free human induced pluripotent stem cells with an excisable single polycistronic vector. Nat Protoc. 2011;6(9):1251–1273. doi: 10.1038/nprot.2011.374. [DOI] [PubMed] [Google Scholar]
- 39.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 43.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alvarez-Dominguez JR, et al. De novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab. 2015;21(5):764–776. doi: 10.1016/j.cmet.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.