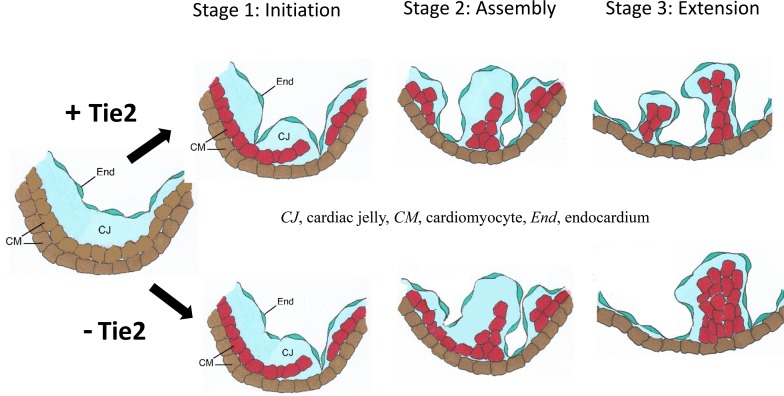
Abstract
The ang1-tyrosine kinase with immunoglobulin-like and EGF-like domains 2 (Tie2) pathway is required for normal vascular development, but its molecular effectors are not well-defined during cardiac ontogeny. Here, we show that endocardial-specific attenuation of Tie2 results in midgestation lethality due to heart defects associated with a hyperplastic but simplified trabecular meshwork (fewer but thicker trabeculae). Reduced proliferation and production of endocardial cells following endocardial loss of Tie2 results in decreased endocardial sprouting required for trabecular assembly and extension. The hyperplastic trabeculae result from enhanced proliferation of trabecular cardiomyocytes, which is associated with upregulation of bone morphogenetic protein 10, increased retinoic acid (RA) signaling, and extracellular signal-regulated protein kinases 1 and 2 hyperphosphorylation in the myocardium. Intriguingly, myocardial phenotypes in conditional knockout hearts could be partially rescued by inhibiting in utero RA signaling with pan-RA receptor antagonist BMS493. These findings reveal 2 complementary functions of endocardial Tie2 during ventricular chamber formation: ensuring normal trabeculation by supporting endocardial cell proliferation and sprouting and preventing hypertrabeculation via suppression of RA signaling in trabecular cardiomyocytes.
Keywords: Cardiology, Development
Keywords: Cardiovascular disease, Embryonic development, Mouse models
Endocardial Tie2 ensures normal ventricular trabeculation by supporting proliferation and sprouting of endocardial cells but prevents hypertrabeculation via suppression of trabecular cardiomyocyte proliferation.
Introduction
In vertebrates, the heart is the first organ to form and function during embryogenesis. Ventricular chamber formation is one of the key development processes in cardiac development, and alterations in this process can cause severe human heart defects. One of the first signs of chamber development is the formation of trabeculae. Trabeculation is the process by which ventricular cardiomyocytes (CMs) grow toward the ventricular lumen, forming a complex sponge-like meshwork (1, 2). Trabeculation begins in the mouse around E9.0 when clusters of myocardial cells delaminate from the outer curvature of the ventricle and invaginate into the cardiac jelly or extracellular matrix (ECM), separating the myocardium from the endocardium (3). It is believed that trabeculae increase cardiac output and facilitate oxygen and nutrient exchange in the embryonic myocardium prior to the development of the coronary circulation (1). Progressively, the ventricular myocardium differentiates into 2 distinct layers: the outer proliferative compact myocardium and the inner, more differentiated, trabecular myocardium. Around E14.5, trabeculae growth subsides with subsequent remodeling or “compaction” (3). Lack of trabeculation frequently causes embryonic lethality in mice, and excess trabeculation causes cardiomyopathy and heart failure in humans (4). The molecular and cellular mechanisms controlling trabeculation are not fully understood, however.
A number of signaling molecules from myocardium, endocardium, and cardiac ECM have been implicated in trabeculation, and an intimate communication between endocardium and myocardium promotes CM proliferation and differentiation, leading to trabeculae formation (2, 4). The Notch signaling pathway has been the most intensely investigated in this process. Global or endothelial-specific abrogation of Notch1, or the Notch1 receptor Delta-like ligand 4 (Dll4), causes ventricular hypoplasia and trabeculation defects (5, 6). Alternatively, accentuated expression of Notch1 via suppression of Numb (7) or Fkbp1a (8) leads to ventricular hypertrabeculation. Notch signaling abrogation affects the expression of 3 signaling pathways required for trabeculation: bone morphogenetic protein 10 (Bmp10), neuregulin 1 (Nrg1)-ErbB2/4, and EphrinB2/EphB4. Bmp10, Nrg1, and EphrinB2 signaling are downstream targets of Notch signaling during trabeculation (5). Bmp10 is transiently enriched in the trabecular CMs between E9.0 and E13.5. Bmp10-deficient mice have hypoplastic ventricular walls and form only primitive trabeculation, consequently dying at around E10.5 (9). Nrg1 is produced in endocardial cells (ECs) and acts through myocardial receptors ErbB2 and ErbB4. Null mutations of Nrg1, ErbB2, and ErbB4 all display similar phenotypes of a hypoplastic ventricular wall, lacking normal trabeculation and lethality between E10.5 and E11.5 in mice (10). Mice null for Efnb2, encoding EphrinB2, and its specific receptor EphB4 both display failure to form ventricular trabeculae and lethality between E10.5 and E11.0 (11).
Retinoic acid (RA) signaling represents an additional key contributor to ventricular trabeculation. RA is a vitamin A–derived, nonpeptidic, small lipophilic molecule that acts as a ligand for nuclear RA receptors on the nuclear membrane, converting them from transcriptional repressors to activators. The distribution and levels of RA in embryonic tissues are tightly controlled by regulated synthesis through the action of specific retinol and retinaldehyde dehydrogenases and by degradation via specific cytochrome P450s. Both vitamin A deficiency and exposure of embryos to excess vitamin A lead to cardiac defects (12, 13). Gene knockout studies confirmed the crucial functions of RA signaling in mouse development. For example, Rxra–/– mice died at E13.5–E16.5 with cardiac outflow tract, large vessel abnormalities, and myocardial hypoplasia (14). Retinol dehydrogenase 10–mutant (Rdh10-mutant) mice died at E10.5–E14.5 with abnormal heart tube formation (15). Raldh2-mutant mice died at E9.5–E10.5 with impaired heart looping and chamber differentiation (16).
Tyrosine kinase with immunoglobulin-like and EGF-like domains 2 (Tie2) is an endothelial-specific type 1 transmembrane protein receptor tyrosine kinase that mediates angiopoietin signaling for endothelial cell survival, vascular remodeling, and integrity (17, 18). Tie2-deficient mouse embryos die at E10.5 due to vessel remodeling defects and lack of trabeculation (19, 20). Deficiency of angiopoietin-1 (Ang1), a Tie2 agonist, results in marked simplification of the cardiac trabeculation and lethality at E11–E12.5 (21, 22), which is a phenotype similar to the phenotype of Tie2-deficient mice, although not as severe. Myocardial overexpression of Ang1 under the control of the tetracycline promoter also results in reduced trabeculation and lethality at E12.5–E15.5 (23). Thus, Ang1 from the myocardium likely signals via Tie2 on the endocardium to regulate ventricular trabeculation, although it is not clear how Ang1/Tie2 signaling controls this process. A major impediment to defining the role of Tie2 in cardiac trabeculation has been early lethality in mutant mice due to diffuse vascular defects. Thus, any cardiac abnormalities seen could be merely secondary to vascular pathology. To circumvent this problem and to allow temporal and tissue-specific gene inactivation, we have developed a conditional allele of the mouse Tie2 gene and utilized an endocardial-specific nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 1; endonuclease-mediated mutation 1, Bin Zhou (Nfatc1Cre) allele to delete Tie2 exclusively in the endocardium, thus bypassing the vascular requirement for Tie2 (24). We find that loss of endocardial Tie2 results in embryonic heart failure characterized by impaired endocardial growth and hyperplastic trabeculation. The endocardial phenotype is caused by decreased EC proliferation and impaired migration, which are associated with a dramatic change of expression of several endocardial genes, including antibodies to Notch1 intracellular domain (N1ICD) and vascular endothelial growth factor receptor 2 (Flk1/VEGFR2). The thicker trabeculae phenotype results from enhanced trabecular CM proliferation, which is caused by downregulation of cyclin-dependent kinase inhibitor 1C (p57) in the endocardium, upregulation of Bmp10, increased RA signaling, and hyperphosphorylation of extracellular signal–regulated kinase 1/2 (Erk1/2) in trabecular myocardium. Thus, these studies reveal 2 complementary functions of Tie2 in regulating trabeculation: autocrine support of EC proliferation and sprouting and paracrine prevention of hypertrabeculation through inhibition of trabecular CM proliferation. These studies provide further experimental evidence that endocardial sprouting and touchdown are critical for ventricular trabeculation.
Results
Tie2 is essential for ventricular chamber development.
To circumvent the potential early requirement for Tie2 signaling in systemic vascular endothelium and investigate the role of Tie2 signaling specifically in the heart, mice with a floxed Tie2 allele (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.96002DS1) were crossed with the Nfatc1Cre line to drive expression of Cre throughout the embryonic atrial and ventricular endocardium from E9.0 but not in the endothelial cells of the peripheral vasculature (refs. 24, 25, and data not shown). We refer to Nfatc1Cre Tie2fl/fl mice as Tie2 conditional knockout (Tie2-cko). Immunostaining of E9.5, E10.5 (data not shown), and E11.5 heart sections detected only minimal and sporadic Tie2 staining in the endocardium and confirmed the specific deletion of Tie2 in the endocardium of Tie2-cko without a change of normal expression of Tie2 in epicardial blood vessel endothelial cells (Supplemental Figure 2, A and B). Consistently, loss of Tie2 expression in E9.5, E10.5, and E11.5 Tie2-cko hearts was 75.5%, 84.4%, and 85.1%, respectively as determined by quantitative PCR (qPCR) (Supplemental Figure 2C), indicating that the efficiency of Tie2 deletion via Nfatc1Cre reached a peak from E10.5 and persisted throughout the period of investigation.
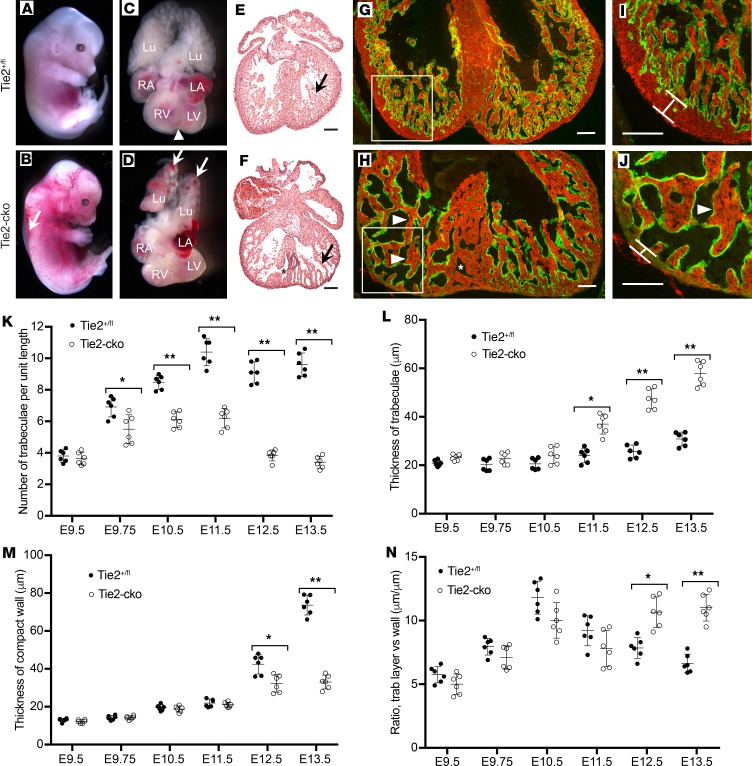
Similar to global deletion of Tie2, none of the endocardial Tie2-cko animals from 22 litters survived to birth. Tie2-cko embryos were grossly indistinguishable from their littermates until E12.5, at which point most began to show dorsal edema that increased until demise (Figure 1, A and B). In addition, enlarged right atria, widespread hemorrhage (69%, 22 out of 32), and severe pulmonary congestion (56%, 18 out of 32) as well as liver congestion were frequently seen, suggesting heart failure and hemodynamic dysfunction. All mutants developed pericardial effusions and died at E13.0–E13.5. Hearts isolated from these E13.5 Tie2-cko embryos lacked the normal ventricular groove, an indication of abnormal ventricle chamber development (Figure 1, C and D). Histological analysis (Figure 1, E and F) and dual-fluorescence immunostaining of heart sections (E9.5–E13.5) with antibodies for troponin T (myocardial marker) and endomucin (which identifies ECs) (Figure 1, G–J and Supplemental Figure 2D) confirmed abnormal trabeculation, thin compact myocardium, and ventricular septum defects compared with littermate controls (Tie2+/fl or Tie2fl/fl). Quantification of trabecular complexity and compact myocardium thickness revealed that, most strikingly, Tie2-cko hearts displayed fewer trabeculae per unit length as early as E9.75, but thicker trabeculae from E11.5 when compared with controls (increased by 53.3%) (Figure 1, K and L). These changes were more apparent at E12.5 (increased by 84.4%) and at E13.5 (increased by 87.4%). There was no significant difference in overall trabecular area or the average thickness of the trabecular layer between control and Tie2-cko, because the compact zone of Tie2-cko became thinner than that of the control from E12.5 (Figure 1M). However, the ratio of overall trabecular area versus the compact wall thickness (data not shown) and the ratio of trabecular layer thickness versus the compact wall thickness were significantly higher in Tie2-cko hearts than those in control hearts (Figure 1N). Thus, endocardial attenuation of Tie2 leads to hyperplastic trabeculation.
Figure 1. Endocardial-specific deletion of Tie2 results in abnormal trabeculation.
(A–D) Gross images of Tie2+/fl (A and C) and Tie2-cko (B and D) embryos, lungs, and hearts at E13.5, showing the perivascular hemorrhage, edema (arrow), and pulmonary congestion (arrows) in the mutant embryos. Arrowhead indicates formation of the ventricular groove in the control heart (C), which is usually not obvious in the mutants (D). (E and F) H&E–stained heart sections of E13.5 Tie2+/fl (E) and Tie2-cko (F) embryos. Arrows indicate trabeculae. (G and H) Tie2+/fl and Tie2-cko ventricular sections were stained with troponin T (myocardial marker) and endomucin (endocardial marker) antibodies. Compared with the control (Tie2+/fl), the mutant heart had fewer but thicker trabeculae (arrowheads), thinner compact myocardium, and ventricular septation defects (*). The boxed regions are enlarged in I and J. The width of the ventricular compact wall is indicated. LA, left atrium; Lu, lung; LV, left ventricle; RA, right atrium; RV, right ventricle. Scale bars: I and J, 50 μm; others, 100 μm. A representative of more than 10 images was chosen for each panel. (K–N) Quantification of trabecular myocardium complexity (trabeculae density, trabeculae thickness, and ratio of trabecular layer and compact zone) and compact myocardium thickness from E9.5–E13.5 (n = 6 per group). Data are expressed as mean ± SEM. *P < 0.05; **P < 0.01, 2-way ANOVA.
Endocardial-specific deletion of Tie2 results in an abnormal endocardial network.
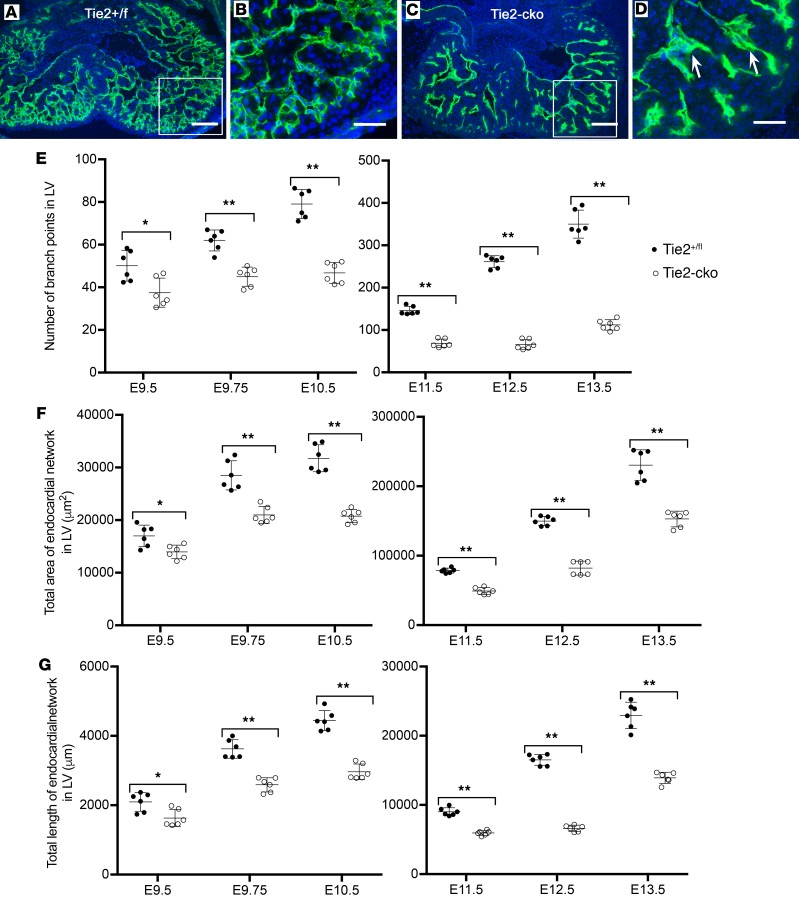
One of the most striking phenotypes of Tie2-cko embryos was their abnormal endocardial network. The endocardial network, visualized by immunostaining for endomucin, was simplified in Tie2-cko ventricles compared with controls (Figure 2, A–D and Supplemental Figure 3). Quantification of endocardial complexity in the left ventricles revealed that the number of endocardial branch points, the total area covered by the endocardial network, and the total length of endocardial network in Tie2-cko were all significantly reduced compared with controls at E9.5–E13.5 (Figure 2, E–G). Most strikingly, in the controls, the number of endocardial branch points increased 7-fold from E9.5–E13.5, but in the mutants, there was only a 2.8-fold increase. Immunostaining of Tie2-cko and control heart sections with antibodies against other endocardial markers such as CD31, CD34, CD54, and intercellular adhesion molecule 2 (data not shown) further validated the abnormal endocardium phenotype in Tie2-cko mice. In addition, the endocardial sproutings in Tie2-cko ventricles were thickened (Figure 2D). Therefore, endocardial network complexity in Tie2-cko is significantly reduced, and the endocardial defects (at least from E9.5) appear to precede the major trabecular defects.
Figure 2. Endocardial-specific deletion of Tie2 results in abnormal endocardial network.
(A and B) Tie2+/fl and Tie2-cko heart sections at E13.0 were immunostained with anti-endomucin antibody (green), showing simplified endocardial networks but thickened endocardial sproutings (arrows) in mutant ventricles. The boxed regions are enlarged in C and D. Nuclei are counterstained with DAPI (blue). Scale bars: A and B, 100 μm; C and D, 50 μm. (C–E) Quantification of endocardial network complexity (branch points, total area and total length of endocardial network) of Tie2-cko embryos and their littermates at E9.5–E13.5 (n = 6 per group). *P < 0.05; **P < 0.01, 2-way ANOVA.
A subset of ECs undergo endothelial to mesenchymal transformation (EnMT) and contribute to the formation of the atrioventricular canal and outflow track endocardial cushions. Thus, a primary endocardial defect could result in altered endocardial cushion function resulting in secondary adverse effects on myocardial function and trabeculation. Although we occasionally observed developmental delay of cushion formations in Tie2-cko hearts, the Tie2-cko embryonic hearts have normal atrioventricular canals and outflow tracts during the subsequent critical stages seen at E11.5 (Supplemental Figure 4, A and B). Furthermore, when floxed Tie2 mice were crossed with a valvular endothelial-specific line, Nfatc1enCre (26), the resulting mutant mice (Nfatc1enCre Tie2fl/fl) had completely normal heart valves (data not shown) with efficient Tie2 deletion in valvular endothelium (Supplemental Figure 4, C and D). Therefore, it is unlikely that the impairment of the endocardial network and myocardial trabeculation in Tie2-cko hearts was secondary to endocardial cushion abnormalities.
Endocardial loss of Tie2 impairs trabecular assembly and extension.
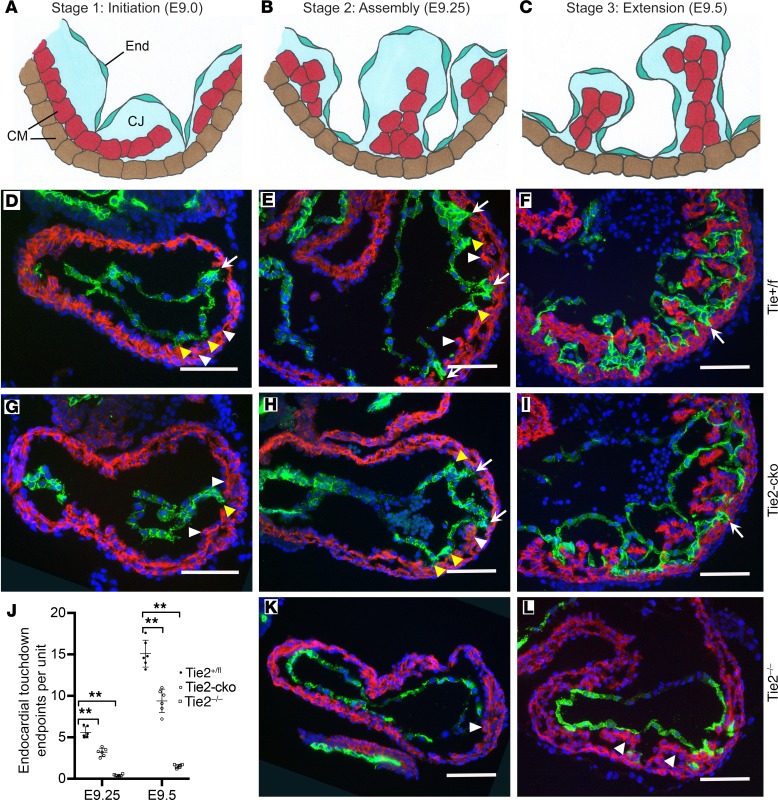
To investigate the initiation as well as sequential morphological phases of trabecular development, we utilized immunofluorescent detection of troponin T (myocardium) and endomucin (endocardium) in WT E9.0–E9.5 murine heart sections to facilitate visualization of ventricular structures (Figure 3, D–F). Based on both previously published work as well as our observations, the anatomical changes associated with trabeculation can be divided into 3 distinct steps (Figure 3, A–C). At E9.0, the endocardium sends out angiogenic sprout-like protrusions similar to those detected in other vascular beds. These sprouts penetrate the thick ECM, traverse the inner or luminal layer of myocardium, and make direct contact or “touchdown” with the outer layer of myocardium. Simultaneously, the myocardial wall thickens to become multicellular, and CMs in specific regions along the inner myocardial layer delaminate into the lumen and form sheet-like protrusions. Thus, the first stage or initiation of trabeculation is characterized by endocardial sprout formation and touchdown coincident with myocardial delamination (Figure 3, A and D). Next, endocardial sproutings progress laterally beneath the myocardial lamina, resulting in isolated clusters of myocardial cells within a bubble of cardiac ECM, covered by endocardium. By E9.25, the endocardial sprouting-enveloped myocardial lamina assemble to isolated short trabecular clusters (stage 2: assembly) (Figure 3, B and E), which is followed by trabecular extension (stage 3) at E9.5, leading to finger-like long trabecular structures (Figure 3, C and F). We detected no endocardial touchdown endpoints in ventricles of Tie2–/– mutants (data not shown) and a decreased number of touchdown endpoints in Tie2-cko embryos (Figure 3G) at E9.0, compared with controls. The myocardial wall became multicellular in the all 3 genotypes, however, and delaminated myocardial lamina were present in even Tie2–/– ventricles (Figure 3J and Supplemental Figure 1F), indicating that trabeculation was initiated in the mutants. At E9.25 and E9.5, the number of endocardial touchdown points gradually increased in both control and Tie2-cko ventricles, although fewer were detected in Tie2-cko animals. The number of endocardial touchdown points continued to be markedly attenuated in Tie2–/–, even at E9.5 (Figure 3, K and L). At E9.5, Tie2-cko embryos had bigger but fewer isolated trabecular clusters than control, whereas no such trabecular structures were detected in Tie2–/– embryos, indicating defective assembly/extension of trabeculae (stages 2 and 3) in the null mutants. Thus, although delamination occurs in the absence of Tie2, trabecular assembly and extension appear to be Tie2 dosage–dependent processes.
Figure 3. Endocardial loss of Tie2 impairs endocardial sprouting and trabecular assembly and extension.
(A–C) The 3 basic stages of trabeculation (initiation, assembly, and extension) and their corresponding sections of Tie2+/fl (D–F), Tie2-cko (G–I), and Tie2–/– (K and L) embryos dual immunostained with troponin T (red) and endomucin (green) antibodies. Nuclei are counterstained with DAPI (blue). CJ, cardiac jelly; CM, cardiomyocytes; End, endocardium. At stage 1 (initiation, around E9.0), as the inner layer CMs delaminate into the lumen and form sheet-like protrusions (myocardial lamina, white arrowheads), the endocardium sends out sproutings to penetrate the thick cardiac jelly and make direct touchdown (arrows) with the outer layer of the myocardium in the control embryo. In Tie2-cko, however, the endocardial sproutings have not yet reached the outer layer of the myocardium (yellow arrowheads). At stage 2 (assembly, E9.25), as endocardial sproutings progress laterally beneath the myocardial lamina, they later assemble to isolated short trabecular clusters. At stage 3 (extension, E9.5), finger-like, long trabecular structures are formed by extension. More endocardial touchdown endpoints were detected in the control than in Tie2-cko and Tie2–/– ventricles. Although the myocardial wall in Tie2–/– ventricles (K and L) was able to thicken to become multicellular and form sheet-like protrusions (myocardial lamina, white arrowheads), assembly and extension of trabeculae were impaired. Scale bars: 100 μm. A representative of more than 8 images was chosen for each panel. (J) Quantification of endocardial touchdown endpoints in control, Tie2-cko, and Tie2–/– embryos at E9.25 and E9.5 (n = 6 per group). **P < 0.01, 1-way ANOVA.
Endocardial attenuation of Tie2 results in decreased EC proliferation and Notch signaling.
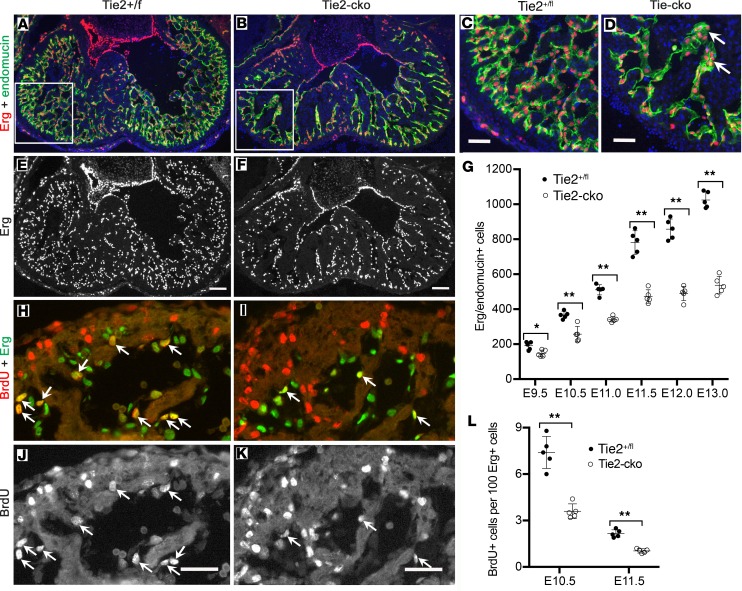
To analyze whether the reduced endocardial area was accompanied by a decreased number of ECs in Tie2-cko mice, the total EC number was determined by counting cells positive for the endothelial-specific ETS transcription factor Erg. The Erg immunostaining revealed a gradual loss of ECs in the mutant endocardium, significant from E9.5 and more severe at E12.0 and E13.0 (Figure 4, A–G). In addition, aggregations of ECs in thickened sproutings were often observed in Tie2-cko endocardium (Figure 4D and Figure 2D), suggesting defective EC migration. EC proliferation of endocardium was analyzed in Tie2-cko and control embryos by costaining for Erg and BrdU. This analysis revealed that EC proliferation in Tie2-cko endocardium was reduced by nearly 50% when compared with control mice at E10.5 and E11.5 (Figure 4, H–L). Because staining of heart sections with antibodies against endomucin and cleaved caspase-3 detected no significant difference in EC apoptosis between Tie2-cko and littermate controls (data not shown and Supplemental Figure 5), the reduced EC number in Tie2-cko mice is not due to decreased EC survival but results from impaired EC proliferation.
Figure 4. Endocardial attenuation of Tie2 results in decreased EC number and proliferation.
(A–F) Dual immunostaining of Tie2+/fl (A and C) and Tie2-cko (B and D) heart sections for Erg (red) and endomucin (green) at E13.0 reveals a significant decrease in Erg-positive EC number in the mutant ventricles following endocardial loss of Tie2. (C and D) Boxed regions in A and B are enlarged, and aggregations of ECs in dilated sproutings (arrow) are often observed in Tie2-cko endocardium. Nuclei are counterstained with DAPI (blue). (G) Quantification of total Erg/endomucin–positive cells indicated a gradual loss of ECs in the mutant endocardium, significant from E9.5. (H–K) Dual immunostaining of control (H and J) and Tie2-cko (I and K) heart sections for BrdU (red) and Erg (green) at E10.5. The cells positive for both BrdU and Erg represent proliferating ECs (arrows), which are decreased in the mutant ventricles. (L) Quantification of BrdU/Erg–positive cells as a percentage of total Erg-positive cells in endocardium indicated that Tie2-cko displayed lower proliferation rates of ECs at E10.5 and at E11.5. Scale bars: C and D, 100 μm; others, 50 μm. A representative of more than 10 images was chosen for each panel (n ≥ 5 per group). *P < 0.05; **P < 0.01, 1-way ANOVA.
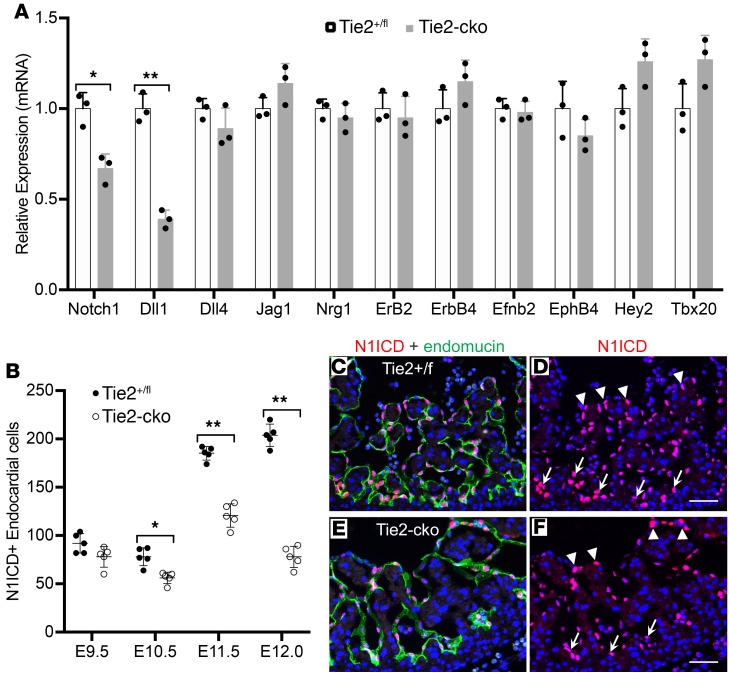
Because it has been elegantly shown that Notch signaling is a key signaling pathway in trabeculation (5, 6), we examined Notch1 signaling using qPCR analysis, which detected reduced expression of Notch1 and Dll1 in Tie2-cko hearts at E11.5. Although there was no significant change in expression of other components or downstream targets, including Dll4, Jag1, Nrg1, Erb2, Erb4, Efnb2, EphB4, Hey2, and Tbx20 (Figure 5A), immunostaining for N1ICD (active Notch1) confirmed a reduction of Notch1 activity in Tie2-cko endocardium (Supplemental Figure 6). Quantification of N1ICD/endomucin double-positive ECs in left ventricles revealed that N1ICD+ ECs in Tie2-cko hearts were reduced by 28%, 35%, and 62%, respectively at E10.5, E11.5, and E12.0 (Figure 5B). Furthermore, compared with the control, the reduction of N1ICD expression in the mutant endocardium was often more pronounced in the distal endocardial “tip” cells at the base of the forming trabeculae than in the more proximal luminal endocardium (Figure 5, C–F). Thus, endocardial distribution of N1ICD expression was altered by Tie2 attenuation.
Figure 5. Endocardial attenuation of Tie2 results in decreased Notch signaling.
(A) qPCR analysis of genes associated with Notch signaling pathway detected reduced expression of Notch1 and Dll1 in E11.5 Tie2-cko hearts, although there was no significant change in expression of other components or downstream targets. (B) Quantification of N1ICD/endomucin double-immunostained hearts revealed that N1ICD+ ECs in Tie2-cko left ventricles were significantly reduced at E10.5, E11.5, and E12.0 when compared with the controls. (C–F) Dual immunostaining of control and Tie2-cko heart sections at E11.5 for NICD1 (red) and endomucin (green). In comparison with the control, the reduction of N1ICD staining in the mutant endocardium is more significant at the base of the forming trabeculae (arrows) than in distal endocardium (arrowheads). Scale bars: 50 μm. A representative of more than 10 images was chosen for each panel; n = 3 (A) or 5 (B) per group. *P < 0.05; **P < 0.01, Student’s t test (A) and 2-way ANOVA (B).
To identify the possible downstream targets by which Tie2 regulates EC proliferation, we characterized expression of endocardial-specific genes in whole E11.5 Tie2-cko and littermate control hearts using RNA-Seq and confirmed candidate gene expression by qPCR analysis. Following endocardial loss of Tie2, the expression levels of several endocardial genes were remarkably reduced, while only a few were strikingly upregulated, including Pdgfb, Robo4, and Edn1 (Supplemental Figure 7A). Reduced expression levels of Sox7, Gata2, Flk1, VE-Cad, and Nos3 in the mutant endocardium were also confirmed with immunostaining (Supplemental Figure 7, B–K).
Global disruption of Sox7 (27), Tmem100 (28), or Arap3 (29) caused embryonic lethality around E11.0 with abnormal vascular development and defective myocardial growth and trabeculation. Therefore, we speculated that a reduction in the expression of these endocardial-specific genes in our Tie2-cko might partially explain the impaired EC proliferation phenotype observed in Tie2-cko embryonic endocardium. However, when we made endocardial-specific deletions of each of these genes using available mice with a floxed allele (29–31) by crossing with Nfatc1Cre, all the conditional mutant mice had normal development of the endocardium and normal trabeculation (data not shown). Therefore, it is unlikely that these 3 genes are downstream targets by which Tie2 regulates EC proliferation or trabeculation.
Endocardial attenuation of Tie2 results in enhanced trabecular CM proliferation.
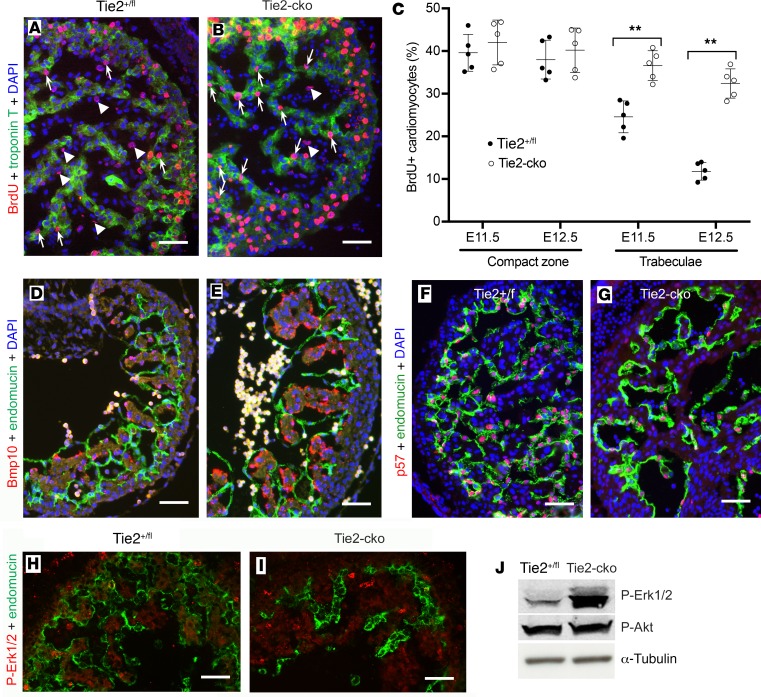
To determine if the thicker trabeculae in Tie2-cko hearts were due to an enhanced proliferation rate in myocardium, we assessed the CM proliferation rate by BrdU pulse labeling, which revealed that the percentage of BrdU-positive CMs in the trabecular zone of the Tie2-cko hearts was significantly higher than that in the control, for example, 36.6% versus 24.6% at E11.5 (increase by 48.8%) and 32.4% versus 11.8% at E12.5 (increase by 174.6%) (Figure 6, A–C). Interestingly, there was no significant difference in the percentage of BrdU-positive CMs in the compact zone between the control and Tie2-cko hearts. This increase in trabecular myocardial cell proliferation in Tie2-cko mice was further confirmed by immunohistochemistry with another cell proliferation marker, phospho-histone H3 (Supplemental Figure 8). In addition, similar to endocardium staining, immunostaining with anti–active caspase 3 revealed little staining within the heart wall and trabeculae in both mutants and littermate controls (Supplemental Figure 5), suggesting that apoptosis did not contribute to the abnormal trabeculation phenotype in the mutants.
Figure 6. Endocardial attenuation of Tie2 results in enhanced CM proliferation.
(A and B) BrdU pulse labeling and coimmunostaining for BrdU (red) and troponin T (green) showing more BrdU-positive CMs (arrows) but fewer BrdU-positive ECs (arrowheads) in the trabecular zone of Tie2-cko embryos at E11.5 than those in the control group. (C) Quantification of BrdU-positive nuclei as a percentage of total nuclei in myocardium indicated that Tie2-cko displayed higher proliferation rates of CMs in the trabecular zone at E11.5 and E12.5 (n = 5 per group). **P < 0.01, 2-way ANOVA. (D and E) Compared with the control littermates (D), Bmp10 expression (red, in situ hybridization) in the trabecular CMs of Tie2-cko embryos (E) at E10.5 was enhanced. Endocardium was labeled with endomucin antibody (green). (F and G) Dual immunostaining of control and Tie2-cko heart sections for the cell cycle inhibitor p57 (red) and endomucin (green) at E11.5 showing significantly reduced p57 expression in the mutant endocardium. (H and I) Dual immunostaining of control and Tie2-cko heart sections for p-Erk1/2 (red) and CD31 (green) at E11.5 showing Erk1/2 hyperphosphorylation in the mutant trabecular myocardium. (J) Western blot analysis of E12.5 control and Tie2-cko hearts confirming significantly enhanced phosphorylation of Erk1/2 in the mutants. α-Tubulin was used as a loading control. Scale bars: 50 μm. For the studies in A–I, more than 5 embryos per genotype collected from at least 3 independent litters were analyzed. A representative of more than 10 images was chosen for each panel. For the studies in J, 58 embryonic hearts per genotype harvested from 23 independent litters were analyzed, and data are expressed as a representative of 3 independent experiments.
Bmp10, enriched in trabecular myocardium, has been identified as a critical growth factor for maintaining CM proliferation associated with attenuation of p57 (Cdkn1c or p57kip2) (9). Consistent with increased CM proliferation in Tie2-cko trabeculae, our in situ hybridization detected an increase in Bmp10 mRNA in the mutant trabeculae when compared with the control at E10.5–E12.5 (Figure 6, D and E, and data not shown). qPCR analysis of E11.5 hearts detected increased expression of Bmp10 (by 47%) and decreased expression of p57 (by 40%) (Supplemental Figure 9). Although some reports have previously suggested that p57 is restricted to myocardium (8, 32), dual immunostaining of heart sections with antibodies against p57 and 2 different endothelial/endocardial markers (endomucin and Erg) documents that p57 is predominantly expressed in endocardium with lower expression in myocardium in the mouse embryo (Supplemental Figure 10). Compared with the control, expression levels of p57 in Tie2-cko ECs were reduced at all stages (E9.5–E13.5) examined (Figures 6, F and G, and data not shown). Thus, our work provides further confirmation of accentuated expression of p57 in the endocardium compared with the myocardium and supports an inverse relationship between endocardial p57 expression, myocardial Bmp10 expression, and myocardial proliferation.
Previous studies showed that Erk1/2 hyperactivation was associated with enhanced CM proliferation in embryonic ventricles (33), and myocardial trabecular Erk1/2 activation could be regulated by Nrg1-ErbB1/4 signaling (10), secondary to endocardial Notch1 activation. We detected an increase in Erk1/2 phosphorylation in Tie2-cko ventricular myocardium at E11.5 with immunostaining (Figure 6, H and I), which was confirmed by Western blot using control and Tie2-cko hearts at E12.5 (Figure 6J; see complete unedited blots in the supplemental material). These results suggest that endocardial Tie2 signaling regulates CM proliferation at least partially via Erk1/2 hyperphosphorylation.
The acellular cardiac jelly or ECM is also essential for normal trabeculation and regulates CM proliferation (25, 34, 35). However, our Alcian blue staining (Supplemental Figure 11, A and B) and immunohistochemistry with antibodies that detect major components of the cardiac ECM such as versican, hyaluronic acid (data not shown), fibronectin, heparan sulfate proteoglycan, and laminin (Supplemental Figure 11, C–H) detected no significant difference between Tie2-cko embryos and their littermate controls at E10.5 and E11.5. Therefore, normal cardiac ECM in Tie2-cko embryos suggests that abnormal CM proliferation in Tie2-cko hearts is not secondary to alteration in the cardiac ECM.
Tie2 regulates trabecular CM proliferation via RA signaling.
To address other possible mechanisms that might contribute to the hyperplastic trabeculation (few but thicker trabeculae) and simplified endocardial networks observed in Tie2-cko hearts, we investigated gene expression in whole E11.5 Tie2-cko and littermate control hearts using RNA-Seq and qPCR analysis. Analysis of genes associated with CM differentiation revealed elevated levels of Myh11, Isl1, Vcam1, and Pdgfc, and no change in expression of Nkx2.5, HOP, PEG1, and Irx3 following endocardial Tie2 loss (Supplemental Figure 12). Upregulation of Myh11, Isl1, and Vcam1 suggests a cardiac differentiation phenotype in Tie2-cko embryos. Interestingly, RNA-Seq and qPCR analysis of E11.5 hearts revealed elevated RA signaling following endocardial loss of Tie2. Numerous important components of the RA signaling pathway, including cellular RA-binding proteins (Crabp1, Crabp2), Rdh10, encoding cytochrome P450 1B1 and retinol-binding protein 4, a number of known RA-inducible genes, such as Stra6, encoding α-fetoprotein (Figure 7A), and several members of the Hox gene family, including Hoxa4, Hox4, and Hoxb5 (data not shown) were upregulated. The endocardial-specific fatty acid–binding protein 5 was the only RA signaling component that was downregulated, while Raldh2 was not changed in Tie2-cko hearts. Immunofluorescence of control and Tie2-cko heart sections at E9.5–E11.5 (Figures 7, B and C) (data not shown) for Crabp2 further confirmed elevated Crabp2 expression in mutant CMs.
Figure 7. Myocardial phenotypes in Tie2-cko hearts were partially rescued by inhibiting in utero RA signaling with BMS493 treatment.
(A) qPCR analysis of genes associated with the RA signaling pathway in E11.5 hearts revealed elevated RA signaling following endocardial loss of Tie2. (B and C) Immunostaining of control and Tie2-cko heart sections at E9.5 for Crabp2 showing elevated Crabp2 expression in mutant CMs. (D–F) Gross images of in utero BMS493 (pan-RA receptor antagonist)-treated Tie2+/fl (D), Tie2-cko/R (E, rescued group) and Tie2-cko/P (F, poorly rescued group) embryos at E14.0. Note that dorsal edema (arrow) was common in all mutant embryos, but pericardial effusions (arrowhead) were usually not obvious in the rescued group. (G–I) Heart sections of BMS493-treated Tie2+/fl (G), Tie2-cko/R (H), and Tie2-cko/P (I) embryos at E14.0 were stained with troponin T (red) and endomucin (green) antibodies. (J–L) Quantification of trabecular myocardium complexity (trabeculae density and thickness) and compact myocardium thickness from E11.5–E14.0. The trabeculae in the Tie2-cko rescued group were still thicker than those of BMS493-treated controls but much thinner than those of the Tie2-cko poorly rescued embryos, which were similar to the untreated Tie2-cko embryos. However, the defects on the density of trabeculae and thickness of compact wall and simplification of endocardium in the Tie2-cko rescued group were not significantly improved. (M and N) BrdU pulse labeling and immunostaining showed a little more BrdU-positive (red) CMs, stained with troponin T (green) in the trabecular zone of BMS493-treated Tie2-cko embryos (N) at E11.5 than those in the treated controls (M). (O) Quantification of BrdU-positive nuclei as a percentage of total nuclei in myocardium indicated that BMS493-treated Tie2-cko embryos displayed slightly higher proliferation rates of CMs in the trabecular zone at E11.5. Scale bars: G–I, 100 μm; others, 50 μm. A representative of more than 10 images was chosen for each panel; n = 3 (A) or 6 (J–L and O) per group. *P < 0.05; **P < 0.01, Student’s t test (A) and 2-way ANOVA (J–L and O).
It is known that a precisely regulated supply of RA is essential for normal cardiogenesis (12, 13). To determine whether upregulation of components of RA signaling pathway in mouse embryonic hearts observed in Tie2-cko embryos was consistent with enhanced RA signaling, we injected WT pregnant mice at E8.5 with all-trans RA and analyzed the RA-treated embryos 2 days later. qPCR analysis of RA-treated WT embryonic hearts recapitulated the elevation of components in the RA signaling pathway observed in the Tie2-cko mutants (Supplemental Figure 13A). As seen in Tie2-mutant embryos, expression levels of Crabp1/2 were increased 2.6- to 3.2-fold, and Rdh10 was increased 1.6-fold, while there was no change in expression of Raldh2. In addition, immunostaining of WT control and RA-treated heart sections at E10.5 for Crabp2 detected elevated myocardial Crabp2 staining following treatment, as seen in Tie2 mutants (Supplemental Figure 13, B and C). Finally, similar to the results documented in the Tie2-cko mutants, immunostaining of WT control and RA-treated heart sections at E10.5 for p-Erk1/2 detected significant myocardial Erk1/2 hyperphosphorylation following RA treatment when compared with controls (Supplemental Figure 13, D and E).
In order to rescue cardiac phenotypes in Tie2-cko by inhibiting RA signaling, we then supplied pregnant Tie2fl/fl mice (bred with Nfatc1Cre Tie2+/fl male) with the pan-RA receptor antagonist BMS493 (36). Treatment started at E9.75 and was terminated 6 to 12 hours before mice were sacrificed at E11.5–E14.5. Initially, we used 5 mg/kg as previously described (36), but this dosage caused maternal toxicity and early lethality of most control and Tie2-cko embryos. We found that 3.5 mg/kg was the highest dosage tolerated by dams and control pups and initiated the rescue experiments at E9.75. Even with this reduced dosage, however, some control embryos still developed dorsal edema at E14.0, indicating that RA signaling inhibition by BMS493 is toxic to early lymphatic development (37). We treated the pregnant mice with BMS493 by oral gavages twice a day with 10- to 12-hour intervals. In contrast to untreated Tie2-cko embryos, which began to show dorsal edema and pericardial effusions from E12.5 and died at E13.0–E13.5, most (82.4%, 28 out of 34) treated Tie2-cko embryos were grossly indistinguishable from their control littermates until E13.75 and survived to E14.0 (we refer to these embryos as the “rescued group”), approximately 12 hours longer than untreated Tie2-cko embryos. Six out of 34 (17.6%) of treated Tie2-cko embryos began to show dorsal edema and pericardial effusions from E13.0 or died at E13.0–E14.0 (we refer to these as the “poorly rescued group”) (Figure 7, D–F). Accordingly, immunostaining (Figure 7, G–I) and quantification of trabecular complexity detected an improvement of the increased trabecular thickness phenotype in the Tie2-cko rescued group. Thickness of trabeculae in the Tie2-cko rescued group was respectively 21.7%, 37.6%, and 27.8% higher than that of BMS493 treated controls at E11.5, E12.5, and E13.5 (Supplemental Figure 14, A–H and Figure 7J), compared with trabecular thickness increase in untreated Tie2-cko embryos of 53.3%, 84.4%, and 87.4% at E11.5, E12.5, and E13.5 (Figure 1L). At E13.5 and E14.0, the trabeculae in the Tie2-cko rescued group were still thicker than those of BMS493 treated controls but much thinner than those of the Tie2-cko poorly rescued embryos or the untreated Tie2-cko embryos (Figure 1). However, the defects detected in the density of trabeculae and thickness of compact wall and simplification of endocardium in the Tie2-cko rescued group were not significantly improved (Figure 7, K and L). Cell proliferation analyses revealed that the percentage of BrdU-positive CMs in the trabecular zone of the BMS493-treated Tie2-cko hearts was only slightly higher than that in the BMS493-treated controls, that is, 25.0% versus 20.1% at E11.5 (increase by 24.4%) (Figure 7, M–O), compared to the increase of 48.8% seen in CMs of untreated Tie2-cko embryos (Figure 6 C). The reduced CM proliferation with BMS493 treatment is consistent with partially rescued trabecular phenotypes. However, as anticipated, the reduced proliferation rate in ECs was not improved (Supplemental Figure 14, I–K); this finding was consistent with the lack of rescue in the endocardial phenotypes. Therefore, myocardial phenotypes in Tie2-cko hearts were partially rescued by inhibiting in utero RA signaling with BMS493 treatment. These results suggest that Tie2 may regulate CM proliferation at least partially via suppression of RA signaling.
Discussion
Previous reports have suggested a role for Tie2 signaling in trabeculation because Tie2-deficient embryos have no trabeculae (19, 20) and Ang1 deficiency results in a marked reduction in cardiac trabeculation (21, 22). However, the molecular basis for these phenotypes has remained elusive. In this report, we have used endocardial-specific deletion of Tie2 in the developing mouse to show that during ventricular trabeculation, Tie2 regulates proliferation, migration, and sprouting of ECs to ensure normal endocardial growth required for normal trabeculation. In addition, endocardial Tie2 is also instrumental in the generation of paracrine signaling that inhibits CM proliferation to prevent hypertrabeculation through downregulation of RA signaling, Bmp10 signaling, and Erk1/2 phosphorylation in the myocardium.
During ventricular trabeculation, myocardium grows out from the inner ventral surface in the apical region of the presumptive ventricle, expands rapidly either by myocyte recruitment or by proliferation, and subsequently differentiates. However, mechanisms regulating CM extension into the luminal part of the ventricle remain largely unknown. Based on the anatomical changes during trabeculation, we suggest a 3-step model for trabecular formation: stage 1 at E9.0, initiation of trabeculation; stage 2 at E9.25, trabecular assembly; and stage 3 at E9.5, trabecular extension (Figure 3). Although CM delamination occurs in both Tie2-cko and Tie2–/– mutants, trabecular assembly and extension are impaired. In Tie2–/– embryonic ventricles, trabecular assembly and extension are totally absent due to complete loss of endocardial touchdown sproutings. Thus, the endocardium is not merely responsible for regulating myocyte proliferation in the process of trabeculation but also plays a critical role in initiating and orchestrating trabecular formation. The endocardial defects preceded the trabecular defects in the mutants, and endocardial sprouting is critical for trabecular assembly and extension. This is consistent with our previous report that Tie2 plays a critical role in sprouting tip cells during postnatal angiogenesis (38). Interestingly, although a role for Tie1 in modulating Tie2 function has been described (38–40), the role of Tie2 in ventricular trabeculation appears to be independent of Tie1, as attenuation of Tie1 with the same endocardial-specific Cre deletor mouse line showed none of the defects described in this article (41). This finding further supports the context-dependent nature of Tie RTK signaling (17, 39). In addition, our observations on the importance of endocardial sprouting and myocardial touchdown in regulation of the trabecular process are supported by a recent publication by Del Monte-Nieto et al. (35) published after the original submission of this article.
It is difficult to determine the specific downstream targets of Tie2 activation in the developing endocardium. Following endocardial loss of Tie2, the expression levels of several endocardial-specific genes were altered. We were able to show that several of the downregulated endocardial genes previously implicated in cardiac development and trabeculation (Sox7, Tmem100, and Arap3) did not phenocopy the defects in the endocardial network formation of the Tie2-cko embryos. It is possible that the reduced expression of N1ICD and Vegfr2 (Flk1) might impair endocardial sprouting and touchdown formation. Both VEGFA and Notch pathways are key modulators in angiogenic sprouting in developing vascular beds (42) and ventricular trabeculation (35). However, because the reduction of N1ICD-positive ECs is proportionate to the reduction of total ECs in Tie2-cko endocardium, this reduction might be due to a loss of ECs in mutants, and it is unlikely that Tie2 specifically targets Notch1 signaling in trabeculation. Furthermore, we did not see the dramatic alteration in trabecular ECM production characteristic of Notch signaling (35), suggesting the defects we observed were not primarily a result of altered Notch signaling.
It has been reported that roundabout guidance receptor 4 (Robo4) is an endothelial-specific receptor that inhibits endothelial cell migration (43), and as noted, Robo4 was increased in our mutants. Similarly, deficiency in Plexin D1 (44), Rasip1 (45), and Acap1 (46) all impair endothelial cell migration, and all were downregulated in Tie2-cko mutants. Mice lacking Rasip1 even fail to form patent lumens in all blood vessels, including the early endocardial tube. Therefore, a 2.1-fold increase of Robo4 and downregulation of Plexin D1, Rasip1, and Acap1 would at least partially explain the EC migration/sprouting defects in Tie2-cko embryonic endocardium.
RA is required for normal embryonic heart development. An excess of RA has also been found to cause teratogenic effects during early heart development, indicating that a precisely regulated supply of RA is essential for normal cardiogenesis (12). We show that enhanced trabecular cardiomyocyte proliferation in Tie2-cko hearts is associated with elevated expression of numerous components of the RA signaling pathway including Rdh10, Crabp1, Crabp2, encoding cytochrome P450 1B1, retinol-binding protein 4, and Stra6. Crabp1 and -2 are proteins of approximately 16–17 kDa, which bind RA with high affinity. It is currently thought that the expression and location of Crabp1 and -2 in the developing heart may reflect the timeframe and areas of RA activity (47). We also identified increased expression of α-fetoprotein and several members of the Hox gene family. Interestingly, promoters of these Hox genes (Stra6 and both Crabp genes) all contain functional and/or evolutionarily conserved RA-response elements (13, 47). It has been reported that a single dose of RA administered at the critical time of cardiogenesis (E8.5) may affect production of endogenous RA within the developing heart (47). In addition, a rapid activation of Erk1/2 by RA has been widely detected in cell culture systems (48). To decipher the possible interplay between retinoid and Tie2 signaling pathways in myocardial trabeculation, we treated mouse embryos with RA and confirmed that RA treatment at E8.5 increased expression of Crabp1 and Crabp2 in the heart in the 48-hour period. Furthermore, trabecular myocardial Erk1/2 hyperphosphorylation also occurs within the same time window. Erk1/2 not only mediates cell survival but also regulates cell differentiation and proliferation (49). Classically, phosphorylation and activation of Erk1/2 has been associated with a positive effect on cellular proliferation.
Furthermore, to rescue cardiac phenotypes in Tie2-cko by inhibiting RA signaling, we treated pregnant Tie2fl/fl mice (bred with Nfatc1Cre Tie2+/fl male) with pan-RA receptor antagonist BMS493. This treatment allowed the majority (82.4%) of Tie2-cko embryos to survive to E14.0; three-quarters lived about 12 hours longer than the untreated mutants. The enhanced proliferation rate in CMs of Tie2-cko embryos was consistently reduced by BMS493 treatment, thus the defect of the thickened trabeculae in the Tie2-cko rescued group was significantly improved when compared with the untreated mutants. However, because BMS493 treatment did not affect the reduced proliferation rate in ECs, endocardial sprouting and touchdown formation in BMS493-treated Tie2-cko embryos were still abnormal. Thus, other components of the cardiac phenotype (low density of trabeculae and thin compact wall) of the Tie2-cko mutant embryos were not rescued by inhibiting RA signaling with BMS493 treatment. Therefore, myocardial phenotypes in Tie2-cko hearts were partially rescued by inhibiting in utero RA signaling with BMS493 treatment. These results suggest that Tie2 may regulate CM proliferation at least partially via suppression of RA signaling. Experiments are ongoing to further determine how Tie2 signaling affects the RA signaling pathway in embryonic trabecular CMs.
In summary, these studies reveal a critical role for the endocardium in the orchestration of ventricular trabeculation which is dependent on endocardial RTK Tie2 signaling. Tie2 plays a primary autocrine function in support of EC proliferation and angiogenic sprouting and touchdown and is also instrumental in the generation of paracrine signaling that results in inhibition of trabecular cardiomyocyte proliferation via suppression of RA signaling that prevents hypertrabeculation. These insights will be important in further defining mechanisms of disease pathology related to ventricular chamber morphogenesis and function.
Methods
Mouse models and related methods.
Nfatc1Cre mice have been previously described (24). Tie2fl/fl mice were generated as shown in Supplemental Figure 1. For BrdU incorporation, pregnant females were injected i.p. using a BrdU cell proliferation kit (RPN20, Amersham Biosciences) at a dosage of 1 ml/100 g body weight 2 hours prior to sacrifice. For RA treatment, at E8.5, half of the pregnant females were injected i.p. with all-trans RA (Sigma-Aldrich) dissolved in 100 μl of DMSO, at a dose of 7 mg/kg body weight. After 2 days, mice were sacrificed. For rescue experiments with BMS493 treatment, as described previously (36), we supplied pregnant Tie2fl/fl mice (bred with Nfatc1Cre/+ Tie2fl/+ male) with pan-RA receptor antagonist BMS493 (Tocris Bioscience; 3.5 mg/kg) or vehicle (DMSO) 1:10 in sunflower oil by oral gavages twice a day at 10- to 12-hour intervals. Treatment started at E9.75 and was terminated 6 to 12 hours before mice were sacrificed at E11.5 to E14.5.
Generation of conditional Tie2 allele.
A BAC recombineering approach was used to generate a floxed Tie2 allele, with loxP sites flanked at the Tie2 minimal promoter (50) and exon 1 (containing the initial ATG codon and encoding the first 17 amino acids) (Supplemental Figure 1A). Correctly targeted embryonic stem (ES) cell clones were identified by Southern blot analysis with probes outside the region of homology and PCR genotyping (Supplemental Figure 1B). After injection of the G4 ES cells into C57BL/6 blastocysts and breeding of chimeras, heterozygous floxed Tie2 mice were bred to mice expressing flippase recombinase to remove the neomycin cassette. Heterozygous Tie2loxP mice (Tie2+/fl) were intercrossed to generate homozygous Tie2fl/fl mice. These mice developed with no obvious abnormalities and were fertile. To confirm that excision of the floxed allele results in a null Tie2 allele, Tie2+/fl mice were crossed with E2A-Cre transgenic mice (51) to produce germline deletion of Tie2 (Tie2–/+) (Supplemental Figure 1C). Breeding to homozygosity (Tie2–/–) resulted in embryonic lethality at E10.5–E11.0, similar to the conventional null mutants previously described (19, 20). At E10.5, Tie2–/– embryos were developmentally delayed (Supplemental Figure 1D) and showed heart defects with loss of trabeculations (Supplemental Figure 1F), identical to those of the conventional knockout. Heterozygous embryos showed no phenotype. Thus, the Tie2del allele is functionally a null allele. The sequences of the 3 primers for PCR genotyping are: 5′-CATGGGAGGTGCACATTTTCAGG-3′, 5′-TATACGCATGCTACTGGCTACAGC-3′, and 5′-CTTCAAAACCGTTGCCATGTGTA-3′.
Histology.
For H&E staining, fixed tissues were put through an ethanol gradient, embedded in paraffin, and sectioned at 6 μm. Sections were deparaffinized and stained with Mayer’s H&E following standard protocols. Alcian blue staining was performed using a 1% solution of Alcian blue (pH 2.5) in acetic acid followed by counterstaining with 0.1% Nuclear Fast Red (Vector Laboratories).
Western blotting.
Lysates from E12.5 hearts of each genotype were processed for Western blotting as previously described (52). The Western blots were probed with the primary antibody followed by incubation with an IRDye 680– or IRDye 800CW–conjugated secondary antibody (LI-COR). Membranes were imaged with the Odyssey infrared imaging system (LI-COR), and quantitative densitometric analysis was performed with Odyssey version 1.2 infrared imaging software. For normalization of signals, mouse anti–α-tubulin was used as a loading control.
Immunostaining.
Paraffin (6 μm) or frozen (10 μm) sections were incubated overnight with primary antibodies (Supplemental Table 2), followed by 1-hour incubation with a fluorescent dye–conjugated secondary antibody. N1ICD staining was performed using tyramide signal amplification (6). For fluorescence staining, Alexa Fluor 488, 555, 594, and 647 fluorochrome-conjugated secondary antibodies (Invitrogen) were used for signal detection. Images were acquired with a Leica TCS SP2 confocal system or a Nikon Eclipse E800 microscope and processed using Adobe Photoshop.
In situ hybridization.
Samples were prepared for RNAscope (Advanced Cell Diagnostics, ACD) according to manufacturer’s guidelines. In situ hybridization was performed on 6-μm paraffin sections with a Mm-Bmp10 probe (ACD, no. 415921) according to the protocol for RNAscope 2.5 HD Red Detection Reagent (ACD, no. 322360). The target region of the RNA probe is a 1.05-kb mouse Bmp10 fragment corresponding to nucleotides 78-1124 of NM_009756.2. The probe was used to hybridize the samples for 2 hours at 40°C. Slides were then washed twice with 1× Wash Buffer (ACD, no. 310091) for 2 minutes each with agitation. After the probe amplification steps, 120 μL Red Working Solution was placed onto each section for 3 minutes at room temperature in the humidity chamber. Slides were then washed twice in double-distilled water, immunostained with anti-endomucin, and mounted in Prolong Gold anti-fade reagent with DAPI (Life Technologies, P36935).
qPCR.
Control and mutant or RA-treated embryos at different stages were dissected in ice-cold PBS. Whole hearts of E9.5–11.5 embryos were separated from the rest of the body, and RNA was extracted. Total RNA was purified using TRIzol Reagent (Invitrogen) with additional DNase treatment (Promega). cDNA was then generated using the SuperScript III Reverse Transcriptase kit (Invitrogen). qPCR was performed on the CFX96 Touch thermal cycler (Bio-Rad) using iQTM SYBR Green Supermix (Bio-Rad). All assays were repeated at least twice, and all samples were run in triplicate. Analysis of gene expression was carried out using the comparative Ct (ΔΔCt) method as described by the manufacturer. Relative quantification of gene expression was normalized to 18S mRNA expression level. Sequences of the PCR primers used are listed in Supplemental Table 1.
RNA-Seq.
RNA was isolated at E11.5 from whole hearts of Tie2+/fl and Tie2-cko embryos (18 per genotype) and then pooled into 3 replicates. RNA was prepared using the standard Illumina TrueSeq RNA-Seq library preparation kit. Libraries were sequenced in a HiSeq 2500 Illumina sequencer using a 76-bp single-end elongation protocol. The resulting raw data were in FASTQ format, QC, and preprocessed. Resulting reads were aligned and gene expression quantified using TopHat v2.0.9 over mouse reference mm10 (GRCm38) and Ensembl genebuild Mus_musculus.GRCm38.75. Gene differential expression was analyzed using the EdgeR R package63. The gene expression was quantified as FPKM value, and the gene differential expression as log2 (fold change) value, using Cufflinks/Cuffdiff v2.2.1 for the comparison of the control group and mutant group. The cluster analysis and heatmap, based on the gene FPKM value of the samples, were plotted in R with the heatmap3 package. Genes showing altered expression with adjusted P < 0.05 were considered differentially expressed. All data files can be accessed at NCBI’s Gene Expression Omnibus (GEO GSE130741).
Quantification of trabecular complexity and compact myocardial thickness.
The number of trabeculae, the thickness of individual trabeculae and the compact zone, the thickness of total trabecular layer, and total trabecular area were quantified as described (6, 7, 53). Transverse heart sections from E9.5–E13.5 control and mutant embryos were stained with anti-endomucin (endocardial marker) and anti-Tnnt2 (recognizes myocardial marker troponin T) to facilitate visualization of ventricular structures. Images were used for the measurements with ImageJ software (NIH). In E12.5 and E13.5 heart sections, the thickness of the compact myocardium in the left ventricle was measured.
Quantification of endocardial complexity and sprouting activity.
To measure endocardial branch points, the total area covered by endocardial network, total length of endocardial network, and transverse heart sections of E9.5–E13.5 control and mutant embryos were stained with anti-endomucin and anti-Erg or N1ICD for visualization of endocardial networks. To measure the endocardial sprouting touchdown points in E9.0–E9.5 endocardium, transverse sections were stained with anti-endomucin and anti-Tnnt2. Images were used for the measurements with AngioTool (54).
Statistics.
Data are reported as means ± SEM. Multiple groups were compared by 1-way ANOVA, and multiple groups with multiple time points were compared by 2-way ANOVA. Student’s t test was used when comparing 2 experimental groups. A P value of less than 0.05 was considered significant. All analyses were performed with GraphPad Prism 7.
Study approval.
The animals were handled in accordance with institutional guidelines with the approval of the Institutional Animal Care and Use Committee of Vanderbilt University School of Medicine.
Author contributions
XQ and HSB designed the research. XQ and CH performed the experiments. XQ and HSB analyzed data and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Jeremy Nathans (Johns Hopkins Medical School) for providing the floxed Sox7 mice, S. Paul Oh (St. Joseph’s Hospital and Medical Center) for providing the floxed Tmem100 mice, and Sonja Vermeren (The Queen’s Medical Research Institute, United Kingdom) for providing the floxed Arap3 mice used in these studies. We thank Alice Qu for the illustration depicting trabeculation in Figure 4. This work was supported by NIH grant R01 HL118386 (to HSB).
Version 1. 05/21/2019
In-Press Preview
Version 2. 07/11/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(13):e96002. https://doi.org/10.1172/jci.insight.96002.
Contributor Information
Xianghu Qu, Email: xianghu.qu@vanderbilt.edu.
Cristina Harmelink, Email: cristina.harmelink@vanderbilt.edu.
H. Scott Baldwin, Email: scott.baldwin@vanderbilt.edu.
References
- 1.Sedmera D, Pexieder T, Vuillemin M, Thompson RP, Anderson RH. Developmental patterning of the myocardium. Anat Rec. 2000;258(4):319–337. doi: 10.1002/(SICI)1097-0185(20000401)258:4<319::AID-AR1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 2.Captur G, et al. Morphogenesis of myocardial trabeculae in the mouse embryo. J Anat. 2016;229(2):314–325. doi: 10.1111/joa.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samsa LA, Yang B, Liu J. Embryonic cardiac chamber maturation: trabeculation, conduction, and cardiomyocyte proliferation. Am J Med Genet C Semin Med Genet. 2013;163C(3):157–168. doi: 10.1002/ajmg.c.31366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang W, Chen H, Qu X, Chang CP, Shou W. Molecular mechanism of ventricular trabeculation/compaction and the pathogenesis of the left ventricular noncompaction cardiomyopathy (LVNC) Am J Med Genet C Semin Med Genet. 2013;163C(3):144–156. doi: 10.1002/ajmg.c.31369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grego-Bessa J, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12(3):415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Amato G, et al. Sequential Notch activation regulates ventricular chamber development. Nat Cell Biol. 2016;18(1):7–20. doi: 10.1038/ncb3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao C, et al. Numb family proteins are essential for cardiac morphogenesis and progenitor differentiation. Development. 2014;141(2):281–295. doi: 10.1242/dev.093690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, et al. Fkbp1a controls ventricular myocardium trabeculation and compaction by regulating endocardial Notch1 activity. Development. 2013;140(9):1946–1957. doi: 10.1242/dev.089920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H, et al. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development. 2004;131(9):2219–2231. doi: 10.1242/dev.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai D, et al. Neuregulin 1 sustains the gene regulatory network in both trabecular and nontrabecular myocardium. Circ Res. 2010;107(6):715–727. doi: 10.1161/CIRCRESAHA.110.218693. [DOI] [PubMed] [Google Scholar]
- 11.Gerety SS, Wang HU, Chen ZF, Anderson DJ. Symmetrical mutant phenotypes of the receptor EphB4 and its specific transmembrane ligand ephrin-B2 in cardiovascular development. Mol Cell. 1999;4(3):403–414. doi: 10.1016/S1097-2765(00)80342-1. [DOI] [PubMed] [Google Scholar]
- 12.Pan J, Baker KM. Retinoic acid and the heart. Vitam Horm. 2007;75:257–283. doi: 10.1016/S0083-6729(06)75010-5. [DOI] [PubMed] [Google Scholar]
- 13.Rhinn M, Dollé P. Retinoic acid signalling during development. Development. 2012;139(5):843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 14.Sucov HM, Dyson E, Gumeringer CL, Price J, Chien KR, Evans RM. RXR alpha mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev. 1994;8(9):1007–1018. doi: 10.1101/gad.8.9.1007. [DOI] [PubMed] [Google Scholar]
- 15.Sandell LL, et al. RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes Dev. 2007;21(9):1113–1124. doi: 10.1101/gad.1533407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niederreither K, Vermot J, Messaddeq N, Schuhbaur B, Chambon P, Dollé P. Embryonic retinoic acid synthesis is essential for heart morphogenesis in the mouse. Development. 2001;128(7):1019–1031. doi: 10.1242/dev.128.7.1019. [DOI] [PubMed] [Google Scholar]
- 17.Augustin HG, Koh GY, Thurston G, Alitalo K. Control of vascular morphogenesis and homeostasis through the angiopoietin-Tie system. Nat Rev Mol Cell Biol. 2009;10(3):165–177. doi: 10.1038/nrm2639. [DOI] [PubMed] [Google Scholar]
- 18.Eklund L, Kangas J, Saharinen P. Angiopoietin-Tie signalling in the cardiovascular and lymphatic systems. Clin Sci. 2017;131(1):87–103. doi: 10.1042/CS20160129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dumont DJ, et al. Dominant-negative and targeted null mutations in the endothelial receptor tyrosine kinase, tek, reveal a critical role in vasculogenesis of the embryo. Genes Dev. 1994;8(16):1897–1909. doi: 10.1101/gad.8.16.1897. [DOI] [PubMed] [Google Scholar]
- 20.Sato TN, et al. Distinct roles of the receptor tyrosine kinases Tie-1 and Tie-2 in blood vessel formation. Nature. 1995;376(6535):70–74. doi: 10.1038/376070a0. [DOI] [PubMed] [Google Scholar]
- 21.Suri C, et al. Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell. 1996;87(7):1171–1180. doi: 10.1016/S0092-8674(00)81813-9. [DOI] [PubMed] [Google Scholar]
- 22.Jeansson M, et al. Angiopoietin-1 is essential in mouse vasculature during development and in response to injury. J Clin Invest. 2011;121(6):2278–2289. doi: 10.1172/JCI46322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward NL, Van Slyke P, Sturk C, Cruz M, Dumont DJ. Angiopoietin 1 expression levels in the myocardium direct coronary vessel development. Dev Dyn. 2004;229(3):500–509. doi: 10.1002/dvdy.10479. [DOI] [PubMed] [Google Scholar]
- 24.Wu B, et al. Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling. Cell. 2012;151(5):1083–1096. doi: 10.1016/j.cell.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Z, et al. The cerebral cavernous malformation pathway controls cardiac development via regulation of endocardial MEKK3 signaling and KLF expression. Dev Cell. 2015;32(2):168–180. doi: 10.1016/j.devcel.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu B, et al. Nfatc1 coordinates valve endocardial cell lineage development required for heart valve formation. Circ Res. 2011;109(2):183–192. doi: 10.1161/CIRCRESAHA.111.245035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wat MJ, et al. Mouse model reveals the role of SOX7 in the development of congenital diaphragmatic hernia associated with recurrent deletions of 8p23.1. Hum Mol Genet. 2012;21(18):4115–4125. doi: 10.1093/hmg/dds241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Somekawa S, et al. Tmem100, an ALK1 receptor signaling-dependent gene essential for arterial endothelium differentiation and vascular morphogenesis. Proc Natl Acad Sci U S A. 2012;109(30):12064–12069. doi: 10.1073/pnas.1207210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gambardella L, Hemberger M, Hughes B, Zudaire E, Andrews S, Vermeren S. PI3K signaling through the dual GTPase-activating protein ARAP3 is essential for developmental angiogenesis. Sci Signal. 2010;3(145):ra76. doi: 10.1126/scisignal.2001026. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, Williams J, Smallwood PM, Nathans J. Sox7, Sox17, and Sox18 cooperatively regulate vascular development in the mouse retina. PLoS One. 2015;10(12):e0143650. doi: 10.1371/journal.pone.0143650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon EH, et al. Generation of mice with a conditional and reporter allele for Tmem100. Genesis. 2010;48(11):673–678. doi: 10.1002/dvg.20674. [DOI] [PubMed] [Google Scholar]
- 32.Kochilas LK, Li J, Jin F, Buck CA, Epstein JA. p57Kip2 expression is enhanced during mid-cardiac murine development and is restricted to trabecular myocardium. Pediatr Res. 1999;45(5 pt 1):635–642. doi: 10.1203/00006450-199905010-00004. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura T, et al. Mediating ERK 1/2 signaling rescues congenital heart defects in a mouse model of Noonan syndrome. J Clin Invest. 2007;117(8):2123–2132. doi: 10.1172/JCI30756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stankunas K, et al. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell. 2008;14(2):298–311. doi: 10.1016/j.devcel.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Del Monte-Nieto G, et al. Control of cardiac jelly dynamics by NOTCH1 and NRG1 defines the building plan for trabeculation. Nature. 2018;557(7705):439–445. doi: 10.1038/s41586-018-0110-6. [DOI] [PubMed] [Google Scholar]
- 36.van de Pavert SA, et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature. 2014;508(7494):123–127. doi: 10.1038/nature13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burger NB, et al. Involvement of neurons and retinoic acid in lymphatic development: new insights in increased nuchal translucency. Prenat Diagn. 2014;34(13):1312–1319. doi: 10.1002/pd.4473. [DOI] [PubMed] [Google Scholar]
- 38.Savant S, et al. The orphan receptor Tie1 controls angiogenesis and vascular remodeling by differentially regulating Tie2 in tip and stalk cells. Cell Rep. 2015;12(11):1761–1773. doi: 10.1016/j.celrep.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leppänen VM, Saharinen P, Alitalo K. Structural basis of Tie2 activation and Tie2/Tie1 heterodimerization. Proc Natl Acad Sci U S A. 2017;114(17):4376–4381. doi: 10.1073/pnas.1616166114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seegar TC, et al. Tie1-Tie2 interactions mediate functional differences between angiopoietin ligands. Mol Cell. 2010;37(5):643–655. doi: 10.1016/j.molcel.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qu X, Zhou B, Scott Baldwin H. Tie1 is required for lymphatic valve and collecting vessel development. Dev Biol. 2015;399(1):117–128. doi: 10.1016/j.ydbio.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shalaby F, et al. A requirement for Flk1 in primitive and definitive hematopoiesis and vasculogenesis. Cell. 1997;89(6):981–990. doi: 10.1016/S0092-8674(00)80283-4. [DOI] [PubMed] [Google Scholar]
- 43.Park KW, et al. Robo4 is a vascular-specific receptor that inhibits endothelial migration. Dev Biol. 2003;261(1):251–267. doi: 10.1016/S0012-1606(03)00258-6. [DOI] [PubMed] [Google Scholar]
- 44.Gitler AD, Lu MM, Epstein JA. PlexinD1 and semaphorin signaling are required in endothelial cells for cardiovascular development. Dev Cell. 2004;7(1):107–116. doi: 10.1016/j.devcel.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 45.Xu K, et al. Blood vessel tubulogenesis requires Rasip1 regulation of GTPase signaling. Dev Cell. 2011;20(4):526–539. doi: 10.1016/j.devcel.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Ballif BA, Powelka AM, Dai J, Gygi SP, Hsu VW. Phosphorylation of ACAP1 by Akt regulates the stimulation-dependent recycling of integrin beta1 to control cell migration. Dev Cell. 2005;9(5):663–673. doi: 10.1016/j.devcel.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 47.Stachurska E, et al. Expression of cellular retinoic acid-binding protein I and II (CRABP I and II) in embryonic mouse hearts treated with retinoic acid. Acta Biochim Pol. 2011;58(1):19–29. [PubMed] [Google Scholar]
- 48.Persaud SD, Lin YW, Wu CY, Kagechika H, Wei LN. Cellular retinoic acid binding protein I mediates rapid non-canonical activation of ERK1/2 by all-trans retinoic acid. Cell Signal. 2013;25(1):19–25. doi: 10.1016/j.cellsig.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ebisuya M, Kondoh K, Nishida E. The duration, magnitude and compartmentalization of ERK MAP kinase activity: mechanisms for providing signaling specificity. J Cell Sci. 2005;118(pt 14):2997–3002. doi: 10.1242/jcs.02505. [DOI] [PubMed] [Google Scholar]
- 50.Fadel BM, Boutet SC, Quertermous T. Functional analysis of the endothelial cell-specific Tie2/Tek promoter identifies unique protein-binding elements. Biochem J. 1998;330(pt 1):335–343. doi: 10.1042/bj3300335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lakso M, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93(12):5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qu X, Tompkins K, Batts LE, Puri M, Baldwin HS, Baldwin S. Abnormal embryonic lymphatic vessel development in Tie1 hypomorphic mice. Development. 2010;137(8):1285–1295. doi: 10.1242/dev.043380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen H, Zhang W, Li D, Cordes TM, Mark Payne R, Shou W. Analysis of ventricular hypertrabeculation and noncompaction using genetically engineered mouse models. Pediatr Cardiol. 2009;30(5):626–634. doi: 10.1007/s00246-009-9406-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zudaire E, Gambardella L, Kurcz C, Vermeren S. A computational tool for quantitative analysis of vascular networks. PLoS One. 2011;6(11):e27385. doi: 10.1371/journal.pone.0027385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.