Abstract
Acute rejection of human allografts has been viewed mostly through the lens of adaptive immunity, and the intragraft landscape of innate immunity genes has not been characterized in an unbiased fashion. We performed RNA sequencing of 34 kidney allograft biopsy specimens from 34 adult recipients; 16 were categorized as Banff acute T cell–mediated rejection (TCMR) and 18 as normal. Computational analysis of intragraft mRNA transcriptome identified significantly higher abundance of mRNA for pattern recognition receptors in TCMR compared with normal biopsies, as well as increased expression of mRNAs for cytokines, chemokines, interferons, and caspases. Intragraft levels of calcineurin mRNA were higher in TCMR biopsies, suggesting underimmunosuppression compared with normal biopsies. Cell-type-enrichment analysis revealed higher abundance of dendritic cells and macrophages in TCMR biopsies. Damage-associated molecular patterns, the endogenous ligands for pattern recognition receptors, as well markers of DNA damage were higher in TCMR. mRNA expression patterns supported increased calcium flux and indices of endoplasmic, cellular oxidative, and mitochondrial stress were higher in TCMR. Expression of mRNAs in major metabolic pathways was decreased in TCMR. Our global and unbiased transcriptome profiling identified heightened expression of innate immune system genes during an episode of TCMR in human kidney allografts.
Keywords: Immunology, Transplantation
Keywords: Innate immunity
Global and unbiased transcriptome profiling of human kidney allograft biopsies reveals heightened expression of innate immune system genes during acute T cell–mediated rejection.
Introduction
Kidney transplantation is a life restorative procedure (1, 2), but multiple challenges still exist (3). Transplantation is a unique situation from an immunological perspective in that 2 diploid genomes are brought together at the time of transplant surgery (4). Immunological injury to the allograft results from the interactions between the donor and recipient genomes. Acute allograft rejection (AR) is a major risk factor for long-term graft failure (5, 6). While the clinical application of potent immunosuppressive and anti-infective prophylactic drugs is a major reason for the current success rates, there is continuing need for better understanding of the alloimmune response to further optimize patient outcomes.
The host defense responses are often categorized into innate and adaptive components (7). Alloimmunity is considered an adaptive immune response, as it represents an adaptation that occurs during the lifetime of an individual. Adaptive immunity is antigen specific and reciprocal cognate interactions between T cells and B cells play key roles in the generation of alloimmune responses. Our current armamentarium of immunosuppressive drugs is designed primarily to keep the adaptive immunity in check.
After remaining in the background for decades, the role of innate immunity as a significant driver of alloimmune response is increasingly recognized (7–9). However, interrogation of innate immunity after transplantation is mostly in the context of ischemia-reperfusion injury. There is an unmet need for the investigation of innate immunity during an episode of AR, especially in human organ transplants (10, 11). The activation of innate immunity in the immediate posttransplant period in the context of ischemia-reperfusion injury does not fully explain its role in AR, which typically happens weeks to months after transplantation.
To fill gaps in our knowledge of the innate immune system’s role in AR, we performed RNA sequencing of human kidney allograft biopsies and computational analysis of gene expression pattern and strengths. Our identification of heightened intragraft expression of innate immune system genes in biopsies categorized as acute T cell–mediated rejection (TCMR) compared with normal biopsies (Normal) brings into sharp focus the association between innate immunity and TCMR, a process hitherto viewed mostly through the lens of adaptive immunity.
Results
Study groups, kidney allograft biopsies, RNA sequencing, and transcriptome analysis.
We studied the mRNA transcriptome of 34 kidney allograft biopsies from 34 adult kidney transplant recipients; 16 biopsies were categorized as TCMR per the Banff 2017 update of the Banff 1997 classification of allograft pathology, a standardized international nomenclature and classification schema for kidney allograft pathology (Supplemental Figure 1A, Supplemental Table 1, and ref. 12; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.128014DS1). These 16 biopsies were done in recipients with graft dysfunction (for-cause biopsies) prior to treatment with antirejection therapy. The remaining 18 biopsies were categorized as normal/nonspecific (Normal) and were done in recipients with stable graft function while on maintenance immunosuppression (surveillance biopsies). Immunostaining of biopsies confirmed the higher abundance of CD3+ T lymphocytes in TCMR. Biopsies were reported independently by 2 transplant pathologists at our center.
We isolated total RNA from the stored allograft biopsy specimens and used 400 ng of total RNA from each biopsy tissue (RNA integrity number 7.8 [7.0–8.5], median [IQR], in TCMR and 8.2 [7.4–8.7] in Normal) to sequence the mRNA transcriptome. In this report, the terms mRNA expression and mRNA abundance are used interchangeably. A gene was considered expressed in the TCMR or Normal group when its normalized mRNA count was ≥1 count per million mapped reads (CPM) in at least 1 sample in that group.
We identified 16,381 protein-coding genes including mitochondrial genes; 6441 (39.3%) were differentially expressed between TCMR and Normal biopsies at P adjusted for FDR (P-FDR) <0.05. Among the 6441, the abundance of 3622 (56%) was higher and that of 2819 (44%) was lower in TCMR compared with Normal biopsies (Supplemental Figure 1, B–D). The difference in mRNA abundance between TCMR and Normal was expressed as fold change (FC); a value of 1 corresponds to no difference in expression between TCMR and Normal (TCMR/Normal = 1). We cataloged the genes of the innate immune system based on published literature and public databases (13–15). We followed the Human Genome Organization (HUGO) Gene Nomenclature Committee (HGNC) approved names for each gene (16).
Transmembrane and cytosolic pattern recognition receptors in TCMR.
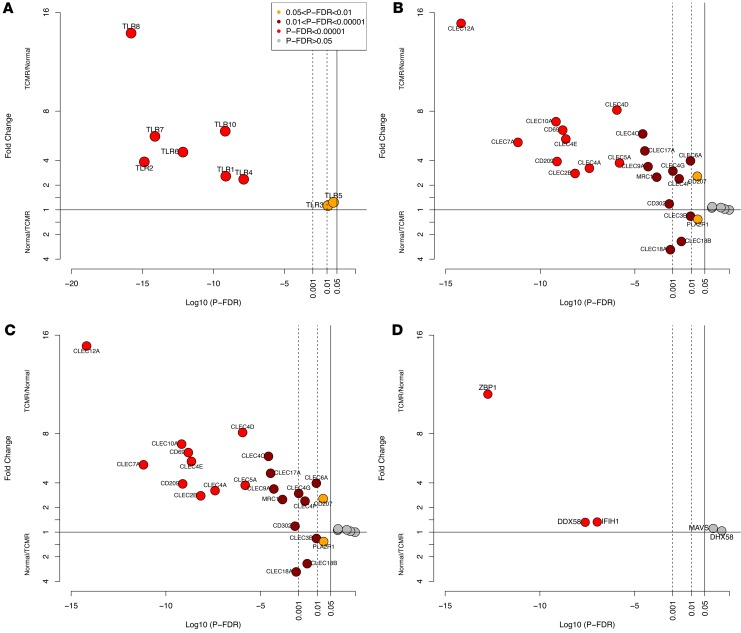
We first analyzed the expression of mRNAs for pattern recognition receptors (PRRs), the cornerstone of the innate immune system, in TCMR and Normal biopsies. PRRs (which include membrane-bound TLRs) are distinct sensors of cells of the innate immune system programmed to detect invariant molecular patterns (17–21). Among the 10 TLRs previously reported in humans, 9 were expressed in the kidney allografts. The abundance of all 9 mRNAs encoding TLRs was significantly higher in TCMR compared with Normal biopsies (Figure 1A and Supplemental Table 2A). TLR9 was not expressed in the kidney allograft. mRNA expression of myeloid differentiation primary response 88 (MYD88), a key signal transducer of TLRs that leads to the activation of nuclear factor kappa-light-chain-enhancer of activated B cells (NFKB), interferon (IFN) regulatory factor (IRF), and nuclear factor of activated T cells (NFATC), was higher in TCMR (FC 2.1, P-FDR < 0.0001). The associations between TLR and its key signal adapters/transducers are shown in Supplemental Table 2B and Supplemental Figure 2, A–C).
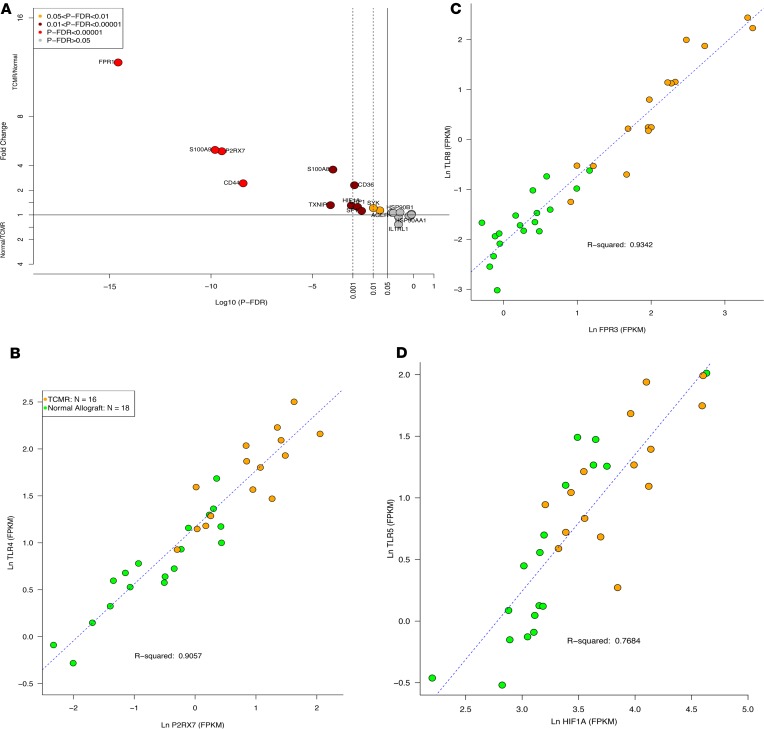
Figure 1. Differential intragraft abundance of mRNAs encoding transmembrane and cytosolic PRRs in human kidney allografts.
On the y axis, a fold change value of 1 (horizontal line) corresponds to no difference in expression levels between TCMR and Normal biopsies, TCMR/Normal = 1. For values plotted above 1, mRNA abundance in TCMR is higher than in Normal. For values plotted below 1, mRNA abundance in Normal is higher than in TCMR. Fold change was calculated using edgeR. The P-FDR is the probability value adjusted for FDR using the Benjamini-Hochberg method. The x axis is the log10P-FDR values; for example, a value of –5 corresponds to P-FDR = 0.00001. The 3 vertical lines correspond to P-FDR < 0.05, < 0.01, and < 0.001, respectively. (A) mRNAs for transmembrane TLRs. (B) mRNAs for transmembrane CLECs, CLRs, and mannose receptors.(C) mRNAs for transmembrane NLRs. (D) mRNAs for cytosolic RIGI-I–like receptors.
Transmembrane C-type lectins (CLECs), C-type lectin receptors (CLRs), and mannose receptors are expressed by macrophages and dendritic cells (DCs). Of the 38 CLRs and mannose receptors previously identified in humans, mRNAs for 28 of 38 were expressed in the kidney allograft biopsy and 23 of the 28 were differentially expressed between TCMR and Normal (Figure 1B and Supplemental Table 3).
Of the 25 transmembrane nucleotide-binding oligomerization domain–containing (NOD) proteins and NOD-like receptors (NLRs) found in humans, 19 NLRs were expressed in the 34 kidney biopsy samples. Among them, 13 were different between TCMR and Normal biopsies (Figure 1C and Supplemental Table 4). Except NLRP6, the other NLRs and Pyrin family members that constitute the inflammasome were higher in TCMR compared with Normal.
Cytosolic retinoic acid–inducible gene I (RIG-I)–like receptors (RLRs) consist of 3 members, DDX58, IFIH1 (MDA5), and DHX58 (LGP2), and recognize viral RNAs and DNAs in the cytoplasm. They associate with an adaptor protein MAVS (IFN-β promoter stimulator 1). DDX58 and IFIH1 were higher in TCMR biopsies (Figure 1D).
By gene-set testing, a differential expression analysis in which a P value is assigned to a set of genes as a unit, there was upregulation of the TLRs, CLRs, NLRs, and RLRs in TCMR, and by pathway enrichment analysis, the signaling pathways of TLRs, CLRs, NLRs, and RLRs were enriched in TCMR biopsies (Supplemental Table 5).
Our unbiased deep sequencing of human kidney allografts and data analysis revealed higher abundance of PRRs in TCMR compared with Normal at the single-gene level, gene-set level, and at the level of pathway enrichment.
Caspases, chemokines, cytokines, IFNs, and TNF in TCMR.
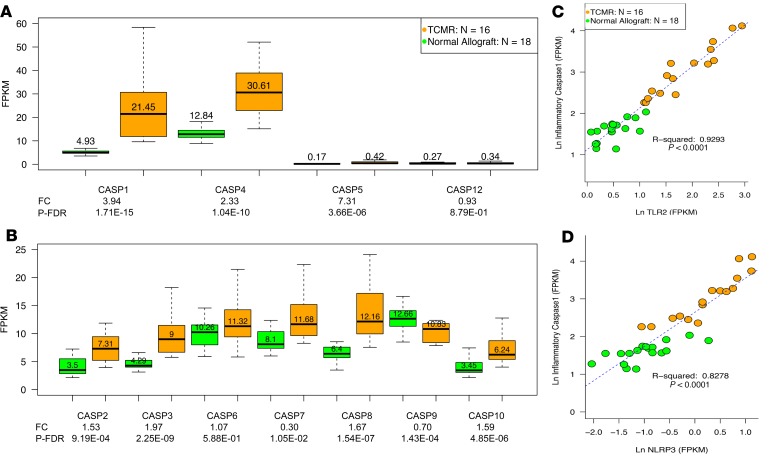
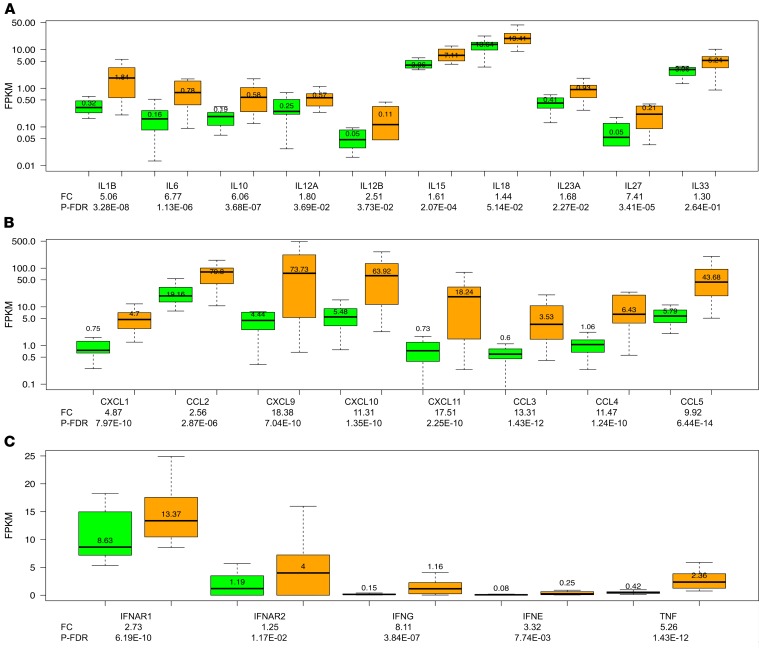
Signaling through PRRs activates inflammatory caspases, NF-κB, IRF, and NFATC transcription factors (22, 23) and triggers the expression of cytokines, chemokines, IFNs, and TNF. We therefore analyzed their expression in biopsies classified as TCMR or Normal. All 4 inflammatory caspases including caspase 1, the proinflammatory IL-1–converting enzyme that cleaves pro-IL-1β into active IL-1β, were higher in TCMR (Figure 2, A and B). There was a positive association between caspase 1 and TLR2 or NLRP3 (Figure 2, C and D). Of the apoptotic caspases, caspase 9, the essential initiator caspase required for apoptosis signaling through the mitochondrial pathway, was significantly decreased in TCMR (FC –1.45, P-FDR < 0.001), whereas caspase 3, the apoptosis-executioner caspase that is directly activated by granzyme B and FasR, was significantly increased in TCMR (FC 1.94, P < 0.0001). The associations between the NLRs and the caspases are shown in Supplemental Table 6. T cell differentiation chemokines CXCL1 and CCL2; T cell chemotactic chemokines CXCL9, CXCL10, and CXCL11; NK cell chemotactic chemokines CCL3, CCL4, and CCL5; inflammatory cytokines including IL1B and IL6; IFN-α receptors, IFN-γ, IFN-ε, and TNF-α, were all higher in TCMR biopsies compared with Normal biopsies (Figure 3, A–C).
Figure 2. Intragraft abundance of mRNAs encoding caspases in human kidney allograft biopsies.
mRNA abundance of inflammatory (A) and apoptosis caspases (B) in TCMR and Normal. The number in each box plot is the median value of mRNA abundance as fragments per kilobase of exon per million reads mapped (FPKM), which normalizes the RNA sequencing data for sequencing depth (per million scaling factor) and the gene length (per kilobase scaling factor) and was assessed with Cufflinks. The fold change (FC) values shown beneath the x axis and the P-FDR were calculated using edgeR. Signaling through PRRs activates caspases. There were strong associations between several PRRs and caspases. As an example, the association between mRNA expression levels of the key inflammatory caspase, caspase 1, and TLR2 (C) or NLRP3 (D) are shown. The strength of the association is expressed as the coefficient of determination (r2). NLRP3 is a key component of the inflammasome, which is a multiprotein complex of receptors and sensors that regulate the activation of caspase 1 and induce inflammation.
Figure 3. Intragraft abundance of mRNAs encoding cytokines, chemokines, IFNs, and TNF in human kidney allograft biopsies.
mRNA abundance of cytokines (A), chemokines (B), IFNs and TNF (C) in TCMR and Normal. Signaling through the PRRs results in the activation of NF-κB, IFN regulatory factors, and NFATC transcription factors, which together with caspases trigger the expression of cytokines, chemokines, IFNs, and TNF.
Our computational analysis identifying increased expression of genes downstream of PRR signaling suggests a mechanism for the inflammatory milieu and T cell activation conducive to TCMR.
Secreted PRRs and complement system proteins in TCMR.
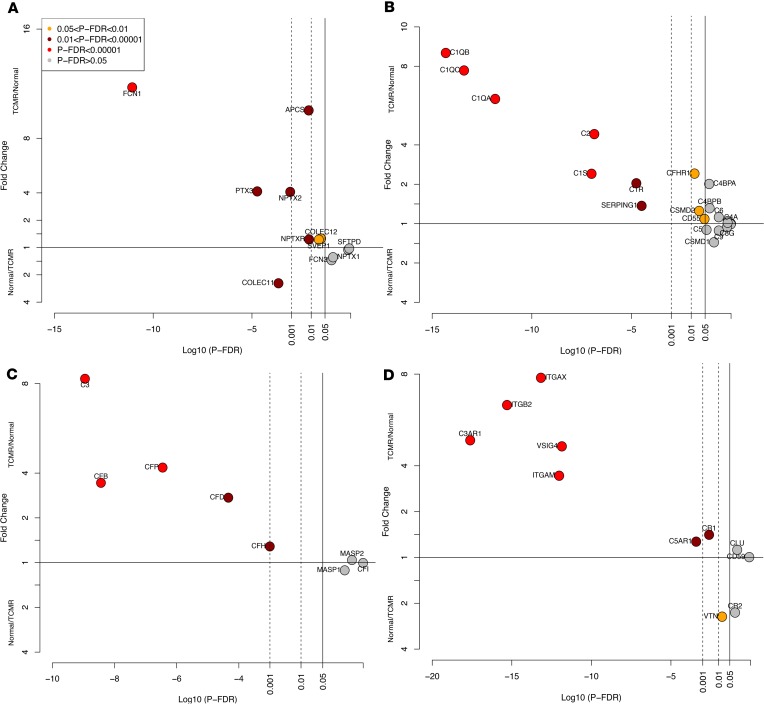
Secreted PRRs activate complements, especially the classical and lectin pathways (24). We therefore analyzed the expression of mRNAs for secreted PRRs and complements in TCMR and Normal biopsies. Among the 12 mRNAs encoding secreted PRRs expressed in the kidney allografts, the intragraft abundance of 8 mRNAs was significantly different between TCMR and Normal biopsies. Of the 8 mRNAs, 7 were higher in TCMR (Supplemental Table 7A).
Abundance of mRNA for C1Q, the key activator of the classical complement pathway, was higher in TCMR compared with Normal biopsies. All 3 complement activation pathways (classical, alternative, and lectin) culminate in C3, the central step in complement activation. mRNA levels of C3 were higher in TCMR (FC 8.2, P-FDR < 0.0001). The receptor for C3, C3AR1, was significantly higher in TCMR (FC 5.11, P-FDR < 0.0001). Of the classical pathway C3 convertase proteins C4 and C2, C2 was expressed at very low levels, while C4 was significantly higher in TCMR than Normal. mRNA levels of alternative pathway complement proteins CFB, CFD, CFH, and CFP, were higher in TCMR but the key lectin pathway activating proteins, MASP1 and MASP2, were not different between TCMR and Normal. Abundance of C3 receptor (C3AR1), C5 receptor (C5AR1), as well as C3 regulators CR1 (CD35), ITGAX (CD11c), ITGB2 (CD18), ITGAM (CD11b), and VSIG4 were higher in TCMR. Downstream of C3, the mRNAs levels of C5, C6, C7, and C8, all components of the membrane attack complex, were not different between TCMR and Normal, and all except C7 were expressed at very low levels. The terminal component C9 was not expressed in the kidney allograft (Figure 4, A–D, and Supplemental Table 7, B–D).
Figure 4. Differential intragraft abundance of mRNAs encoding secreted PRRs and the complement system proteins in human kidney allografts.
mRNA abundance of secreted PRRs (A), classical complement pathway proteins (B), alternative and lectin pathway proteins (C), and complement inhibitors and receptors (D) in TCMR and Normal.
Our analyses reveal the higher abundance of secreted PRRs and several key classical and alternative pathways complement components in TCMR. However, the low level of expression and lack of significant difference between TCMR and Normal of the downstream complement components C5 through C9 suggests the absence of canonical cascade reaction of the complement system in TCMR.
Pathogen-associated molecular patterns in TCMR.
As PRRs were higher in TCMR, we investigated whether the ligand for PRRs, pathogen-associated molecular patterns (PAMPs), the microbe-specific small-molecule motifs recognized by PRRs, were also higher in TCMR. None of the 34 kidney recipients had symptoms/signs of active infection at the time of kidney allograft biopsy or in the immediate pre- and postbiopsy period. Our analysis using Kraken (25), a bioinformatics tool that aligns the RNA sequencing data with metagenomic annotation databases, identified 287 microbiota (235 bacteria and 52 viruses) in the 34 kidney biopsies. The total counts of the 287 microbiota transcripts were lower in TCMR biopsies compared with Normal biopsies (log2CPM 5.12 vs. 5.69, P < 0.0001, Mann-Whitney test).
Our analyses showing that the pathogen counts were not increased in TCMR suggest that PAMPs were unlikely to be responsible for the increased expression of PRRs during an episode of TCMR.
Damage/danger–associated molecular patterns in TCMR.
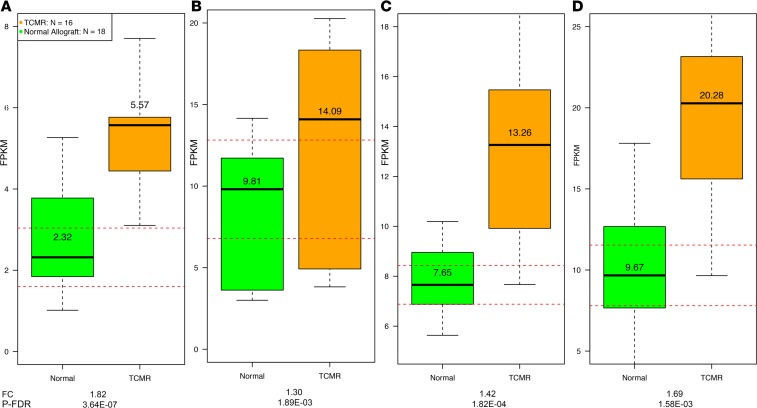
Damage/danger–associated molecular patterns (DAMPs) are ligands for PRRs (26). We analyzed the expression pattern of mRNAs encoding DAMPs in the biopsies. Among the 27 mRNAs for the DAMPs expressed in the kidney allograft, the abundance of 17 mRNAs was significantly different between TCMR and Normal biopsies (Supplemental Table 8). Sensors of cellular and mitochondrial damage such as FPR1 (FC 12.4, P-FDR < 0.0001), a high-affinity receptor for N-formyl-methionyl peptides, and its 2 low-affinity receptor isoforms FPR2 (FC 9.3, P-FDR < 0.0001) and FPR3 (FC 6.2, P-FDR < 0.0001) were both higher in TCMR biopsies. Bacteria and mitochondria are the only source for N-formyl peptides in nature (27). CD44, a receptor for the extracellular matrix component hyaluronan (HA), was higher (FC 2.6, P-FDR < 0.0001) in TCMR. Tissue injury leads to extracellular matrix breakdown, including the degradation of HA, which signals through a receptor complex of TLR4, CD44, and LY96, to activate NF-κB. P2RX7, a potent activator of NLRP3 inflammasome and a receptor for extracellular ATP, was higher in TCMR (FC 5.2, P-FDR < 0.0001). Sensing of extracellular ATP by P2X7R promotes calcium influx and activation of the intracellular ATP export channel PANX1 (28). mRNA expression levels of PANX1 were higher (FC 1.5, P-FDR < 0.01) in TCMR. Low intracellular ATP together with sustained calcium influx activates AMP-activated protein kinase (AMPK), which acts as a sensor of cellular energy status (29). Pathway enrichment analysis showed that the AMPK signaling pathway was enriched in TCMR (P-FDR < 0.0001). Abundance of hypoxia-inducible factor 1α (HIF-1α), a transcription factor that functions as a master regulator of oxygen homeostasis, was higher (FC 1.8, P-FDR < 0.0001) in TCMR together with CYBB (NOX2; FC 6.4, P-FDR < 0.0001), a critical component of the membrane-bound oxidase of phagocytes that generates superoxide and is activated by hypoxia. Thioredoxin is a thiol-oxidoreductase that is a major regulator of cellular redox signaling protecting cells from oxidative stress. TXNIP, thioredoxin-interacting protein, is a DAMP that was higher in TCMR (FC 1.8, P-FDR < 0.0001). It inhibits the antioxidative function of thioredoxin, resulting in the accumulation of reactive oxygen species (ROS) and cellular stress (Figure 5, A–D).
Figure 5. Differential intragraft abundance of mRNAs encoding DAMPs in human kidney allografts.
(A) mRNA abundance of DAMPs and DAMP sensors in TCMR and Normal. (B–D) Scatterplots depict the relation between the DAMPs, P2RX7 (a receptor that detects extracellular ATP), and a PRR, TLR4 (B); FPR3, a receptor for N-formyl-methionyl peptides, and a PRR, TLR8 (C); and HIF-1α, a transcription factor that functions as a master regulator of oxygen homeostasis, and a PRR, TLR5 (D).
Taken together, our analysis suggests that the ligands for signaling through PRRs in TCMR are more likely to be DAMPs rather than PAMPs, and the higher DAMPs in TCMR compared with Normal is likely due to cellular and mitochondrial stress.
Indicators of DNA damage in TCMR.
As the source of DAMPs is cellular stress and damage, we investigated for markers of DNA damage in TCMR. The 3 key DNA damage sensors, ATM and PRKDC (DNA-PK), which recognize DNA double-strand breaks, and ATR, which recognizes DNA replication stress (30), were higher in TCMR (Figure 6, A and B). Changes in chromatin structure can lead to activation of ATM at the damage site and recruitment of proteins involved in DNA repair. Of the 14 proteins that are described to participate in repair of double-strand breaks and expressed in the kidney allograft, 9 were higher in TCMR. Of the 8 proteins that participate in the repair of damage caused by replication stress and expressed in the kidney allograft, 3 were higher in TCMR (Supplemental Table 9, A–C). Of the 9 cytosolic nucleic acid sensors, 7 were higher in TCMR. TP53 (p53), a transcription factor that is involved in DNA repair (31), and CDKN1A (P21), which is activated by p53 and which inhibits cellular proliferation in response to DNA damage, were higher in TCMR (FC 1.4, P-FDR < 0.001, and FC 1.7, P-FDR < 0.01, respectively; Figure 6, C and D). By gene-set testing, DAMPs and DNA damage and nucleic acid sensors were upregulated in TCMR (Supplemental Table 10).
Figure 6. Kidney allograft expression of mRNAs encoding DNA damage sensors in TCMR and Normal.
Expression of 2 key DNA damage sensors, ataxia telangiectasia mutated (ATM) (A), a serine-threonine kinase that recognizes DNA double strand breaks, and ataxia telangiectasia and Rad3 related (ATR) (B), a serine-threonine kinase that recognizes DNA replication stress, in TCMR and Normal. Another key sensor, PRKDC (DNA-PK), was also increased in TCMR (FC 1.22, P < 0.05). All 3 DNA damage checkpoint proteins activate p53 (C), a tumor suppressor protein that activates either the DNA repair pathway or apoptosis pathway. CDKN1A (P21) (D) is a cyclin-dependent kinase inhibitor that is activated by p53, thus linking DNA damage to cell cycle arrest.
Our analysis reveals increased abundance of markers of DNA damage in TCMR, suggesting cell injury and death as the source of DAMPs in TCMR.
DCs and macrophages in TCMR.
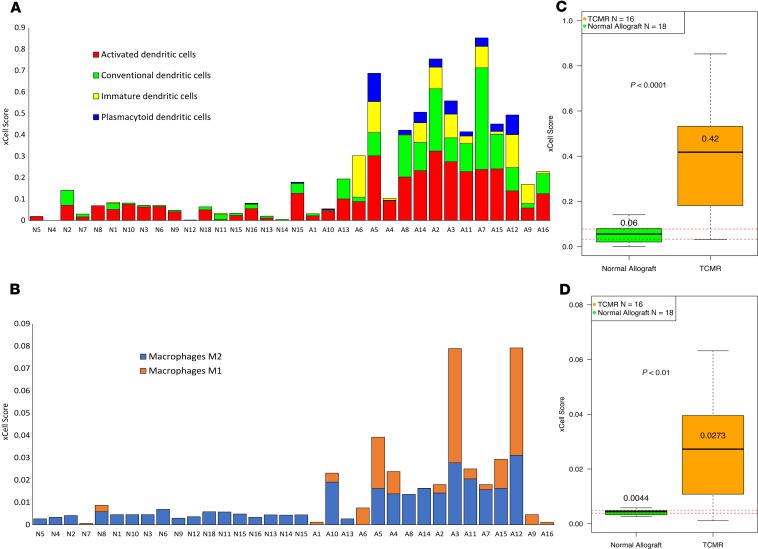
Because DCs and macrophages are the commonest cell types that express PRRs, we evaluated intragraft abundance of genes related to DCs and macrophages. The abundance of mRNA for CD209, a transmembrane receptor expressed on the surface of DCs and macrophages, was significantly higher in TCMR biopsies than Normal biopsies (FC 3.9, P-FDR < 0.0001). Cell-type-enrichment analysis using xCell (32) showed high scores for activated DCs in TCMR compared with Normal (median 0.44 vs. 0.06, P < 0.0001, Mann-Whitney test). Similarly, the total macrophage score was higher in TCMR (0.0273 vs. 0.0044, P < 0.01; Figure 7, A and B).
Figure 7. Cell-type enrichment scores for DCs and macrophages in TCMR and Normal.
Distribution of cell-type enrichment scores for DCs (A) and macrophages (B) in individual TCMR and Normal samples. We used xCell, a bioinformatics tool, to provide an enrichment score for different cell types that allow comparison of cell types across samples (not across cell types within a sample). The y axis represents the xCell enrichment score (not a percentage). The height of each stacked bar column represents the total DC or macrophage xCell score for that sample. The figure also depicts box plots of total DC score (C) and total macrophage score (D) in TCMR and Normal. The difference in total DC score and the total macrophage score between TCMR and Normal were statistically significant by Mann-Whitney test.
Our analysis reveals that the expression of genes encoding DCs and macrophages, the commonest cell types that express PRRs, are increased in TCMR compared with Normal.
Immunosuppressive status in TCMR.
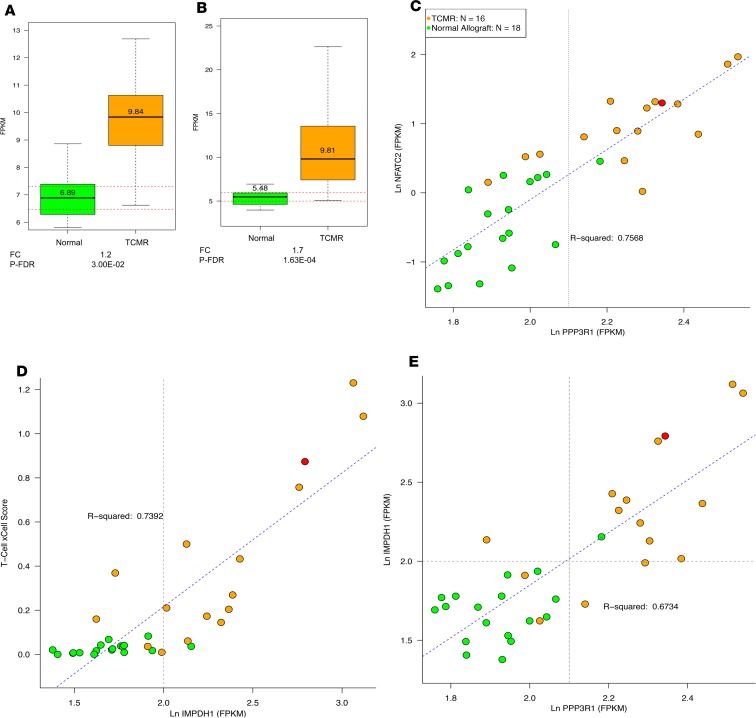
Our unbiased characterization of the intragraft transcriptome of human kidney allografts revealed innate immune system genes to be overexpressed in TCMR compared with Normal biopsies. Immunosuppressive drugs are essential for the prevention of rejection of allografts. Because recipients in the Normal group had stable graft function and normal/nonspecific histopathological findings, we considered them to have an immune system that was adequately suppressed and wondered whether an undersuppressed immune system could be the basis for activation of the innate immune system in TCMR. In order to assess whether the immune system is differentially suppressed during TCMR compared with Normal, we analyzed abundance of mRNA for calcineurin, the primary target of commonly used immunosuppressant tacrolimus. Four of the 5 reported isoforms of calcineurin were expressed in the kidney and mRNA levels of PPP3R1 (regulatory subunit α) were higher (FC 1.2, P-FDR < 0.05) in TCMR compared with Normal biopsies (Figure 8A). The other 3 calcineurin isoforms, PPP3CA (catalytic subunit α), PPP3CB (catalytic subunit β), and PPP3CC (catalytic subunit γ), were also higher in TCMR compared with Normal biopsies but the differences were not statistically significant (Supplemental Figure 3 A). There was a strong positive association (r2 = 0.76) between calcineurin (PPP3R1) and NFATC2 (Figure 8C). The transcription factor NFATC2 regulates T cell activation, differentiation, and development. We examined whether the tacrolimus trough level at the time of biopsy was different between TCMR and Normal groups. The whole-blood tacrolimus trough level was 5.2 ng/mL (median, IQR 4.2–6.7) in TCMR and 7.0 ng/mL (6.0–8.7) in Normal (P < 0.01, Mann-Whitney test).
Figure 8. Kidney allograft expression of mRNAs encoding calcineurin PPP3R1 and inosine-5′-monophosphate dehydrogenase 1 (IMPDH1) in TCMR and Normal.
Intragraft mRNA abundance of calcineurin isoform PPP3R1 (protein phosphatase 3 regulatory subunit B, α) (A), and IMPDH1 (B) in TCMR and Normal. In the box plots, the 2 dotted red lines represent the 95% confidence interval of the Normal group. Calcineurin, a phosphatase made of 2 proteins, CNA and CNB, cleaves phosphate from serine and threonine in proteins. CNA possesses the phosphatase enzymatic activity and binds calmodulin. CNB binds Ca2+ directly and regulates the activity of CNA. Tacrolimus acts as an immunosuppressive drug by binding to and inhibiting calcineurin. We used PPP3R1 mRNA abundance as a marker of calcineurin inhibition. Mycophenolate acts by inhibiting IMPDH, an enzyme that catalyzes the oxidation of inosine monophosphate to xanthosine monophosphate, which is the rate-limiting step in the de novo biosynthesis of guanine nucleotide. We used IMPDH1 mRNA abundance as a marker of mycophenolate action. Scatter plots depict the association between calcineurin isoform PPP3R1 and NFATC2, a key transcription factor necessary to produce the T cell proliferative cytokine IL-2 (C), between IMPDH1 and a T cell score derived by xCell (D), and between PPP3R1 and IMPDH1 (E). We used NFATC2 as a marker of the effect of calcineurin inhibition on T cells. We used T cell xCell score as a marker of the effect of mycophenolate on T cells. One patient in the TCMR group who did not receive tacrolimus is shown in red in the scatter plot.
The commonly used immunosuppressive drug mycophenolate acts by blocking IMPDH, an enzyme that catalyzes the rate-limiting step in the de novo biosynthesis of guanine nucleotide. The mRNA level of IMPDH1 isoform was higher (FC 1.7, P-FDR < 0.001) in TCMR (Figure 8B). IMPDH2 mRNA was not significantly different (Supplemental Figure 3A). There was a strong positive association (r2 = 0.74) between IMPDH1 mRNA and xCell T cell score (Figure 8D), but not between IMPDH2 and T cell score or between IMPDH1 and IMPDH2 (Supplemental Figure 3B). A positive linear association, however, was observed between PPP3R1 and IMPDH1 (r2 = 0.67; Figure 8E).
Our results demonstrating that mRNAs for key targets of clinically used immunosuppressive drugs are in higher abundance in Normal biopsies compared with TCMR biopsies provide a mechanism for the heightened expression of innate immune system genes in biopsies manifesting TCMR.
Intracellular Ca2+ homeostasis in TCMR.
Because Ca2+ and calcineurin/NFAT signaling is mechanistically linked, we evaluated whether Ca2+ homeostasis was altered in TCMR. To maintain cytosolic calcium concentration in the low nM range, the low-affinity, high-capacity sarcoplasmic reticular Ca2+ ATPases (SERCAs) pump Ca2+ into the ER and plasma membrane Ca2+ ATPases (PMCAs) pump Ca2+ out of cells (33). Expression of the SERCA ATP2A3 and the PMCAs ATP2B1 and ATP2B4 was higher in TCMR (FC 2.8, P-FDR < 0.0001, FC 1.3, P-FDR < 0.01, and FC 1.5, P-FDR < 0.001, respectively). Similarly, the expression of Na+/Ca2+ exchangers SLC24A4 and SLC8A3, which pump Ca2+ out of the cell, was higher in TCMR compared with Normal. (FC 3.8, P-FDR < 0.0001 and FC 2.2, P-FDR < 0.05, respectively; Supplemental Table 11).
Among the store-operated Ca2+ channels, which are activated by depletion of ER Ca2+ stores and facilitate Ca2 entry from the extracellular milieu into the cytoplasm, ORAI1 and ORAI2 were higher (FC 1.29, P-FDR < 0.05, and FC 1.35, P-FDR < 0.01, respectively), whereas ORAI3 was reduced in TCMR (FC –1.4, P-FDR < 0.05; Supplemental Table 11). Intragraft abundance of STIM1 and STIM2, the membrane sensors of Ca2+ depletion, was higher in TCMR but the P-FDR was >0.05. The mRNA expression of ITPR3, which facilitates release of ER-stored Ca2+ into the cytoplasm, was increased (FC 1.9, P < 0.0001) in TCMR compared with Normal (Supplemental Table 11).
Mitochondrial calcium uniporter (MCU), the primary source of mitochondrial uptake of Ca2+ from the cytosol, was increased in TCMR but the difference was not statistically significant (FC 1.2, P-FDR = 0.08). However, its dominant-negative paralog MCUB was significantly increased in TCMR (FC 3.94, P-FDR < 0.0001). The regulators of MCU, MICU1 and MICU2, were reduced in TCMR (FC –1.7, P-FDR < 0.0001, and FC –1.3, P-FDR < 0.001; Supplemental Table 12). The mRNA expression of TLR2 and MCUB was strongly associated (r2 = 0.86). There was also a positive association between mRNA expression of calcineurin PPP3R1 and Ca2+ pumps/channels (Supplemental Figure 4, A–F).
Our analyses demonstrating that mRNAs encoding key intracellular, ER, and mitochondrial Ca2+ channels/pumps are increased in TCMR compared with Normal biopsies indicate calcium-dependent immune activation during TCMR.
Increased ER stress and oxidative stress in TCMR.
As increased Ca2+ flux can cause ER stress and oxidative stress, we evaluated whether the abundance of mRNAs encoding proteins implicated in cellular stress responses is increased in TCMR. ER stress activates the unfolded protein response (UPR). The major protein sensors on the ER membrane that initiate UPR, ERN1 (IRE1) and EIF2AK3 (PERK), were higher in TCMR (FC 1.5, P-FDR < 0.001, and FC 1.6, P-FDR < 0.0001, respectively). ERO1A (ERO1L), an oxidoreductase involved in the production of ROS, was higher in TCMR (FC 1.5, P-FDR < 0.0001). The UPR activates both the adaptive and proapoptotic pathways simultaneously. Interestingly, DDIT3 (CHOP), the master regulator of ER stress–induced apoptosis, was reduced in TCMR (FC –1.3, P-FDR < 0.05). PPP1R15A, a phosphatase that reverses the shutoff of protein synthesis initiated by stress-inducible kinases and facilitates recovery of cells from stress, was increased in TCMR compared with Normal (FC 1.2, P-FDR < 0.05; Supplemental Table 13).
NADPH oxidases (NOX) are the key producers of ROS within cells (34). Phagocytic NOX2 (CYBB) together with neutrophil cytosolic factor 1 (NCF1), NCF2, and NCF4, which form the NADPH oxidase complex, were higher in TCMR. The infiltrating macrophages are likely to be the predominant source of NOX2, though it is also expressed in kidney cells. Expression levels of the cytochrome c oxidase COX4I1, the terminal oxidase in mitochondrial electron transport, were reduced together with downregulation of the electron transport chain. SOD1, the cytosolic isoform of superoxide dismutase which converts O2– to O2 and H2O2, and CAT which converts H2O2 to O2 and H2O, were low in TCMR compared with Normal (FC –1.7, P-FDR < 0.001 and FC –1.7, P-FDR < 0.001, respectively). Interestingly, NOX4, the NOX isoform with highest expression levels in the kidney and produces H2O2 constitutively and does not require other cytosolic oxidases for its activation, was lower in TCMR compared with Normal (FC –2.5, P-FDR < 0.01; Supplemental Table 14).
Our analysis revealing that ER stress and cellular oxidative responses are increased in TCMR compared with Normal biopsies is consistent with increased abundance of mRNAs encoding channels/receptors conducive to an increased intracellular Ca2+ turnover during TCMR.
Mitochondrial stress and energy metabolism in TCMR.
Because ER stress is linked to mitochondrial stress and because mitochondrial products were the major DAMPs in TCMR, we investigated mitochondrial stress and energy metabolism during TCMR.
Complex 1 of the electron transport chain consists of 46 subunits and catalyzes a 2-electron oxidation of NADH by ubiquinone. Of the 39 subunits expressed in the kidney, abundance was significantly low in 34. Abundance of all 4 subunits of complex II, which contains a bound flavin adenine dinucleotide (FAD) cofactor, was lower. mRNA levels of all subunits of complex III, which catalyzes the oxidation of 1 molecule of ubiquinol and 2 molecules of cytochrome c, were low. Similarly, the abundance of complex IV, which transfers electrons to oxygen while pumping protons across the membrane, was lower in TCMR compared with Normal.
Of the 39 differentially expressed glycolysis and gluconeogenesis genes, 30 were reduced in TCMR. mRNA levels of all 3 hexokinase isoenzymes (HK1, HK2, and HK3), which catalyze the first step of glycolysis to produce glucose-6-phophate, were higher in TCMR (FC 1.4, P-FDR < 0.05, FC 4.7, P-FDR < 0.0001, and FC 8, P-FDR < 0.0001, respectively). However, the abundance of mRNA for glucose-6-phosphate dehydrogenase (G6PD), a major producer of the electron donor NADPH in defense of oxidizing agents and the rate-limiting enzyme of the oxidative pentose-phosphate pathway, was not different between the 2 groups. All 3 key proteins of the pyruvate dehydrogenase complex were reduced in TCMR. Lactate dehydrogenase A (LDHA), which converts pyruvate to lactate, was higher (FC 1.38, P-FDR < 0.05), while LDHB, which converts lactate to pyruvate, was reduced (FC –1.64, P-FDR < 0.01) in TCMR (Supplemental Table 15).
All major citric acid cycle enzymes and enzyme complexes were reduced in TCMR (Supplemental Table 16). Only carnitine shuttle isoform CPT1B, which moves fatty acids (acyl-CoA) into the mitochondria, was significantly increased in TCMR (FC 1.7, P-FDR < 0.05), while nuclear protein CPT2, a mitochondrial membrane protein that is transported to the mitochondrial inner membrane and oxidizes long-chain fatty acids, was lower in TCMR (FC –2, P-FDR < 0.001). MLYCD, which inhibits the transport of fatty acyl CoAs into mitochondria, and PANK1, a key regulator of coenzyme A, were reduced in TCMR. Enzymes involved in β-oxidation (initial breakdown of free fatty acids to acetyl-CoA) that include acyl-CoA dehydrogenase, enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase, and acetyl-coenzyme A acetyltransferases, were reduced in TCMR.
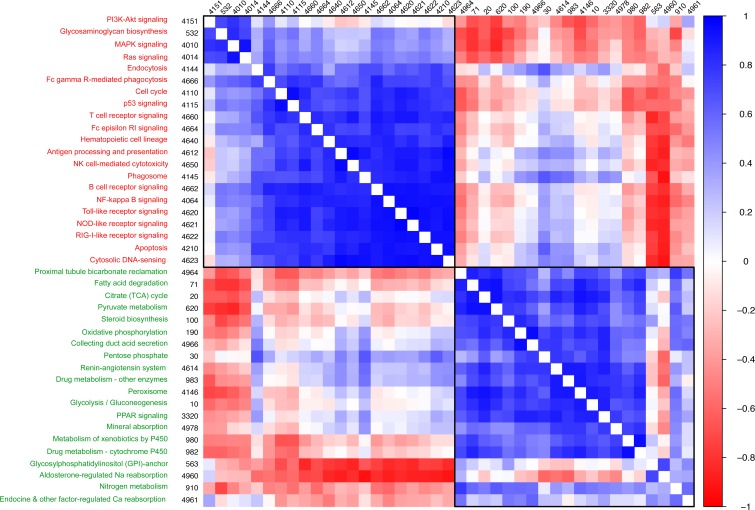
Of the 2 mitochondrial uncoupling proteins expressed in the kidney, UCP2 was significantly increased in TCMR compared with Normal biopsies (FC 3.7, P-FDR < 0.0001), while UCP3 was not different (FC 1.2, P-FDR = 0.3). Furthermore, mRNA expression of SIRT3, SIRT4, and SITR5, which are localized in the mitochondria and regulate the function of the TCA cycle, fatty acid oxidation, and electron transport chain, were significantly decreased in TCMR (FC –1.4, P < 0.0001, FC –1.9, P < 0.0001, and logFC –1.3, P < 0.01, respectively). We identified 102 mitochondrial genes not directly involved in oxidative phosphorylation or fatty acid β-oxidation: 77 ribosomal proteins, 18 mitochondria-encoded tRNAs, 2 mitochondrial encoded rRNAs, and 5 translocases of outer mitochondrial membrane complex. mRNA expression was low in 55 ribosomal proteins, 7 tRNAs, 2 rRNAs, and 5 translocases in TCMR. A clustering analysis of 41 significant energy metabolism and immune response pathways in the 16 TCMR samples is shown in Figure 9.
Figure 9. Correlation clustering of enriched energy metabolism and immune response pathways in TCMR.
Hierarchical correlation clustering of 41 statistically significant enriched immune response and energy metabolism pathways in TCMR. Pathway names (red, immune response pathways; green, energy metabolism pathways) and numbers correspond to the KEGG pathway database. We used GAGE for pathway enrichment analysis. For a given pathway, the GAGE algorithm tests whether specific gene sets are significantly differentially expressed (each TCMR vs. all Normal) relative to the background whole gene set of 16,381 genes (each TCMR vs. all Normal) that we identified, using a rank-based 2-sample t test. For each gene set, a global P value is derived in a meta-test on the negative log sum of all P values from the individual TCMR vs. Normal comparisons. The stat mean is the mean of the individual statistics from multiple gene-set tests. Among the TCMR samples, the relationship of the stat mean between any 2 pathways is represented by the correlation coefficient (r) value. In the heatmap above, each square is the correlation coefficient, adjusted for multiple comparisons, of the 2 pathways. If the correlation coefficient is positive (blue), then both pathways are concordant (both are either up- or downregulated). If the correlation coefficient is negative (red), then both pathways are discordant (one is up- and the other is downregulated).
Our analyses showing reduced glycolysis with reduced pyruvate substrate available for mitochondrial respiration in TCMR, and the downregulation of the TCA cycle, oxidative phosphorylation, and fatty acid β-oxidation together with reduced ribosomal proteins, tRNAs, rRNAs, and translocases are all indicative of mitochondrial dysfunction in TCMR compared with Normal biopsies.
PRRs and T cell costimulatory molecules in TCMR.
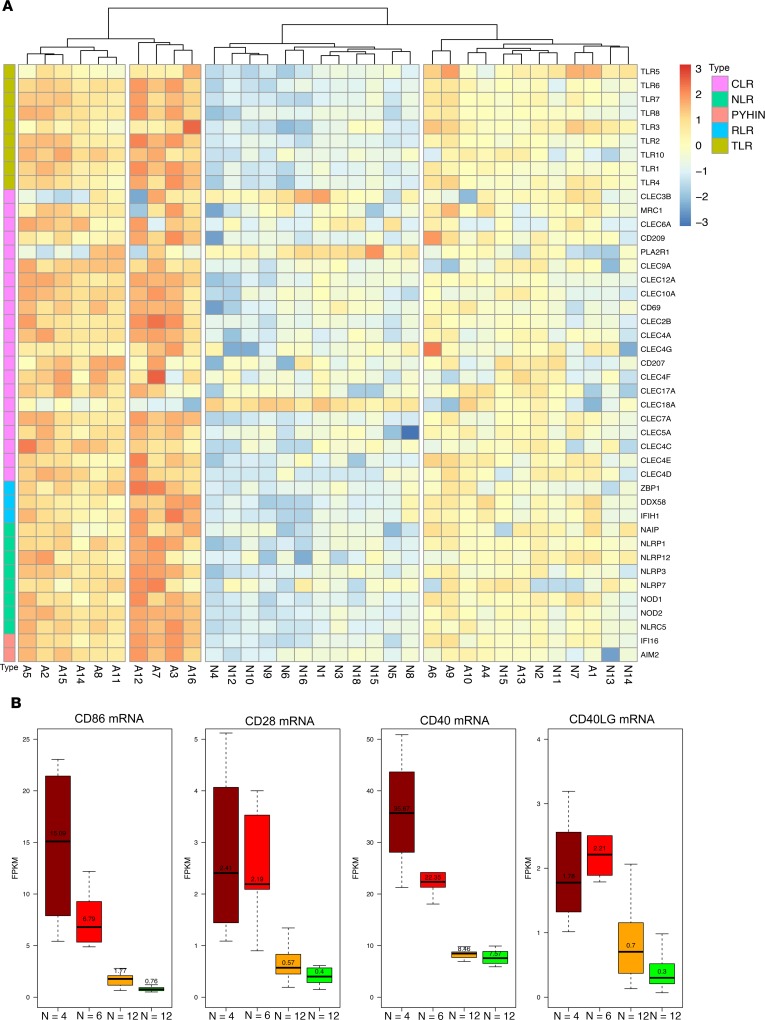
Because T lymphocytes are the primary effector cells in TCMR, and because signaling through PRRs can culminate in an inflammatory microenvironment conducive to the costimulatory signaling of T cells by the cells of the innate immune system, we assessed the relationship between the intragraft abundance of mRNAs for PRRs and the intragraft abundance of T cell costimulatory molecules CD86, CD28, CD40, and CD40LG. Clustering analysis based on the expression strength of mRNA for PRRs showed that the 34 kidney allograft samples clustered into 4 subgroups (Figure 10A). We compared the expression of T cell costimulatory molecules CD86, CD28, CD40, and CD40LG among these 4 subgroups. The group with the highest expression of PRRs had the highest expression of costimulatory molecules, and the group with the lowest expression of PRRs had the lowest expression of costimulatory molecules (Figure 10B).
Figure 10. Relationship between mRNA abundance of PRRs and T cell costimulatory molecules in TCMR and Normal.
By hierarchical clustering analysis of the differentially expressed PRRs (A), the 34 kidney allografts (TCMR: A1–A16, Normal: N1–N18) separate into 4 groups. We labeled the groups as dark red (highest gene expression, middle left, N = 4), red (left, N = 6), orange (right, N = 12), and green (lowest gene expression, middle right, N = 12). Signaling through PRRs culminates in costimulation of T cells. Box plots (B) depict the expression of T cell costimulatory molecules CD86 (B7-2, expressed on APCs and serves as a ligand for 2 T cell proteins, CD28 and CTLA4), CD28 (T cell surface protein that provides signals needed for T cell activation and survival), CD40 (present on APCs and required for its activation), and CD40LG (present on T cells and serves as a ligand for CD40) in the 4 groups.
Our data demonstrating that the expression of PRRs is positively associated with the expression of T cell costimulatory molecules suggest a molecular basis for plenary T cell activation during an episode of TCMR.
Cell death in TCMR.
Because T cell activation eventually results in target cell death, we assessed the major pathways of cell death in TCMR.
We studied the apoptosis, necroptosis, autophagy, and mitophagy pathways. Apoptosis and necroptosis pathways were significantly enriched in the upward direction (increased) in TCMR (P-FDR 3.46 × 10–15 and 9.20 × 10–15, respectively). The autophagy pathway was not statistically different between TCMR and Normal. The mitophagy pathway was significantly enriched in the downward direction (decreased) in TCMR (P-FDR 1.146 × 10–2). The key regulators of mitophagy, PINK1 and PRKN, were lower in TCMR (FC –1.66, P-FDR < 0.0001, and FC –1.59, P-FDR < 0.001, respectively).
Our analysis reveals that both apoptosis and necroptosis pathways of regulated cell death are active in TCMR. However, cytoprotective autophagy and mitophagy pathways are not increased in TCMR.
Validation of the increased abundance of innate immunity genes in TCMR.
We selected a panel of key innate immune system mRNAs that were increased in TCMR and sought to confirm the increased abundance of their respective proteins. Immunostaining of TCMR and Normal biopsies for CD68, TLR4, NLRP1, and CASP1 proteins confirmed their increased abundance in TCMR (Supplemental Figure 5, A–H).
Gene expression and time from transplantation to biopsy.
As the median time from transplantation to biopsy was different in TCMR and Normal, we evaluated whether the higher abundance of key mRNAs of the innate immune system in TCMR was a reflection of the disease phenotype (TCMR vs. Normal) or the time from transplantation to biopsy (early vs. late).
There was no association between time from transplantation to biopsy and the abundance of a select panel of mRNAs including TLR2 (r2 = 0.05), TLR8 (r2 = 0.06), MYD88 (r2 = 0.03), CLEC7A (r2 = 0.04), CLEC10A (r2 = 0.07), NLRP1 (r2 = 0.04), or CASP1 (r2 = 0.03; Supplemental Figure 6, A–G), as well as the macrophage score (r2 = 0.03) or DC score (r2 = 0.02; Supplemental Figure 6, H–I).
We next analyzed the RNA sequencing data of 4 native nontransplant kidneys from a public database (proteinatlas.org) and compared the expression of the above mRNAs with that of TCMR and Normal. The mRNA expression of TLR2, TLR8, MYD88, CLEC7A, CLEC10A, NLRP1, and CASP1, as well as the macrophage and DC scores in the Normal group, were similar to the native nontransplant kidneys and were lower in abundance compared with TCMR (Supplemental Figure 7, A–I).
Our analysis suggests that the intragraft expression of innate immune system genes is independent of time from transplantation. The differential intragraft expression is due to their higher abundance in TCMR rather than lower abundance in Normal, compared with native nontransplant kidneys.
Discussion
In this report, the first of its kind to our knowledge, we used RNA sequencing of kidney allograft biopsies and found that the mRNAs of key players of the innate immune system are in higher abundance in human kidney allograft biopsies displaying TCMR compared with biopsies categorized as Normal. Figure 11 is a formulation of the contributions and consequences of innate immune system activation during TCMR. Research elucidating the role of innate immunity in organ transplantation is primarily focused on the inflammatory milieu during the peritransplant period, including cytokine release and complement activation during brain death, mechanical injury during organ removal, and ischemia-reperfusion injury (8, 10, 35). AR seldom occurs in the immediate posttransplant period; the activation of the innate immune system in AR vis-à-vis normal/stable graft function remains unclear. Understanding the role of innate immunity in AR is essential to better define the molecular events that lead to alloimmune inflammation and injury and facilitate the development of therapies that may improve responses to currently available therapies that primarily target the adaptive immune system.
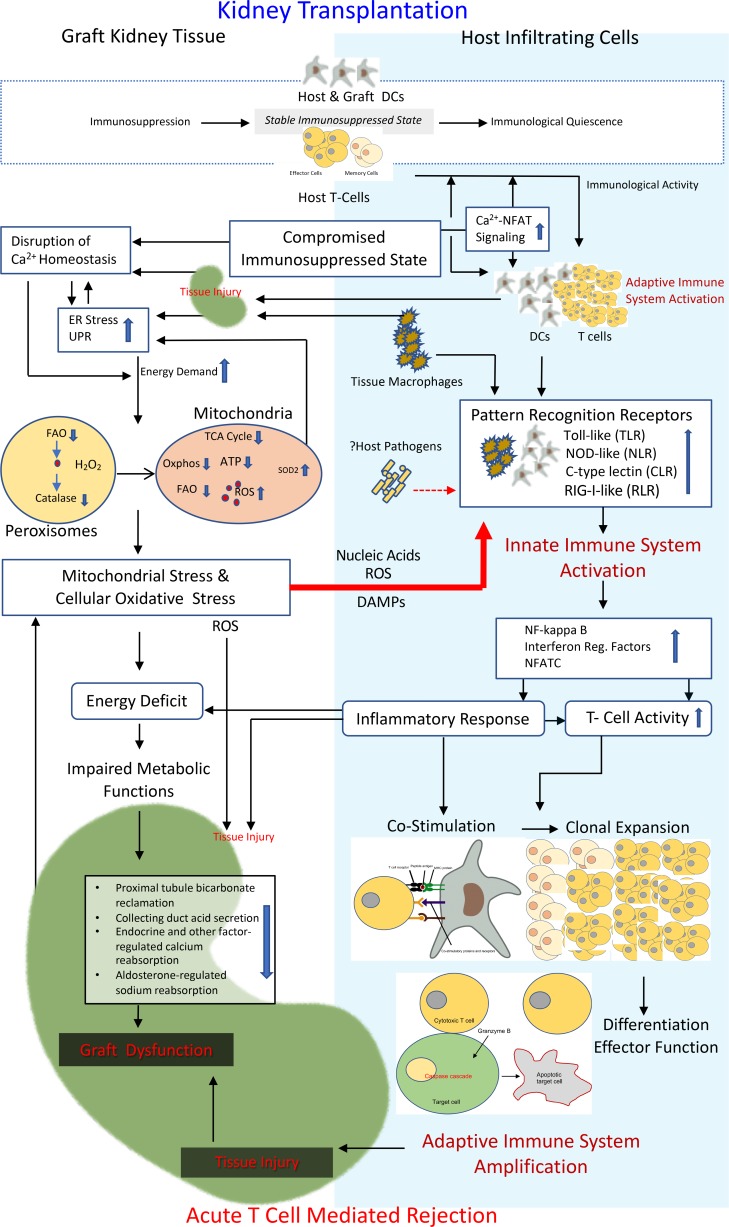
Figure 11. A model for innate immune system activation in TCMR.
At transplantation, allograft resident donor DCs migrate to host secondary lymphoid organs and function as antigen-transporting cells. Host T cells recognize intact donor HLA on donor DCs (direct allorecognition) or host DCs (semidirect allorecognition). Donor HLA can also be internalized by host DCs, processed, and presented as peptide fragments along with host HLA; self-MHC-restricted host T cells then recognize the peptide fragments presented by the host DCs (indirect allorecognition). Before antigen presentation, DCs, the key component of innate immune system, need to transform from an immature antigen-capture phenotype to a mature T cell–activation phenotype. Peritransplant tissue injury and ensuing inflammatory microenvironment cause DC maturation and priming of the adaptive immune system, resulting in clonal expansion and differentiation of T cells. Immunosuppressive drug therapy keeps the immune system relatively quiescent. Any compromise to this stable immunosuppressed state (Figure 8) activates alloreactive T cells that cause tissue injury, probably at a low level. In this context, increased cellular calcium flux (Supplemental Tables 11 and 12) results in increased ER stress (Supplemental Table 13). The high energy demand leads to increased cellular oxidative stress (Supplemental Table 14) and mitochondrial stress, resulting in reduced cellular peroxisomes and mitochondrial energy production (Supplemental Tables 15 and 16), and generation of reactive oxygen species. Products of cellular damage (DAMPs; Supplemental Tables 8–10) are sensed by PRRs (Supplemental Tables 2–7), predominantly expressed on DCs and macrophages, leading to a full-fledged activation of the innate immune system. The resultant inflammatory response generates costimulatory signals (Figure 10) for T cell clonal expansion and differentiation. The energy deficit compromises several key metabolic pathways within the graft (Figure 9), resulting in graft dysfunction. Tissue injury and graft dysfunction further increases the cellular stress, thus amplifying the feed-forward loop.
We first studied PRRs, the primary sensors of danger/damage that enable maturation of antigen-presenting cells (APCs) and subsequent activation of the adaptive immune response. Our findings of increased expression of PRRs in TCMR compared with Normal, as well as the strong association between TLRs and MYD88, a key intracellular signaling molecule for TLRs, are consistent and extend the findings in preclinical models (7, 8, 35). Skin grafts transplanted between mice mismatched for the male minor antigen are not rejected if both donor and recipient lack MYD88 (36). TLR3 signaling does not require MYD88 but involves MAPK (37). We observed a statistically significant association between TLR3 and MAPK but not with MYD88. NLRP3, the best-characterized inflammasome, was increased in TCMR. PRRs are also expressed in kidney parenchymal cells and play a role in a variety of kidney diseases (38). In a recent study of kidney recipients, an NLRP3 gain-of-function single-nucleotide variant in donors was associated with an increased risk of AR, while a loss-of-function variant in the recipient was associated with a decreased risk of AR in the first year after kidney transplantation (39).
As signaling through PRRs culminates in an inflammatory microenvironment, we assessed cytokines, chemokines, IFNs, and TNF. Our findings of increased CXCL10 and CXCL9 in TCMR is supported by clinical data from multicenter studies of increased urinary CXCL9 and CXCL10 mRNA and protein as biomarkers diagnostic of TCMR (40, 41).
We evaluated complements, which are activated by secreted PRRs. The liver is the main source of circulating complement proteins. Our findings of increased intragraft expression of complements suggest their production in the allograft itself. Extrahepatic production of complement is increasingly recognized, especially by the kidney cells and the immune cells (42). In human kidneys, complement component C4 mRNA amount has been found comparable to that of the liver (43). C1q, C1r, and C1s are synthesized in cells of myeloid origin, including macrophages and DCs, and are regulated by IFN-γ. C3 is produced by a variety of cells including the proximal tubular cells. Various immune cells secrete complements in their own microenvironment (42). Emerging data indicate that immune cell–derived complement activation physiologically regulates immune cell survival and proliferation, modulating the strength and phenotype of adaptive T cell responses (44).
We assessed if PAMPs are the primary drivers of the increased PRRs. Our observation of the lack of clinical symptoms or signs of active infection and the absence of significant increase in tissue microbiota in TCMR compared to Normal is consistent with literature showing that the principle triggers of inflammation after organ transplantation are likely to be endogenous rather than microbial in origin (45). However, the role of human microbiota in modulating the immune system is emerging and gut, urinary, or tissue microbiota as initiators or modulators of the immune response directed against the allograft need to be further explored (46).
We evaluated whether DAMPs are the primary drivers of increased PRRs in TCMR. Our analyses show that activation of the innate immune system through PRR signaling is not predominantly microbe driven but DAMPs driven. The major source of DAMPs are DNA and nucleic acid fragments, both of nuclear and mitochondrial origin (47). We found evidence for DNA damage and repair in TCMR. This was consistent with clinical studies of TCMR that show positive staining for DNA fragmentation by TUNEL, tissue transglutaminase II, a marker of DNA damage, and 8-hydroxy-2′-deoxyguanosine, an indicator of oxidative injury of nucleic acids (48). We did not find HMGB1, a well characterized nuclear protein DAMP that plays an important role in ischemia-reperfusion injury of the kidney, to be higher in TCMR compared to Normal biopsy group (49). Similarly, HSP, another well-characterized DAMP, was not higher in TCMR than in the Normal biopsy group. Together, our data suggest that the inciting event for TCMR is less likely to be the immune activation in the peritransplant period due to ischemia-reperfusion injury.
We assessed the primary cells of the innate immune system that express PRRs, the DCs, and found increased DCs in TCMR. At the time of transplantation, the donor DCs that accompany the graft migrate to secondary lymphoid organs of the host and function as APCs (50). That the median time from transplantation to TCMR was 12 months in our study raises interesting questions regarding the origin of intragraft DCs during an episode of TCMR. Host and recipient DCs prime the adaptive immune response by 3 ways: direct allorecognition, semidirect allorecognition, and indirect allorecognition (51). In an ear-skin-graft model, intravital imaging found donor DCs to emigrate rapidly from the graft and were replaced by host CD11b+ mononuclear cells (52). The current finding of increased DCs in TCMR is supported by clinical data from 105 kidney recipients, including 53 with rejection, that showed independent association between densities of DCs and T cells in allograft biopsies, and poor allograft outcomes (53). The role of DCs in priming the immune response to the transplanted organ is well recognized. Recent preclinical studies have extended this fundamental paradigm by showing that DCs also sustain the alloimmune response outside the secondary lymphoid tissues in the graft (54). Our observation of immature DCs in human TCMR compared with Normal biopsies provides support and extends the elegant preclinical observation that priming of T cells occurs in the allograft as well. The role of APCs is also played by macrophages (8, 11). Our finding of increased macrophages in TCMR is consistent with immunohistochemical data demonstrating increased CD68+ macrophages in allograft biopsies of kidney recipients with TCMR (4, 55). We found increased M1 macrophages in TCMR, which are activated by IFN-γ, are proinflammatory, and act as an APC (56). Interestingly, all biopsies had M2 macrophages that are involved in tissue repair and remodeling (57).
Because the immune system must be suppressed to prevent rejection and drugs are the common means of achieving this, we asked the question whether the drug-induced immunosuppressed state was compromised in TCMR. In this study, whole-blood tacrolimus trough levels were significantly lower in the patients with TCMR than in patients with Normal biopsies. We found that the abundance of mRNA for calcineurin regulatory subunit, PPP3R1, is higher in TCMR compared with Normal. Although mononuclear cell NFAT-regulated gene expression as a surrogate of calcineurin inhibition has been studied as a pharmacodynamics monitoring tool (58), calcineurin mRNA has not been measured in kidney recipients. Calcineurin is expressed in various tissues in humans, with maximal expression in the brain (59). However, as the effect of tacrolimus on calcineurin mRNA is not known, we compared the calcineurin mRNA expression of the 4 native nontransplant kidneys. The levels of 4 calcineurin-isoform mRNAs in native nontransplant kidneys were higher compared with kidney graft recipients with normal biopsies. This provided reassurance that kidney tissue calcineurin mRNA in transplant recipients in the Normal group was indeed decreased compared with native nontransplant kidneys. Similarly, we found IMPDH1 mRNA to be increased in TCMR compared with Normal and the lowest levels were found in the native nontransplant kidneys. Both isoforms of IMPDH are expressed in various tissues in humans (59). In peripheral blood mononuclear cells, induction of both mRNAs was observed after mitogenic stimulation and mycophenolic acid did not significantly alter the induction of mRNAs (60). Clinical studies have shown an increase in levels of mononuclear cell IMPDH1 mRNA in stable recipients during the first 3 to 4 months of transplantation compared with pretransplant levels. There was an increase in IMPDH1 mRNA levels during AR compared with other time points within the first 2 years of transplantation (61, 62).
Our analysis revealed increased expression of Ca2+ pumps conducive to increased cellular Ca2+ turnover. We found evidence for ER stress, oxidative stress, and mitochondrial stress and reduced energy metabolism in TCMR, and all point toward severe cellular stress in TCMR. Mitochondria and ER interact both physiologically and functionally, and Ca2+ signaling is the critical link between the 2 organelles. Mitochondria-ER cross talk is mediated by protein misfolding within the ER, which causes Ca2+ release into the cytoplasm that is taken up by the mitochondria and results in Ca2+ overload and mitochondrial stress (63). Oxidative phosphorylation was reduced in TCMR. Mitochondrial dysfunction is crucial for PRR signaling and mitochondrial products including mitochondrial DNA are an important source of DAMPs (64). Importantly, TCMR was associated with increased mitochondrial uncoupling, as evidenced by mRNA abundance of UCP2, the inner mitochondrial membrane uncoupling protein whose physiological role is to mediate regulated proton leak (65). In a rat model of kidney transplantation, UCP2-mediated mitochondrial uncoupling was accompanied by increased kidney oxygen consumption, and decreased kidney pO2 was observed as an early event (66).
We found both apoptosis and necroptosis pathways to be upregulated in TCMR. Both pathways of regulated cell death have been shown to be involved in preclinical models of acute kidney injury (67). These 2 forms are not mutually exclusive and can coexist. Our observation that caspase 9, the mitochondrial apoptosis-initiator caspase, is not high in TCMR compared to Normal suggests that cellular stress–activated apoptosis is not the major apoptotic pathway of cell death in TCMR. This is consistent with evidence showing that the perforin/granzyme pathway utilized by cytotoxic T cells activates caspase 3 directly and does not require activation of caspase 9 (68). The lack of activation of caspase 9 also suggests resilience of target cells and could partly explain the immediate improvement in kidney function following quenching of inflammation with antirejection treatment. Because autophagy and mitophagy are critical cytoprotective processes to scavenge ROS-damaged cellular organelles and reduce further ROS production (69), the mechanistic basis for tissue injury in TCMR could be explained by our finding of lack of activation of autophagy and mitophagy and hence a dysfunction in damaged-organelle clearance in TCMR. While the precise reasons for lack of induction of autophagy and downregulation of mitophagy in TCMR are not known, preclinical studies suggest a role for tacrolimus in the induction of autophagy (70). The potential for pharmacological manipulation of these pathway as a damage-limiting strategy in TCMR needs further exploration.
Our analysis confirmed that the increased expression of genes in TCMR was independent of time from transplantation. Also, comparison with native nontransplant kidneys revealed that the innate immune system was upregulated in TCMR rather than downregulated in Normal. Accordingly, our working model for the activation of the innate immune system in TCMR begins with a breach in the stability of drug-induced suppression of the immune system (Figure 11). Because the initial interaction between the DCs and naive T cells happens immediately after the transplantation (71) and the activated alloreactive T cells are maintained in a quiescent state by immunosuppressive drugs, it is likely that underimmunosuppression triggers a low-level tissue injury by the alloreactive T cells. This tissue injury initiates a cascade of events that activates the innate immune system, which culminates in costimulatory signals for the full activation of T cells. Interestingly, calcineurin/NFAT signaling is not related to T cells alone and recent studies have shown that it also contributes to innate immunity and regulates the homeostasis of innate cells (22, 72). Similarly, mycophenolate inhibits the differentiation, maturation, and allostimulatory function of human DCs (73). The resulting cellular injury increases DAMPs that further activates the innate immune system in a feed-forward loop. That such a compromised immunosuppressed state can cause rejection is supported by clinical studies documenting increased risk of rejection in immunosuppression minimization/withdrawal protocols (74).
Our study has limitations. Not every differentially abundant mRNA needs to be different at the protein level. However, the coordinated expression of multiple genes in multiple pathways as well as the differences in the abundance of proteins of key innate immune system mRNAs suggests this to be an unlikely scenario. Moreover, the cascade of events in the activation of the innate immune system that we report here in humans has been elegantly developed in animal models. Biopsy is typically a small piece of tissue and need not represent the processes within the entire allograft. Also, we provide a cross-sectional view of the transcriptome; thus, an association rather than causality can be claimed. An unresolved issue, in view of the cross-sectional nature of our study, is when the increase in mRNA abundance witnessed in TCMR occurs. In view of the clinical studies of urinary chemokines showing molecular quiescence in those who subsequently develop AR (41, 75), it is possible that the increased expression is proximate to TCMR. We are unable to assess if any or all innate immune pathways are essential for TCMR. In animal models, skin and cardiac allografts without evidence of inflammation are vigorously rejected by transferred T cells or when recipients are reconstituted with T cells at a physiologic rate by nude bone graft transplantation (76). In contrast, emerging studies in animal models have provided compelling evidence that innate cells engage in allorecognition and this form of nonmicrobial, non–self-recognition, referred to as innate allorecognition, plays an important role in rejection (77).
In summary, we utilized the high resolution of RNA sequencing to investigate human kidney allografts and provide a landscape of innate immune system gene expression patterns and strength in kidney recipients with biopsies manifesting TCMR or Normal features. Our analyses suggest an active innate immune system during an episode of TCMR in human kidney recipients. Therapies targeting the innate immune system are extensively used for autoimmune diseases. Our data provide a compelling argument for repurposing such drugs to prevent or treat allograft rejection in the clinical setting.
Methods
Study group, kidney allograft biopsies, and RNA sequencing.
We performed kidney allograft biopsies under ultrasound guidance. A portion of the biopsy tissue was immediately immersed in RNAlater RNA stabilization solution (Life Technologies) and stored at –80°C. From the stored samples, we isolated total RNA using the miRNeasy Mini Kit (Qiagen). We used the TruSeq sample preparation kit v2 (Illumina) to prepare individual cDNA libraries from 400 ng total RNA from each sample. Briefly, this consists of poly-A selection of mRNAs and conversion to single-stranded cDNA using random hexamer primers followed by second-strand generation to create double-stranded cDNA. Sequencing adapters were then ligated to the fragmented cDNA. This was followed by PCR amplification and pooling of the libraries. We sequenced the biopsy samples in 2 different batches as part of a larger RNA sequencing study. The first batch (10 TCMR and 5 Normal) was sequenced as single-end, 6 pooled libraries per lane of a flow cell, on a HiSeq 2500 (Illumina) sequencer. The second batch (6 TCMR and 13 Normal) was sequenced as paired-end, 10 pooled libraries per lane of a flow cell, on a HiSeq 4000 sequencer. The raw sequencing data were stored in FASTQ format.
Sequencing data analysis.
Data processing of all samples was done using the same analysis pipeline. Quality control of FASTQ data was performed using FastQC:Read QC (78) and adapter sequences, if any, were removed with trimmomatic (79). The 101-bp single- and paired-end reads were then aligned to human genome (GRCh37/hg19 assembly) using Tophat2 (80). The human UCSC reference annotation was used to align and quantitate the transcripts. The overall mapping rate was 91.8% ± 3.9% (mean ± SD). Transcript assembly and abundance were determined using Cufflinks (81). The hg19 genes file was used as reference annotation guide to determine the FPKM for each transcript with the Cufflinks and raw counts with HTseq (82). We further established that normalization by library size, TMM (weighted trimmed mean), and log2 transformation of raw counts, as described in the edgeR user guide (83), were sufficient to remove the potential batch effects. After removal of duplicate transcripts and retaining genes with at least 1 CPM in one sample, the initial transcript size of 22,582 was reduced to 16,381. Differential RNA sequencing analysis was performed with edgeR (83). We performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis using the R packages GAGE (84), limma (85), and pathview (86). We have deposited the RNA sequence data in NCBI’s Gene Expression Omnibus (accession number GSE131179).
Statistical analysis.
We used edgeR to determine FCs (83). edgeR fits a negative binomial model of the sequence data and derives probability values for differential expression using an exact test. Probability values were adjusted for FDR; P-FDR < 0.05 was considered statistically significant.
Study approval.
Kidney recipients reported herein provided written informed consent to participate in the study, prior to their inclusion in the study. Our Institutional Review Board approved the study. The clinical and research activities that we report here are consistent with the principles of the Declaration of Helsinki (87).
Author contributions
MS and TM designed the research. HY, JZX, and TM conducted experiments. FBM, HY, ML, AV, JRL, DMD, JZX, SPS, SVS, VKS, OE, and TM acquired data. FBM, HY, AV, SVS, and TM analyzed data. JZX, OE, and MS provided reagents. FBM, MS, and TM wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported in part by awards from the NIH to M. Suthanthiran (MERIT Award, R37-AI051652), T. Muthukumar (K08-DK087824), and Weill Cornell Medical College (Clinical and Translational Science Center Award UL1TR000457). We thank Bing He (Research Administrator) and Yigfang Liu (Research Specialist), both at the Translational Research Core Laboratory in the Department of Pathology and Laboratory Medicine, Catherine Snopkowski (Research Specialist), Emmanuel Edusei (Research Coordinator), Mila Lagman (Staff Associate), and ThuTrang Du (Division Administrator), all in the Division of Nephrology, for their help with the execution of this project.
Version 1. 07/11/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(13):e128014. https://doi.org/10.1172/jci.insight.128014.
Contributor Information
Hua Yang, Email: huy2001@med.cornell.edu.
Michelle Lubetzky, Email: mil9044@med.cornell.edu.
Akanksha Verma, Email: vermaakanksha22@gmail.com.
Darshana M. Dadhania, Email: dmd2001@med.cornell.edu.
Olivier Elemento, Email: ole2001@med.cornell.edu.
Manikkam Suthanthiran, Email: msuthan@med.cornell.edu.
Thangamani Muthukumar, Email: mut9002@med.cornell.edu.
References
- 1.Orandi BJ, et al. Survival benefit with kidney transplants from HLA-incompatible live donors. N Engl J Med. 2016;374(10):940–950. doi: 10.1056/NEJMoa1508380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolfe RA, et al. Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. N Engl J Med. 1999;341(23):1725–1730. doi: 10.1056/NEJM199912023412303. [DOI] [PubMed] [Google Scholar]
- 3.Legendre C, Canaud G, Martinez F. Factors influencing long-term outcome after kidney transplantation. Transpl Int. 2014;27(1):19–27. doi: 10.1111/tri.12217. [DOI] [PubMed] [Google Scholar]
- 4.Thareja G, et al. Single nucleotide variant counts computed from RNA sequencing and cellular traffic into human kidney allografts. Am J Transplant. 2018;18(10):2429–2442. doi: 10.1111/ajt.14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole EH, Johnston O, Rose CL, Gill JS. Impact of acute rejection and new-onset diabetes on long-term transplant graft and patient survival. Clin J Am Soc Nephrol. 2008;3(3):814–821. doi: 10.2215/CJN.04681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alkadi MM, et al. Kidney allograft failure in the steroid-free immunosuppression era: A matched case-control study. Clin Transplant. 2017;31(11) doi: 10.1111/ctr.13117. [DOI] [PubMed] [Google Scholar]
- 7.Oberbarnscheidt MH, Zecher D, Lakkis FG. The innate immune system in transplantation. Semin Immunol. 2011;23(4):264–272. doi: 10.1016/j.smim.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cucchiari D, Podestà MA, Ponticelli C. The critical role of innate immunity in kidney transplantation. Nephron. 2016;132(3):227–237. doi: 10.1159/000444267. [DOI] [PubMed] [Google Scholar]
- 9.Farrar CA, Kupiec-Weglinski JW, Sacks SH. The innate immune system and transplantation. Cold Spring Harb Perspect Med. 2013;3(10):a015479. doi: 10.1101/cshperspect.a015479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy SP, Porrett PM, Turka LA. Innate immunity in transplant tolerance and rejection. Immunol Rev. 2011;241(1):39–48. doi: 10.1111/j.1600-065X.2011.01009.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhuang Q, Lakkis FG. Dendritic cells and innate immunity in kidney transplantation. Kidney Int. 2015;87(4):712–718. doi: 10.1038/ki.2014.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haas M, et al. The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant. 2018;18(2):293–307. doi: 10.1111/ajt.14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lata S, Raghava GP. PRRDB: a comprehensive database of pattern-recognition receptors and their ligands. BMC Genomics. 2008;9:180. doi: 10.1186/1471-2164-9-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardy JL, Lynn DJ, Brinkman FS, Hancock RE. Enabling a systems biology approach to immunology: focus on innate immunity. Trends Immunol. 2009;30(6):249–262. doi: 10.1016/j.it.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Breuer K, et al. InnateDB: systems biology of innate immunity and beyond--recent updates and continuing curation. Nucleic Acids Res. 2013;41(Database issue):D1228–D1233. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yates B, Braschi B, Gray KA, Seal RL, Tweedie S, Bruford EA. Genenames.org: the HGNC and VGNC resources in 2017. Nucleic Acids Res. 2017;45(D1):D619–D625. doi: 10.1093/nar/gkw1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17(1):1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 18.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327(5963):291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figdor CG, van Kooyk Y, Adema GJ. C-type lectin receptors on dendritic cells and Langerhans cells. Nat Rev Immunol. 2002;2(2):77–84. doi: 10.1038/nri723. [DOI] [PubMed] [Google Scholar]
- 20.Inohara N, Nuñez G. The NOD: a signaling module that regulates apoptosis and host defense against pathogens. Oncogene. 2001;20(44):6473–6481. doi: 10.1038/sj.onc.1204787. [DOI] [PubMed] [Google Scholar]
- 21.Amarante-Mendes GP, Adjemian S, Branco LM, Zanetti LC, Weinlich R, Bortoluci KR. Pattern recognition receptors and the host cell death molecular machinery. Front Immunol. 2018;9:2379. doi: 10.3389/fimmu.2018.02379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fric J, Zelante T, Wong AY, Mertes A, Yu HB, Ricciardi-Castagnoli P. NFAT control of innate immunity. Blood. 2012;120(7):1380–1389. doi: 10.1182/blood-2012-02-404475. [DOI] [PubMed] [Google Scholar]
- 23.Feerick CL, McKernan DP. Understanding the regulation of pattern recognition receptors in inflammatory diseases - a ‘Nod’ in the right direction. Immunology. 2017;150(3):237–247. doi: 10.1111/imm.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Degn SE, Thiel S. Humoral pattern recognition and the complement system. Scand J Immunol. 2013;78(2):181–193. doi: 10.1111/sji.12070. [DOI] [PubMed] [Google Scholar]
- 25.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen GY, Nuñez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol. 2010;10(12):826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabiet MJ, Huet E, Boulay F. Human mitochondria-derived N-formylated peptides are novel agonists equally active on FPR and FPRL1, while Listeria monocytogenes-derived peptides preferentially activate FPR. Eur J Immunol. 2005;35(8):2486–2495. doi: 10.1002/eji.200526338. [DOI] [PubMed] [Google Scholar]
- 28.Di Virgilio F, Dal Ben D, Sarti AC, Giuliani AL, Falzoni S. The P2X7 Receptor in Infection and Inflammation. Immunity. 2017;47(1):15–31. doi: 10.1016/j.immuni.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 29.Marin TL, et al. AMPK promotes mitochondrial biogenesis and function by phosphorylating the epigenetic factors DNMT1, RBBP7, and HAT1. Sci Signal. 2017;10(464):eaaf7478. doi: 10.1126/scisignal.aaf7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: The trinity at the heart of the DNA damage response. Mol Cell. 2017;66(6):801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 31.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16(7):393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 32.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clapham DE. Calcium signaling. Cell. 2007;131(6):1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 34.Panday A, Sahoo MK, Osorio D, Batra S. NADPH oxidases: an overview from structure to innate immunity-associated pathologies. Cell Mol Immunol. 2015;12(1):5–23. doi: 10.1038/cmi.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrade CF, Waddell TK, Keshavjee S, Liu M. Innate immunity and organ transplantation: the potential role of Toll-like receptors. Am J Transplant. 2005;5(5):969–975. doi: 10.1111/j.1600-6143.2005.00829.x. [DOI] [PubMed] [Google Scholar]
- 36.Goldstein DR, Tesar BM, Akira S, Lakkis FG. Critical role of the Toll-like receptor signal adaptor protein MyD88 in acute allograft rejection. J Clin Invest. 2003;111(10):1571–1578. doi: 10.1172/JCI17573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnsen IB, Nguyen TT, Bergstrøm B, Lien E, Anthonsen MW. Toll-like receptor 3-elicited MAPK activation induces stabilization of interferon-β mRNA. Cytokine. 2012;57(3):337–346. doi: 10.1016/j.cyto.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 38.Schroppel B, He JC. Expression of Toll-like receptors in the kidney: their potential role beyond infection. Kidney Int. 2006;69(5):785–787. doi: 10.1038/sj.ki.5000190. [DOI] [PubMed] [Google Scholar]
- 39.Dessing MC, Kers J, Damman J, Navis GJ, Florquin S, Leemans JC. Donor and recipient genetic variants in NLRP3 associate with early acute rejection following kidney transplantation. Sci Rep. 2016;6:36315. doi: 10.1038/srep36315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hricik DE, et al. Multicenter validation of urinary CXCL9 as a risk-stratifying biomarker for kidney transplant injury. Am J Transplant. 2013;13(10):2634–2644. doi: 10.1111/ajt.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suthanthiran M, et al. Urinary-cell mRNA profile and acute cellular rejection in kidney allografts. N Engl J Med. 2013;369(1):20–31. doi: 10.1056/NEJMoa1215555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lubbers R, van Essen MF, van Kooten C, Trouw LA. Production of complement components by cells of the immune system. Clin Exp Immunol. 2017;188(2):183–194. doi: 10.1111/cei.12952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feucht HE, et al. Biosynthesis of complement C4 messenger RNA in normal human kidney. Nephron. 1989;53(4):338–342. doi: 10.1159/000185778. [DOI] [PubMed] [Google Scholar]
- 44.Kwan WH, van der Touw W, Heeger PS. Complement regulation of T cell immunity. Immunol Res. 2012;54(1-3):247–253. doi: 10.1007/s12026-012-8327-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mori DN, Kreisel D, Fullerton JN, Gilroy DW, Goldstein DR. Inflammatory triggers of acute rejection of organ allografts. Immunol Rev. 2014;258(1):132–144. doi: 10.1111/imr.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JR, et al. Gut microbial community structure and complications after kidney transplantation: a pilot study. Transplantation. 2014;98(7):697–705. doi: 10.1097/TP.0000000000000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grazioli S, Pugin J. Mitochondrial damage-associated molecular patterns: from inflammatory signaling to human diseases. Front Immunol. 2018;9:832. doi: 10.3389/fimmu.2018.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ott U, Aschoff A, Fünfstück R, Jirikowski G, Wolf G. DNA fragmentation in acute and chronic rejection after renal transplantation. Transplant Proc. 2007;39(1):73–77. doi: 10.1016/j.transproceed.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 49.Wu H, et al. HMGB1 contributes to kidney ischemia reperfusion injury. J Am Soc Nephrol. 2010;21(11):1878–1890. doi: 10.1681/ASN.2009101048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lechler RI, Batchelor JR. Restoration of immunogenicity to passenger cell-depleted kidney allografts by the addition of donor strain dendritic cells. J Exp Med. 1982;155(1):31–41. doi: 10.1084/jem.155.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lakkis FG, Lechler RI. Origin and biology of the allogeneic response. Cold Spring Harb Perspect Med. 2013;3(8):a014993. doi: 10.1101/cshperspect.a014993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Celli S, Albert ML, Bousso P. Visualizing the innate and adaptive immune responses underlying allograft rejection by two-photon microscopy. Nat Med. 2011;17(6):744–749. doi: 10.1038/nm.2376. [DOI] [PubMed] [Google Scholar]
- 53.Batal I, et al. Dendritic cells in kidney transplant biopsy samples are associated with T cell infiltration and poor allograft survival. J Am Soc Nephrol. 2015;26(12):3102–3113. doi: 10.1681/ASN.2014080804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhuang Q, et al. Graft-infiltrating host dendritic cells play a key role in organ transplant rejection. Nat Commun. 2016;7:12623. doi: 10.1038/ncomms12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bergler T, et al. Infiltration of macrophages correlates with severity of allograft rejection and outcome in human kidney transplantation. PLoS ONE. 2016;11(6):e0156900. doi: 10.1371/journal.pone.0156900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mills CD. Anatomy of a discovery: M1 and M2 macrophages. Front Immunol. 2015;6:212. doi: 10.3389/fimmu.2015.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25(12):677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 58.Sommerer C, Giese T, Meuer S, Zeier M. New concepts to individualize calcineurin inhibitor therapy in renal allograft recipients. Saudi J Kidney Dis Transpl. 2010;21(6):1030–1037. [PubMed] [Google Scholar]
- 59.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 60.Jain J, et al. Regulation of inosine monophosphate dehydrogenase type I and type II isoforms in human lymphocytes. Biochem Pharmacol. 2004;67(4):767–776. doi: 10.1016/j.bcp.2003.09.043. [DOI] [PubMed] [Google Scholar]
- 61.Sombogaard F, et al. Inosine monophosphate dehydrogenase messenger RNA expression is correlated to clinical outcomes in mycophenolate mofetil-treated kidney transplant patients, whereas inosine monophosphate dehydrogenase activity is not. Ther Drug Monit. 2009;31(5):549–556. doi: 10.1097/FTD.0b013e3181b7a9d0. [DOI] [PubMed] [Google Scholar]
- 62.Sanquer S, et al. Expression of inosine monophosphate dehydrogenase type I and type II after mycophenolate mofetil treatment: a 2-year follow-up in kidney transplantation. Clin Pharmacol Ther. 2008;83(2):328–335. doi: 10.1038/sj.clpt.6100300. [DOI] [PubMed] [Google Scholar]
- 63.Malhotra JD, Kaufman RJ. ER stress and its functional link to mitochondria: role in cell survival and death. Cold Spring Harb Perspect Biol. 2011;3(9):a004424. doi: 10.1101/cshperspect.a004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sandhir R, Halder A, Sunkaria A. Mitochondria as a centrally positioned hub in the innate immune response. Biochim Biophys Acta Mol Basis Dis. 2017;1863(5):1090–1097. doi: 10.1016/j.bbadis.2016.10.020. [DOI] [PubMed] [Google Scholar]
- 65.Krauss S, Zhang CY, Lowell BB. The mitochondrial uncoupling-protein homologues. Nat Rev Mol Cell Biol. 2005;6(3):248–261. doi: 10.1038/nrm1592. [DOI] [PubMed] [Google Scholar]
- 66.Papazova DA, Friederich-Persson M, Joles JA, Verhaar MC. Renal transplantation induces mitochondrial uncoupling, increased kidney oxygen consumption, and decreased kidney oxygen tension. Am J Physiol Renal Physiol. 2015;308(1):F22–F28. doi: 10.1152/ajprenal.00278.2014. [DOI] [PubMed] [Google Scholar]
- 67.Linkermann A. Nonapoptotic cell death in acute kidney injury and transplantation. Kidney Int. 2016;89(1):46–57. doi: 10.1016/j.kint.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 68.Goping IS, et al. Granzyme B-induced apoptosis requires both direct caspase activation and relief of caspase inhibition. Immunity. 2003;18(3):355–365. doi: 10.1016/S1074-7613(03)00032-3. [DOI] [PubMed] [Google Scholar]
- 69.Anding AL, Baehrecke EH. Cleaning house: selective autophagy of organelles. Dev Cell. 2017;41(1):10–22. doi: 10.1016/j.devcel.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakagaki T, et al. FK506 reduces abnormal prion protein through the activation of autolysosomal degradation and prolongs survival in prion-infected mice. Autophagy. 2013;9(9):1386–1394. doi: 10.4161/auto.25381. [DOI] [PubMed] [Google Scholar]
- 71.Hommel M, Kyewski B. Dynamic changes during the immune response in T cell-antigen-presenting cell clusters isolated from lymph nodes. J Exp Med. 2003;197(3):269–280. doi: 10.1084/jem.20021512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Müller MR, Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol. 2010;10(9):645–656. doi: 10.1038/nri2818. [DOI] [PubMed] [Google Scholar]
- 73.Colic M, Stojic-Vukanic Z, Pavlovic B, Jandric D, Stefanoska I. Mycophenolate mofetil inhibits differentiation, maturation and allostimulatory function of human monocyte-derived dendritic cells. Clin Exp Immunol. 2003;134(1):63–69. doi: 10.1046/j.1365-2249.2003.02269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prashar R, Venkat KK. Immunosuppression minimization and avoidance protocols: when less is not more. Adv Chronic Kidney Dis. 2016;23(5):295–300. doi: 10.1053/j.ackd.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 75.Rabant M, et al. Early low urinary CXCL9 and CXCL10 might predict immunological quiescence in clinically and histologically stable kidney recipients. Am J Transplant. 2016;16(6):1868–1881. doi: 10.1111/ajt.13677. [DOI] [PubMed] [Google Scholar]
- 76.Bingaman AW, et al. Vigorous allograft rejection in the absence of danger. J Immunol. 2000;164(6):3065–3071. doi: 10.4049/jimmunol.164.6.3065. [DOI] [PubMed] [Google Scholar]
- 77.Lakkis FG, Li XC. Innate allorecognition by monocytic cells and its role in graft rejection. Am J Transplant. 2018;18(2):289–292. doi: 10.1111/ajt.14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Andrews S. Babraham Bioinformatics. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc Accessed: September 1st, 2018.
- 79.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29(14):1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.