Supplemental Digital Content is available in the text
Keywords: Fusion genes, Endometrial cancer, Drug resistance, Prognosis
Abstract
Background:
Fusion genes may play an important role in tumorigenesis, prognosis, and drug resistance; however, studies on fusion genes in endometrial cancer (EC) are rare. This study aimed to identify new fusion genes and to explore their clinical significance in EC.
Methods:
A total of 28 patients diagnosed with EC were enrolled in this study. RNA sequencing was used to obtain entire genomes and transcriptomes. STAR-comparison and STAR-fusion prediction were applied to predict the fusion genes. Chi-square tests and Student t tests were used to verify the clinical significance with SPSS 13.0 software.
Results:
New fusion genes were found, and the number of fusion genes varied from 3 to 110 among all patients with EC. The type of fusion genes varied and included messenger RNA (mRNA)-mRNA, long non-coding RNA (lncRNA)-lncRNA, and lncRNA-mRNA. There were six fusion genes with high fusion rates, namely, RP11–123O10.4–GRIP1, RP11–444D3.1–SOX5, RP11–680G10.1–GSE1, NRIP1–AF127936.7, RP11–96H19.1–RP11–446N19.1, and DPH7–PTP4A3. Further studies showed that these fusion genes are related to stage, grade, and recurrence, in which NRIP1–AF127936.7 and DPH7–PTP4A3 were found only in stage III patients with EC. DPH7–PTP4A3 was found in grades 2 and 3, and recurrent patients with EC.
Conclusion:
Fusion genes play an essential role in EC. Six genes that are overexpressed with high fusion rates are identified. NRIP1–AF127936.7 and DPH7–PTP4A3 might be related to stage, and DPH7–PTP4A3 be related to grade and recurrence.
Introduction
Endometrial cancer (EC) is one of the most common gynecological tumors in developed and developing countries, and its prevalence has recently increased.[1] Environmental factors, including estrogen, an abnormal mismatch repair system, genetic abnormalities, and the aberrant methylation of DNA and microRNA, are currently proposed as major mechanisms of carcinogenesis in EC.[2] Nevertheless, the management of EC has become more complex during the past 5 to 10 years because the carcinogenic mechanisms of EC remain unclear. Various classifications are used to assess the risks of recurrence and to determine optimum postoperative management.[3] However, there are few methods that can be used to analyze the prognosis of patients with EC.
Fusion genes are hybrid genes that combine parts of two or more original genes formed as the result of either structural chromosomal rearrangement, including primarily translocation, inversion, amplification and deletion, or non-structural aberrations caused by cis- and trans-splicing or transcriptional read-through. Such events are known to play important roles during the initial steps of tumorigenesis[4] and have been recognized in leukemic tumors,[5] neuroblastoma,[6] and endometrial stromal sarcoma.[7,8] These studies have also indicated that fusion genes such as MBTD1-CXorf67,[9]JAZF1-SUZ12, JAZF1-PHF1, EPC1-PHF1,[10] and TSNAX-DISC1[11] play an essential role in cell proliferation, apoptosis, and drug sensitivity.[12,5,13–16] However, research on the fusion genes of EC is rare.
In this study, we aimed to identify and verify new fusion genes in EC using RNA sequencing and analyzed the clinical significance of these fusion genes in EC.
Methods
Ethical approval
This study was approved by the Research Ethics Committee of Peking University People's Hospital. All patients were fully informed of the purpose and methods of the present study, and written informed consent was obtained from the patients themselves or their guardians.
Patients
Twenty-eight patients were enrolled in this study. The patients’ characteristics are listed in Table 1.
Table 1.
Clinical characteristics of 28 patients with endometrial cancer.

RNA sequence analysis
RNA quantification and qualification
RNA degradation and contamination were monitored on 1% agarose gels. RNA purity was checked using a NanoPhotometer® spectrophotometer NP80 (IMPLEN, CA, USA). RNA concentration was measured using a Qubit® RNA Assay Kit on a Qubit® 2.0 Fluorometer (Life Technologies, CA, USA). RNA integrity was assessed using an RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
Library preparation for transcriptome sequencing
A total amount of 3 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer's recommendations, and index codes were added to attribute sequences to each sample. Briefly, messenger RNA (mRNA) was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEB Next First Strand Synthesis Reaction Buffer (5×). First-strand complementary DNA (cDNA) was synthesized using a random hexamer primer and M-MuLV Reverse Transcriptase (RNaseH–). Second-strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H. The remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of the 3′ ends of the DNA fragments, NEB Next Adaptor with a hairpin loop structure was ligated to prepare for hybridization. To select cDNA fragments of preferentially 250 to 300 bp in length, the library fragments were purified with an AMPure XP system (Beckman Coulter, Beverly, MA, USA). Then, 3 μL of USER Enzyme (NEB) was used with the size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before polymerase chain reaction (PCR). PCR was then performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. Finally, PCR products were purified (AMPure XP system), and library quality was assessed on the Agilent Bioanalyzer 2100 system.
Clustering and sequencing
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using a TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer's instructions. After cluster generation, the library preparations were sequenced on an Illumina HiSeq platform, and 125 bp/150 bp paired-end reads were generated.
Fusion analysis
A fusion gene refers to two genes (either in whole or in part) that undergo fusion resulting in a chimeric gene, which is usually caused by reasons such as chromosome translocation and associated problems. STAR-fusion (1.2.0) software analysis and the detection of fusion genes were used to identify fusion genes. Fusion gene lists were filtered with STAR-fusion with the standard filter method and other parameters (--annotate; --examine_coding_effect; -FusionInspectorinspect; --denovo_reconstruct; --min_junction_reads1; -min_sum_frags2).
Statistical analysis
The software SPSS version 13.0 (SPSS Inc., Chicago, IL, USA) was used for the statistical analysis. The chi-square test and Student's t test were used to calculate the associations between clinical signatures and fusion gene distribution. P values less than 0.05 were considered statistically significant.
Results
Fusion gene types in EC
Fusion genes were identified in all 28 cases, as shown in Figure 1 and supplementary material. The number of fusion genes varied from 3 to 110. The type of fusion gene included mRNA-mRNA, long non-coding RNA (lncRNA)-lncRNA, and lncRNA-mRNA. The results indicated that fusion genes are present in EC, while the type distribution of the fusion genes varied.
Figure 1.
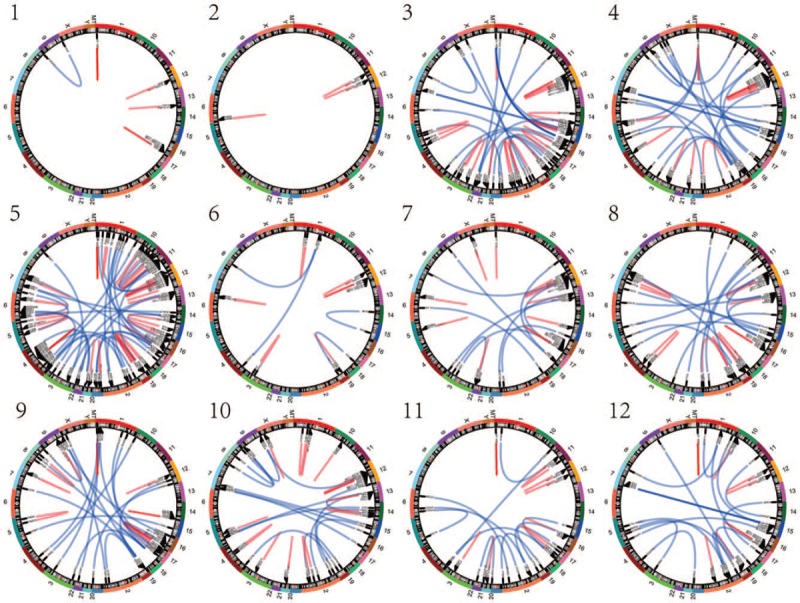
The fusion genes in 28 endometrial cancer patients: 1 to 28 in the figure show the fusion gene distributions of 28 patients with endometrial cancer.
Fusion gene expression in patients with EC
There were six fusion genes with a high fusion rate (fusion rate = the number of patients with fusion gene(+)/28). RP11–123O10.4–GRIP1 had the highest fusion rate of approximately 89.28% (25/28). The expression of RP11–444D3.1–SOX5 was 60.71% (17/28). The fusion rate of RP11–680G10.1–GSE1 was 50.00% (14/28). The fusion rate of NRIP1–AF127936.7 was 42.86% (12/28). The fusion rate of RP11–96H19.1--RP11–446N19.1 was 39.29% (11/28). The fusion rate of DPH7--PTP4A3 was 32.29% (9/28). The mean values of the junction reads of RP11–123O10.4--GRIP1, RP11–444D3.1--SOX5, RP11–680G10.1--GSE1, NRIP1--AF127936.7, RP11–96H19.1--RP11–446N19.1, and DPH7--PTP4A3 were 16.36, 4.71, 7.04, 1.14, 3.25, and 10.82, respectively. These results indicate that the fusion rate and junction reads of these fusion genes are both high.
Relationship Between Fusion Gene Expression and Clinical Characteristics
Fusion gene distribution in stage I-II and stage III EC
The fusion gene distribution was different between stage I-II and stage III patients with EC, as shown in Figure 2 and Table 1. The fusion genes of stage I-II patients with EC, including AC024084.1--AC009784.3, BMPR1B--PDLIM5, and C15orf57--CBX3, are shown in Figure 2A. The fusion genes of stage III patients with EC, including REV3L--RPL28, RPL18--MT-ATP6, and SLC39A11--CPSF4L, are shown in Figure 2B. These results indicate that the fusion genes RP11–123O10.4--GRIP1, RP11–444D3.1--SOX5, RP11–680G10.1--GSE1, and RP11–96H19.1--RP11–446N19.1 are found in all stages of EC, while NRIP1--AF127936.7 and DPH7--PTP4A3 are found only in stage III EC, as shown in Table 2.
Figure 2.
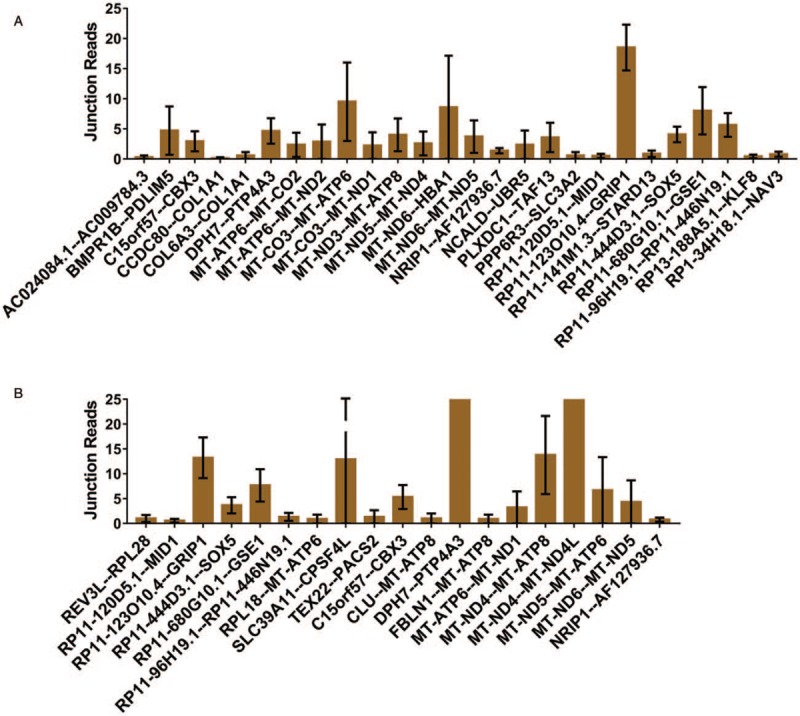
Fusion genes in different stages of endometrial cancer: (A) The junction reads of fusion genes in stage I-II patients with EC; (B) the junction reads of fusion genes in stage III patients with EC (note: only fusion genes that appeared in more than one patient or had more than one variant are listed). EC: Endometrial cancer.
Table 2.
Fusion genes in different stages of endometrial cancer.
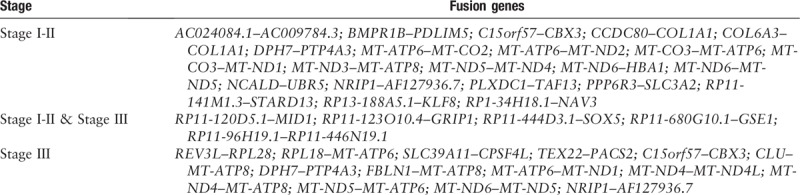
Fusion gene distribution among grades 1, 2, and 3 patients with EC
The fusion gene distribution among different grades of patients with EC is shown in Figure 3. The fusion genes of grade 1 patients with EC, including AC024084.1--AC009784.3, BMPR1B--PDLIM5, and MT-ATP6--MT-CO2, are shown in Figure 3A. The fusion genes of grade 2 patients with EC, including C15orf57--CBX3, CCDC80--COL1A1, and DPH7--PTP4A3, are shown in Figure 3B. The fusion genes of grade 3 patients with EC, including BMPR1B--PDLIM5, C15orf57--CBX3, and COL6A3--COL1A1, are shown in Figure 3C. These results indicate that the fusion genes RP11–123O10.4--GRIP1, RP11–444D3.1--SOX5, RP11–680G10.1--GSE1, RP11–96H19.1--RP11–446N19.1, and NRIP1--AF127936.7 were found in all grades of EC, while DPH7--PTP4A3 is found only in grades 2 and 3 EC.
Figure 3.
Fusion genes in different grades of EC: (A) The junction reads of fusion genes in grade 1 patients with EC; (B) the junction reads of fusion genes in grade 2 patients with EC; (C) the junction reads of fusion genes in grade 3 patients with EC (only fusion genes that appeared in more than one patient or had more than one variant are listed). EC: Endometrial cancer.
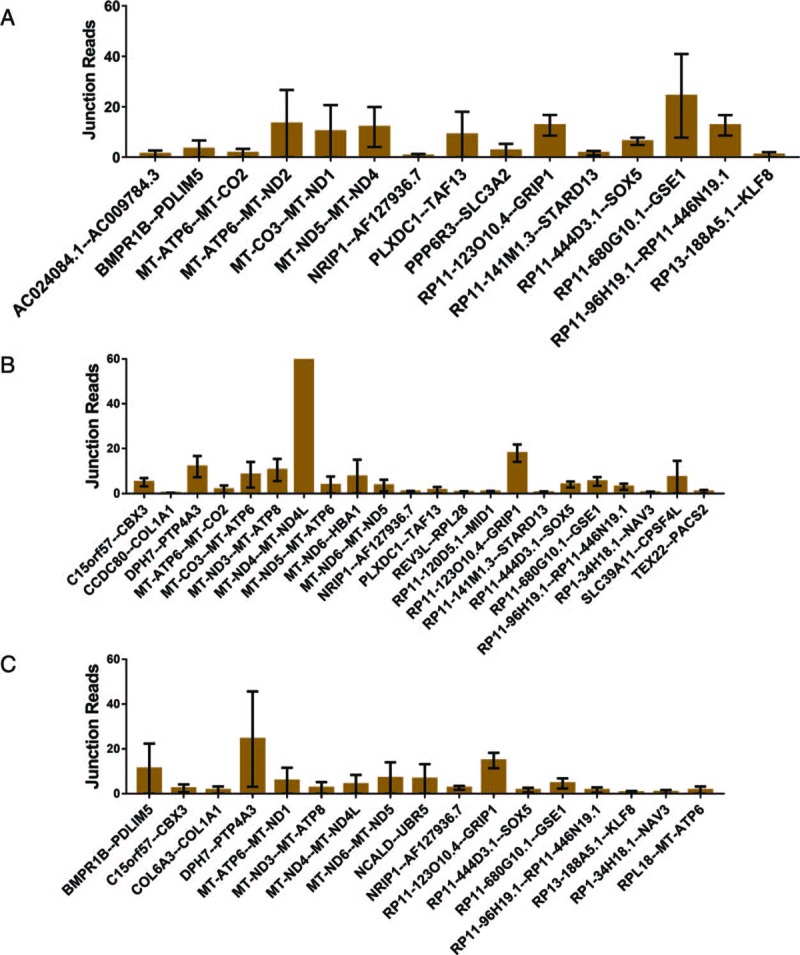
Fusion gene distribution in recurrent and non-recurrent EC
The fusion gene distributions were different between recurrent and non-recurrent EC, as shown in Figure 4. The diagnosis of recurrence should be adjusted by abdominal, pelvic, and/or chest computed tomography or a physical exam based on the National Comprehensive Cancer Network (NCCN) guidelines. The fusion genes in patients with recurrent EC, including AC024084.1--AC009784.3, BMPR1B--PDLIM5, and MT-ATP6--MT-CO2, are shown in Figure 4A. The fusion genes in patients with non-recurrent EC, including BMPR1B--PDLIM5, C15orf57--CBX3, and COL6A3--COL1A1, are shown in Figure 4B. These results indicate that the fusion genes RP11–123O10.4--GRIP1, RP11–444D3.1--SOX5, RP11–680G10.1--GSE1, RP11–96H19.1--RP11–446N19.1, and NRIP1--AF127936.7 are over-expressed in non-recurrent EC, while DPH7--PTP4A3 is found in recurrent EC.
Figure 4.
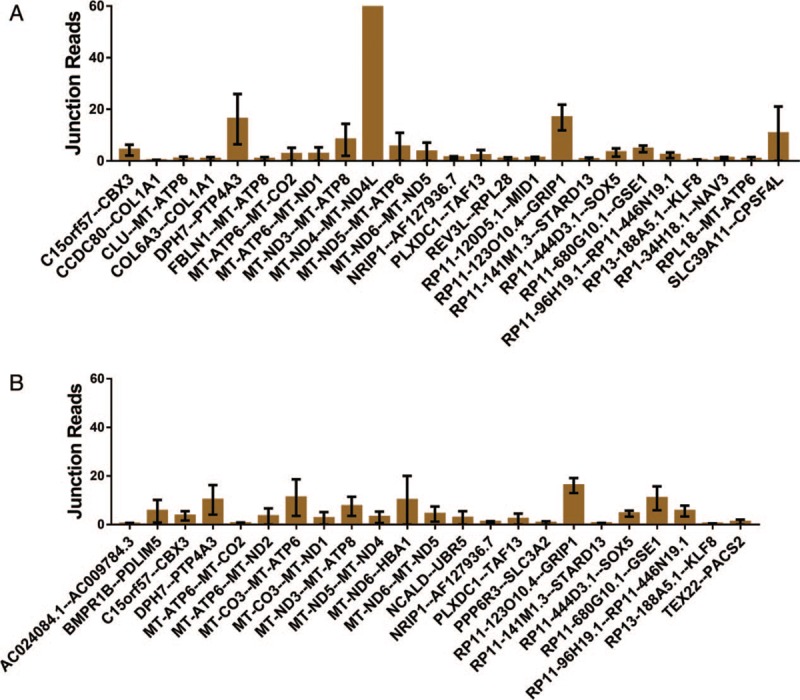
Fusion genes in recurrent and non-recurrent endometrial cancer: (A) The junction reads of fusion genes in recurrent patients with EC; (B) the junction reads of fusion genes in non-recurrent patients with EC (only fusion genes that appeared in more than one patient or had more than one variant are listed). EC: Endometrial cancer.
Discussion
Fusion genes are hybrid genes that combine parts of two or more original genes, which are formed as a result of chromosomal rearrangements or abnormal transcription and have been shown to act as drivers of malignant transformation and progression in leukaemic tumors,[5] neuroblastoma,[6] and endometrial stromal sarcoma.[7,8] Fusion genes are also used as diagnostic and prognostic markers to confirm cancer diagnosis and to monitor the response to molecular therapies.[17] Thus, to identify and verify the fusion genes of EC is of great importance.
To date, the study of fusion genes of uterine tumors have focused on endometrial stromal sarcoma-associated genomic rearrangements, including BRD8-PHF1,[7]MBTD1-CXorf67,[9]JAZF1-SUZ12, JAZF1-PHF1, EPC1-PHF1,[10] and TSNAX-DISC1,[11] which proved that fusion genes play essential roles in diagnosis, treatment, and prognosis.[7,10,18,19] In fact, there are few studies on fusion genes of endometrioid adenocarcinoma.
In this study, we proved that fusion genes were found in endometrioid adenocarcinoma and that the distribution of the fusion genes of endometrioid adenocarcinoma varied. We first showed that RP11–123O10.4--GRIP1, RP11–444D3.1--SOX5, RP11–680G10.1--GSE1, RP11–96H19.1--RP11–446N19.1, NRIP1--AF127936.7, and DPH7--PTP4A3 had high fusion rates and were over-expressed at high levels in patients with EC, suggesting that these fusion genes might be related to tumorigenesis and prognosis.
Further study of these six fusion genes with high fusion rates revealed that these fusion genes can be detected, while their biological functions remain unclear. RP11–123O10.4--GRIP1 was the fusion gene with the highest fusion rate (approximately 89.29%). RP11–123O10.4 is a lncRNA located on chromosome 12, and only one variant was found. lncRNAs are a set of non-protein-coding transcripts longer than 200 nucleotides[20] that play vital roles in cancer proliferation and drug resistance. GRIP1 belongs to the p160 steroid receptor co-activator family that plays essential roles in nuclear receptor-dependent transcriptional regulation.[21] GRIP1 is an mRNA located on chromosome 12 and contains two variants. Previous studies have shown that GRIP1 plays an essential role in cell proliferation.[22] However, the biological function of RP11–123O10.4--GRIP1 remains unclear.
The fusion rate of RP11–444D3.1--SOX5 was 61%. RP11–444D3.1 is a lncRNA located on chromosome 12, and two variants were found. SOX5 is an mRNA located on chromosome 12 and contains three variants. SOX5 encodes a member of the SRY-related HMG-box family of transcription factors involved in the determination of cell fate and the regulation of embryonic development.[23] As a nuclear transcription factor, the high expression of SOX5 is detected in many malignant tumors. The aberrant expression of SOX5 can promote the epithelial-mesenchymal transition, proliferation, invasion, and migration of cancers by targeting different downstream genes, such as Twist1, Snail, secreted protein acidic and rich in cysteine, and microphthalmia-associated transcription factor, as well as up-regulating the protein abundance of p27, β-catenin, etc.[23,24]
The fusion rate of RP11–680G10.1--GSE1 was 50%. RP11–680G10.1 is a lncRNA located on chromosome 16, and one variant was found. GSE1 is an mRNA located on chromosome 16 and contains two variants. GSE1, also known as KIAA0182, is a novel protein that has been isolated and identified by ion trap mass spectrometry. GSE1 is a proline-rich protein, and its estimated molecular mass is 136,000 kDa.[25] GSE1 can mediate the progression of breast cancer in vitro. GSE1 overexpression in breast cancer and the knockdown of GSE1 significantly suppressed breast cancer cell proliferation, migration, and invasion.[26] Furthermore, GSE1 was identified as a direct target of miR-489-5p, which is significantly reduced in breast cancer tissues. In addition, the forced expression of miR-489-5p suppressed breast cancer cell proliferation, migration, and invasion.[26] Moreover, the depletion of GSE1 by siRNAs significantly abrogated the enhanced proliferation, migration, and invasion of breast cancer cells consequent to miR-489-5p depletion.[26]
The fusion rate of NRIP1--AF127936.7 was 43%. NRIP1 is an mRNA located on chromosome 21, and two variants were found. AF127936.7 is located on chromosome 21 and contains two variants. NRIP1 is a transcriptional co-regulator (also known as RIP140) that plays very important physiological roles by finely tuning the activity of a large number of transcription factors. Noticeably, the RIP140 gene has been shown to be involved in the regulation of energy expenditure, mammary gland development, and intestinal homeostasis as well as behavior and cognition. RIP140 is also involved in the regulation of various oncogenic signaling pathways and participates in the development and progression of solid tumors.[27] However, there are few studies on the AF127936.7 gene.
The fusion rate of RP11–96H19.1--RP11–446N19.1 was 39%. RP11–96H19.1 is a lncRNA located on chromosome 12, and two variants were found. RP11–446N19.1 is a lncRNA located on chromosome 12 and contains two variants. However, the functions of RP11–96H19.1 and RP11–446N19.1 remain unclear.
The fusion rate of DPH7--PTP4A3 was 32%. DPH7 is an mRNA located on chromosome 9, and one variant was found. PTP4A3 is an mRNA located on chromosome 8 and contains one variant. The membrane-associated intracellular protein tyrosine phosphatase PTP4A3 is over-expressed in human colorectal cancer and contributes to cell migration and invasion.[28] However, few studies have illustrated the biological functions of DPH7.
In general, the detection of fusion genes might be recognized as a promising clinical tool for the diagnosis and staging classification of EC. Our present study indicates that fusion genes can be found in EC and play a key role in contributing to the progression of EC. We suggest that NRIP1--AF127936.7 and DPH7--PTP4A3 may be potential candidates for a new genetic and an epigenetic marker, respectively, in EC.
In this study, the fusion genes RP11–123O10.4--GRIP1, RP11–444D3.1--SOX5, RP11–680G10.1--GSE1, NRIP1--AF127936.7, RP11–96H19.1--RP11–446N19.1, and DPH7--PTP4A3 were proved to have a high fusion rate in EC. RP11–123O10.4--GRIP1, RP11–444D3.1--SOX5, RP11–680G10.1--GSE1, and RP11–96H19.1--RP11–446N19.1 might be related to tumorigenesis, while NRIP1--AF127936.7 and DPH7--PTP4A3 might be related to stage and DPH7--PTP4A3 might be related to grade and a poor prognosis. The biological functions of these fusion genes need to be further studied.
Funding
This work was supported by grants from the National Key R&D programme of China (No. YFC1303100 and No. YFC1303103).
Conflicts of interest
None.
Supplementary Material
Footnotes
How to cite this article: Yao T, Liu JJ, Zhao LJ, Zhou JY, Wang JQ, Wang Y, Wang ZQ, Wei LH, Wang JL, Li XP. Identification of new fusion genes and their clinical significance in endometrial cancer. Chin Med J 2019;00:00–00. doi: 10.1097/CM9.0000000000000203
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017; 67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Banno K, Yanokura M, Iida M, Masuda K, Aoki D. Carcinogenic mechanisms of endometrial cancer: involvement of genetics and epigenetics. J Obstet Gynaecol Res 2014; 40:1957–1967. doi: 10.1111/jog.12442. [DOI] [PubMed] [Google Scholar]
- 3.Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet 2016; 387:1094–1108. doi: 10.1016/s0140-6736(15)00130-0. [DOI] [PubMed] [Google Scholar]
- 4.Dai X, Theobard R, Cheng H, Xing M, Zhang J. Fusion genes: a promising tool combating against cancer. Biochim Biophys Acta Rev Cancer 2018; 1869:149–160. doi: 10.1016/j.bbcan.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Lyu X, Wang X, Zhang L, Chen Z, Zhao Y, Hu J, et al. Detection of 22 common leukemic fusion genes using a single-step multiplex qRT-PCR-based assay. Diagn Pathol 2017; 12:55.doi: 10.1186/s13000-017-0634-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schleiermacher G, Raynal V, Janoueix-Lerosey I, Combaret V, Aurias A, Delattre O. Variety and complexity of chromosome 17 translocations in neuroblastoma. Genes Chromosomes Cancer 2004; 39:143–150. doi: 10.1002/gcc.10313. [DOI] [PubMed] [Google Scholar]
- 7.Micci F, Brunetti M, Dal Cin P, Nucci MR, Gorunova L, Heim S, et al. Fusion of the genes BRD8 and PHF1 in endometrial stromal sarcoma. Genes Chromosomes Cancer 2017; 56:841–845. doi: 10.1002/gcc.22485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang B, Lu LX, Tu XY, Cheng YF, Bi R, Yang WT. Endometrial stromal sarcoma: morphologic features and detection of JAZF1-SUZ12 and YWHAE FAM22 fusion genes (in Chinese). Chin J Pathol 2016; 45:308–313. doi: 10.3760/cma.j.issn.0529-5807.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Dewaele B, Przybyl J, Quattrone A, Finalet Ferreiro J, Vanspauwen V, Geerdens V, et al. Identification of a novel, recurrent MBTD1-CXorf67 fusion in low-grade endometrial stromal sarcoma. Int J Cancer 2014; 134:1112–1122. doi: 10.1002/ijc.28440. [DOI] [PubMed] [Google Scholar]
- 10.Dickson BC, Lum A, Swanson D, Bernardini MQ, Colgan TJ, Shaw PA, et al. Novel EPC1 gene fusions in endometrial stromal sarcoma. Genes Chromosomes Cancer 2018; 57:598–603. doi: 10.1002/gcc.22649. [DOI] [PubMed] [Google Scholar]
- 11.Li N, Zheng J, Li H, Deng J, Hu M, Wu H, et al. Identification of chimeric TSNAX-DISC1 resulting from intergenic splicing in endometrial carcinoma through high-throughput RNA sequencing. Carcinogenesis 2014; 35:2687–2697. doi: 10.1093/carcin/bgu201. [DOI] [PubMed] [Google Scholar]
- 12.Xu H, Wu X, Sun D, Shijun L, Siwen Z, Miao T, et al. SEGF: a novel method for gene fusion detection from single-end next-generation sequencing data. Genes Basel 2018; 9:7.doi: 10.3390/genes9070331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandran RK, Geetha N, Sakthivel KM, Aswathy CG, Gopinath P, Raj TVA, et al. Genomic amplification of BCR-ABL1 fusion gene and its impact on the disease progression mechanism in patients with chronic myelogenous leukemia. Gene 2019; 686:85–91. doi: 10.1016/j.gene.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Fimereli D, Fumagalli D, Brown D, Gacquer D, Rothé F, Salgado R, et al. Genomic hotspots but few recurrent fusion genes in breast cancer. Genes Chromosomes Cancer 2018; 57:331–338. doi: 10.1002/gcc.22533. [DOI] [PubMed] [Google Scholar]
- 15.Komuro A, Raja E, Iwata C, Soda M, Isogaya K, Yuki K, et al. Identification of a novel fusion gene HMGA2-EGFR in glioblastoma. Int J Cancer 2018; 142:1627–1639. doi: 10.1002/ijc.31179. [DOI] [PubMed] [Google Scholar]
- 16.Choi Y, Kwon CH, Lee SJ, Park J, Shin JY, Park DY. Integrative analysis of oncogenic fusion genes and their functional impact in colorectal cancer. Br J Cancer 2018; 119:230–240. doi: 10.1038/s41416-018-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Annala MJ, Parker BC, Zhang W, Nykter M. Fusion genes and their discovery using high throughput sequencing. Cancer Lett 2013; 340:192–200. doi: 10.1016/j.canlet.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Micci F, Gorunova L, Gatius S, Matias-Guiu X, Davidson B, Heim S, et al. MEAF6/PHF1 is a recurrent gene fusion in endometrial stromal sarcoma. Cancer Lett 2014; 347:75–78. doi: 10.1016/j.canlet.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 19.Tamura R, Yoshihara K, Yamawaki K, Suda K, Ishiguro T, Adachi S, et al. Novel kinase fusion transcripts found in endometrial cancer. Sci Rep 2015; 5:18657.doi: 10.1038/srep18657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perkel JM. Visiting “noncodarnia”. Biotechniques 2013; 54:303–304. doi: 10.2144/000114037. [DOI] [PubMed] [Google Scholar]
- 21.Hoang T, Fenne IS, Madsen A, Bozickovic O, Johannessen M, Bergsvåg M, et al. cAMP response element-binding protein interacts with and stimulates the proteasomal degradation of the nuclear receptor coactivator GRIP1. Endocrinology 2013; 154:1513–1527. doi: 10.1210/en.2012-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta P, Park SW, Farooqui M, Wei LN. Orphan nuclear receptor TR2, a mediator of preadipocyte proliferation, is differentially regulated by RA through exchange of coactivator PCAF with corepressor RIP140 on a platform molecule GRIP1. Nucleic Acids Res 2007; 35:2269–2282. doi: 10.1093/nar/gkl1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Zheng Q, Li W, Lu Y, Ni Y, Ma L, et al. SOX5 induces lung adenocarcinoma angiogenesis by inducing the expression of VEGF through STAT3 signaling. Onco Targets Ther 2018; 11:5733–5741. doi: 10.2147/ott.s176533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou H, Wang S, Wang S, Wu H, Yu J, Chen Q, et al. SOX5 interacts with YAP1 to drive malignant potential of non-small cell lung cancer cells. Am J Cancer Res 2018; 8:866–878. [PMC free article] [PubMed] [Google Scholar]
- 25.Hakimi MA, Dong Y, Lane WS, Speicher DW, Shiekhattar R. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J Biol Chem 2003; 278:7234–7239. doi: 10.1074/jbc.M208992200. [DOI] [PubMed] [Google Scholar]
- 26.Chai P, Tian J, Zhao D, Zhang H, Cui J, Ding K, et al. GSE1 negative regulation by miR-489-5p promotes breast cancer cell proliferation and invasion. Biochem Biophys Res Commun 2016; 471:123–128. doi: 10.1016/j.bbrc.2016.01.168. [DOI] [PubMed] [Google Scholar]
- 27.Lapierre M, Docquier A, Castet-Nicolas A, Gitenay D, Jalaguier S, Teyssier C, et al. The emerging role of the transcriptional coregulator RIP140 in solid tumors. Biochim Biophys Acta 2015; 1856:144–150. doi: 10.1016/j.bbcan.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 28.McQueeney KE, Salamoun JM, Ahn JG, Pekic P, Blanco IK, Struckman HL, et al. A chemical genetics approach identifies PTP4A3 as a regulator of colon cancer cell adhesion. FASEB J 2018; 32:5661–5673. doi: 10.1096/fj.201701446R. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


