Abstract
Biofilms are surface-associated bacterial communities that play both beneficial and harmful roles in nature, in medicine, and in industry. Tolerant and persister cells are thought to underlie biofilm-related bacterial recurrence in medical and industrial contexts. Here, we review recent progress aimed at understanding the mechanical features that drive biofilm resilience and the biofilm formation process at single-cell resolution. We discuss findings regarding mechanisms underlying bacterial tolerance and persistence in biofilms and how these phenotypes are linked to antibiotic resistance. New strategies for combatting tolerance and persistence in biofilms and possible methods for biofilm eradication are highlighted to inspire future development.
Introduction
We live in societies made of individuals with interacting social connections and enduring architectural infrastructures. On a six order of magnitude smaller scale, bacterial cells also build microbial cities called biofilms in which individual cells and groups of cells interact and a global infrastructure is assembled. Biofilms are surface-attached communities of bacteria embedded in an extracellular matrix (Hall-Stoodley et al., 2004). Biofilms can be beneficial for health, for example, as normal components of plant, animal, and human microbiomes and they can be crucial for effective industrial processes such as wastewater treatment. However, often, biofilms cause major problems: in medicine, biofilms underlie chronic infections, and in industry, biofilms foul surfaces of pipes and clog filtration devices.
Biofilm eradication, whether in medicine or industry, is remarkably difficult. One feature thought to underlie biofilm tenacity is that biofilm communities can harbor tolerant and persister cells (Lewis, 2005): cells that can survive transient antibiotic treatment and that regrow when the antibiotic is withdrawn (Brauner et al., 2016). Indeed, both hyper-biofilm-forming mutants and mutants exhibiting enhanced persistence are isolated from patients with chronic infections (Hall-Stoodley et al., 2004; Lewis, 2010). In this Mini Review, we summarize recent progress in the understanding of biofilm formation, focusing on the mechanical attributes of biofilms that endow them with their remarkable resilience. We highlight progress aimed at defining mechanisms underlying the tolerant and persister phenotypes. Finally, we provide an overview of exciting new strategies for combatting harmful bacterial biofilms.
Biofilm matrices: mechanical shelters for bacterial cells
A defining feature of a biofilm is the presence of the extracellular matrix, made up of extracellular polymeric substances (EPS) secreted by the cells dwelling inside (Hall-Stoodley et al., 2004). The EPS is usually a mixture of polysaccharides, proteins, extracellular DNA (eDNA), and other minor components. The physical and chemical properties of the biofilm matrix constituents coupled with their particular interactions give rise to the global biofilm mechanical properties. These properties allow the matrix to shield the resident cells from desiccation, chemical perturbation, invasion by other bacteria, and killing by predators. The matrix also provides the mechanical properties necessary to protect the cells from external forces such as fluid shear and to ensure the biofilm community remains attached to a surface. In the context of infectious biofilms, neutrophils can only ingest pathogens smaller than 10 μm, thus, participating in the biofilm lifestyle protects individual bacteria and small bacterial clusters from neutrophil attack. Moreover, to access biofilm-dwelling bacteria, neutrophils need to first break biofilms (~100 μm) into smaller pieces. However, neutrophils can only exert stress up to ~1 kPa during phagocytosis (Kovach et al., 2017), so biofilm mechanics could potentially prevent neutrophils from making biofilm cells available for killing.
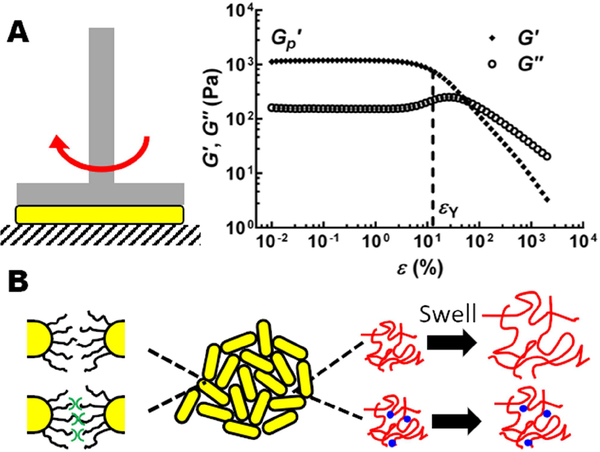
Tools and concepts from the rheology field have been adapted to quantitatively define biofilm mechanical properties. Rheology is the study of viscoelastic materials: materials that have both solid and liquid properties (Billings et al., 2015). For rheologic measurements, biofilms are sandwiched between parallel plates and subjected to shearing (Figure 1A, Left). These analyses define the elastic modulus, which is the stiffness of the biofilm at small deformation, and the yield strain, which is how much deformation a biofilm can sustain before it fails (Figure 1A, Right) (Kovach et al., 2017). The product of the elastic modulus and the yield strain defines the yield stress, which is the minimum force needed to cause a biofilm to fail. Below we summarize insight gained from recent rheological measurements of three model biofilm forming species: Vibrio cholerae, Pseudomonas aeruginosa, and Staphylococcus epidermidis.
Figure 1.
Biofilm mechanics. (A) Left: schematic of a rheometer setup for measuring biofilm mechanics. Biofilms (yellow) are sandwiched between a rotating (red arrow) shaft and a stationary plate (gray stripes). Right: Representative storage modulus G’ and loss modulus G” curves as a function of shear strain ε measured for a V. cholerae biofilm. G’ and G” correspond to the elastic and viscous responses of the biofilm, respectively. From the curve, the elastic modulus G’p and the yield strain εY are extracted. (B) Schematic of V. cholerae biofilm matrix components and how they contribute to biofilm mechanical properties. Cells (yellow cylinders) interact through surface lipopolysaccharides (black curvy lines) or by crosslinking via RbmA (green symbols). VPS (red wavy lines) is crosslinked by RbmC and Bap1, both depicted as blue dots. Removal of RbmC and Bap1 causes the VPS to swell. Images are adapted from Yan et al., 2018.
V. cholerae is the causative agent of the pandemic disease cholerae. The major V. cholerae biofilm matrix component is the Vibrio polysaccharide (VPS) and there are three matrix proteins RbmA, Bap1, and RbmC (Teschler et al., 2015). Deletion of genes encoding matrix components, followed by rheological measurements, enabled the mechanical properties of V. cholerae biofilms to be defined (Yan et al., 2018). The V. cholerae biofilm can be described as a double-networked hydrogel with an elastic modulus of ~1kPa. One network is formed by the VPS polysaccharide reinforced by RbmC and Bap1, and the second network is formed by the cells connected by RbmA (Figure 1B). Elimination of RbmA or RbmC/Bap1 weakens the dual network and reduces the elastic modulus. Elimination of all three matrix proteins causes the VPS to swell, resulting in an increased yield strain but at the expense of a highly reduced elastic modulus. Only when all the matrix components are present do V. cholerae biofilms possess a large enough yield stress (~100 Pa) to withstand the flow regimes they experience in their natural habitat, for example, on sinking marine snow.
P. aeruginosa is an opportunistic pathogen that forms chronic biofilm infections in patients with compromised immune systems, burns, in-dwelling devices, and cystic fibrosis (CF). Combinations of three polysaccharides can be present in the P. aeruginosa biofilm matrix: Psl, Pel, and alginate (Kovach et al., 2017). Rheological measurements using the model virulent strain PAO1 and isolates from CF lungs show that Psl, together with its cross-linking protein CdrA, are the main contributors to the biofilm elastic modulus (Kovach et al., 2017). Overproduction of Pel increases the biofilm yield strain but does not alter the elastic modulus. Pel is positively changed and it binds eDNA (Jennings et al., 2015). Pel-eDNA interactions could be instrumental in driving overall biofilm mechanics, but this aspect has not yet been studied. Mucoid P. aeruginosa biofilms that overproduce alginate are fluid-like, possessing reduced elastic modulus and reduced yield stress compared to PAO1 biofilms (Gloag et al., 2018). In CF patients, cells in P. aeruginosa biofilms tend to increase both alginate and Psl production. Alginate overproduction causes a decrease in yield stress that is compensated for via overproduction of Psl. Together, these alterations enable the biofilm to preserve its original yield stress (Kovach et al., 2017). It is possible that maintaining a minimum yield stress is required for P. aeruginosa biofilm cells to avoid immune clearance. The unique combinations of matrix components displayed by different P. aeruginosa strains suggest that such blends promote distinct biofilm mechanical properties, each presumably optimized for a particular environmental condition.
S. epidermidis is a member of the human skin microbiome but also occurs in medical device and hospital acquired infections. The main S. epidermidis matrix component is called polysaccharide intercellular adhesin (PIA), a positively charged polymer (Otto, 2009). At low pH, purified PIA in solution possesses concentration-dependent viscoelasticity that is well described by the classical model of associative polymers: polymers that can both physically entangle and chemically interact through hydrogen bonding (Ganesan et al., 2016). The main contribution to biofilm rheology, however, stems from chemical interactions, as the concentration of PIA in biofilms is too low for physical entanglement. At pH = 7 (or lower), PIA associates, becomes unstable in solution, and it phase separates together with the S. epidermidis cells to form biofilm-like structures with rheological properties similar to native S. epidermidis biofilms (Stewart et al., 2015). Indeed, simply increasing the pH above 7 was sufficient to stabilize PIA and make S. epidermidis biofilms more malleable. The strong pH-dependent phase behavior exhibited by PIA suggests S. epidermidis biofilms may possess distinct mechanical properties in particular local infection environments.
Biofilm architectures: from individual cells to macroscopic communities
Until recently, there was little understanding of how cells are arranged within biofilms and how 3D biofilm structures are built cell by cell. Custom high-resolution confocal microscopy technologies and companion imaging analysis algorithms were developed that enabled investigation of biofilms at single cell resolution. Initially, high-resolution confocal laser scanning microscopy was used to extract spatial information regarding fixed cells in S. epidermidis biofilms (Stewart et al., 2013). By tracking the centers of the spherical cells and analyzing the local cell density and cluster distribution, local biofilm compactness parameters were defined and were discovered to vary within an S. epidermidis biofilm. In regions with high and medium cell density, nearly all of the cells were present in a single cluster that exhibited characteristics of a dense disordered fluid. In regions of sparse cell density, cell clusters displayed open, fractal features similar to colloidal gels. Upon osmotic stresses (high salt concentration) or antibiotic challenge (vancomycin), however, the S. epidermidis biofilm structure only exhibited the low-density phenotype. The mechanism(s) driving these regional packing differences is unclear. We hypothesize that, as highlighted in the preceding section, local variations in pH or in PIA concentration that alter PIA solution behavior could lead to distinct biofilm packing density phenotypes.
Images of fixed V. cholerae cells obtained at different times during biofilm formation were acquired to learn how cell arrangement changes as biofilms mature (Drescher et al., 2016). The community transitions from a 2D branched morphology to a dense 3D cluster. In the mature V. cholerae biofilm cluster, vertical cells reside at the biofilm center and radially orientated cells are at the periphery. This entire sequence of structural transitions was subsequently visualized in living, growing V. cholerae biofilms (Figure 2A–B) (Yan et al., 2016). Mutagenesis coupled with matrix labeling showed that V. cholerae biofilms lacking cell-surface adhesion due to deletion of rbmC and bap1 exhibit normal cell density but show no cell ordering. By contrast, biofilms lacking cell-cell connections due to deletion of rbmA display reduced cell packing density and enhanced vertical cell alignment.
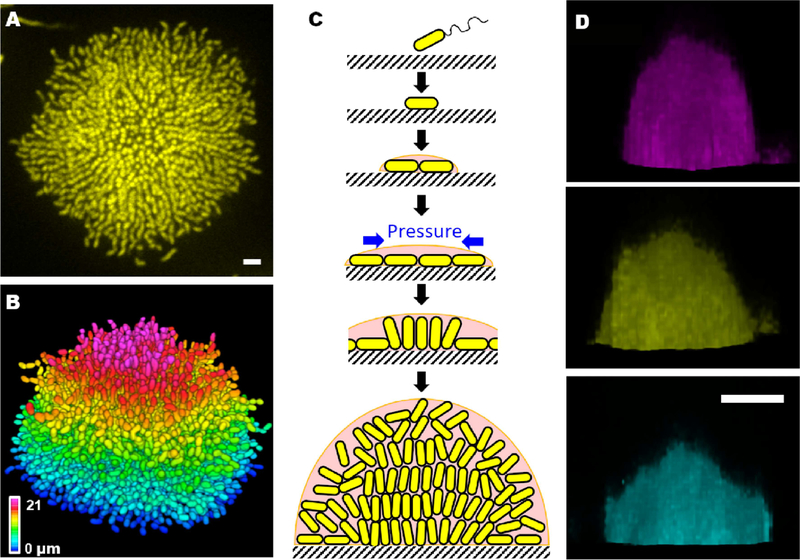
Figure 2.
Biofilm formation process. (A) Cross-sectional image of the bottom cell layer of a growing V. cholerae biofilm cluster at 18 h and (B) the corresponding segmented image with color-coding according to z position. Scale bar: 3 μm. (C) Schematic representation of the steps in the V. cholerae biofilm formation process. Cells are in yellow and the matrix is in pink. (D) Side views of 7 h old biofilms grown with 0.4 μg/mL A22 (magenta), without treatment (yellow) and with 4 μg/mL cefalexin (cyan). A22 and cefalexin cause the cells to become shorter and longer, respectively. Scale bar, 10 μm. Images in A-C are adapted from Yan et al., 2016. Images in D are adapted from Beroz et al., 2018.
To explore the forces driving structural transitions in V. cholerae biofilms, agent-based simulations were developed to investigate cell-surface interactions (Beroz et al., 2018). When a biofilm begins to form on a surface, it expands outward from the founder cell, as a one-cell-layer thick 2D film. During expansion, cells experience increasing mechanical pressure as they divide and push against their neighbors. These neighboring cells, in turn, resist the pushing force via surface adhesion. Ultimately, the pressure from pushing exceeds the cell-to-surface adhesion force and causes individual cells to reorient at the center of the biofilm where the pressure is the greatest. Cells transition from aligning parallel to aligning perpendicular to the substrate. When verticalized cells divide, they place their offspring further into the third dimension, thus the biofilm gradually transitions from a 2D surface layer to a mature 3D community (Figure 2C). To bolster these theoretical arguments, V. cholerae cell lengths were manipulated using chemicals. The timing of verticalization was altered: biofilms with shorter (longer) cells transitioned from 2D to 3D earlier (later) than cells of normal length, because shorter (longer) cells required lower (higher) critical forces to drive verticalization. A consequence of altering cell length was to change the overall width to height ratio of the resulting biofilm (Figure 2D).
Tolerance and persistence: how bacterial cells survive antibiotic challenge
Antibiotic resistance is caused by mutations that make a bacterial cell impervious to the toxic effect of the antibiotic, endowing that cell and its descendants with a selective growth advantage over non-resistant cells. Beyond classic resistance mechanisms, bacteria can display “tolerance”, the ability to survive transient exposure to high concentrations of an antibiotic (Brauner et al., 2016). Tolerant bacteria grow slower or have longer non-growing lag times when they exit stationary phase than their non-tolerant counterparts. Common targets of antibiotics, e.g., RNA polymerase, cell-wall biosynthetic enzymes, exhibit low activity in non-growing cells, and thus, slow-growing or non-growing cells can evade killing. In this respect, tolerance differs fundamentally from resistance, as resistance is usually specific to one antibiotic or one class of antibiotics.
Tolerant cells display a longer minimum duration of killing by an antibiotic than non-tolerant cells, enabling tolerant cells to enjoy a selective advantage during transient or periodic antibiotic treatment (Brauner et al., 2016). Indeed, tolerant Escherichia coli cells spontaneously arose after repeated cycles of ampicillin treatment (Fridman et al., 2014). The increased lag time the E. coli cells exhibited matched the duration of ampicillin exposure. Because antibiotic treatment usually occurs in timed doses, patients experience periodic fluctuations in antibiotic concentration, likely favoring the emergence of tolerant cells.
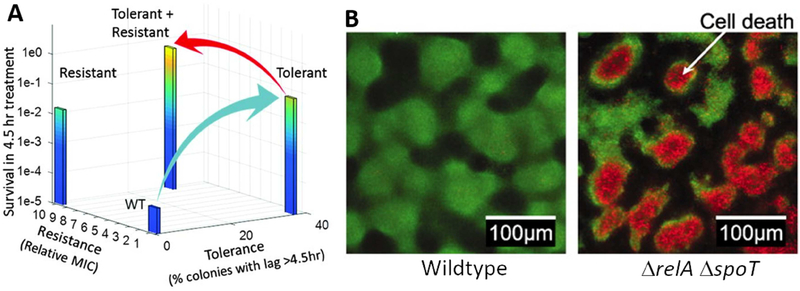
Mutations in genes encoding a methionyl-tRNA synthetase, ribose-phosphate diphosphokinase, and toxin-antitoxin (TA) modules all promote tolerance by extending lag phase (Fridman et al., 2014). It is hypothesized that a sequential relationship exists between bacterial tolerance and bacterial resistance. Indeed, in an experiment probing periodic ampicillin treatment that mimicked medical practice, resistant E. coli strains isolated at the end of the experiment all arose from ancestral, tolerant strains (Figure 3A) (Levin-Reisman et al., 2017). The logic is that tolerance mutations occur more frequently than resistance mutations due to a larger target size of the former: there are many genes that when mutated confer tolerance while mutations in only a few genes confer resistance to a particular antibiotic. Once a tolerant mutant becomes established in the population, its presence gives the rarer, resistance mutations more opportunities to occur (Brauner et al., 2016; Levin-Reisman et al., 2017).
Figure 3.
Tolerance and persistence in bacterial biofilms. (A) Experimental trajectory of E. coli cells from tolerant to resistant in response to periodic treatment with 50 μg/mL ampicillin. Wildtype E. coli cells transition to tolerant (blue arrow), and subsequently, to resistant (red arrow). MIC is defined as the minimum inhibitory antibiotic concentration that prevents bacterial growth. See Levin-Reisman et al., 2017. (B) Confocal images of P. aeruginosa biofilms following viability staining. Live cells are green and dead cells are red. Spontaneous cell death in the absence of antibiotics as shown in the figure, occurs in the interior, nutrient limited region of of ΔrelA ΔspoT biofilms but not in wildtype biofilms. The image in A is adapted from Levin-Reisman et al., 2017. Images in B are adapted from Nguyen et al., 2011.
Another form of tolerance, not obtained through heritable mutations but rather through phenotypic differentiation, is called persistence (Balaban et al., 2004; Lewis, 2005). Originally observed by Bigger (Bigger, 1944), bacterial persistence is receiving renewed interest due to its medical relevance, in particular, in the context of biofilms (Lewis, 2005). Time dependent antibiotic killing of a bacterial population shows that actively growing cells are killed first whereas persister cells are killed in a second phase at a much lower rate. Visualization of individual bacterial cells established that, prior to antibiotic treatment, an exponentially growing bacterial population contains a pre-existing fraction of non-growing cells (Balaban et al., 2004). It is this sub-population that survives antibiotic treatment and regrows after the antibiotic is withdrawn. Another source of persister cells are those that have become dormant during stationary phase. Such cells are simply carried over to the new culture upon sub-culturing.
The mechanisms driving subpopulations of cells to enter the persistent state are the subject of intense research and debate. One mechanism involves toxin-antitoxin (TA) modules (Lewis, 2005; Rotem et al., 2010). Indeed, the first identified high-persistence E. coli strain harbors a mutation in hipAB encoding a TA module (Moyed and Bertrand, 1983). The HipA toxin is a serine-protein kinase that phosphorylates GltX, a glutamyl-tRNA synthetase. HipA is inactivated by the companion antitoxin HipB (Schumacher et al., 2015). When HipA levels exceed a threshold in a cell due to stochastic fluctuations, protein synthesis is inhibited, and as a consequence, cell growth is arrested (Rotem et al., 2010). Growth-arrested cells can become persisters. The originally isolated high-persister strain possesses a mutation that impairs HipA-HipB binding (Schumacher et al., 2015), which increases the chances of cells of this strain entering the growth-arrested state. TA modules do not appear to underlie persistence in Staphylococcus aureus as elimination of all TAs had no effect on persister cell formation (Conlon et al., 2016). Rather, some S. aureus cells stochastically enter into stationary phase earlier than others to become persister cells. In this case, stationary phase entry is accompanied by a decrease in intracellular ATP levels, which, in turn, reduces the activity of ATP-dependent antibiotic targets (DNA gyrase, DNA topoisomerase, RNA polymerases, etc.). Therefore, stationary phase S. aureus cells are naturally prone to becoming persister cells. Lastly, genomic studies of E. coli and P. aeruginosa have identified many metabolic genes connected to persister cell formation (Amato et al., 2014).
Normally, persister cells make up from 10−2 to 10−5 of a population, so such cells might seemingly not be clinically relevant given that the goal of antibiotic treatment is to eliminate the majority of actively growing pathogens and to expect the immune system to clear the remainder (Lewis, 2010). However, persister cells may be dangerous to particular patient populations. In immunocompromised individuals, persister cells can likely regrow. In some diseases such as tuberculosis, antibiotic treatment must drive pathogens to very low numbers to achieve a clinical outcome. In such cases, persister cells could be problematic. In chronic infections, such as those in CF patients, high-persister mutants can be isolated after prolonged antibiotic treatment (Lewis, 2010). In these real-life cases, persister cells could be present and not eliminated by current drug regimes.
Tolerance and persistence in bacterial biofilms
The ability of biofilms to house tolerant and persister cells is proposed to underlie the difficulties encountered in eliminating biofilms during chronic infections (Lewis, 2005). Impeded antibiotic penetration into biofilms was initially proposed to be responsible, however it is now known that the matrix mesh size is much larger than antibiotic molecules (Ganesan et al., 2016; Yan et al., 2018), and most antibiotics do not interact strongly with biofilm matrix components (Spoering and Lewis, 2001). Rather, increased antibiotic tolerance and persistence in biofilms likely arises from altered physiology of biofilm cells. Cells buried inside thick biofilms could be in stationary phase, as penetration of nutrients and oxygen are known to be limited due to consumption by peripherally-located cells (Walters et al., 2003). Indeed, increasing evidence supports similarities between the physiological states of biofilm-dwelling cells and stationary phase planktonic cells. For example, the levels of persister cell formation by P. aeruginosa are comparable in the biofilm state and in stationary phase (Spoering and Lewis, 2001). Likewise, antibiotic tolerance phenotypes of S. aureus biofilm cells, stationary phase planktonic cells, and persister cells are strikingly similar. (Waters et al., 2016). Nutrient starvation, a common environmental situation encountered during both biofilm formation and entrance into stationary phase could promote antibiotic tolerance and persister cell formation by triggering the stringent response (Nguyen et al., 2011). In P. aeruginosa biofilms, disabling the stringent response via deletion of both relA and spoT leads to a 1,000-fold reduction in cell survival upon antibiotic treatment. The ΔrelA ΔspoT mutant cells possess impaired antioxidant defenses and increased oxidant production, which, together, sensitize the cells to antibiotic treatment and to nutrient limitation. Indeed, even in the absence of antibiotics, spontaneous death of the ΔrelA ΔspoT P. aeruginosa cells occurs in the nutrient-limited interior regions of biofilm clusters (Figure 3B). Although there are some results suggesting specific genes drive persister cell formation exclusively in biofilms (Harrison et al., 2009), the current notion is that mechanisms underlying persister cell formation under planktonic conditions apply to persister cell formation in biofilms.
Even if the mechanisms giving rise to persister cells in biofilms and in planktonic environments are similar or identical, persister cells in in vivo biofilms could be particularly tenacious because the biofilm matrix provides a physical barrier that protects the persister cells from immune components (Lewis, 2005). As mentioned above, in vivo biofilm stiffness could exceed the maximum mechanical stress neutrophils are capable of exerting, in the present context, preventing neutrophils from accessing persister cells buried deep in the interior of a biofilm. Moreover, the rigidity of the biofilm matrix scaffold remains even if the majority of the biofilm cells have been killed by antibiotics (Zrelli et al., 2013). Such residual structures could harbor tolerant or persister cells that can regrow and cause recurrent infections.
New strategies to target tolerant and persister cells in biofilms
The notion that biofilms provide a “safe haven” for persister cells to arise and evade antibiotics and immune components suggests that entire biofilm structures must be removed from infection sites for successful elimination of pathogens. Mechanical debridement (scraping of biofilms from wounds) is the standard-of-care for chronic wounds (Gordon et al., 2017). However, it is difficult to completely remove all cells once a biofilm is sheared into pieces. Moreover, this arduous process can only be applied to infected regions that are exposed and thus amenable to mechanical manipulation. To address this challenge, a capillary peeling method has been developed in which liquid is slowly applied to a biofilm grown at an air-solid interface and capillary forces gently peel the biofilm off in its entirety (Yan et al., 2018). This method applies to a variety of surfaces including metals, hydrogels, and membranes and to biofilms formed by different bacterial species. This new method does not yet address situations in which biofilms are submerged or reside internally in patients.
In instances of internal biofilms, chemicals that induce biofilm dispersal have been pursued alone or in combination with antibiotic treatment. Examples include Dispersin B that degrades poly-N-acetylglucosamine, a common biofilm matrix component (McDougald et al., 2012). An unsaturated fatty acid produced by P. aeruginosa, cis-2-decenoic acid, can trigger the dispersal of cells from biofilms formed by a range of bacteria including P. aeruginosa itself (Davies and Marques, 2009). Once the bacterial cells are dispersed into the solution, they become vulnerable to clearance by the immune system and/or killing by antibiotics. To date, Dispersin B is marketed as an ingredient in a wound care gel and as a medical device coating. In both cases, when Dispersin B is combined with antibiotics, the compound shows efficacy in prevention of bacterial infections (Kaplan, 2010).
Regarding targeting and eliminating tolerant or persistent cells, E. coli and S. aureus persister cells in biofilms can be re-sensitized to an aminoglycoside antibiotic by providing metabolites that generate a proton-motive force facilitating aminoglycoside uptake (Allison et al., 2011). Interestingly, the above dispersal promoting molecule cis-2-decenoic acid can also transform P. aeruginosa and E. coli persister cells from dormant to metabolically active (Marques et al., 2014). Again, this change renders the cells susceptible to antibiotics. Another good example is provided by the acyldepsipeptide antibiotic ADEP4, which kills persister cells in S. aureus biofilms via activation of ClpP (Conlon et al., 2013). ADEP4-activated ClpP exhibits promiscuous protease activity, cleaves essential proteins, and causes persister cells to die. Together with rifampicin, ADEP4 treatment successfully eradicated S. aureus biofilms both in vitro and in a mouse model.
Other clever strategies to eliminate chronic infections are being developed and have potential for use to combat persister cells in biofilms. Here, we provide one new example as a representative to highlight these emerging applications. The membranes of S. aureus and other Gram-positive bacteria contain functional membrane microdomains (FMM) similar to lipid rafts in eukaryotic membranes. The FMM of methicillin-resistant S. aureus (MRSA) contain a high level of staphyloxanthin, an essential membrane-bound antioxidant. Illuminating MRSA with blue light promotes degradation of staphyloxanthin and sensitizes MRSA to reactive oxygen attack, both in the planktonic and biofilm states. Treatment with light was effective in a mouse wound infection model (Dong et al., 2019). Whether such a mechanism is generalizable remains to be tested, and the delivery method (light in this case) might also restrict its use due to penetration issues for thick tissues.
Perspectives
Biofilm formation and persister cell formation can be viewed as two types of collective bacterial behaviors. In the case of biofilm formation, bacterial cells collectively produce extracellular matrices, a public good that profits the entire community. In the case of persister cell formation, the entire population benefits when, a subpopulation of cells survives a hostile environment in which the majority of cells perish. Combining these two collective lifestyles endows the bacteria with powerful mechanisms to survive harsh perturbations, including mechanical stress and antibiotic treatment. This good news for the bacteria presents humanity with a serious challenge regarding chronic infections.
To address this challenge, a deeper understanding of the biofilm formation process, biofilm mechanics, tolerance, and persistence is necessary. We expect new technologies to provide insight into how biofilm mechanics arise from particular steps in biofilm development and the features mechanics provide to these living structures. Simultaneous single-cell resolution biofilm imaging and rheological measurements will allow interesting questions to be answered including: what happens at the single-cell level during biofilm failure or detachment? How does the biofilm internal structure evolve as the biofilm relaxes stress? How does local cellular configuration determine the local biofilm stiffness and do weak regions exist in biofilms that could be exploited to drive failure? Such measurements could provide a comprehensive understanding of biofilms as dynamic living materials.
Regarding tolerant and persister cells, additional evidence is required to confirm their presence in biofilms, especially in vivo during disease. Single-cell visualization of the process of tolerant/persister cell birth to non-growth to rejuvenation in a biofilm will yield valuable information. Questions that can be addressed include: Where and when do tolerant/persister cells arise in spatially-structured biofilms and what is their spatial distribution? Which cells enjoy the mechanical protection provided by the matrix, what selects them, and are their locations pre-defined or random? How do persistent/tolerant cells deal with debris left by cells that have been killed by antibiotics? Ultimately, a deep understanding of the behaviors of all cells in biofilms, both at the individual and collective levels, could lead to currently unimaginable strategies to combat harmful biofilms and to promote beneficial biofilms.
Biofilms are surface-associated bacterial communities that play both beneficial and harmful roles in nature, in medicine, and in industry. Here, we review recent progress aimed at understanding the mechanical features that drive biofilm resilience, and the mechanisms underlying why biofilm-dwelling cells are tolerant and persistent to antibiotic treatment.
Acknowledgements
This work was supported by the Howard Hughes Medical Institute, National Science Foundation Grant MCB-1713731, NIH Grant 2R37GM065859, and the Max Planck Society-Alexander von Humboldt Foundation (B.L.B.). J.Y. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allison KR, Brynildsen MP, and Collins JJ (2011). Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473, 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato S,M, Fazen CH, Henry TC, Mok WWK, Orman MA, Sandvik EL, Volzing KG, and Brynildsen MP (2014). The role of metabolism in bacterial persistence. Front. Microbiol 5, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, and Leibler S (2004). Bacterial persistence as a phenotypic switch. Science 305, 1622–1625. [DOI] [PubMed] [Google Scholar]
- Beroz F, Yan J, Meir Y, Sabass B, Stone HA, Bassler BL, and Wingreen NS (2018). Verticalization of bacterial biofilms. Nat. Phys 14, 954–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger JW (1944). Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244, 497–500. [Google Scholar]
- Billings N, Birjiniuk A, Samad TS, Doyle PS, and Ribbeck K (2015). Material properties of biofilms — a review of methods for understanding permeability and mechanics. Rep. Prog. Phys 78, 036601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauner A, Fridman O, Gefen O, and Balaban NQ (2016). Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol 14, 320–330. [DOI] [PubMed] [Google Scholar]
- Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, and Lewis K (2013). Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, and Lewis K (2016). Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol 1, 16051. [DOI] [PubMed] [Google Scholar]
- Davies DG, and Marques CNH (2009). A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol 191, 1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P-T, Mohammad H, Hui J, Leanse LG, Li J, Liang L, Dai T, Seleem MN, and Cheng J-X (2019). Photolysis of staphyloxanthin in methicillin-resistant Staphylococcus aureus potentiates killing by reactive oxygen species. Adv. Sci 1900030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher K, Dunkel J, Nadell CD, van Teeffelen S, Grnja I, Wingreen NS, Stone HA, and Bassler BL (2016). Architectural transitions in Vibrio cholerae biofilms at single-cell resolution. Proc. Natl. Acad. Sci. USA 113, E2066–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman O, Goldberg A, Ronin I, Shoresh N, and Balaban NQ (2014). Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513, 418–421. [DOI] [PubMed] [Google Scholar]
- Ganesan M, Knier S, Younger JG, and Solomon MJ (2016). Associative and entanglement contributions to the solution rheology of a bacterial polysaccharide. Macromolecules 49, 8313–8321. [Google Scholar]
- Gloag ES, German GK, Stoodley P, and Wozniak DJ (2018). Viscoelastic properties of Pseudomonas aeruginosa variant biofilms. Sci. Rep 8, 9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon VD, Davis-Fields M, Kovach K, and Rodesney Christopher.A. (2017). Biofilms and mechanics: a review of experimental techniques and findings. J. Phys. D 50, 223002. [Google Scholar]
- Hall-Stoodley L, Costerton JW, and Stoodley P (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol 2, 95–108. [DOI] [PubMed] [Google Scholar]
- Harrison J,J, Wade W,D, Akierman S, Vacchi-Suzzi C, Stremick CA, Turner R,J, and Ceri H (2009). The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob. Agents Chemother 53, 2253–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, Secor PR, Tseng BS, Scian M, Filloux A, et al. (2015). Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 112, 11353–11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JB (2010). Biofilm Dispersal: Mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res 89, 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach K, Davis-Fields M, Irie Y, Jain K, Doorwar S, Vuong K, Dhamani N, Mohanty K, Touhami A, and Gordon VD (2017). Evolutionary adaptations of biofilms infecting cystic fibrosis lungs promote mechanical toughness by adjusting polysaccharide production. NPJ Biofilms Microbiomes 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, and Balaban NQ (2017). Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830. [DOI] [PubMed] [Google Scholar]
- Lewis K (2005). Persister cells and the riddle of biofilm survival. Biochemistry (Mosc) 70, 267–274. [DOI] [PubMed] [Google Scholar]
- Lewis K (2010). Persister cells and the paradox of chronic infections. Microbe 5, 429–437. [Google Scholar]
- Marques CNH, Morozov A, Planzos P, and Zelaya HM (2014). The fatty acid signaling molecule cis-2-decenoic acid increases metabolic activity and reverts persister cells to an antimicrobial-susceptible state. Appl. Environ. Microbiol 80, 6976–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougald D, Rice SA, Barraud N, Steinberg PD, and Kjelleberg S (2012). Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol 10, 39–50. [DOI] [PubMed] [Google Scholar]
- Moyed HS, and Bertrand KP (1983). hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol 155, 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, et al. (2011). Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M (2009). Staphylococcus epidermidis — the “accidental” pathogen. Nat. Rev. Microbiol 7, 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem E, Loinger A, Ronin I, Levin-Reisman I, Gabay C, Shoresh N, Biham O, and Balaban NQ (2010). Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc. Natl. Acad. Sci. USA 107, 12541–12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Balani P, Min J, Chinnam NB, Hansen S, Vulić M, Lewis K, and Brennan RG (2015). HipBA–promoter structures reveal the basis of heritable multidrug tolerance. Nature 524, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoering AL, and Lewis K (2001). Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol 183, 6746–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart EJ, Satorius AE, Younger JG, and Solomon MJ (2013). Role of environmental and antibiotic stress on Staphylococcus epidermidis biofilm microstructure. Langmuir 29, 7017–7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart EJ, Ganesan M, Younger JG, and Solomon MJ (2015). Artificial biofilms establish the role of matrix interactions in staphylococcal biofilm assembly and disassembly. Sci. Rep 5, 13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschler JK, Zamorano-Sanchez D, Utada AS, Warner CJA, Wong GCL, Linington RG, and Yildiz FH (2015). Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat. Rev. Microbiol 13, 255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters MC, Roe F, Bugnicourt A, Franklin MJ, and Stewart PS (2003). Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother 47, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters EM, Rowe SE, O’Gara JP, and Conlon BP (2016). Convergence of Staphylococcus aureus persister and biofilm research: can biofilms be defined as communities of adherent persister cells? PLOS Pathog. 12, e1006012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Sharo AG, Stone HA, Wingreen NS, and Bassler BL (2016). Vibrio cholerae biofilm growth program and architecture revealed by single-cell live imaging. Proc. Natl. Acad. Sci. USA 113, E5337–E5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Moreau A, Khodaparast S, Perazzo A, Feng J, Fei C, Mao S, Mukherjee S, Košmrlj A, Wingreen NS, et al. (2018). Bacterial biofilm material properties enable removal and transfer by capillary peeling. Adv. Mater 30, 1804153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrelli K, Galy O, Latour-Lambert P, Kirwan L, Ghigo JM, Beloin C, and Henry N (2013). Bacterial biofilm mechanical properties persist upon antibiotic treatment and survive cell death. New J. Phys 15, 125026. [Google Scholar]