Abstract
Nano-titanium dioxide (nano-TiO2), though one of the most utilized and produced engineered nanomaterials (ENMs), diminishes cardiovascular function through dysregulation of metabolism and mitochondrial bioenergetics following inhalation exposure. The molecular mechanisms governing this cardiac dysfunction remain largely unknown. The purpose of this study was to elucidate molecular mediators that connect nano-TiO2 exposure with impaired cardiac function. Specifically, we were interested in the role of microRNA (miRNA) expression in the resulting dysfunction. Not only are miRNA global regulators of gene expression, but miRNA-based therapeutics provide a realistic treatment modality. Wild type and MiRNA-378a knockout mice were exposed to nano-TiO2 with an aerodynamic diameter of 182 ± 1.70 nm and a mass concentration of 11.09 mg/m3 for four hours. Cardiac function, utilizing the Vevo 2100 Imaging System, electron transport chain complex activities, and mitochondrial respiration assessed cardiac and mitochondrial function. Immunoblotting and qPCR examined molecular targets of miRNA-378a. MiRNA-378a-3p expression was increased 48 hours post inhalation exposure to nano-TiO2. Knockout of miRNA-378a preserved cardiac function following exposure as revealed by preserved E/A ratio and E/SR ratio. In knockout animals, complex I, III, and IV activities (~2–6 fold) and fatty acid respiration (~5 fold) were significantly increased. MiRNA-378a regulated proteins involved in mitochondrial fusion, transcription, and fatty acid metabolism. MiRNA-378a-3p acts as a negative regulator of mitochondrial metabolic and biogenesis pathways. MiRNA-378a knockout animals provide a protective effect against nano-TiO2 inhalation exposure by altering mitochondrial structure and function. This is the first study to manipulate a miRNA to attenuate the effects of ENM exposure.
Keywords: Cardiovascular, Mitochondria, Inhalation Exposure
Introduction
The advent and advancement of clinical applications continues to drive utilization of engineered nanomaterials (ENMs) in consumer goods (Dong et al., 2014, Besinis et al., 2015). As such, titanium dioxide (TiO2) has become one of the most prolifically implemented ENMs (Hendren et al., 2011, Gottschalk et al., 2015). While the beneficial manufacturing properties of ENMs are clear, the rate of incorporation in new products and devices outpaces our understanding of the toxicology of these materials. Though progress has been made outlining the consequences on organ systems and the physiological impact of ENM exposure, the molecular mechanisms governing their toxicity are poorly defined. Understanding how to manage the untoward consequences of ENM inhalation exposure therapeutically involves the exploitation of molecular pathways central to the etiology.
Systemic health effects following ENM exposure are linked to mitochondrial dysfunction, not only in the lungs (Ruenraroengsak and Tetley, 2015) but also in the cardiovascular system (Stapleton et al., 2015, Hathaway et al., 2017a, Nichols et al., 2018). The heart has been established as a significant contributor to the overall pathophysiological outcomes elicited by ENM exposure (Li et al., 2007, Kan et al., 2012, Stapleton et al., 2012, Holland et al., 2016). Cardiac mitochondrial function is crucial in maintaining/restoring cardiac homeostasis after ENM insult, as it is a central determinant of metabolic and bioenergetic capacity. Recently, particulate matter (PM2.5) air pollution has been shown to alter microRNA (miRNA) expression in the serum of a human cohort in Shanghai, China (Chen et al., 2018). The correlation between serum miRNA expression and inflammatory cytokines involved in cardiovascular health reveals the clinical relevance of miRNAs in shaping the effects of inhalation exposure. Because miRNAs can influence numerous biological target axes, therapeutic interventions targeting their regulation offer distinct advantages to approaches designed to restore a single molecular target as they provide the potential for a coordinated response to pathological insults at numerous loci.
MiRNAs are transcribed as pri-miRNA species within the nucleus (processed through Drosha) and travel to the cytoplasm as pre-miRNA (processed through Dicer) where they eventually assume their mature miRNA conformation (Shukla et al., 2011). The mature, miRNA species (~22 nt) that originate in both intragenic and intergenic regions of the genome are known to individually function in regulating multiple genetic and epigenetic pathways (Sato et al., 2011). The manipulation of a miRNA that controls metabolic and mitochondrial function provides a potentially novel therapeutic strategy for controlling cellular insults precipitated by ENM exposure. In the heart, and other mitochondrially dense tissue, peroxisome proliferator-activated receptor gamma coactivator 1-beta (Ppargc1b or PGC-1β) is highly expressed (St-Pierre et al., 2003), and is central in the control of metabolism (Carrer et al., 2012). MiRNA-378a, which encodes both the 3p and 5p mature variants, resides within the first intron of the PGC-1β gene.
MiRNA-378a is involved in regulating apoptosis (Kim et al., 2013), insulin sensitivity (Knezevic et al., 2012), angiogenesis (Hua et al., 2006), metabolism (Carrer et al., 2012), and other cellular processes (Gagan et al., 2011, Wang et al., 2012) implicating it in the coordination of cellular responses. Though multiple miRNAs regulate heart function (Divakaran and Mann, 2008, Espinoza-Lewis and Wang, 2012), the involvement of miRNA-378a with shifting substrate metabolism and regulation of mitochondrial bioenergetics (Jagannathan et al., 2015) provides an intimate, and novel, connection between its expression and mitochondrial health. Carrer et al. reported on the propensity of miRNA-378a knockout mice to protect against metabolic insult (Carrer et al., 2012). These researchers found that a high-fat diet induced obesity in wild type mice, but not miRNA-378a knockout mice, suggesting miRNA-378a may repress metabolic genes in response to environmental stimuli. The role of miRNA-378a in regulating metabolism and its high abundance in cardiac tissue (Krist et al., 2015) indicates that this small RNA may represent a therapeutic target for attenuation of metabolic disturbances and mitochondrial dysfunction resulting from pathological insult.
The objective of the current study was to determine whether manipulation of miRNA-378a expression could alter adverse cardiovascular and metabolic outcomes associated with ENM inhalation exposure. Our findings indicate that knockout, and knockdown to a lesser extent, of miRNA-378a improves cardiac and mitochondrial function by altering global bioenergetic and respiration capacity following nano-TiO2 inhalation exposure. This is the first study to determine the impact of altered miRNA expression on the response to ENM exposure in vivo.
Materials and Methods
MiRNA-378a Knockout Mouse Model
The West Virginia University Animal Care and Use Committee approved all animal studies, which conformed to the most current National Institutes of Health (NIH) Guidelines for the Care and Use of Laboratory Animals manual. Specifics about the animal model have been previously published (Carrer et al., 2012). Briefly, miRNA-378a is located within the first intron of the PGC-1β gene (Figure S1). Removal of the miRNA-378a loci was achieved by replacing the region with a neomycin cassette and LoxP sites. The recombination removed the miRNA-378a transcribed region but retained PGC-1β gene function. MiRNA-378a animals have been shown to be more metabolically active, specifically through mitochondrial fatty acid oxidation (Carrer et al., 2012). Sperm, which was cryopreserved from the miRNA-378a knockout 129SvEv/C57BL/6 mouse line, was used for in vitro fertilization of FVB mice. The progeny heterozygous miRNA-378a FVB mice were bred for greater than 10 generations, establishing the congenic line.
Experimental Model
Mice were housed in the West Virginia University Health Sciences Center Animal Facility and given access to a rodent diet and water ad libitum. To verify presence of the knockout allele, traditional PCR was performed amplifying the first intronic region of the PGC-1β gene (Table S1). The WT allele amplifies a 400 bp product while the transgenic allele amplifies a 470 bp product. Between 14–18 weeks of age, mice were subjected to a single exposure of either control, filtered air (Sham exposure) or nano-TiO2 aerosol (Ex exposure). Animals used in the analyses, unless stated, included Con Sham (n = 6), Con Ex (n = 6), Het Sham (n = 7), Het Ex (n = 8), KO Sham (n = 4), and KO Ex (n = 4). Both male and female mice were used in the analyses.
Engineered Nanomaterial
Nano-TiO2 P25 powder, purchased from Evonik (Aeroxide TiO2, Parsippany, NJ), was prepared through drying, sieving, and storing, as previously described (Nurkiewicz et al., 2008, Knuckles et al., 2012), and was composed of anatase (80%) and rutile (20%) TiO2 with a primary particle size of 21 nm. This particle has been previously characterized: Zeta potential = −56.6 mV (Nichols et al., 2018) and specific surface area = 48.08 m2/g (Nurkiewicz et al., 2008, Sager et al., 2008) determined.
Inhalation Exposure System
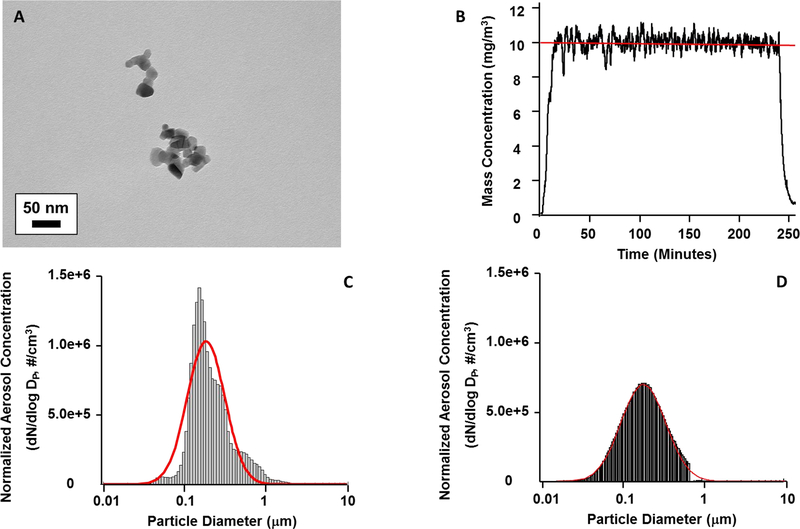
Nano-TiO2 aerosols were created with a customized high-pressure acoustical generator (HPAG). TiO2 bulk nano-powders were aerosolized with the HPAG and fed into the whole-body exposure chamber. Figure 1A shows a representative transmission electron micrograph of nano-TiO2 aerosols sampled from the chamber, imaged with the JEOL 1400 transmission electron microscope (Tokyo, Japan). A personal DataRAM (pDR-1500; Thermo Environmental Instruments Inc., Franklin, MA) sampled the exposure chamber air and utilized light scattering techniques to estimate the mass concentration in real-time. Feedback loops within the software automatically adjusted the acoustic energy to maintain a stable mass concentration during the exposure. A target DataRAM concentration of 10 mg/m3 for a period of 240 minutes was chosen in order to achieve a similar lung burden (7–10 μg/exposure) as that in previous studies (Nichols et al., 2018, Stapleton et al., 2018). The DataRAM signal during the exposure is shown in Figure 1B. A concurrent gravimetric measurement indicated an average mass concentration of 11.09 mg/m3 during the 240 minute period. Aerosol size distributions were examined from the exposure chamber with: 1) a high-resolution electrical low-pressure impactor (ELPI+; Dekati, Tampere, Finland), (2) a scanning particle mobility sizer (SMPS 3938; TSI Inc., St. Paul, MN), and (3) an aerodynamic particle sizer (APS 3321; TSI Inc., St. Paul, MN). The ELPI+ data (Figure 1C) and the combined SMPS-APS data (Figure 1D) both indicated log-normal aerosol size distributions with Count Median Diameters of 182 nm and 190 nm, respectively. Bedding material soaked with water was used in the exposure chamber to maintain a comfortable humidity during the exposure. Control animals were exposed to HEPA filtered air with similar chamber conditions in terms of temperature and humidity.
Figure 1:
Nano-TiO2 aerosol characterizations. (A) Representative transmission electron micrograph of aerosolized agglomerates collected on a filter. Bar is 50 nm. (B) Real-time estimation of the aerosol mass concentration measured with a light scattering device. The red line represents the target concentration of 10 mg/m3. The black line is the measured concentration during the exposure period. Note that computerized feedback system (see Methods) maintains a stable concentration. Mean DataRAM aerosol concentration during this exposure was 9.88 ± 0.03 mg/m3. Data was sampled every 2 seconds. (C) High resolution ELPI aerodynamic diameter measurements. The red line designates the log normal distribution obtained with the log probability plot method. This produced a count median diameter of 182 nm with a geometric standard deviation of 1.70. (D) SMPS (light gray bars) and APS (dark gray bars) mobility diameter measurements. The red line designates the log normal distribution obtained with the log probability plot method. This produced a count median diameter of 190 nm with a geometric standard deviation of 1.88. Note: APS bar values are nominal and difficult to visualize.
Echocardiography–M-mode, B-Mode and PW Doppler Imaging
Both one week prior to exposure and within 24 hours following exposure to control filtered air or nano-TiO2, ultrasound imaging was performed. Animals were initially anesthetized using 2.5% isoflurane, with a sustained rate of 1% isoflurane during imaging, in order to more appropriately replicate physiological conditions (James et al., 1998). To monitor the condition of the mice, a rectal temperature probe was used. Motion-mode (M-mode), Brightness-mode (B-mode), and Pulse Wave (PW) Doppler imaging acquisition and analysis have been previously described by our laboratory (Nichols et al., 2015, Thapa et al., 2015, Shepherd et al., 2016, Hathaway et al., 2017a, Nichols et al., 2018). Briefly, images were procured with a linear array transducer at 32–40 MHz, with a frame rate of 233–401 frames/second. Ultrasound images were taken using the Vevo 2100 Imaging System (Visual Sonics, Toronto, Canada). Briefly, imaging of the left ventricle in long-axis B-mode was achieved through placing the transducer left of the sternum. Rotation 90°, perpendicular to the animal, allowed for short-axis B-Mode images. Gating midway between the short-axis B-Mode images allowed for acquisition of M-mode parameters. Pulse Wave Doppler echocardiography measured mitral valve function, determining early (E-wave) and late (A-wave) diastolic filling of the left ventricle. Ejection Fraction is considered the percent of blood leaving the heart following each contraction and fractional shortening the change in left ventricular diameter. Total cardiac output was measured as the heart rate times the stroke volume per beat (volume of blood leaving the heart). All analyses were performed by one analyst. Three replicate analyses were performed for each echocardiographic measure per animal.
Stress Strain-Speckle Tracking
In both the short and long axis, left ventricular visualization B-Mode images were used for speckle-tracking-based strain analyses. The procedure for speckle-tracking-based strain analysis have been previously described by our laboratory (Shepherd et al., 2016, Hathaway et al., 2017a, Nichols et al., 2018). As a summary, strain was measured as a function of total deformation length divided by the original length of a segment, computed for each cardiac cycle (Pavlopoulos and Nihoyannopoulos, 2008, Blessberger and Binder, 2010). Peak strain and strain rates were measured across radial, circumferential, and longitudinal dimensions. Displacement length, velocity, strain, and strain rate were measured in the endocardium in the short (radial and circumferential dimensions) and long (radial and longitudinal) axes over a minimum of three cycles. Short-axis images of the left ventricle were split into anterior free, lateral, posterior, inferior free, posterior septum, and anterior septum while long-axis images were split into anterior base, anterior mid, anterior apex, posterior apex, posterior mid, and posterior base. The average of segmental values was used for analyses in this study. Assessment of Global Longitudinal Strain (GLS) in the left ventricle provides a speckle tracking-based valuation of myocardium contraction which correlates with left ventricular function (Ersboll et al., 2013, Biering-Sorensen et al., 2017), and can be combined with PW Doppler E-wave (E/SR) to make clinical prognostics of left ventricular health (Wang et al., 2007).
Mitochondrial Isolations
48 hours post-exposure to control filtered air or nano-TiO2, mice were euthanized, the heart, lungs, and liver were excised, and cardiac and lung mitochondrial subpopulations were isolated for analyses as previously described (Palmer et al., 1977), with modifications by our laboratory (Dabkowski et al., 2010, Baseler et al., 2011, Baseler et al., 2013). Samples were prepared through differential centrifugation, allowing for the compartmentalization of nuclear, cytoplasmic, and mitochondrial fractions. Mitochondrial subpopulations, subsarcolemmal (SSM) and interfibrillar (IFM) were combined to form a total mitochondrial population.
Mitochondrial Respiration
State 3 and state 4 respiration rates were analyzed in isolated mitochondria as previously described (Croston et al., 2014, Thapa et al., 2015) with adaptations. The Bradford method (Bradford, 1976), with standardization to bovine serum albumin, provided normalization of protein concentrations for cardiac mitochondria loading. To measure oxygen consumption of mitochondria, a multi-unit (8 channel) Oxytherm Peltier Electrode apparatus (Hansatech Instruments, Norfolk, England) was utilized. Calibration of the machine was performed through maximal oxygen concentration in respiration buffer (80mM KCl, 50mM MOPS, 1 mmol/l EGTA, 5 mmol/l KH2PO4 and 1 mg/ml BSA) and zero oxygen concentration through NaHSO3. Oxygen concentration measurements were assessed using the S1 Clark type polarographic oxygen electrode disc, with a central platinum cathode and a concentric silver anode. Mitochondria were loaded into 500 mL of respiration buffer in the Peltier Chamber. Changes in oxygen levels were measured in real-time, providing both nmol O2/mL consumption and rates per 10s, 30s, and 1min. Maximal complex I-mediated respiration was initiated by the addition of glutamate (5 mM) and malate (5 mM) for glucose mediated pathways or palmitoyl-carnitine (50 μM) and malate (5mM) for fatty-acid mediated pathways. Data for state 3 (250mM ADP) and state 4 (ADP-limited) respiration were expressed as nmol of oxygen consumed/mL/min, standardized to protein concentration.
Electron Transport Chain Complex Activities
Maximal activities of electron transport chain (ETC) complexes I, III, IV and V were measured as previously described (Hathaway et al., 2017a, Shepherd et al., 2017). The ETC is comprised of five protein complexes which transport electrons, in a redox fashion, to create a proton (H+) gradient which contributes toward the production of ATP. Complexes I (ubiquinone oxidoreductase), III (succinate dehydrogenase), and IV (cytochrome c reductase) can complex together to form a respirasome. Complex V (ATP Synthase) produces ATP from ADP using the proton gradient. The Bradford method (Bradford, 1976), with standardization to bovine serum albumin, provided normalization of protein concentrations for cardiac and lung mitochondria loading. Activity was measured in complex I (reduction of decyclubiquinone), complex III (reduction of cytochrome c), complex IV (oxidation of reduced cytochrome c), and complex V (oligomycin-sensitive ATPase activity through pyruvate kinase and phosphoenolpyruvate.).
Quantitative PCR
RNA was isolated from 20 mg of cardiac, lung, and liver tissue or from isolated cardiac mitochondria, using the miRNeasy Mini Kit (product no.: 217004, Qiagen, Hilden, Germany ) per manufacturer’s instructions, with minor modifications. For tissue, 700 mL QIAzol was used for homogenization with a Polytron PowerGen 500 S1 tissue homogenizer (Fisher Scientific, Hampton, NH). Total RNA was isolated from both the whole tissue and mitochondrial samples and used in subsequent analyses. RNA was converted to cDNA with the First-strand cDNA Synthesis kit for miRNA (product no.: HP100042, Origene, Rockville, MD), per manufacturer’s instructions. Differential expression of miRNA-378a-3p, miRNA-378a-5p, PGC-1β, mitofusin 1 (Mfn1), and mediator complex subunit 19 (Med19) were measured. Primer design for qPCR is provided (Table S1). Briefly, primers for mRNA transcripts were derived using Primer-BLAST through NCBI, while miRNA forward primers were designed to span the 5’ region of the sequence with a standard reverse to flank the poly-A tail following cDNA synthesis. Expression was normalized to GAPDH in whole tissue and U6 in isolated mitochondria. Experiments were performed on the Applied Biosystems 7900HT Fast Real-Time PCR system (Applied Biosystems, Foster City, CA), using 2X SYBR Green Master Mix. Quantification was achieved using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Western Blot Analyses
Immunoblotting was performed on 4 –12% gradient gels through MES SDS-PAGE, as previously described (Dabkowski et al., 2010, Baseler et al., 2011, Baseler et al., 2013, Nichols et al., 2015, Thapa et al., 2015). The Bradford method (Bradford, 1976), with standardization to bovine serum albumin, provided normalization of protein concentrations for the cytoplasmic portion of cardiac tissue for loading. Primary antibodies used in the study included: anti-GAPDH (product no.: ab8245, Abcam, Cambridge, MA), anti-optic atrophy 1 (OPA1, product no.: ab42364, Abcam, Cambridge, MA), and anti-peroxisome proliferator-activated receptor alpha (PPARα, product no.: ab2779, Abcam, Cambridge, MA). All primary antibodies were used at a 1:1000 concentration. Secondary antibodies used included the following: goat anti-mouse IgG (H&L) horseradish peroxidase (HRP) conjugate (product no. 31430, Thermo Fisher, Waltham, MA) and goat anti-rabbit IgG (H&L) HRP conjugate (product no. ab6721, Abcam, Cambridge, MA). Standardization was determined through GAPDH expression. Pierce® ECL Western Blotting Substrate (product no.: 32106, Thermo Fisher) was used per manufacturer’s instructions to detect tagged proteins through a G:Box Bioimaging system (Syngene, Frederick, MD). Data were captured using GeneSnap/GeneTools software (Syngene). Densitometry was analyzed using Image J Software (NIH, Bethesda, MD). All values were expressed as optical density with arbitrary units.
Mitochondrial Size, Internal Complexity, and Membrane Potential
Flow cytometry was used to assess structural and functional parameters of mitochondria as previously described (Dabkowski et al., 2009, Dabkowski et al., 2010, Williamson et al., 2010, Croston et al., 2014), with modifications. Briefly, the LSRFortessa (BD Biosciences, Franklin Lakes, NJ) was used to perform flow cytometric analyses. To stain viable mitochondria, MitoTracker™ Deep Red FM/633 (Thermo Fisher) was used to produce a threshold for gating. Measurements of size were assessed through Sphero AccuCount Blank Particles, 2.0 μm (Spherotech Inc., Lake Forest, IL) sizing beads. Forward Scatter (FSC PMT-H) measured the absolute size of the mitochondria while the Side Scatter (SSC-H) measured the granularity of the mitochondria. The ratiometric mitochondrial dye JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazol carbocyanine iodide, Molecular Probes, Carlsbad, CA) was used to measure mitochondrial membrane potential. Changes in membrane potential were assessed as the ratio of orange to green fluorescence of JC-1. All flow cytometric measures were performed at the West Virginia University Flow Cytometry & Single Cell Core Facility. FSC files were processed using the FCS Express Flow Research Edition (De Novo Software, Glendale, CA).
Transmission Electron Microscopy of Mitochondria
After excision of cardiac tissue, ~1–2 mm3 pieces were cut and sent to the WVU Electron Microscopy Shared Facilities where they were processed and further imaged at the WVU Electron Microscopy Histopathology and Tissue Bank. Briefly, cardiac tissue was fixed in 2.5% glutaraldehyde in 0.1 M PBS (pH 7.4) for 60 minutes. After washing in PBS, samples were incubated in 1% osmium tetroxide containing 1% potassium ferricyanide for 60 minutes. Following additional PBS washes, tissue was dehydrated using a graded series of alcohol dilutions and propylene oxide. Samples were then incubated overnight in a 1:1 mix of propylene oxide and EPON. Samples were incubated in pure EPON for 4 washes, each for 60 minutes. Samples were cured in pure EPON inside the BEEM® Embedding Capsules Size 3 (Electron Microscopy Sciences, Hatfield, PA) starting at 37°C for 24 hours and then 60°C for 48 hours. Tissue was sectioned using an Ultramicrotome Leica EM UC7 (Leica Microsystems, Wetzler, Germany) and stained with uranyl acetate and lead citrate. Sections were imaged using the JEOL JEM-2100 Transmission Electron Microscope (JOEL, Akishima, Tokyo, Japan). Semi-quantitative analyses of mitochondrial size was processed through Fiji (NIH, Bethesda, MD) and the Trainable Weka Segmentation (TWS) plugin by providing a probability map by which particle size could be determined. Selection of 5 mitochondria per group were used to perform analyses.
In Situ Hybridization (ISH) for miRNA-378a
After excision of cardiac tissue, the apex of the heart was removed (~5 mm3) and prepared per manufacturer’s instructions (Qiagen) (Schaeren-Wiemers and Gerfin-Moser, 1993, Heller et al., 1998), with slight modifications. Tissue was treated with freshly prepared 4% paraformaldehyde in PBS and was incubated at 4°C overnight. Tissue was then directly transferred to 0.5M Sucrose/PBS and was incubated at 4°C overnight. Following incubation, tissue was flash frozen and samples taken to the WVU Pathology Laboratory of Translational Medicine to embed and section tissue. Tissue was cut in 12–14 μM sections and mounted on Superfrost/Plus slides. Sections were further fixed in 4% paraformaldehyde in PBS and was incubated at 4°C for 10 minutes and then washed with PBS 3 times for 5 minutes each. For epitope retrieval, slides were immersed in a citric acid bath (10 mM citric acid, pH 6.0) at 95°C for 15 minutes and then washed in PBS for 5 minutes at room temperature. Sections were acetylated (2.33 mL of triethanolamine and 500 μL acetic anhydride in 197 mL water) for 10 minutes. Slides were washed 3 times with PBS for 5 minutes each. Slides were hybridized using a hybridization buffer (50% deionized formamide, 0.3M NaCl, 20 mM Tris HCl, pH 8.0, 5 mM EDTA, 10 mM Na3PO4, pH 8.0, 10% Dextran Sulfate, 1X Denhardt’s solution, and 0.5 mg/mL of yeast RNA) (Wilkinson, 1999) and 40 nM of miRNA-378a-3p 3’-DIG labeled probe (Qiagen).
Total hybridization mixture was heated to 65°C to linearize the probe and 100 μL of solution was added to slides, cover slipped, and hybridized at 56°C for 18 hours in a HybEZ™ Hybridization System (Advanced Cell Diagnostics, Newark, CA). Coverslips were removed in 5X SSC buffer at room temperature for 20 minutes. Slides were then washed twice in 50% formamide, 0.1% Tween-20, and 1X SSC at 56°C. Following two washes in PBS for 15 minutes each, blocking solution (3% Donkey serum, 0.1% Titron-100X, in PBS) was applied for 1 hour at room temperature. AP conjugated ant-DIG Fab fragments (1:1000) (Sigma-Aldrich, St. Louis, MO) were then applied to the blocking solution for 2 hours at room temperature. Following two washes in 0.1% Tween/PBS for 30 minutes each and 1X PBS for 10 minutes each, BM Purple AP Substrate (Sigma-Aldrich) and 2mM Levamisole were applied to slides for 6 days. Staining was quantified through light microscopy using the EVOS™ FL Auto Imaging System (Thermo Fisher). Briefly, using images derived with the same light intensity, Fiji (NIH, Bethesda, MD) was applied to measure intensity of BM Purple staining by using the log(max intensity/mean intensity) of Purple (R: 0.4643, G: 0.8303, B: 0.3083).
Citrate Synthase Activity
Citrate synthase is key to metabolism through the production of citrate from acetyl-CoA and oxaloacetate, initiating the tricarboxylic acid (TCA) cycle. To determine the number of mitochondria in heart tissue, a colorimetric Citrate Synthase Assay (Sciencell, San Diego, CA) was employed per manufacturer’s instructions (Jagannathan et al., 2015). The Bradford method (Bradford, 1976), with standardization to bovine serum albumin, provided normalization of protein concentrations for cardiac mitochondria. Briefly, 1 μg of mitochondria was used to measure the colorimetric change produced from 5’, 5’-Dithiobis 2-nitrobenzoic acid (DTNB) and CoA-SH forming of TNB at an absorbance maximum of 412 nm.
Hydrogen Peroxide (H2O2) Assay
To measure the concentration of H2O2 in the heart we used an Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit (Thermo Fisher, Waltham, MA) per manufacturer’s instructions and as previously described (Nichols et al., 2018), with modifications. Heart tissue was homogenized in NP-40 buffer (150 mM NaCl, 50 mM Tris-HCl, 1% Triton-100X, and pH adjusted to 7.4) and 50 μg of protein, determined through the Bradford method (Bradford, 1976), was used in the assay. The reaction of the Amplex® Red reagent (10-acetyl-3,7-dihydroxyphenoxazine) with H2O2 occurs at a stoichiometric ratio of 1:1. Sample H2O2 concentrations were determined through running a standard curve (0 to 20 μM H2O2), with absorbance measured at 560 nm. Data was normalized to the number of mitochondria for each sample, using citrate synthase activity (described above).
TargetScan
The miRNA-mRNA binding prediction software TargetScan 7.1 Server was implemented in order to determine seed-sequence binding affinities (Lewis et al., 2005, Agarwal et al., 2015). Briefly, binding affinities were examined in the 3’ untranslated region (UTR) of Mfn1 and Med19, where context++ scores (CSs) predicted the propensity for binding. CSs are derived from a variety of variables including 3’ UTR length, supplementary pairing, local AU content, and conservation of target sequence. Seed-sequence regions begin with the second base pair on the miRNA, starting from the 5’ end, and can identically bind to eight (8mer), seven (7mer), or six (6mer) consecutive base pairs on the mRNA strand. http://www.targetscan.org/vert_71/
IntaRNA
RNA-RNA interaction software IntaRNA 2.0 was used to assess miRNA binding in the 3’ UTR of Mfn1 (mouse), MFN1 (human), Med19 (mouse), and MED19 (human) (Busch et al., 2008, Wright et al., 2014, Mann et al., 2017). Unlike TargetScan 7.1, IntaRNA considers free energy binding as well as intramolecular forces between base pairs. Ensembl was used to obtain the 3’ UTR sequence for Mfn1 and Med19. The 3’ UTR sequences were then run against the miRNA-378a-3p sequence (5’ – ACUGGACUUGGAGUCAGAAGG – 3’) to determine complementarity. The interaction energy is a measure of binding potential.
Mfold
The UNAFold Web Server was used to evaluate miRNA folding, specifically through using the mfold software (Zuker, 2003). The pre-miRNA-378a (5’ – AGGGCUCCUGACUCCAGGUCCUGUGUGUUACCUCGAAAUAGCACUGGACUUGGAGUCAGAAGGCCU – 3’), mature miRNA-378a-3p (5’ – ACUGGACUUGGAGUCAGAAGG – 3’), and mature miRNA-378a-5p (5’ – CUCCUGACUCCAGGUCCUGUGU – 3’) sequences from miRBase release 21 were used.
Ingenuity Pathway Analysis (IPA)
QIAGEN’s IPA Server (December 2017 update) provides protein ontology and the capacity to correlate gene expression data through molecular pathways experimentally verified. The “PPARα/RXRα Activation” pathway and graphical illustrations were used in the analysis www.qiagen.com/ingenuity.
Statistics
Significance was determined using either a two-tailed Student’s t-test or one-way analysis of variance (ANOVA), where appropriate. Tukey’s multiple comparisons test was implemented following the ANOVA to derive significance between multiple groups. Differences between groups were considered statistically different (P ≤ 0.05, denoted by *). All data are presented as the mean ± standard error of the mean (SEM).
Results
MiRNA-378a Expression following Nano-TiO2 Exposure
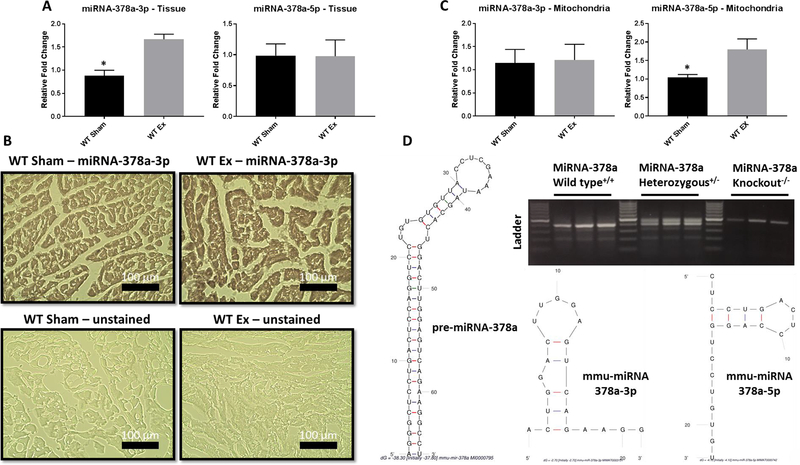
Because the pre-miRNA-378a gene actually contains both a 3’ (miRNA-378a-3p) and a 5’ (miRNA-378a-5p) arm mature miRNA (Krist et al., 2015), expression of both miRNAs needed to be assessed in total heart homogenates and within isolated mitochondria. This tissue and organellar-specific approach allowed for the determination of miRNA-378a’s involvement in regulating a variety of pathways (whole tissue) or those specifically related to mitochondrial bioenergetics and mitochondrial transcription (mitochondria). In whole heart tissue (Figure 2A), the expression of miRNA-378a-3p was significantly increased following exposure, while the 5p strand was unchanged. Using in situ hybridization, miRNA-378a-3p was also shown to be significantly elevated in cardiac tissue, displaying universal, not localized, expression of the miRNA throughout the cell (Figure 2B). The gene that encodes these two mature miRNAs, PGC-1β, was also shown to have increased expression following exposure (Figure S2A). The heart, in comparison to the lung and liver, was the only tissue where miRNA-378a-3p was increased, with a trend toward decreased expression in both the lung and liver following exposure (Figure S2B).
Figure 2:
Expression of miRNA-378a following nano-TiO2 inhalation and miRNA-378a animal models. Following inhalation exposure to control filtered air or nano-TiO2 qPCR was implemented to assess miRNA-378a-3p and miRNA-378a-5p expression in (A) whole heart tissue. (B) Representative images of in situ hybridization of miRNA-378a-3p in cardiac tissue performed under light microscopy (n = 4 for both groups). (C) qPCR was also used to measure miRNA-378a-3p and miRNA-378a-5p expression in isolated mitochondria (D) Genotyping through PCR amplification and agarose gel electrophoresis for the miRNA-378a loci in WT, Het, and KO animals. Additionally, RNA folding software Mfold evaluated the likely folding characteristics of the miRNA-378a species. Groups are considered significantly different if P ≤ 0.05 = *. All data are presented as the mean ± standard error of the mean (SEM). WT = wild type, Sham = control filtered air exposed, Ex = nano-TiO2 exposed.
In mitochondria (Figure 2C), the 3p strand remained unchanged, while miRNA-378a-5p was increased after nano-TiO2 exposure. Though alterations in mitochondrial miRNA-378a-5p could potentially change dynamics surrounding mitochondrial metabolism (Jagannathan et al., 2015), the relative abundance of the 5p strand to the 3p strand was significantly lower (Figure S2C) in both the whole heart tissue and the mitochondrion, likely contributing very little to systemic cellular mechanisms. Genotyping was performed using PCR and gel agarose electrophoresis for the genic region surrounding miRNA-378a for wild type (WT), heterozygous knockout (Het) and total knockout (KO) animals; the size and characteristic folding of the pre-miRNA and mature strands are also represented (Figure 2D). The expression of miRNA-378a-3p and 5p were then confirmed both in whole heart tissue and mitochondria through qPCR in transgenic animals (Figure S2D). The increased expression of miRNA-378a-3p in the whole heart homogenates following exposure provided a pathway for examining how miRNAs can alter cardiac function following exposure to nano-TiO2 through a transgenic model.
Cardiac Contractile Function and miRNA-378a
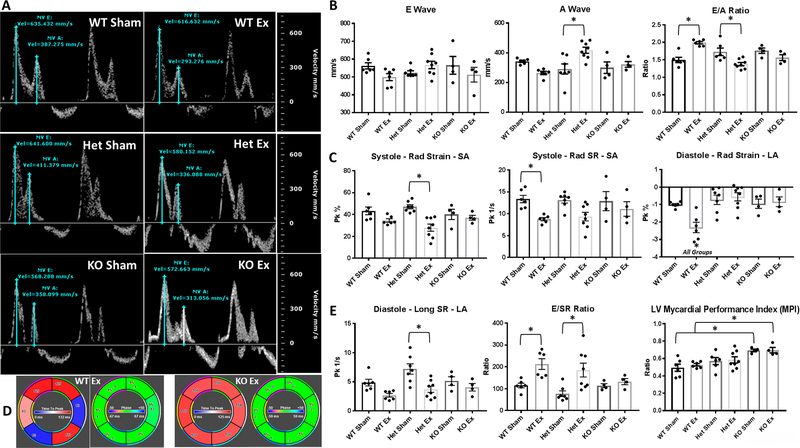
A list of all M-mode parameters are provided in Table 1. Of note, a decrease in the ejection fraction (percentage of total blood leaving the heart) and fractional shortening (change in left ventricular diameter) between the WT Sham and WT Ex group was observed, though preserved in the Het and KO groups. The WT groups also had larger systolic volumes than both the Het and KO groups. M-mode characteristics are also provided for Het and KO mice pre-exposure, validating the sham exposure model (Table S2). PW Doppler was implemented to measure mitral valve function and filling parameters (Figure 3A). The E wave (early diastolic filling of the left ventricle) was shown to not be significantly altered between groups, with the Het group exhibiting a higher A wave (late diastolic filling of the left ventricle) following exposure (Figure 3B). As seen in other studies following nano-TiO2 exposure (Hathaway et al., 2017a, Nichols et al., 2018), the WT group had an E/A ratio of ~2. When the E/A ratio increases, it indicates either more passive (early) filling or less active (late) filling of the left ventricle; this signifies a pathological shift in blood flow in the left ventricle and indicates diastolic dysfunction and increased left atrial pressure.
Table 1: M-Mode Cardiac Function.
Ultrasound imaging following nano-TiO2 inhalation exposure. Measurements were taken for at least three consecutive systolic and diastolic peaks and troughs, respectively, for each animal. Significance (P ≤ 0.05) is denoted for a specific category through bold text and reference to the sample number that it is significantly different from. Sample numbers are listed in the column heading. WT = wild type, Het = heterozygous for the miRNA-378a allele, KO = knockout for the miRNA-378a allele, Sham = control filtered air exposed, Ex = nano-TiO2 exposed, CO = cardiac output, Diameter;d = diastolic diameter, Diameter;s = systolic diameter, EF = ejection fraction, FS = fractional shortening, LV Mass = left ventricular mass, LV Mass Corr = left ventricular mass corrected, SV = stroke volume, V;d = volume during diastole, V;s = volume during systole.
| Calculation | Units | WT Sham1 | WT Ex2 | Het Sham3 | Het Ex4 | KO Sham5 | KO Ex6 |
|---|---|---|---|---|---|---|---|
| CO | mL/min | 13.04 ± 1.15 | 11.00 ± 1.41 | 12.11 ± 2.22 | 12.79 ± 1.97 | 12.34 ± 0.82 | 14.35 ± 0.62 |
| Diameter;d | mm | 2.33 ± 0.06 | 2.15 ± 0.13 | 2.52 ± 0.15 | 2.58 ± 0.14 | 2.62 ± 0.07 | 2.82 ± 0.29 |
| Diameter;s | mm | 1.01 ± 0.03 | 1.03 ± 0.08 | 0.83 ± 0.09 | 0.85 ± 0.08 | 0.79 ± 0.08 | 0.99 ± 0.21 |
| EF | % | 87.39 ± 1.16 |
84.17 ± 1.75 6 |
89.07 ± 1.54 | 87.43 ± 1.89 | 90.05 ± 0.79 | 91.60 ± 0.45 |
| FS | % | 56.44 ± 1.62 |
52.14 ± 2.11 6 |
57.88 ± 2.06 | 55.86 ± 2.50 | 59.86 ± 0.80 | 62.96 ± 2.74 |
| LV Mass | mg | 108.88 ± 10.24 | 111.87 ± 10.05 | 100.21 ± 6.94 | 132.61 ± 8.47 | 122.29 ± 10.26 | 108.32 ± 9.97 |
| LV Mass Corr | mg | 87.10 ± 8.20 | 89.50 ± 8.04 | 80.17 ± 5.55 | 106.08 ± 6.78 | 97.84 ± 8.21 | 86.66 ± 7.97 |
| SV | uL | 27.16 ± 1.38 | 22.06 ± 2.62 | 22.24 ± 3.19 | 23.62 ± 3.26 | 24.22 ± 1.46 | 31.49 ± 4.82 |
| V;d | uL | 30.86 ± 1.44 | 29.65 ± 1.92 | 25.13 ± 3.13 | 25.18 ± 2.13 | 25.41 ± 1.66 | 29.66 ± 2.82 |
| V;s | uL |
2.63 ± 0.36 3,4,5,6 |
2.93 ± 0.29 3,4,5,6 |
1.45 ± 0.40 | 1.55 ± 0.28 | 1.19 ± 0.31 | 1.68 ± 0.14 |
Figure 3:
Cardiac function following nano-TiO2 inhalation exposure. (A) PW Doppler images illustrate changes in E and A wave velocities following exposure. (B) E and A wave velocities, as well as E/A wave ratios, are represented from PW Doppler imaging. Speckle-tracking analyses from B-Mode short and long axis images for (C) systolic and diastolic strain and strain rate. (D) An illustration of dyssynchrony of the endocardium during short axis systolic radial stain (indicated by color) in the WT nano-TiO2 exposed animals compared to conserved strain parameters in the KO exposed animals. (E) The diastolic longitudinal strain rate coupled with the E wave velocities provided a prognostic cardiac health index for the left ventricle (LV), while the LV myocardial performance index (MPI) further provided a prognostic index of the left ventricular ejection parameters. Groups are considered significantly different if P ≤ 0.05 = *. All data are presented as the mean ± standard error of the mean (SEM). WT = wild type, Het = heterozygous for the miRNA-378a allele, KO = knockout for the miRNA-378a allele, Sham = control filtered air exposed, Ex = nano-TiO2 exposed, SA = left ventricular short axis, LA = left ventricular long axis, Long = longitudinal, Rad = radial.
While the KO group preserved E/A ratio, the Het group had a significant decrease in E/A ratio. Short axis radial strain and strain rate were significantly decreased following exposure to nano-TiO2 in the WT and Het groups, respectively, while no changes were shown in the KO (Figure 3C). Also, long axis diastolic radial strain was increased in the WT group after exposure. Generally, decreases in systolic measures of strain infer impaired contractile properties of the left ventricle, while increases in diastolic measures of strain could suggest aberrant relaxation of the left ventricle. Strain measures represent the contraction and relaxation of the myocardium and subsequent cardiomyocyte contractility. The segmental changes in short axis systolic radial strain are illustrated as an example of the segmental analysis of speckle tracking stress-strain (Figure 3D).
In the long axis, global longitudinal strain (GLS) during end-diastole provides significant prognostic value in assessing cardiac health (Ersboll et al., 2013) (Figure 3E). By combining both the E wave and end-diastolic GLS (E/SR ratio), it provides both cardiac strain and mitral valve function in a prognostic assessment (Wang et al., 2007). An increased E/SR ratio in the WT and Het indicates increased susceptibility to adverse cardiac events, while the KO group remained unchanged (Figure 3E). Cardiac function following exposure is further captured in the left ventricle myocardial performance index (), revealing increased function in the KO group compared to the WT (Figure 3E). All other measures of diastolic (Table S3 and S4) and systolic (Table S5 and S6) stress-strain are included. Decreases in miRNA-378a expression attenuates the effect of nano-TiO2 exposure to the heart, preserving cardiac function following acute nano-TiO2 exposure.
Mitochondrial Bioenergetics and miRNA-378a
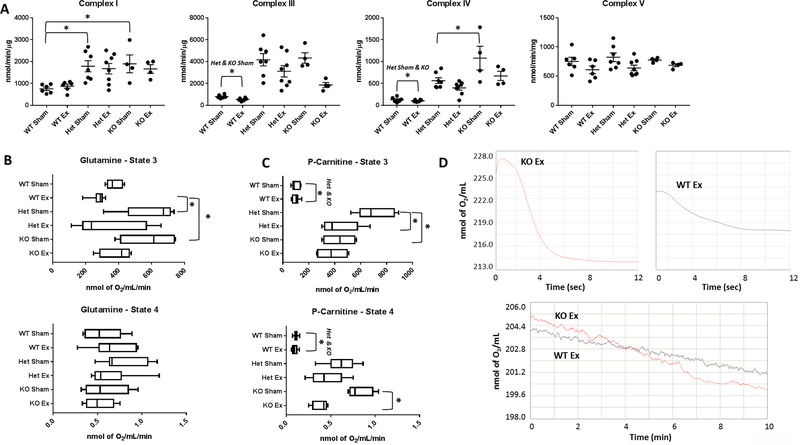
In the WT, changes to ETC activities and mitochondrial respiratory capacity were observed following exposure. Complex III activity and glucose-mediated state 3 respiration were decreased following nano-TiO2 exposure (Figure S3A and S3B). When examining bioenergetic capacity of all the groups, a distinctive metabolic profile was observed. Irrespective of exposure, complex I, III, and IV activities were shown to be significantly increased in the Het and KO groups (Figure 4A), potentially contributing to the cardioprotection observed after exposure. Unlike in the WT mice, glucose-mediated metabolism was not the most significantly impacted metabolic pathway in the Het or KO animals (Figure 4B); the main pathway affected was fatty-acid metabolism (Figure 4C). Both the Het and KO groups displayed a more robust fatty acid metabolic profile, which is illustrated in Figure 4D.
Figure 4:
Bioenergetic function of cardiac mitochondria following nano-TiO2 inhalation exposure. (A) Electron transport chain complex activities were measured for complexes I, III, IV, and V. Mitochondrial State 3 and State 4 respiration were measured through (B) glucose mediated pathways and (C) fatty acid mediated pathways. (D) State 3 and State 4 respiration changes between a representative WT and KO animal illustrates changes in oxygen consumption when using p-carnitine for fatty acid mediated respiration. Groups are considered significantly different if P ≤ 0.05 = *. All data are presented as the mean ± standard error of the mean (SEM). WT = wild type, Het = heterozygous for the miRNA-378a allele, KO = knockout for the miRNA-378a allele, Sham = control filtered air exposed, Ex = nano-TiO2 exposed, P-Carnitine = palmitoyl carnitine.
While exposure primarily affected glucose substrate utilization pathways in WT animals (Figure S3B), the Het and KO animals displayed decrements in fatty acid substrate utilization in state 3 and state 4 respiration, respectively. The increased utilization of fatty acid metabolites has been previously shown in the liver of miRNA-378a knockout animals (Carrer et al., 2012), supporting the parallels described in the heart and suggesting a metabolic contribution toward attenuating the pathology. Lung tissue of the Het and KO animals showed conserved ETC complex activity following exposure (Figure S4A and S4B), indicating preservation of cardiac function following inhalation exposure may be a function of protection to multiple organ systems in the miRNA-378a knockout/knockdown animals. The increase in complex activities and fatty acid-driven state 3 and state 4 respiration in Het and KO animals revealed a bioenergetic role for cardioprotection following nano-TiO2 exposure.
Mitochondrial Ultrastructure
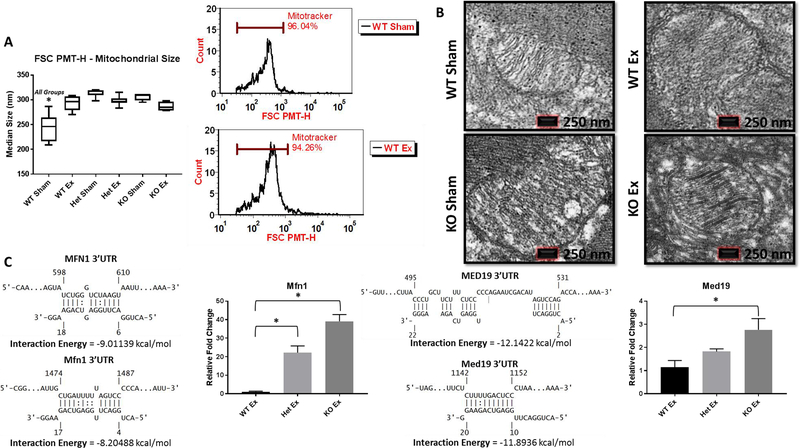
Mitochondrial size, through flow cytometry (Figure S5A), was shown to increase following exposure in the WT group (Figure 5A), as well as in both the Het and KO groups, irrespective of exposure. The shape and size of mitochondria were also examined through transmission electron microscopy (TEM), confirming the increased size of mitochondria following ENM inhalation exposure in the WT group using a semi-quantitative approach (Figure 5B). Citrate synthase activity, used as a common marker of mitochondrial number/content and shown to be highly correlative (Larsen et al., 2012), revealed decreased mitochondrial number in the WT group after exposure, while the Het and KO were protected from the effect (Figure S5B). In the WT group following exposure, though the size of the mitochondria increased, decreased mitochondrial number suggests a role in contributing to the dysfunctional respiratory capacity of the mitochondria (Johnson et al., 2013). Alternatively, in the Het and KO groups, mitochondrial size was shown increased while mitochondrial size and number were unaffected by the exposure paradigm. The increased size and unchanging number of mitochondria in the Het and KO groups could relate to the increased bioenergetic capacity and potential protective mechanism observed following nano-TiO2 inhalation exposure. Other measures of mitochondrial ultrastructure, such as internal complexity (Figure S5C) and membrane potential (Figure S5D) were measured and found to be unchanged. Measurements of H2O2 were performed to further understand mitochondrial stress/dysfunction. In the WT group following exposure, the concentration of H2O2 in cardiac tissue was significantly increased (Figure S5E). Following inhalation exposure to nano-TiO2, the density, size, and impact of ROS is altered, but knockout, or even knockdown, of miRNA-378a produces a protective phenotype.
Figure 5:
Mechanisms governing mitochondrial function and molecular targets of miRNA-378a following inhalation exposure. (A) Mitochondrial size was determined through forward scatter, gating mitochondria positively stained with MitoTracker™ Deep Red FM/633; representative images of the WT sham and nano-TiO2 exposure are illustrated. (B) Representative Transmission Electron Microscopy (TEM) images depicting changes to mitochondrial size (n = 1 for each group). (C) Binding interactions and qPCR for both human (MFN1 and MED19) and mouse (Mfn1 and Med19) using IntaRNA 2.0. Interaction energies are calculated as binding propensity through free energy binding near the seed sequence region. Groups are considered significantly different if P ≤ 0.05 = *. All data are presented as the mean ± standard error of the mean (SEM). WT = wild type, Het = heterozygous for the miRNA-378a allele, KO = knockout for the miRNA-378a allele, Sham = control filtered air exposed, Ex = nano-TiO2 exposed, FSC PMT-H = forward scatter, Mfn1 = mitofusin 1, Med19 = mediator complex subunit 19, 3’ UTR = 3’ untranslated region.
Targets of miRNA-378a and Pathways Affected
Gene transcripts, which were related to mitochondrial function and revealed highest 3’ UTR binding scores by miRNA-378a, were examined for further in-silico binding and expression. Mfn1 and Med19 were identified to be directly targeted by miRNA-378a-3p using bioinformatics software IntaRNA and TargetScan. MiRNA-378a-3p binding to the 3’ untranslated region (UTR) was evaluated through both free energy binding affinity (IntaRNA) (Figure 5D) and seed sequence complementarity (TargetScan) (Figure S6A). The mRNA of Mfn1 and Med19 were found to be significantly increased following exposure in the Het and KO groups (Figure 5D), suggesting that the negative regulation of miRNA-378a-3p was degrading transcripts and limiting translation of these proteins. While Med19 is involved in general transcriptional activation of RNA Polymerase, Mfn1 is involved prolifically in mitochondrial fusion dynamics and the health of mitochondria.
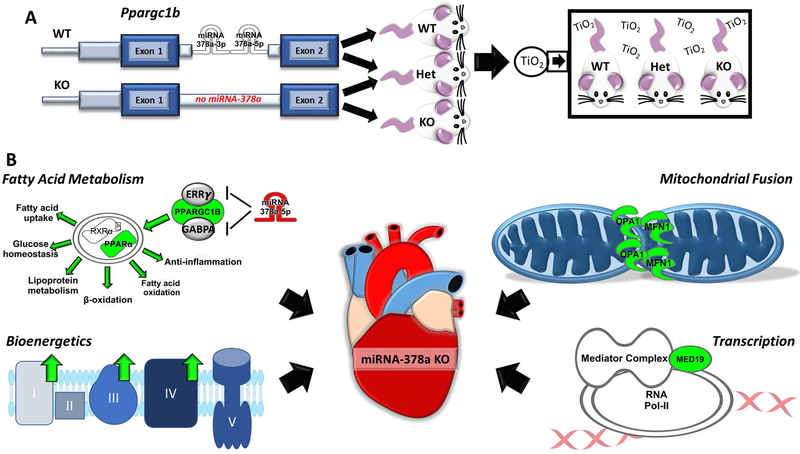
Opa1, a direct mediator with Mfn1 of inducing mitochondrial fusion, was shown to be significantly increased in the Het and KO groups following inhalation exposure; likewise, expression of the fatty acid metabolism protein Pparα was also increased (Figure S6B). The knockdown/knockout of miRNA-378a-5p, which acts as a negative regulator of estrogen-related receptor gamma (EERγ) and GA-binding protein alpha chain (GABPA), could promote the expression of PGC-1β and subsequently interact with Pparα (Eichner et al., 2010). In the KO animal, PGC-1β is found to be significantly increased compared to the WT group, following inhalation exposure (Figure S6C). Following maternal nano-TiO2 exposure, PGC-1β displayed increased histone 3 lysine 4 tri-methylation (H3K4me3) association at its promoter region, suggesting an increased expression of both the gene and miRNA-378a through epigenetic mechanisms (Figure S6D) (Stapleton et al., 2018). Suppressing miRNA-378a during inhalation exposure could diminish cardiac dysfunction by offering cardioprotection through pathways including transcription, bioenergetics, fatty acid metabolism, and mitochondrial fusion (Figure 6A and 6B).
Figure 6:
General schema of miRNA-378a interactions following inhalation exposure. (A) Transgenic mouse model, illustrating miRNA-378a wild type and knockout alleles. Mice were exposed to a total nano-TiO2 deposition of 9.88 ± 0.03 mg/m3. Cardiac issue was excised for subsequent experimental procedures. (B) Molecular changes in the KO nano-TiO2 exposed heart compared to WT exposed. Med19 was shown to be significantly elevated, an important cofactor in regulating cellular transcription. Metabolically, fatty acid metabolism and electron transport chain complex activities were elevated, likely through mediators such as Pparα and Ppargc1b and enhanced mitochondrial ultrastructure. Increased mitochondrial fusion (Mfn1 and Opa1) contributes to the integrity of mitochondria following exposure. These molecular pathways altered with the repression of miRNA-378a contribute to elevated cardiac function in KO compared to the WT mice. WT = wild type, Het = heterozygous for the miRNA-378a allele, KO = knockout for the miRNA-378a allele, Mfn1 = mitofusin 1, Med19 = mediator complex subunit 19, 3’ UTR = 3’ untranslated region, OPA1 = optic atrophy 1, PPARα = peroxisome proliferator-activated receptor alpha, Ppargc1b or PGC-1β = peroxisome proliferator-activated receptor gamma coactivator 1-beta.
Discussion
The scope and impact of research examining the potential human consequences of ENM exposure continues to grow, with considerations into the dose, physiochemical characteristics, route, and time course of exposure and the impact on human health and safety adding multiple layers of complexity. Nano-TiO2 remains a significant contributor to total ENM production, affecting both those involved in manufacturing and utilization of the material. The life cycle of ENMs, including byproducts of manufacturing, disposal through waste and recycling, use by consumers, and other mediums, eventually carry ENMs into the environment (Gottschalk and Nowack, 2011). The environmental sequestration of ENMs and their byproducts could then have secondary or tertiary impacts on human health (Boxall et al., 2007). We provide evidence to support the therapeutic benefit of miRNA-378a downregulation as a means of mediating the ENM toxicological response through metabolism, mitochondrial function, and other cellular processes during ENM inhalation exposure. For the first time, this study illustrates the impact of the manipulation of a miRNA in vivo on controlling toxicity to ENMs.
Under non-pathological states, altered miRNA content may have no overt consequences to basal physiological processes. As a stressor is introduced, such as disease, xenobiotic exposure, or caloric changes, specific miRNAs are needed to mediate the response and attempt to return the cell to homeostasis (Carrer et al., 2012, Mendell and Olson, 2012, Olejniczak et al., 2018). In the transgenic miRNA-378a knockout and knockdown animals, this has been highlighted in both caloric changes (Carrer et al., 2012) and currently with ENM inhalation exposure. Adaptation to a stress has significantly diverse outcomes in transgenic versus wild type animals, though little or no genetic or phenotypic changes exist at a basal condition. Because miRNAs exist in a complicated web of regulatory non-coding RNA, compensatory mechanisms are present which can attenuate loss of function from a gene or genes (Gurtan and Sharp, 2013, Thomson and Dinger, 2016). In the transgenic animals used in this study, it is likely that the loss of miRNA-378a is compensated through increased expression of endogenous miRNAs that regulate similar functional pathways, such as the insulin response and fatty acid metabolism pathways. Additionally, epigenetic remodeling could occur, repressing the expression of previously targeted miRNA-378a transcripts.
Cellular and systemic dysfunction has been attributed to ENM-induced changes in miRNA expression (Halappanavar et al., 2011, Ng et al., 2011, Chew et al., 2012, Balansky et al., 2013, Nagano et al., 2013, Snyder-Talkington et al., 2016). While the contribution of miRNA expression in advancing ENM related insult is beginning to be understood, what lacks clarity are the explicit mechanisms governing these changes in expression. The interplay between the epigenome and miRNA expression provides a new area of research and hierarchy of control, which could potentially explain the regulation of multiple pathways. MiRNAs are known to directly regulate, and to be regulated by, epigenetic function (Sato et al., 2011, Hathaway et al., 2017,b).
In one of the first studies to examine the effects of inhalation of ENM and miRNA expression, Halappanavar et al. showed that following inhalation exposure, miRNA-1 and −449 were elevated (Halappanavar et al., 2011). MiRNA-1 is known to target histone deacetylase 4 (HDAC4) while miRNA-449 has been shown to target HDAC1, resulting in global changes to the epigenetic histone profile in the cell (Chen et al., 2006). Down regulation of HDACs following nano-TiO2 exposure would suggest the activation of genes through open histone conformations, which we have reported in progeny following maternal exposure (Stapleton et al., 2018). Alternatively, the miRNA expression can be altered by epigenetic machinery. The expression of miRNA-378a is regulated through methylation in TGF-β1-treated LX-2 cells (Yu et al., 2016). In the current study, both the expression of the host gene PGC-1β and miRNA-378a-3p were shown to be increased following inhalation exposure to nano-TiO2, with epigenetic mechanisms known to alter the expression of the PGC-1β gene. The epigenetic landscape of miRNA-378a, and other altered miRNAs, could provide valuable information regarding transient and, potentially, sustained effects of ENM exposure on the heart and other organ systems.
The animal models employed in the study examine miRNA-378a across a stratum of normal (WT), knockdown (Het), and knockout (KO) expression levels. Intuitively, one could hypothesize that the health outcomes going from poorest to best prognosis would trend in a linear fashion from WT to Het to KO. While we expect the KO model to have increased mitochondrial fatty acid metabolism (Gerin et al., 2010) and overall better health (Carrer et al., 2012) following ENM exposure, the knockdown expression of miRNA-378a may still be prevalent enough to regulate maladaptive metabolic pathways, meaning that the linearity of functional measures in the three models may not exist in all cases. MiRNA’s are multifaceted transcriptional and translational repressors that target multiple genic groups, making it difficult to speculate the intermediary effects that knockdown expression may have on any one cardiac, mitochondrial, or molecular measure (Melman et al., 2014). That being said, in general, we expected to see overall better function in the Het model compared to the WT control.
The current study proposes the idea that targeting miRNAs can alter cardiac and mitochondrial function and adaptation to ENM inhalation exposure. A noted limitation to the study is the examination of systemic miRNA-378a knockout and knockdown when examining cardiovascular health following ENM inhalation exposure. The current work focuses on cardiac adaptation to ENM inhalation exposure, but does so through systemic knockdown of miRNA-378a; a cardiac-specific knockout model could provide further indication into the absence of localized miRNA-378a expression following insult. We point to miRNA-378a as a target for attenuation of nano-TiO2 inhalation exposure, though it is likely that multiple miRNAs, and other coding and non-coding RNAs, are involved in the response to the insult. Our current work may be limited through the exploration of a single miRNA, but the outlined methodology provides an architype for transgenic animal use in examining ENM toxicology and signifies the importance of non-coding RNA in regulating the cardiovascular response.
The goal of ENM inhalation toxicology is to decrease the severity of biologic effects and/or prevention of exposure, ultimately increasing the positive impact of ENM utilization in human endeavors. The ability to therapeutically target a miRNA, or even group of miRNAs, could beneficially protect those exposed to ENMs. We show through the heterozygous knockout of miRNA-378a that even partial downregulation of a miRNA can beneficially alter the response to ENM inhalation exposure, producing a therapeutic effect. Ultimately, further investigation into the role of miRNAs following ENM exposure is needed as well as understanding their interconnection with the epigenome. Future studies could examine the epigenetic regulation of genes encoding miRNAs, such as the promotor region of PGC-1β, and determining if ENM exposure provides transient or sustained epigenetic control of crucial genes and non-coding RNA.
Understanding the role of miRNAs in regulating the response to ENM inhalation exposure is important in defining the molecular consequences involved in the response. In this study, we discovered that miRNA-378a, a small non-coding RNA, was directly involved in regulating cardiac function following nano-TiO2 inhalation exposure. The knockout, and to a lesser extent knockdown, of miRNA-378a expression was associated with preserved cardiac function following exposure and increased bioenergetic capacity when compared to wild type animals. The mechanisms governing this functional difference are likely through bolstered mitochondrial health, including increased mitochondrial fusion and fatty acid metabolism, as well as changes to cellular transcription. This study investigates for the first time, the in vivo role of a miRNA in attenuating cardiovascular consequences following ENM inhalation exposure, setting miRNA-378a as a potential target for future investigations. Manipulation of miRNA-378a, or other miRNAs found in the heart or systemically, have the potential to improve health outcomes following ENM inhalation exposure.
Supplementary Material
Acknowledgements
We would like to thank Eric Olson and the Transgenic Technology Center at University of Texas Southwestern Medical Center for the miRNA-378a knockout mouse model.
Declaration of Interest
The authors declare that they have no competing interests, financial or otherwise. This work was supported by: R01 HL-128485 (JMH), R01-ES015022 (TRN), AHA-17PRE33660333 (QAH), AHA-13PRE16850066 (CEN), DGE-1144676 (QAH, ABA, TRN), WV-INBRE support by NIH Grant P20GM103434, WVU Flow Cytometry & Single Cell Core supported by MBRCC CoBRE Grant GM103488 and Fortessa S10 Grant OD016165, and the Community Foundation for the Ohio Valley Whipkey Trust.
Footnotes
Publisher's Disclaimer: Disclaimer
Publisher's Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention.
References:
- Agarwal V, Bell GW, Nam JW & Bartel DP, 2015. Predicting effective microRNA target sites in mammalian mRNAs. Elife, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balansky R, Longobardi M, Ganchev G, Iltcheva M, Nedyalkov N, Atanasov P, Toshkova R, De Flora S & Izzotti A, 2013. Transplacental clastogenic and epigenetic effects of gold nanoparticles in mice. Mutat Res, 751–752, 42–8. [DOI] [PubMed] [Google Scholar]
- Baseler WA, Dabkowski ER, Jagannathan R, Thapa D, Nichols CE, Shepherd DL, Croston TL, Powell M, Razunguzwa TT, Lewis SE, Schnell DM & Hollander JM, 2013. Reversal of mitochondrial proteomic loss in Type 1 diabetic heart with overexpression of phospholipid hydroperoxide glutathione peroxidase. Am J Physiol Regul Integr Comp Physiol, 304, R553–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baseler WA, Dabkowski ER, Williamson CL, Croston TL, Thapa D, Powell MJ, Razunguzwa TT & Hollander JM, 2011. Proteomic alterations of distinct mitochondrial subpopulations in the type 1 diabetic heart: contribution of protein import dysfunction. Am J Physiol Regul Integr Comp Physiol, 300, R186–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besinis A, De Peralta T, Tredwin CJ & Handy RD, 2015. Review of nanomaterials in dentistry: interactions with the oral microenvironment, clinical applications, hazards, and benefits. ACS Nano, 9, 2255–89. [DOI] [PubMed] [Google Scholar]
- Biering-Sorensen T, Biering-Sorensen SR, Olsen FJ, Sengelov M, Jorgensen PG, Mogelvang R, Shah AM & Jensen JS, 2017. Global Longitudinal Strain by Echocardiography Predicts Long-Term Risk of Cardiovascular Morbidity and Mortality in a Low-Risk General Population: The Copenhagen City Heart Study. Circ Cardiovasc Imaging, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blessberger H & Binder T, 2010. Two dimensional speckle tracking echocardiography: clinical applications. Heart, 96, 2032–40. [DOI] [PubMed] [Google Scholar]
- Boxall AB, Tiede K & Chaudhry Q, 2007. Engineered nanomaterials in soils and water: how do they behave and could they pose a risk to human health? Nanomedicine (Lond), 2, 919–27. [DOI] [PubMed] [Google Scholar]
- Bradford MM, 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem, 72, 248–54. [DOI] [PubMed] [Google Scholar]
- Busch A, Richter AS & Backofen R, 2008. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics, 24, 2849–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrer M, Liu N, Grueter CE, Williams AH, Frisard MI, Hulver MW, Bassel-Duby R & Olson EN, 2012. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci U S A, 109, 15330–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL & Wang DZ, 2006. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet, 38, 228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Li H, Cai J, Wang C, Lin Z, Liu C, Niu Y, Zhao Z, Li W & Kan H, 2018. Fine Particulate Air Pollution and the Expression of microRNAs and Circulating Cytokines Relevant to Inflammation, Coagulation, and Vasoconstriction. Environ Health Perspect, 126, 017007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew WS, Poh KW, Siddiqi NJ, Alhomida AS, Yu LE & Ong WY, 2012. Short- and long-term changes in blood miRNA levels after nanogold injection in rats--potential biomarkers of nanoparticle exposure. Biomarkers, 17, 750–7. [DOI] [PubMed] [Google Scholar]
- Croston TL, Thapa D, Holden AA, Tveter KJ, Lewis SE, Shepherd DL, Nichols CE, Long DM, Olfert IM, Jagannathan R & Hollander JM, 2014. Functional deficiencies of subsarcolemmal mitochondria in the type 2 diabetic human heart. Am J Physiol Heart Circ Physiol, 307, H54–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabkowski ER, Baseler WA, Williamson CL, Powell M, Razunguzwa TT, Frisbee JC & Hollander JM, 2010. Mitochondrial dysfunction in the type 2 diabetic heart is associated with alterations in spatially distinct mitochondrial proteomes. Am J Physiol Heart Circ Physiol, 299, H529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabkowski ER, Williamson CL, Bukowski VC, Chapman RS, Leonard SS, Peer CJ, Callery PS & Hollander JM, 2009. Diabetic cardiomyopathy-associated dysfunction in spatially distinct mitochondrial subpopulations. Am J Physiol Heart Circ Physiol, 296, H359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaran V & Mann DL, 2008. The emerging role of microRNAs in cardiac remodeling and heart failure. Circ Res, 103, 1072–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Dong C, Ren T, Li Y & Shi D, 2014. Surface-engineered graphene-based nanomaterials for drug delivery. J Biomed Nanotechnol, 10, 2086–106. [DOI] [PubMed] [Google Scholar]
- Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M, St-Pierre J & Giguere V, 2010. miR-378( *) mediates metabolic shift in breast cancer cells via the PGC-1beta/ERRgamma transcriptional pathway. Cell Metab, 12, 352–61. [DOI] [PubMed] [Google Scholar]
- Ersboll M, Valeur N, Mogensen UM, Andersen MJ, Moller JE, Velazquez EJ, Hassager C, Sogaard P & Kober L, 2013. Prediction of all-cause mortality and heart failure admissions from global left ventricular longitudinal strain in patients with acute myocardial infarction and preserved left ventricular ejection fraction. J Am Coll Cardiol, 61, 2365–73. [DOI] [PubMed] [Google Scholar]
- Espinoza-Lewis RA & Wang DZ, 2012. MicroRNAs in heart development. Curr Top Dev Biol, 100, 279–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagan J, Dey BK, Layer R, Yan Z & Dutta A, 2011. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J Biol Chem, 286, 19431–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerin I, Bommer GT, Mccoin CS, Sousa KM, Krishnan V & Macdougald OA, 2010. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab, 299, E198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk F, Lassen C, Kjoelholt J, Christensen F & Nowack B, 2015. Modeling flows and concentrations of nine engineered nanomaterials in the Danish environment. Int J Environ Res Public Health, 12, 5581–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk F & Nowack B, 2011. The release of engineered nanomaterials to the environment. J Environ Monit, 13, 1145–55. [DOI] [PubMed] [Google Scholar]
- Gurtan AM & Sharp PA, 2013. The role of miRNAs in regulating gene expression networks. J Mol Biol, 425, 3582–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halappanavar S, Jackson P, Williams A, Jensen KA, Hougaard KS, Vogel U, Yauk CL & Wallin H, 2011. Pulmonary response to surface-coated nanotitanium dioxide particles includes induction of acute phase response genes, inflammatory cascades, and changes in microRNAs: a toxicogenomic study. Environ Mol Mutagen, 52, 425–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathaway QA, Nichols CE, Shepherd DL, Stapleton PA, Mclaughlin SL, Stricker JC, Rellick SL, Pinti MV, Abukabda AB, Mcbride CR, Yi J, Stine SM, Nurkiewicz TR & Hollander JM, 2017a. Maternal-engineered nanomaterial exposure disrupts progeny cardiac function and bioenergetics. Am J Physiol Heart Circ Physiol, 312, H446–H458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathaway QA, Pinti MV, Durr AJ, Waris S, Shepherd DL & Hollander JM, 2017b. Regulating MicroRNA Expression: At the Heart of Diabetes Mellitus and the Mitochondrion. Am J Physiol Heart Circ Physiol, ajpheart 00520 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller S, Sheane CA, Javed Z & Hudspeth AJ, 1998. Molecular markers for cell types of the inner ear and candidate genes for hearing disorders. Proc Natl Acad Sci U S A, 95, 11400–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendren CO, Mesnard X, Droge J & Wiesner MR, 2011. Estimating production data for five engineered nanomaterials as a basis for exposure assessment. Environ Sci Technol, 45, 2562–9. [DOI] [PubMed] [Google Scholar]
- Holland NA, Thompson LC, Vidanapathirana AK, Urankar RN, Lust RM, Fennell TR & Wingard CJ, 2016. Impact of pulmonary exposure to gold core silver nanoparticles of different size and capping agents on cardiovascular injury. Part Fibre Toxicol, 13, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB & Zhang Y, 2006. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One, 1, e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan R, Thapa D, Nichols CE, Shepherd DL, Stricker JC, Croston TL, Baseler WA, Lewis SE, Martinez I & Hollander JM, 2015. Translational Regulation of the Mitochondrial Genome Following Redistribution of Mitochondrial MicroRNA in the Diabetic Heart. Circ Cardiovasc Genet, 8, 785–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JF, Hewett TE & Robbins J, 1998. Cardiac physiology in transgenic mice. Circ Res, 82, 407–15. [DOI] [PubMed] [Google Scholar]
- Johnson ML, Robinson MM & Nair KS, 2013. Skeletal muscle aging and the mitochondrion. Trends Endocrinol Metab, 24, 247–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan H, Wu Z, Young SH, Chen TH, Cumpston JL, Chen F, Kashon ML & Castranova V, 2012. Pulmonary exposure of rats to ultrafine titanium dioxide enhances cardiac protein phosphorylation and substance P synthesis in nodose ganglia. Nanotoxicology, 6, 736–45. [DOI] [PubMed] [Google Scholar]
- Kim SW, Kim HW, Huang W, Okada M, Welge JA, Wang Y & Ashraf M, 2013. Cardiac stem cells with electrical stimulation improve ischaemic heart function through regulation of connective tissue growth factor and miR-378. Cardiovasc Res, 100, 241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevic I, Patel A, Sundaresan NR, Gupta MP, Solaro RJ, Nagalingam RS & Gupta M, 2012. A novel cardiomyocyte-enriched microRNA, miR-378, targets insulin-like growth factor 1 receptor: implications in postnatal cardiac remodeling and cell survival. J Biol Chem, 287, 12913–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckles TL, Yi J, Frazer DG, Leonard HD, Chen BT, Castranova V & Nurkiewicz TR, 2012. Nanoparticle inhalation alters systemic arteriolar vasoreactivity through sympathetic and cyclooxygenase-mediated pathways. Nanotoxicology, 6, 724–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krist B, Florczyk U, Pietraszek-Gremplewicz K, Jozkowicz A & Dulak J, 2015. The Role of miR-378a in Metabolism, Angiogenesis, and Muscle Biology. Int J Endocrinol, 2015, 281756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen S, Nielsen J, Hansen CN, Nielsen LB, Wibrand F, Stride N, Schroder HD, Boushel R, Helge JW, Dela F & Hey-Mogensen M, 2012. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J Physiol, 590, 3349–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB & Bartel DP, 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120, 15–20. [DOI] [PubMed] [Google Scholar]
- Li Z, Hulderman T, Salmen R, Chapman R, Leonard SS, Young SH, Shvedova A, Luster MI & Simeonova PP, 2007. Cardiovascular effects of pulmonary exposure to single-wall carbon nanotubes. Environ Health Perspect, 115, 377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ & Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25, 402–8. [DOI] [PubMed] [Google Scholar]
- Mann M, Wright PR & Backofen R, 2017. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res, 45, W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melman YF, Shah R & Das S, 2014. MicroRNAs in heart failure: is the picture becoming less miRky? Circ Heart Fail, 7, 203–14. [DOI] [PubMed] [Google Scholar]
- Mendell JT & Olson EN, 2012. MicroRNAs in stress signaling and human disease. Cell, 148, 1172–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Higashisaka K, Kunieda A, Iwahara Y, Tanaka K, Nagano K, Abe Y, Kamada H, Tsunoda S, Nabeshi H, Yoshikawa T, Yoshioka Y & Tsutsumi Y, 2013. Liver-specific microRNAs as biomarkers of nanomaterial-induced liver damage. Nanotechnology, 24, 405102. [DOI] [PubMed] [Google Scholar]
- Ng CT, Dheen ST, Yip WC, Ong CN, Bay BH & Lanry Yung LY, 2011. The induction of epigenetic regulation of PROS1 gene in lung fibroblasts by gold nanoparticles and implications for potential lung injury. Biomaterials, 32, 7609–15. [DOI] [PubMed] [Google Scholar]
- Nichols CE, Shepherd DL, Hathaway QA, Durr AJ, Thapa D, Abukabda A, Yi J, Nurkiewicz TR & Hollander JM, 2018. Reactive oxygen species damage drives cardiac and mitochondrial dysfunction following acute nano-titanium dioxide inhalation exposure. Nanotoxicology, 12, 32–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols CE, Shepherd DL, Knuckles TL, Thapa D, Stricker JC, Stapleton PA, Minarchick VC, Erdely A, Zeidler-Erdely PC, Alway SE, Nurkiewicz TR & Hollander JM, 2015. Cardiac and mitochondrial dysfunction following acute pulmonary exposure to mountaintop removal mining particulate matter. Am J Physiol Heart Circ Physiol, 309, H2017–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurkiewicz TR, Porter DW, Hubbs AF, Cumpston JL, Chen BT, Frazer DG & Castranova V, 2008. Nanoparticle inhalation augments particle-dependent systemic microvascular dysfunction. Part Fibre Toxicol, 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejniczak M, Kotowska-Zimmer A & Krzyzosiak W, 2018. Stress-induced changes in miRNA biogenesis and functioning. Cell Mol Life Sci, 75, 177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JW, Tandler B & Hoppel CL, 1977. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. J Biol Chem, 252, 8731–9. [PubMed] [Google Scholar]
- Pavlopoulos H & Nihoyannopoulos P, 2008. Strain and strain rate deformation parameters: from tissue Doppler to 2D speckle tracking. Int J Cardiovasc Imaging, 24, 479–91. [DOI] [PubMed] [Google Scholar]
- Ruenraroengsak P & Tetley TD, 2015. Differential bioreactivity of neutral, cationic and anionic polystyrene nanoparticles with cells from the human alveolar compartment: robust response of alveolar type 1 epithelial cells. Part Fibre Toxicol, 12, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager TM, Kommineni C & Castranova V, 2008. Pulmonary response to intratracheal instillation of ultrafine versus fine titanium dioxide: role of particle surface area. Part Fibre Toxicol, 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato F, Tsuchiya S, Meltzer SJ & Shimizu K, 2011. MicroRNAs and epigenetics. FEBS J, 278, 1598–609. [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N & Gerfin-Moser A, 1993. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 431–440. [DOI] [PubMed] [Google Scholar]
- Shepherd DL, Hathaway QA, Pinti MV, Nichols CE, Durr AJ, Sreekumar S, Hughes KM, Stine SM, Martinez I & Hollander JM, 2017. Exploring the mitochondrial microRNA import pathway through Polynucleotide Phosphorylase (PNPase). J Mol Cell Cardiol, 110, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd DL, Nichols CE, Croston TL, Mclaughlin SL, Petrone AB, Lewis SE, Thapa D, Long DM, Dick GM & Hollander JM, 2016. Early detection of cardiac dysfunction in the type 1 diabetic heart using speckle-tracking based strain imaging. J Mol Cell Cardiol, 90, 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla GC, Singh J & Barik S, 2011. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol Cell Pharmacol, 3, 83–92. [PMC free article] [PubMed] [Google Scholar]
- Snyder-Talkington BN, Dong C, Sargent LM, Porter DW, Staska LM, Hubbs AF, Raese R, Mckinney W, Chen BT, Battelli L, Lowry DT, Reynolds SH, Castranova V, Qian Y & Guo NL, 2016. mRNAs and miRNAs in whole blood associated with lung hyperplasia, fibrosis, and bronchiolo-alveolar adenoma and adenocarcinoma after multi-walled carbon nanotube inhalation exposure in mice. J Appl Toxicol, 36, 161–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB & Spiegelman BM, 2003. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J Biol Chem, 278, 26597–603. [DOI] [PubMed] [Google Scholar]
- Stapleton PA, Hathaway QA, Nichols CE, Abukabda AB, Pinti MV, Shepherd DL, Mcbride CR, Yi J, Castranova VC, Hollander JM & Nurkiewicz TR, 2018. Maternal engineered nanomaterial inhalation during gestation alters the fetal transcriptome. Part Fibre Toxicol, 15, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton PA, Minarchick VC, Cumpston AM, Mckinney W, Chen BT, Sager TM, Frazer DG, Mercer RR, Scabilloni J, Andrew ME, Castranova V & Nurkiewicz TR, 2012. Impairment of coronary arteriolar endothelium-dependent dilation after multi-walled carbon nanotube inhalation: a time-course study. Int J Mol Sci, 13, 13781–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton PA, Nichols CE, Yi J, Mcbride CR, Minarchick VC, Shepherd DL, Hollander JM & Nurkiewicz TR, 2015. Microvascular and mitochondrial dysfunction in the female F1 generation after gestational TiO2 nanoparticle exposure. Nanotoxicology, 9, 941–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa D, Nichols CE, Lewis SE, Shepherd DL, Jagannathan R, Croston TL, Tveter KJ, Holden AA, Baseler WA & Hollander JM, 2015. Transgenic overexpression of mitofilin attenuates diabetes mellitus-associated cardiac and mitochondria dysfunction. J Mol Cell Cardiol, 79, 212–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson DW & Dinger ME, 2016. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet, 17, 272–83. [DOI] [PubMed] [Google Scholar]
- Wang J, Khoury DS, Thohan V, Torre-Amione G & Nagueh SF, 2007. Global diastolic strain rate for the assessment of left ventricular relaxation and filling pressures. Circulation, 115, 1376–83. [DOI] [PubMed] [Google Scholar]
- Wang P, Gu Y, Zhang Q, Han Y, Hou J, Lin L, Wu C, Bao Y, Su X, Jiang M, Wang Q, Li N & Cao X, 2012. Identification of resting and type I IFN-activated human NK cell miRNomes reveals microRNA-378 and microRNA-30e as negative regulators of NK cell cytotoxicity. J Immunol, 189, 211–21. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, 1999. In Situ Hybridization: A Practical Approach: Oxford University Press. [Google Scholar]
- Williamson CL, Dabkowski ER, Baseler WA, Croston TL, Alway SE & Hollander JM, 2010. Enhanced apoptotic propensity in diabetic cardiac mitochondria: influence of subcellular spatial location. Am J Physiol Heart Circ Physiol, 298, H633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PR, Georg J, Mann M, Sorescu DA, Richter AS, Lott S, Kleinkauf R, Hess WR & Backofen R, 2014. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res, 42, W119–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Yang J, Huang K, Pan X, Chen B, Dong P & Zheng J, 2016. The Epigenetically-Regulated microRNA-378a Targets TGF-beta2 in TGF-beta1-Treated Hepatic Stellate Cells. Cell Physiol Biochem, 40, 183–194. [DOI] [PubMed] [Google Scholar]
- Zuker M, 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res, 31, 3406–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.