Summary
Rotavirus C (RVC) causes enteric disease in multiple species, including humans, swine, bovines, and canines. To date, the evolutionary relationships of RVC populations circulating in different host species are poorly understood, owing to the low availability of genetic sequence data. To address this gap, we sequenced 45 RVC complete genomes from swine samples collected in the United States and Mexico. A phylogenetic analysis of each genome segment indicates that RVC populations have been evolving independently in human, swine, canine, and bovine hosts for at least the last century, with inter-species transmission events occurring deep in the phylogenetic tree, and none in the last 100 years. Bovine and canine RVC populations clustered together 9 of the 11 gene segments, indicating a shared common ancestor centuries ago. The evolutionary relationships of RVC in humans and swine were more complex, due to the extensive genetic diversity and multiple RVC clades identified in pigs, which were not structured geographically. Topological differences between trees inferred for different genome segments occurred frequently, including at nodes deep in the tree, indicating that RVC’s evolutionary history includes multiple reassortment events that occurred a long time ago. Overall, we find that RVC is evolving within host-defined lineages, but the evolutionary history of RVC is more complex than previously recognized due to the high genetic diversity of RVC in swine, with a common ancestor dating back centuries. Pigs may act as a reservoir host for RVC, and a source of the lineages identified in other species, including humans, but additional sequencing is needed to understand the full diversity of this understudied pathogen across multiple host species.
Keywords: Rotavirus C, Phylodynamics, Whole-genome sequences, Surveillance
Introduction
Rotaviruses (RVs) are a major cause of severe gastroenteritis in humans and animals. Globally, most children are exposed to rotaviruses by age five, resulting in over half a million deaths each year (Sadiq, Bostan, Yinda, Naseem, & Sattar, 2018). In pigs, RVs are prevalent in young piglets, causing significant mortality and economic impact through loss of production. RVs, of the family Reoviridae, are non-enveloped double-stranded RNA viruses with a genome consisting of 11 segments that code for six structural proteins (VP1-VP4, VP6-VP7) and five or six non-structural proteins (NSP1-NSP5/NSP6) (Estes & Greenberg, 2013). RVs are categorized into eight phylogenetically distinct species (RVA-RVH), based on the inner capsid protein (VP6) (Matthijnssens et al., 2012). Recently, two putative new RV species I and J were identified (Bányai et al., 2017; Mihalov-Kovacs et al., 2015). RVA, RVB, and RVC are found in mammals, including humans, swine, and cattle. Of all the RV species, RVA is the most important causative agent of diarrhea in humans, and the genetic diversity of RVA has been well characterized by the outer capsid proteins VP7 and VP4 defined as G and P genotypes, respectively (Estes & Greenberg, 2013). Inter-species transmission events of RVA between animals and humans have been well documented and contribute to the genetic diversity of RVA found in humans (Matthijnssens & Van Ranst, 2012). The virus’s segmented genome provides another mechanism for rapid evolution, as entire segments can be exchanged via reassortment.
Rotavirus C (RVC) was first detected in 1980 in a pig in Ohio, US (Saif, Bohl, Theil, Cross, & House, 1980), and subsequently in humans, cattle, ferrets, and recently in dogs (Chang, Nielsen, Ward, & Saif, 1999; Gabbay et al., 2008; T. Mawatari et al., 2004; Otto, Schulze, & Herbst, 1999; Torres-Medina, 1987). RVC in pigs is globally distributed, and associated with epidemic cases of diarrhea (Collins, Martella, & O’Shea, 2008; Jeong et al., 2015; Martella et al., 2007; Marthaler et al., 2013). RVC infections have been associated with acute diarrhea in children and adults in multiple countries (Supplemental Table 1). Notably, two diverse lineages of VP3 have been identified in humans, and human VP6 lineage was identified in swine, providing possible evidence of reassortment of viruses from animal and human reservoirs (Kattoor et al., 2017; Yamamoto et al., 2011). Serological typing of RVC is limited, owing to the difficulty of cultivating RVC strains in cell culture, and RVC genotypes are determined by sequence data (Fujii et al., 2000; Jeong et al., 2015; Marthaler et al., 2013; Moutelikova, Prodelalova, & Dufkova, 2013; Saif, Terrett, Miller, & Cross, 1988; Soma et al., 2013; Suzuki, Hasebe, Miyazaki, & Tsunemitsu, 2014; Tsunemitsu et al., 1991). At the time of this study, the availability of whole-genome sequence (WGS) data for RVC is limited for all hosts: swine (n = 1), canine (n = 1), bovine (n = 7), and human (n = 13) (Baek, Than, Kim, Lim, & Kim, 2013; Chen, Lambden, Lau, Caul, & Clarke, 2002; Doan et al., 2016; Marton et al., 2016; Takahiro Mawatari, Hirano, Tsunemitsu, & Suzuki, 2014; Soma et al., 2013; Yamamoto et al., 2011; Zhirakovskaia et al., 2016).
To address this gap in available RVC WGS data, we sequenced the complete genomes of 45 RVC samples collected from swine in the USA and Mexico (Supplement Table 2) and conducted a phylogenetic analysis including all available RVC sequence data from GenBank and the Virus Pathogen Resource (ViPR). We find that RVC consists of multiple host-specific lineages that have evolved independently, including multiple genetically diverse swine lineages with evidence of multiple reassortment events that occurred deep in the evolutionary history of the virus.
Material and methods
Sample collection and sequencing.
The University of Minnesota Veterinary Diagnostic Laboratory routinely receives swine enteric samples to determine the etiological agents of porcine diseases. Samples were determined to be positive for RVC using a previously described real time RT-PCR method (Homwong, Diaz, Rossow, Ciarlet, & Marthaler, 2016; Marthaler et al., 2014). A total of 64 samples were collected from 16 US states and Mexico between January 2012 and May 2012 and were selected for complete genome sequencing, using primers and thermal cycling conditions as previously described (Yamamoto et al., 2011). Primers were multiplexed to reduce the number of PCR reactions (n = 4) per sample; VP1 and VP4; VP2 and VP3; VP7, VP6 and NSP4; NSP1-NSP3 and NSP5. The amplicons were visualized in an agarose gel. The 4 PCR reactions per sample were combined into a single tube and purified using the MinElute PCR Purification kit (Qiagen). The cDNA was submitted to the University of Minnesota Genomic Center for library preparation and sequencing, using the paired 150 Nextera XT Dual Index Kit for a single run on the Illumina MiSeq platform. In total, from 64 porcine samples, we generated complete WGS for 45 RVC samples and partial genome sequences for 19 RVC samples for which primers failed to amplify every gene segment.
Read processing and genome assembly.
The Illumina paired-end reads were trimmed, and the adapters and primer sequences were removed, using Trimmomatic software with a sliding window of 4 and a quality value cutoff of 16 (Bolger, Lohse, & Usadel, 2014). Reads to swine and various bacterial organisms were mapped and removed using Bowtie2 (Supplemental Material 1) (Langmead & Salzberg, 2012). The number and percentage of RVC, swine, bacterial, and other reads were estimated using Kraken, composed of a custom database containing all the viral sequences, reference bacterial genomes, swine, human, corn, and soybean genomes from GenBank (Supplemental Table 2) (Knutson, Velayudhan, & Marthaler, 2017; Wood & Salzberg, 2014).> Each RVC gene segment was assembled using the A5 assembly software utilizing the IDBA-UD de novo assembler (Tritt, Eisen, Facciotti, & Darling, 2012). Once the RVC genome sequences were assembled, NCBI Blastn was used to identify the RVC gene segments from the contigs generated by the assembly program. After the assembly process, the sequences were trimmed to start from the open reading frame (ORF) for each segment. The sequences were deposited into GenBank and assigned the following accession numbers: MG451081-MG451801. If more than one lineage was identified by de novo assembly, the sequences were arbitrary labeled with the strain name followed by a hyphen and 2, 3 or 4, depending on the number of sequences per sample since determining the relationship of multiple gene segments within a sample is not feasible, and A5 may not identify multiple gene within the same lineage due to the assembly algorithm within the program. Approximately 27.7% (19/64) of the samples identified gene segments from multiple lineages (Supplemental Table 3).
Gene segment analysis
The newly generated sequences were aligned with all publicly available RVC gene segments from Virus Pathogen Resource (ViPR) and GenBank, downloaded on July 31st, 2015, with 80% of the ORF or above for each gene segments (Supplemental Table 4) using MAFFT algorithm available in Geneious (v7.1.2). Recombination was examined for each alignment using the Recombination Detection Program (RDP) v4 (Martin, Murrell, Golden, Khoosal, & Muhire, 2015). There was no evidence of recombination for viruses newly generated for this study. Four previously published strains (KOR/06–144-2, P1; KOR/07–109-12, P4; KOR/2478, P7; and KOR/07–74-11, P7;) were identified as recombinants (Jeong et al., 2015) and removed from the Bayesian analysis. We have also updated our datasets with all publicly available RVC gene segments from Virus Pathogen Resource (ViPR) and GenBank, downloaded on August 7th, 2018, with 80% of the ORF or above for each gene segments (Supplemental Table 4) using MAFFT algorithm available in Geneious (v7.1.2).
Phylogenetic analysis.
The Bayesian framework based on Markov chain Monte Carlo (MCMC) was used to infer a time-scaled phylogeny for each of the RVC gene segments, using BEAST v1.8.4 (Drummond, Nicholls, Rodrigo, & Solomon, 2002; Drummond, Rambaut, Shapiro, & Pybus, 2005; Drummond, Suchard, Xie, & Rambaut, 2012; Drummond & Rambaut, 2007; Drummond & Suchard, 2010). A general-time reversible (GTR) model of nucleotide substitution with gamma distribution among-site rate variation was applied using a relaxed molecular clock and Bayesian Skygrid population prior (Gill et al., 2013). At least 3 MCMC chains were run for 200 million generations each, with sub-sampling every 20,000 iterations since we observed limited temporal signal in the data using TempEst, with correlations in the order of 10−1 to 10−2. A maximum clade credibility (MCC) tree was created for each segment after discarding the first 10% of the chains and summarized in TreeAnnotator (v1.8.4). FigTree (v1.4.3) was used to visualize the tree topologies and key parameters. We also inferred maximum likelihood (ML) trees for the updated dataset using RAxML v7.2.6 (Stamatakis, 2006) with a general time-reversible (GTR) model of nucleotide substitution and gamma-distributed (Γ) rate variation among sites. To assess the robustness of each node, a 500 replicates bootstrap resampling process was performed.
Genetic distance between host clades.
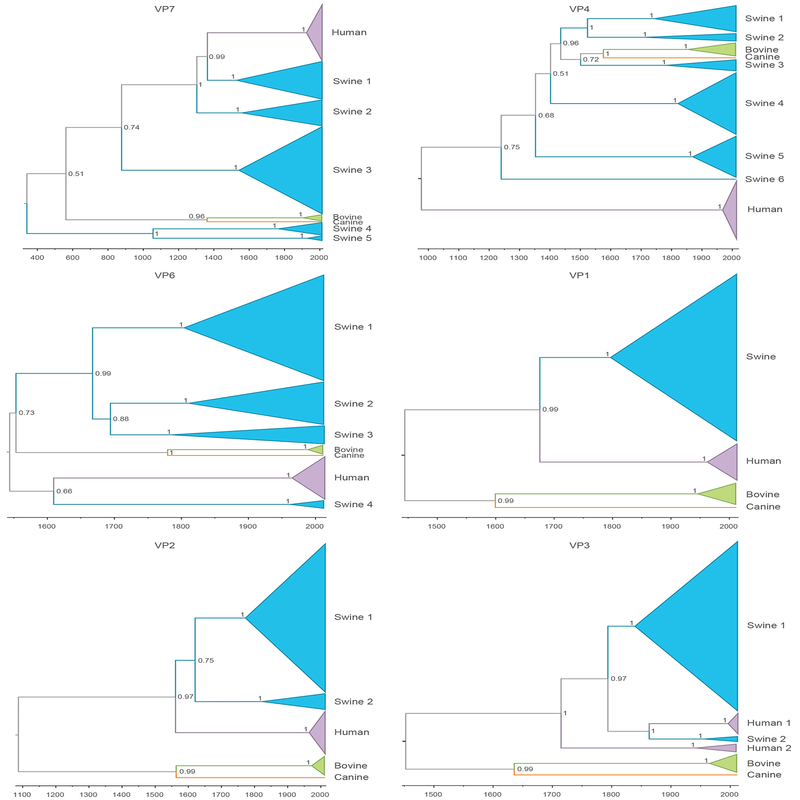
To evaluate genetic divergence between viruses belonging to different host clades of RVC, we estimated the pairwise distance (p-distance) using MEGA v 7.0.2, with 1st+2nd+3rd+non-coding codon positions included and all positions with less than 50% site coverage were eliminated. We calculated the average percentage of genetic similarity [(1−p distance)×100] between clades Bovine, Canine, Human, and Swine clades for the VP7 and VP4 in Figure 1 (Kumar, Stecher, & Tamura, 2016).
Figure 1. MCC trees inferred for 11 dsRNA segments of RVC genome.
Clades of viruses from host are shaded as follows: swine – blue, bovine – green, canine – orange and human – purple. MCC tree is time-scaled, and posterior probabilities for key nodes are provided.
Selection analysis.
Positive selective pressure was assessed using relative rates of non-synonymous and synonymous nucleotide substitutions (dN/dS). We used multiple selection algorithms in Datamokey, including the single-likelihood ancestor counting (SLAC), the fixed effects likelihood (FEL) and the mixed effects model of evolution (MEME) (Delport, Poon, Frost, & Kosakovsky Pond, 2010). Considering the variability between the different computational methods in determining positive selection, only sites verified by all three methods were considered under true positive selection. The analysis was carried out using nucleotide alignments containing all sequences in our data set, as well as specifically for the three-host species for which sufficient data was available (porcine, bovine, and human).
Results
RVC evolves independently in multiple host species.
The evolutionary history of RVC was reconstructed for each of the eleven segments of the RVC genome, using the Bayesian platform implemented in BEAST to infer MCC trees (Supplementary Figure 1). The time-structured MCC trees inferred for each genome segment indicate long-term circulation of RVC as independent lineages in multiple mammalian species, with little evidence of inter-species transmission, at least in the last hundred years. Whereas RVC populations found in bovine and human hosts tend to be monophyletic, multiple highly divergent lineages of RVC were identified in swine as a result of the increased sampling of porcine strains provided by this study. For example, five highly genetically divergent clades of swine viruses were observed on the VP7 tree (Figure 1 and Table 1). These clades diverged from each other centuries -- possibly millennia -- ago. As a result, some porcine RVC segments share less than a 70% nucleotide identity (Table 2 and Figure 1).
Table 1.
Estimated times to the Most Recent Common Ancestor (tMRCA (years, 95% HPD)) in human, swine, and bovine hosts
| Gene Segment | Human (years) | Swine (years) | Bovine (years) |
|---|---|---|---|
| VP7 | 1907.32 (1845.94 – 1956.63) |
† | 1886.02 (1803.69 – 1948.18) |
| VP4 | 1970.79 (1963.57 – 1976.94) |
1299.09 (1080.16 – 1495.86) |
1853.36 (1789.07 – 1909.18) |
| VP6 | 1963.45 (1947.49 – 1976.61) |
1379.22 (1045.84 – 1640.54) |
1972 (1954.69 – 1986.41) |
| VP1 | 1952.59 (1926.01 – 1974.33) |
1815.95 (1710.37 – 1902.59) |
1954.46 (1930.03 – 1974.77) |
| VP2 | 1964.49 (1951.50 – 1975.58) |
1603.29 (1472.08 – 1740.19) |
1971.01 (1959.42 – 1980.61) |
| VP3 | 1721.04 (1542.54 – 1864.66) |
1721.04 (1542.54 – 1864.66) |
1961.46 (1935.14 – 1981.99) |
| NSP1 | 1969.98 (1954.81 – 1982.83) |
1475.54 (1261.86 – 1698.18) |
1972.01 (1953.35 – 1983.77) |
| NSP2 | 1949.83 (1912.24 – 1977.39) |
1571.21 (1228.78 – 1780.01) |
1954.54 (1915.45 – 1981.25) |
| NSP3 | 1976.38 (1966.52 – 1984.70) |
1683.97 (1551.31 – 1805.11) |
1976.02 (1962.24 – 1987.80) |
| NSP4 | † | † | 1703.61 (1465.05 – 1878.10) |
| NSP5 | 1933.06 (1862.66 – 1976.12) |
1755.71 (1496.03 – 1914.99) |
1946.62 (1885.25 – 1986.38) |
Table 2.
Genetic similarity (of most divergent pair of strains for each host) for RVC in human, swine, and bovine hosts
| Gene Segment | Human (% nucleotide identity, 95% HPD) | Swine (% nucleotide identity, 95% HPD) | Bovine (% nucleotide identity, 95% HPD) |
|---|---|---|---|
| VP7 | 93.07–100 | 69.05–100 | 90.96–100 |
| VP4 | 95.09–100 | 70.01–100 | 83.51–100 |
| VP6 | 95.87–100 | 79.50–100 | 95.11–100 |
| VP1 | 92.37–100 | 85.78–100 | 92.87–100 |
| VP2 | 94.27–100 | 78.40–100 | 95.04–99.77 |
| VP3 | 83.42–100 | 83.02–100 | 93.49–100 |
| NSP1 | 92.28–100 | 69.04–100 | 91.94–100 |
| NSP2 | 93.59–100 | 80.77–100 | 94.44–100 |
| NSP3 | 92.37–100 | 74.05–100 | 92.87–100 |
| NSP4 | 70.38–100 | 78.40–100 | 91.90–100 |
| NSP5 | 91.59–100 | 83.02–100 | 94.44–100 |
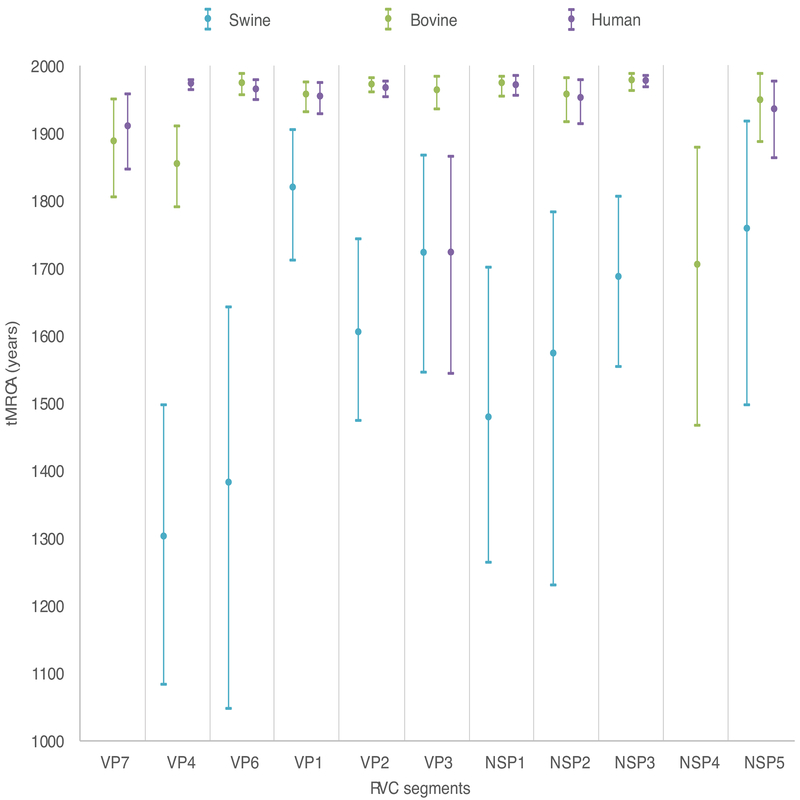
Considerably less genetic diversity was observed among bovine RVCs (>90% genetic similarity for all segments except VP4, Table 2). However, sequences from bovine hosts were only available from Japan (2003–2010) and may not represent the global genetic diversity. Similarly, relatively low genetic diversity was observed among human RVCs, with >90% genetic similarity for all segments except VP3 and NSP4. Human samples were available from multiple countries in Asia and Europe from 1988–2013, and are more likely to be representative of the global diversity of RVC in humans. However, additional RVC populations may be circulating in humans in certain geographical areas that could not be characterized in our study due to the paucity of sequence data, which emphasizes the need of additional sampling from other continents. The time to most recent common ancestor (tMRCA) of the human RVC population fell within the twentieth century for most segments (Figure 2), and it is possible that RVC has emerged more recently in humans. On the VP3 tree, two highly divergent clades of human viruses were identified that share only 83% genetic similarity, as observed previously (Yamamoto et al., 2011). One clade contains viruses collected from humans during 2001–2013 in Asia and Europe; the second clade contains viruses collected from humans during 1988–2010, also from Asia and Europe. These two human clades shared a common ancestor hundreds of years ago, and one clade may represent a separate introduction into humans from an animal reservoir, possibly swine as suggested by the evolutionary relationship displayed in the tree.
Figure 2. Divergence dates for each RVC host.
The time to the Most Recent Common Ancestor (tMRCA) is provided for each RVC host and for each segment of the RVC genome. A tMRCA is not provided for canines because only one canine sequence was available for this analysis. Circles represent the date of divergence (mean), and error bars represent the 95% HPD. Each estimate is shaded according to the host of origin, similar to Figure 1. tMRCA not showed for swine and human NSP4 and for swine VP7 due to uninformative estimate (95% HPD range > 1,000 years).
We also looked at the genetic similarity among different host clades, particularly for VP7 and VP4, as defined in Figure 1. For VP7, we noticed that Swine1 and Swine2 clades are equally close to Human (82%), but have diverged independently, sharing only 80% of their genetic signature (Table 3). For VP4, the highest genetic similarity is shared between the Bovine and Canine clades (76% and 77.50% in VP7), as well as between Swine 2 and Swine 3, and Swine 3 and Swine 4 clades (76%) (Table 4). Despite these clades sharing some degree of genetic similarity, the older tMRCAs along with the lower values of genetic similarity are an indication that these divergence processes took place a long time ago. We have also investigated the phylogenetic relationships of an updated dataset using ML trees (Supplementary Figure 2), and observed similar topologies to those estimated in BEAST, and to a lesser extent for VP4 and VP6. These results reassure that the initial reconstructions represent an accurate representation of RVC evolutionary dynamics.
Table 3.
Genetic similarity (percentage) between pairs of host lineages for VP7.
| VP7 Lineages | Bovine | Canine | Human | Swine 1 | Swine 2 | Swine 3 | Swine 4 | Swine 5 |
|---|---|---|---|---|---|---|---|---|
| Bovine | - | |||||||
| Canine | 77.50 | - | ||||||
| Human | 73.60 | 76.00 | - | |||||
| Swine 1 | 73.00 | 76.00 | 82.00 | - | ||||
| Swine 2 | 73.00 | 76.00 | 82.00 | 80.00 | - | |||
| Swine 3 | 75.00 | 77.00 | 78.00 | 76.00 | 76.00 | - | ||
| Swine 4 | 72.00 | 74.00 | 73.00 | 74.00 | 73.00 | 73.00 | - | |
| Swine 5 | 75.00 | 77.00 | 76.00 | 75.00 | 76.00 | 74.00 | 78.00 | - |
Gradient indicates increase in percentage of genetic similarity, from green (lowest), to yellow, to red (highest).
Table 4.
Genetic similarity (percentage) between pairs of host lineages for VP4.
| VP4 Lineages | Bovine | Canine | Human | Swine 1 | Swine 2 | Swine 3 | Swine 4 | Swine 5 | Swine 6 |
|---|---|---|---|---|---|---|---|---|---|
| Bovine | - | ||||||||
| Canine | 76.00 | - | |||||||
| Human | 71.00 | 71.00 | - | ||||||
| Swine 1 | 74.00 | 73.00 | 70.00 | - | |||||
| Swine 2 | 75.00 | 74.00 | 71.00 | 73.00 | - | ||||
| Swine 3 | 75.00 | 74.00 | 71.00 | 74.00 | 76.00 | - | |||
| Swine 4 | 74.00 | 73.00 | 70.00 | 74.00 | 74.00 | 76.00 | - | ||
| Swine 5 | 74.00 | 74.00 | 72.00 | 74.00 | 74.00 | 74.00 | 74.00 | - | |
| Swine 6 | 73.00 | 73.00 | 70.00 | 72.00 | 72.00 | 73.00 | 73.00 | 74.00 | - |
Gradient indicates increase in percentage of genetic similarity, from green (lowest), to yellow, to red (highest).
Deep evolutionary history of reassortment and inter-species transmission with RVC.
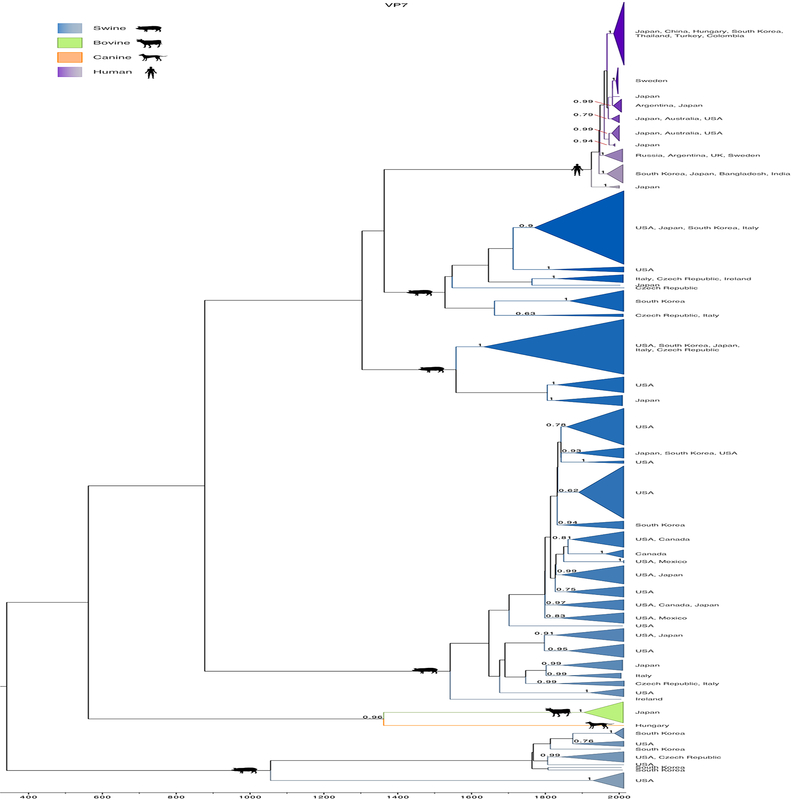
Certain topological patterns were conserved across trees, including the position of bovine and canine clades as sister lineages for all segments except NSP4 and NSP5 (Figure 1). Human viruses also were monophyletic on all trees except for VP3. This may be an oversimplification since understanding the evolutionary history of RVC is complicated by the variable positions of swine viruses in the trees inferred by each gene segment. The swine RVC population was monophyletic for only four of the eleven segments (VP1, NSP4 and NSP5). Up to six highly divergent swine lineages were identified on the VP4 and VP7 trees. These swine lineages were not structured geographically, consistent with long-term circulation in swine hosts (Figure 3 and Supplementary Figure 3). These topological differences are an indication of past reassortment, in which entire genome segments are exchanged between viruses during a co-infection event, producing viruses with segments with different evolutionary histories, and possibly inter-species transmission events that occurred deep in the evolutionary history of these lineages (Figure 1). The extensive genetic diversity observed in swine is consistent with swine being a reservoir host and a possible source of viruses in other host species, including humans. Human and swine lineages often were grouped together on the phylogeny of numerous RV gene segments (VP1, VP2, VP3, VP6, VP7, and NSP2). However, human viruses were positioned basally to all lineages from other hosts on four trees (VP4, NSP1, NSP4, and NSP5) (Figure 3 and Supplementary Figure 2). Without additional sampling, it not possible to determine directionality of RVC transmission between swine and humans.
Figure 3. Detailed phylogeny of VP7 capsid.
Time-scaled Bayesian MCC tree of 301 VP7 sequences, similar to Figure 1. Clades are shaded by host, similar to Figure 1. Clades of viruses from the same host also are labeled by location. Posterior probabilities for key nodes are provided.
Similar evolutionary rates and selection for RVC.
Additional phylogenies inferred for each host-specific lineage revealed no consistent differences in evolutionary rates among human, swine, and bovine lineages (Supplemental Table 5). Rates of RVC evolution varied across segments of the viral genome, ranging from 1.81 × 10−4 substitutions/site/year (95% Highest Posterior Density (HPD) interval: 9.92 × 10−5 - 2.91 × 10−4) for NSP4 to 1.46 × 10−3 substitutions/site/year (95% HPD: 9.90 × 10−4 - 1.96 × 10−3) for NSP3, the fastest evolving segment (Table 5 and Figure 2). Eleven sites spanning six genome segments were identified as under positive selection in swine (Table 6). However, the regions of positive selection did not correspond with significant antigenic regions in the VP7 protein (Eren, Zamuda, & Patton, 2016).
Table 5.
Evolutionary rate for each RVC segment.
| Gene Segment | Evolutionary Rate (substitutions/site/year) (95% HPD interval) |
|---|---|
| VP7 | 4.39 × 10−4 (2.28 × 10−4 – 6.81 × 10−4) |
| VP4 | 9.31 × 10−4 (7.10 × 10−4 – 1.16 × 10−3) |
| VP6 | 6.70 × 10−4 (4.29 × 10−4 – 9.38 × 10−4) |
| VP1 | 7.31 × 10−4 (4.13 × 10−4 – 1.08 × 10−3) |
| VP2 | 7.71 × 10−4 (5.68 × 10−4 – 1.01 × 10−3) |
| VP3 | 9.63 × 10−4 (5.02 × 10−4 – 1.52 × 10−3) |
| NSP1 | 1.19 × 10−3 (7.94 × 10−4 – 1.71 × 10−3) |
| NSP2 | 7.87 × 10−4 (3.93 × 10−4 – 1.17 × 10−3) |
| NSP3 | 1.46 × 10−3 (9.90 × 10−4 – 1.96 × 10−3) |
| NSP4 | 1.81 × 10−4 (9.92 × 10−5 – 2.91 × 10−4) |
| NSP5 | 6.95 × 10−4 (2.32 × 10−4 – 1.24 × 10−3) |
Table 6.
Positively selected sites in the RVC genome.
| Gene Segment | All Host | Swine |
|---|---|---|
| VP7 | 10 | 10 |
| VP2 | None | 127 |
| VP3 | 216, 645, 691 | 9, 300, 645, 691 |
| NSP1 | 386 | 386 |
| NSP4 | None | 70 |
| NSP5 | 130, 131 | 130, 131, 132 |
Discussion
RVC has been greatly undersampled to date, despite the economic importance of RVC in animals and its capacity to infect humans (Tuanthap et al., 2018). While the real time RT-PCR specifically detects porcine strains, the sequencing primers, initially designed for human strains (Yamamoto et al., 2011), did not identify human lineages of RVC and failed to amplify some gene segments in a few porcine RVC strains. However, this study revealed extensive genetic diversity circulating in pigs in the United States and globally, including lineages that diverged hundreds of years ago. We observed a lack of geographical clustering per lineage, which may be a product of several factors, including the long evolutionary relationships of RVC lineages, viral reassortment events, global live swine trade, and passenger air flow.
The extent of RVC diversity identified in US swine has important implications for pathogen control, including the development of porcine RVC vaccines. Although RVC is not considered a clinically important pathogen for human infant mortality due to its milder sporadic and outbreak cases (Bhat et al., 2018; Meleg et al., 2008; Moon et al., 2011), our analysis revealed that the pathogen has been established as an independent lineage in humans for decades, at least, and remains undersampled, as exposed by extremely long branch lengths in the trees. At this time, it is not possible to determine whether the multitude of independently evolving swine lineages arose from viral diversification within pigs, accruing diversity gradually over centuries, or via multiple independent viral introductions into pigs from other RVC hosts that have not been sampled. Although it is clear that RVC exists today as multiple highly divergent host-specific lineages, the lack of RVC sampling in both known and unknown hosts presents an inherent limitation in our analysis, and it is difficult to ascertain the origins of these lineages, or when RVC was introduced from one species to another.
Additional sequence data is also needed to determine if pigs are a major RVC reservoir and a source of RVC for humans and other hosts. The recent identification of human VP6 lineage in swine suggests reverse zoonosis or spill over event from humans to swine (Supplemental Figure 4 (Kattoor et al., 2017). The likelihood that many species infected with RVC remain unidentified was recently underscored by the discovery of RVC in canines. RVA has been detected in a diverse range of host species, including humans, pigs, cows, cats, dogs, horses, birds, rabbits, camelids, and a variety of zoo animals (Badaracco et al., 2013; Bányai et al., 2005; Baumeister, Castro, McGuire-Rodgers, & Ramsay, 1983; Browning, Chalmers, Fitzgerald, & Snodgrass, 1991; Chandler-Bostock et al., 2014; De Grazia et al., 2007; German et al., 2015; Rohwedder, Schütz, Minamoto, & Brüssow, 1995). It is possible that certain rotavirus groups are better at adapting to multiple host species than others, and this question requires additional sampling from multiple species and for non-A rotavirus groups.
The estimated evolutionary rates for RVC had greater variation (1.81 × 10−4 to 1.46 × 10−3) than those estimated for RVA G1P[8] (6.5 – 10 ×10−4) (Zeller et al., 2012). In Zellar’s RVA study, NSP1 and NSP4 had the highest evolutionary rates, whereas in our RVC dataset NSP3 had the highest evolutionary rate among all segments, and NSP1 had the second highest, which is interesting since the RVC and RVA proteins have similar protein motifs (Langland, Pettiford, Jiang, & Jacobs, 1994). Limited research has been conducted on the RVC proteins compared to RVA’s proteins. Additionally, RVC’s VP7 appears to be evolving at approximately half the speed of RVA’s VP7, which may be a contributor to RVA’s higher prevalence and transmissibility (Matthijnssens et al., 2010; Trang et al., 2012; Zeller et al., 2015).
Ultimately, understanding the evolutionary dynamics of RVC in humans and other susceptible animal species is crucial to design better health interventions to minimize the potential impact of RVC strains. However, it is clear that additional whole genome sequences for all host species from a greater geographical range will provide more insight into the spatio-temporal patterns of RVC.
Supplementary Material
Supplementary Figure 1. MCC trees inferred separately for each segment of the RVC genome. Time-scaled Bayesian MCC trees inferred for the 11 RVC genes. The color of each tip indicates the origin of the sample: swine – blue, bovine – green, canine – orange and human – purple. Tip labels and posterior probabilities for host-specific nodes are provided.
Supplementary Figure 2. Maximum likelihood trees inferred for all dsRNA segments of RVC genome in the updated dataset. Clades of viruses from the same host have been collapsed, and shaded as follows: swine – blue, bovine – green, canine – orange and human – purple. Bootstrap support values >30 are provided for key nodes.
Supplementary Figure 3. Additional spatial information in MCC trees inferred for each seg ment. Time-scaled Bayesian MCC trees for all genes, except VP7 (provided Figure 3), with tips annotated with sampling location for each cluster. Each supported location cluster is colored according to host of origin: swine – blue gradient, bovine – green, canine – orange and human – purple gradient. Posterior probabilities for key nodes are provided.
Supplementary Figure 4. Maximum likelihood tree inferred for the VP6 segment of the updated dataset. Clades of viruses from the same host have been collapsed, and shaded as follows: swine – blue, bovine – green, canine – orange and human – purple. Bootstrap support values >30 are provided for key nodes. Porcine strains from India (tip labels in magenta) cluster together with human strains.
Impacts.
RVC strains are limited to host-defined lineages.
Lineages were not geographically defined indicating RVC strains have a long evolutionary history.
Porcine RVC strains are genetically more diverse than RVC strains found in the other mammalian species.
Acknowledgements
This study was partially support by Zoetis (formerly Pfizer Animal Health), the Rapid Agricultural Response Fund, established by the Minnesota legislature and administered by the UM Agricultural Experiment Station, and the USDA National Institute of Food and Agriculture Animal Health project accession number 1013569. Nídia S. Trovão was supported in part by the National Institutes for Health (NIH)/National Institute of Allergy and Infectious Diseases (NIAID) CEIRS contract HHSN272201400008C. The content is solely the responsibility of the authors and does not represent official views of the National Institutes of Health. The authors would like to thank Matthew G. Heffel for his technical assistance.
Footnotes
Conflict of Interest
The authors declare no conflict of interests.
Contributor Information
Nídia S. Trovão, Division of International Epidemiology and Population Studies, Fogarty International Center, National Institutes of Health, Bethesda, Maryland,; Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, United States, Global Health and Emerging Pathogens Institute, Icahn School of Medicine at Mount Sinai, New York, New York, USA
Frances K. Shepherd, Comparative and Molecular Biosciences, College of Veterinary Medicine, University of Minnesota, Minnesota, United States
Katerina Herzberg, Department of Veterinary Population Medicine, College of Veterinary Medicine, University of Minnesota, Minnesota, United States.
Albert Rovira, Department of Veterinary Population Medicine, College of Veterinary Medicine, University of Minnesota, Minnesota, United States.
Marie R. Culhane, Department of Veterinary Population Medicine, College of Veterinary Medicine, University of Minnesota, Minnesota, United States
Ham C. Lam, Department of Veterinary Population Medicine, College of Veterinary Medicine, and Minnesota Supercomputing Institute, University of Minnesota, Minnesota, United States
Matthew C. Jarvis, Department of Veterinary Population Medicine, College of Veterinary Medicine, and Department of Biochemistry, Molecular Biology, and Biophysics, University of Minnesota, Minnesota, United States
Martha I. Nelson, Division of International Epidemiology and Population Studies, Fogarty International Center, National Institutes of Health, Bethesda, Maryland, United States
Douglas G. Marthaler, Veterinary Diagnostic Laboratory, College of Veterinary Medicine, Kansas State University, Manhattan, Kansas, United States Veterinary Diagnostic Laboratory, College of Veterinary Medicine, University of Minnesota, St. Paul, Minnesota.
REFERENCES
- Badaracco A, Matthijnssens J, Romero S, Heylen E, Zeller M, Garaicoechea L, … Parreño V (2013). Discovery and molecular characterization of a group A rotavirus strain detected in an Argentinean vicuña (Vicugna vicugna). Veterinary Microbiology, 161(3–4), 247–254. 10.1016/j.vetmic.2012.07.035 [DOI] [PubMed] [Google Scholar]
- Baek IH, Than VT, Kim H, Lim I, & Kim W (2013). Full genomic characterization of a group C rotavirus isolated from a child in South Korea. Journal of Medical Virology, 85(8), 1478–1484. 10.1002/jmv.23587 [DOI] [PubMed] [Google Scholar]
- Bányai K, Forgách P, Erdélyi K, Martella V, Bogdán A, Hocsák E, … Szucs G (2005). Identification of the novel lapine rotavirus genotype P[22] from an outbreak of enteritis in a Hungarian rabbitry. Virus Research, 113(2), 73–80. 10.1016/j.virusres.2005.03.029 [DOI] [PubMed] [Google Scholar]
- Bányai K, Kemenesi G, Budinski I, Földes F, Zana B, Marton S, … Jakab F (2017). Candidate new rotavirus species in Schreiber’s bats, Serbia. Infection, Genetics and Evolution, 48, 19–26. 10.1016/j.meegid.2016.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister BM, Castro AE, McGuire-Rodgers SJ, & Ramsay EC (1983). Detection and control of rotavirus infections in zoo animals. Journal of the American Veterinary Medical Association, 183(11), 1252–1254. [PubMed] [Google Scholar]
- Bhat S, Kattoor JJ, Malik YS, Sircar S, Deol P, Rawat V, … Kobayashi N (2018). Species C Rotaviruses in Children with Diarrhea in India, 2010–2013: A Potentially Neglected Cause of Acute Gastroenteritis. Pathogens (Basel, Switzerland), 7(1). 10.3390/pathogens7010023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, & Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England), 30(15), 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning GF, Chalmers RM, Fitzgerald TA, & Snodgrass DR (1991). Serological and genomic characterization of L338, a novel equine group A rotavirus G serotype. The Journal of General Virology, 72 ( Pt 5)(Pt 5), 1059–1064. 10.1099/0022-1317-72-5-1059 [DOI] [PubMed] [Google Scholar]
- Chandler-Bostock R, Hancox LR, Nawaz S, Watts O, Iturriza-Gomara M, & Mellits KH (2014). Genetic diversity of porcine group A rotavirus strains in the UK. Veterinary Microbiology, 173(1–2), 27–37. 10.1016/j.vetmic.2014.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KO, Nielsen PR, Ward LA, & Saif LJ (1999). Dual infection of gnotobiotic calves with bovine strains of group A and porcine-like group C rotaviruses influences pathogenesis of the group C rotavirus. Journal of Virology, 73(11), 9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Lambden PR, Lau J, Caul EO, & Clarke IN (2002). Human group C rotavirus: completion of the genome sequence and gene coding assignments of a non-cultivatable rotavirus. Virus Research, 83(1–2), 179–187. [DOI] [PubMed] [Google Scholar]
- Collins PJ, Martella V, & O’Shea H (2008). Detection and characterization of group C rotaviruses in asymptomatic piglets in Ireland. Journal of Clinical Microbiology, 46(9), 2973–2979. 10.1128/JCM.00809-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Grazia S, Martella V, Giammanco GM, Gòmara MI, Ramirez S, Cascio A, … Arista S (2007). Canine-origin G3P[3] rotavirus strain in child with acute gastroenteritis. Emerging Infectious Diseases, 13(7), 1091–1093. 10.3201/eid1307.070239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delport W, Poon AFY, Frost SDW, & Kosakovsky Pond SL (2010). Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics (Oxford, England), 26(19), 2455–2457. 10.1093/bioinformatics/btq429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan YH, Haga K, Fujimoto A, Fujii Y, Takai-Todaka R, Oka T, … Katayama K (2016). Genetic analysis of human rotavirus C: The appearance of Indian-Bangladeshi strain in Far East Asian countries. Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 41, 160–173. 10.1016/j.meegid.2016.03.027 [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Nicholls GK, Rodrigo AG, & Solomon W (2002). Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics, 161(3), 1307–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, & Rambaut A (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7(Journal Article), 214 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, & Pybus OG (2005). Bayesian coalescent inference of past population dynamics from molecular sequences. Molecular Biology and Evolution, 22(5), 1185–1192. 10.1093/molbev/msi103 [DOI] [PubMed] [Google Scholar]
- Drummond AJ, & Suchard MA (2010). Bayesian random local clocks, or one rate to rule them all. BMC Biology, 8(Journal Article), 114–7007–8–114 10.1186/1741-7007-8-114; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, & Rambaut A (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution, 29(8), 1969–1973. 10.1093/molbev/mss075; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren E, Zamuda K, & Patton JT (2016). Modeling of the rotavirus group C capsid predicts a surface topology distinct from other rotavirus species. Virology, 487, 150–162. 10.1016/j.virol.2015.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes M, & Greenberg HB (2013). Rotaviruses In Knipe D & Howley P (Eds.), Fields virology (Vol. 6th, pp. 1347–1395). Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins. Retrieved from [Table of contents only ]--http://www.loc.gov/catdir/toc/ecip072/2006032230.html; [Google Scholar]
- Fujii R, Kuzuya M, Hamano M, Ogura H, Yamada M, & Mori T (2000). Neutralization assay for human group C rotaviruses using a reverse passive hemagglutination test for endpoint determination. Journal of Clinical Microbiology, 38(1), 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbay YB, Borges AA, Oliveira DS, Linhares AC, Mascarenhas JDP, Barardi CRM, … Jiang B (2008). Evidence for zoonotic transmission of group C rotaviruses among children in Belém, Brazil. Journal of Medical Virology, 80(9), 1666–1674. [DOI] [PubMed] [Google Scholar]
- German AC, Iturriza-Gomara M, Dove W, Sandrasegaram M, Nakagomi T, Nakagomi O, … Morgan KL (2015). Molecular epidemiology of rotavirus in cats in the United Kingdom. Journal of Clinical Microbiology, 53(2), 455–464. 10.1128/JCM.02266-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill MS, Lemey P, Faria NR, Rambaut A, Shapiro B, & Suchard MA (2013). Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Molecular Biology and Evolution, 30(3), 713–724. 10.1093/molbev/mss265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homwong N, Diaz A, Rossow S, Ciarlet M, & Marthaler D (2016). Three-Level Mixed-Effects Logistic Regression Analysis Reveals Complex Epidemiology of Swine Rotaviruses in Diagnostic Samples from North America. PloS One, 11(5), e0154734 10.1371/journal.pone.0154734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YJ, Matthijnssens J, Kim DS, Kim JY, Alfajaro MM, Park JG, … Cho KO (2015). Genetic diversity of the VP7, VP4 and VP6 genes of Korean porcine group C rotaviruses. Veterinary Microbiology, 176(1–2), 61–69. 10.1016/j.vetmic.2014.12.024 [DOI] [PubMed] [Google Scholar]
- Kattoor JJ, Saurabh S, Malik YS, Sircar S, Dhama K, Ghosh S, … Singh RK (2017). Unexpected detection of porcine rotavirus C strains carrying human origin VP6 gene. The Veterinary Quarterly, 37(1), 252–261. 10.1080/01652176.2017.1346849 [DOI] [PubMed] [Google Scholar]
- Knutson TP, Velayudhan BT, & Marthaler DG (2017). A porcine enterovirus G associated with enteric disease contains a novel papain-like cysteine protease. The Journal of General Virology, 98(6), 1305–1310. 10.1099/jgv.0.000799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, & Tamura K (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution, 33(7), 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langland JO, Pettiford S, Jiang B, & Jacobs BL (1994). Products of the porcine group C rotavirus NSP3 gene bind specifically to double-stranded RNA and inhibit activation of the interferon-induced protein kinase PKR. Journal of Virology, 68(6), 3821–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, & Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods, 9(4), 357–359. 10.1038/nmeth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V, Banyai K, Lorusso E, Bellacicco AL, Decaro N, Camero M, … Buonavoglia C (2007). Prevalence of group C rotaviruses in weaning and post-weaning pigs with enteritis. Veterinary Microbiology, 123(1–3), 26–33. 10.1016/j.vetmic.2007.03.003 [DOI] [PubMed] [Google Scholar]
- Marthaler D, Homwong N, Rossow K, Culhane M, Goyal S, Collins J, … Ciarlet M (2014). Rapid detection and high occurrence of porcine rotavirus A, B, and C by RT-qPCR in diagnostic samples. Journal of Virological Methods, 209(Journal Article), 30–34. 10.1016/j.jviromet.2014.08.018 [DOI] [PubMed] [Google Scholar]
- Marthaler D, Rossow K, Culhane M, Collins J, Goyal S, Ciarlet M, & Matthijnssens J (2013). Identification, phylogenetic analysis and classification of porcine group C rotavirus VP7 sequences from the United States and Canada. Virology, 446(1–2), 189–198. 10.1016/j.virol.2013.08.001; [DOI] [PubMed] [Google Scholar]
- Martin DP, Murrell B, Golden M, Khoosal A, & Muhire B (2015). RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evolution, 1(1), vev003 10.1093/ve/vev003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton S, Medici MC, Tummolo F, Martella V, Arcangeletti MC, Bányai K, … Fehér E (2016). Analysis of the full genome of human group C rotaviruses reveals lineage diversification and reassortment. Journal of General Virology, 97(8), 1888–1898. 10.1099/jgv.0.000497 [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Otto PH, Ciarlet M, Desselberger U, Van Ranst M, & Johne R (2012). VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Archives of Virology, 157(6), 1177–1182. 10.1007/s00705-012-1273-3 [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, & Van Ranst M (2012). Genotype constellation and evolution of group A rotaviruses infecting humans. Current Opinion in Virology, 2(4), 426–433. 10.1016/j.coviro.2012.04.007; [DOI] [PubMed] [Google Scholar]
- Mawatari T, Taneichi A, Kawagoe T, Hosokawa M, Togashi K, & Tsunemitsu H (2004). Detection of a bovine group C rotavirus from adult cows with diarrhea and reduced milk production. The Journal of Veterinary Medical Science/the Japanese Society of Veterinary Science, 66(7), 887. [DOI] [PubMed] [Google Scholar]
- Mawatari Takahiro, Hirano K, Tsunemitsu H, & Suzuki T (2014). Whole-genome analysis of bovine rotavirus species C isolates obtained in Yamagata, Japan, 2003–2010. The Journal of General Virology, 95(Pt 5), 1117–1125. 10.1099/vir.0.062166-0 [DOI] [PubMed] [Google Scholar]
- Meleg E, Bányai K, Martella V, Jiang B, Kocsis B, Kisfali P, … Szucs G (2008). Detection and quantification of group C rotaviruses in communal sewage. Applied and Environmental Microbiology, 74(11), 3394–3399. 10.1128/AEM.02895-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalov-Kovacs E, Gellert A, Marton S, Farkas SL, Feher E, Oldal M, … Banyai K (2015). Candidate new rotavirus species in sheltered dogs, Hungary. Emerging Infectious Diseases, 21(4), 660–663. 10.3201/eid2104.141370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S, Humphrey CD, Kim JS, Baek LJ, Song J-W, Song K-J, & Jiang B (2011). First detection of group C rotavirus in children with acute gastroenteritis in South Korea. Clinical Microbiology and Infection, 17(2), 244–247. 10.1111/j.1469-0691.2010.03270.x [DOI] [PubMed] [Google Scholar]
- Moutelikova R, Prodelalova J, & Dufkova L (2013). Prevalence study and phylogenetic analysis of group C porcine rotavirus in the Czech Republic revealed a high level of VP6 gene heterogeneity within porcine cluster I1. Archives of Virology, (Journal Article). 10.1007/s00705-013-1903-4 [DOI] [PubMed] [Google Scholar]
- Otto P, Schulze P, & Herbst W (1999). Demonstration of group C rotaviruses in fecal samples of diarrheic dogs in Germany. Archives of Virology, 144(12), 2467–2473. [DOI] [PubMed] [Google Scholar]
- Rohwedder A, Schütz KI, Minamoto N, & Brüssow H (1995). Sequence analysis of pigeon, turkey, and chicken rotavirus VP8* identifies rotavirus 993/83, isolated from calf feces, as a pigeon rotavirus. Virology, 210(1), 231–235. 10.1006/viro.1995.1338 [DOI] [PubMed] [Google Scholar]
- Sadiq A, Bostan N, Yinda KC, Naseem S, & Sattar S (2018). Rotavirus: Genetics, pathogenesis and vaccine advances. Reviews in Medical Virology, e2003 10.1002/rmv.2003 [DOI] [PubMed] [Google Scholar]
- Saif LJ, Bohl EH, Theil KW, Cross RF, & House JA (1980). Rotavirus-like, calicivirus-like, and 23-nm virus-like particles associated with diarrhea in young pigs. Journal of Clinical Microbiology, 12(1), 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif LJ, Terrett LA, Miller KL, & Cross R (1988). Serial propagation of porcine group C rotavirus (pararotavirus) in a continuous cell line and characterization of the passaged virus. Journal of Clinical Microbiology, 26(7), 1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma J, Tsunemitsu H, Miyamoto T, Suzuki G, Sasaki T, & Suzuki T (2013). Whole-genome analysis of two bovine rotavirus C strains: Shintoku and Toyama. The Journal of General Virology, 94(Pt 1), 128–135. 10.1099/vir.0.046763-0; [DOI] [PubMed] [Google Scholar]
- Stamatakis A (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics (Oxford, England), 22(21), 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Hasebe A, Miyazaki A, & Tsunemitsu H (2014). Phylogenetic characterization of VP6 gene (inner capsid) of porcine rotavirus C collected in Japan. Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 26, 223–227. 10.1016/j.meegid.2014.05.024 [DOI] [PubMed] [Google Scholar]
- Torres-Medina A (1987). Isolation of an atypical rotavirus causing diarrhea in neonatal ferrets. Laboratory Animal Science, 37(2), 167–171. [PubMed] [Google Scholar]
- Tritt A, Eisen JA, Facciotti MT, & Darling AE (2012). An integrated pipeline for de novo assembly of microbial genomes. PloS One, 7(9), e42304 10.1371/journal.pone.0042304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemitsu H, Saif LJ, Jiang B, Shimizu M, Hiro M, Yamaguchi H, … Hirai (1991). Isolation, characterization, and serial propagation of a bovine group C rotavirus in a monkey kidney cell line (MA104). Journal of Clinical Microbiology, 29(11), 2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuanthap S, Phupolphan C, Luengyosluechakul S, Duang-In A, Theamboonlers A, Wattanaphansak S, … Poovorawan Y (2018). Porcine rotavirus C in pigs with gastroenteritis on Thai swine farms, 2011–2016. PeerJ, 6, e4724 10.7717/peerj.4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DE, & Salzberg SL (2014). Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biology, 15(3), R46–2014-15–3-r46. 10.1186/gb-2014-15-3-r46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto D, Ghosh S, Kuzuya M, Wang YH, Zhou X, Chawla-Sarkar M, … Kobayashi N (2011). Whole-genome characterization of human group C rotaviruses: identification of two lineages in the VP3 gene. Journal of General Virology, 92(2), 361. [DOI] [PubMed] [Google Scholar]
- Zeller M, Patton JT, Heylen E, De Coster S, Ciarlet M, Van Ranst M, & Matthijnssens J (2012). Genetic Analyses Reveal Differences in the VP7 and VP4 Antigenic Epitopes between Human Rotaviruses Circulating in Belgium and Rotaviruses in Rotarix and RotaTeq. Journal of Clinical Microbiology, 50(3), 966–976. 10.1128/JCM.05590-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhirakovskaia E, Tikunov A, Klemesheva V, Loginovskikh N, Netesov S, & Tikunova N (2016). First genetic characterization of rotavirus C in Russia. Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 39, 1–8. 10.1016/j.meegid.2016.01.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. MCC trees inferred separately for each segment of the RVC genome. Time-scaled Bayesian MCC trees inferred for the 11 RVC genes. The color of each tip indicates the origin of the sample: swine – blue, bovine – green, canine – orange and human – purple. Tip labels and posterior probabilities for host-specific nodes are provided.
Supplementary Figure 2. Maximum likelihood trees inferred for all dsRNA segments of RVC genome in the updated dataset. Clades of viruses from the same host have been collapsed, and shaded as follows: swine – blue, bovine – green, canine – orange and human – purple. Bootstrap support values >30 are provided for key nodes.
Supplementary Figure 3. Additional spatial information in MCC trees inferred for each seg ment. Time-scaled Bayesian MCC trees for all genes, except VP7 (provided Figure 3), with tips annotated with sampling location for each cluster. Each supported location cluster is colored according to host of origin: swine – blue gradient, bovine – green, canine – orange and human – purple gradient. Posterior probabilities for key nodes are provided.
Supplementary Figure 4. Maximum likelihood tree inferred for the VP6 segment of the updated dataset. Clades of viruses from the same host have been collapsed, and shaded as follows: swine – blue, bovine – green, canine – orange and human – purple. Bootstrap support values >30 are provided for key nodes. Porcine strains from India (tip labels in magenta) cluster together with human strains.