Main Text
(iScience 15, 1–15, May 31, 2019)
In the originally published version of this article, the authors found errors in Figures 2, 3, and 4. In Figures 2 and 3, the images for the “scrambled-LNA” control actually represent vehicle-only control. The authors have now provided images showing the scrambled-LNA control alongside the experimental samples processed at the same time. Please note that the scrambled-LNA and vehicle-only controls give similar results. In Figure 4, the original images do represent “scrambled-LNA” and the authors have corrected the label in the figure to state this. The original and corrected versions of the figures and corrected legends appear here and online.
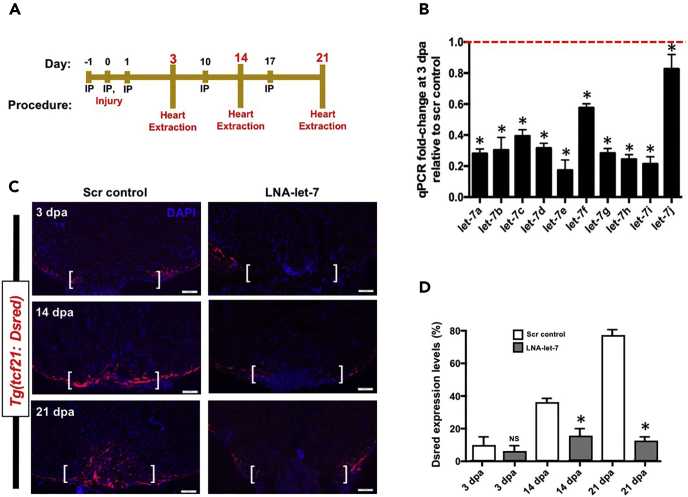
Figure 2. Let-7 Depletion Results in Defects in Wound Closure (Corrected)
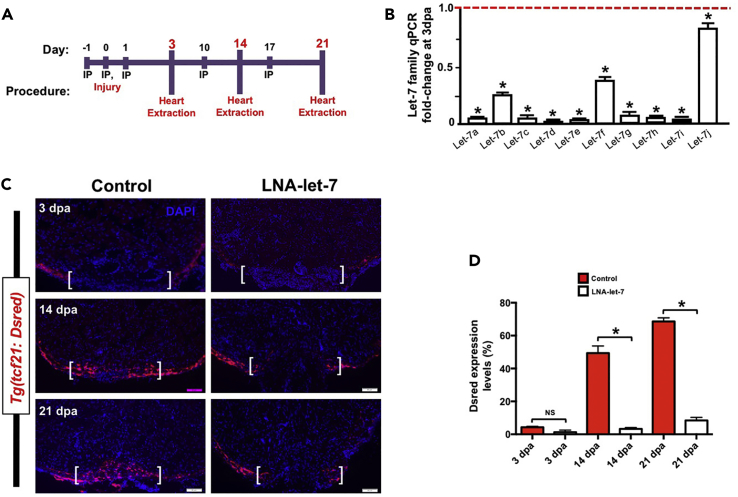
(A) Schematic of locked-nucleic-acid (LNA) microinjection paradigm to deplete let-7 expression.
(B) Real-time qPCR analyses show that LNA treatment results in significant knockdown of mature let-7 expression at 3 dpa, in comparison to scrambled control LNA oligonucleotides. Values are means ± SE. *p < 0.05. (n = 3).
(C) Representative images of tcf21:Dsred expression in scrambled LNA control and LNA-let-7 treated hearts at 3, 14, and 21 dpa showing defects in wound closure. Scale bar represents 50 μm. Brackets indicate approximate resection plane.
(D) Quantification of tcf21:Dsred expression levels within the resection zone. Values are means ± SE. *p < 0.05. (n = 6); IP, intraperitoneal; dpa, days post-amputation; NS, not significant.
Figure 2. Let-7 Depletion Results in Defects in Wound Closure (Original)
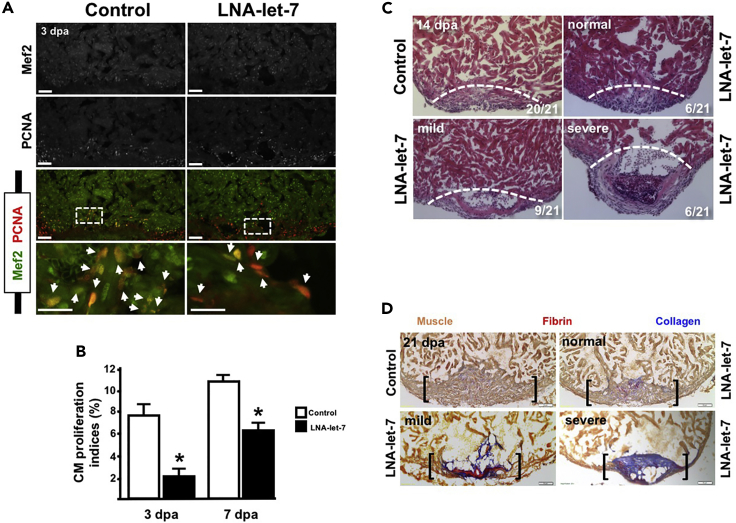
Figure 3. Let-7 Activity Is Required for Heart Regeneration (Corrected)
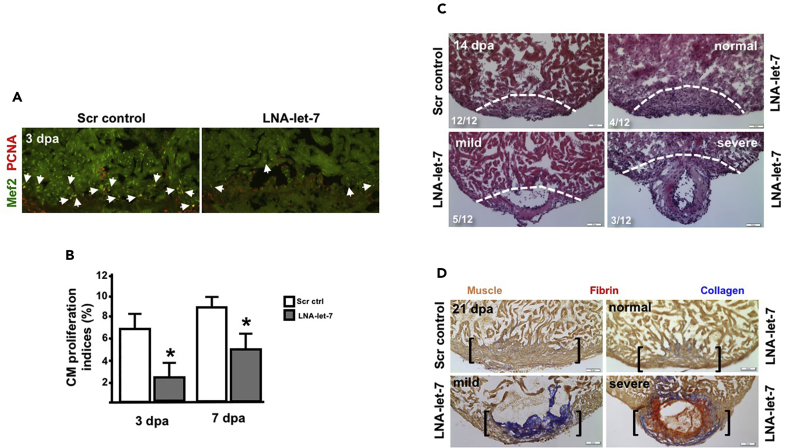
(A) Representative images showing proliferating CMs in scrambled LNA control and LNA-let-7-treated fish. Arrowheads indicate Mef2+PCNA+ cells at the injury zone (n = 7).
(B) CM proliferation indices are suppressed by 68% and 44% in LNA-let-7 hearts at 3 and 7 dpa, respectively, when compared to scrambled LNA control treatment. Values are means ± SE. *p < 0.05.
(C) Representative images of hematoxylin and eosin staining of 14 dpa scrambled LNA control and LNA-let-7-treated hearts reveal a spectrum of heart regeneration defects, categorized as normal, mild, and severe. (N = 12). Dashed lines mark approximate resection plane; dpa, days post-amputation.
(D) Representative images of AFOG stains of scrambled LNA control and LNA-let-7 administered hearts reveal more collagen and fibrin tissue within the injury zone. (N = 10). Brackets mark the injury zone. Brown, muscle; red, fibrin; blue, collagen.
Figure 3. Let-7 Activity Is Required for Heart Regeneration (Original)
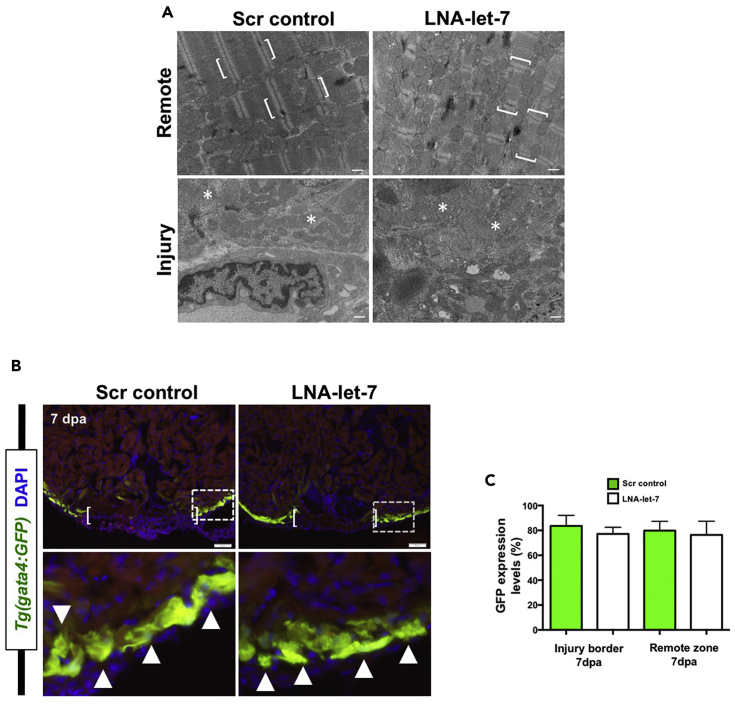
Figure 4. CM Dedifferentiation Is Normal under Conditions of Decreased let-7 Activity (Corrected)
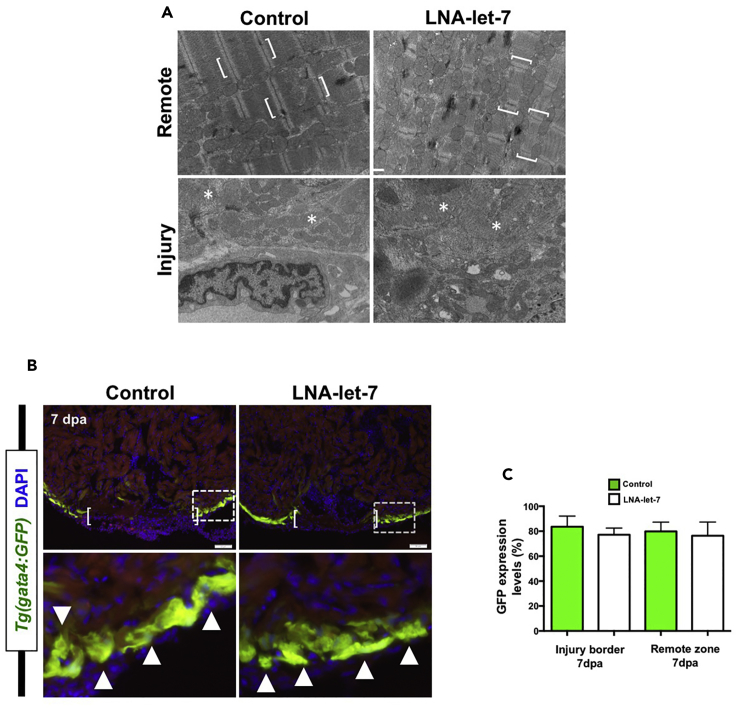
(A) Representative transmission electron microscope images of remote and injury zones of resected 7-dpa hearts from scrambled control and LNA-let-7-treated hearts. Brackets mark intact CM sarcomeres in a remote zone. By contrast, asterisks highlight disorganized and dedifferentiated myosin bundles at the injury zone (n = 4). Scale bar represents 500 nm.
(B) Representative images showing activation of Tg(gata4:GFP) expression in the primordial muscle layer in scrambled control and LNA-let-7-treated hearts at 7 dpa. Brackets in upper panels show approximate resection injury plane. Arrowheads in magnified bottom panels show gata4:GFP expression in both scrambled control and LNA-let-7-treated hearts. (n = 6).
(C) Quantification of gata4:GFP expression levels at the injury border and remote zones show no differences between scrambled control and LNA-let-7-treated hearts. Values are means ± SE. *p < 0.05. (N = 6).
Figure 4. CM Dedifferentiation Is Normal under Conditions of Decreased let-7 Activity (Original)