Abstract
Previous studies have shown that an intronic variant rs780094 of the GCKR gene (glucokinase regulatory protein) is significantly associated with several metabolites, but the associations of this genetic variant with different lipids is largely unknown. Therefore, we applied metabolomics approach to measure metabolites in a large Finnish population sample (METSIM study) to investigate their associations with rs780094 of GCKR. We measured metabolites by mass spectrometry from 5,181 participants. P < 5.8 × 10−5 was considered as statistically significant given 857 metabolites included in statistical analyses. We found novel negative associations of the T allele of GCKR rs780094 with serine and threonine, and positive associations with two metabolites of tryptophan, indolelactate and N-acetyltryptophan. Additionally, we found novel significant positive associations of this genetic variant with 12 glycerolipids and 19 glycerophospholipids. Significant negative associations were found for three glycerophospholipids (all plasmalogen-cholines), and two sphingolipids. Significant novel associations were also found with gamma-glutamylthreonine, taurocholenate sulfate, and retinol. Our study adds new information about the pleiotropy of the GCKR gene, and shows the associations of the T allele of GCKR rs780094 with lipids.
Subject terms: Diabetes complications, Genetic association study
Introduction
Genetic and lifestyle/environmental factors play an important role in the risk of type 2 diabetes. Over 100 genetic variants have been associated with type 2 diabetes in recent genome wide association studies, including glucokinase regulator GCKR gene encoding glucokinase regulatory protein (GKRP), a hepatocyte-specific inhibitor of the glucose-metabolizing enzyme glucokinase in the fasting state. After a meal hepatic glucokinase is released to cytoplasm and stimulates glycogen deposition and de novo lipogenesis1.
Two single nucleotide polymorphisms of GCKR, rs780094 (intronic variant) and rs1260326 (p.P446L) are in a strong linkage disequilibrium. These genetic variants are significantly associated with several metabolites, including amino acids, carbohydrates, ketone bodies, high density lipoprotein, total triacylglycerides (TAG), very low density lipoprotein, and diseases, such as type 2 diabetes, non-alcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH)2–7. Thus, GCKR is a highly pleiotropic gene. The p.P446L GKRP substitution results in destabilization of the GCK binding interface explaining inverse correlation between fasting glucose and triglycerides for this variant8. Increased hepatic GCK activity results in decreased glucose levels and increases in TAG and glycogen synthesis in normoglycemia9.
Previous studies on the associations of genetic variants of GCKR with laboratory measurements have not focused on lipids except for TAG. Given the fact that variants of GCKR have been associated with liver diseases the associations of these variants with lipids, including glycerolipids (GLs), glycerophospholipids (GPLs) and sphingolipids could be of great interest. GLs are composed of mono, di-, and tri-substituted glycerols that are hydrolyzed to free fatty acids (FFAs)10. GPLs are major components of cellular membranes synthesized from phosphatidic acid (PA) and diacylglycerol (DAG)11. PA can also be formed via phosphorylation of DAG11 and dephosphorylated to generate 1,2-DAG or bound with choline, ethanolamine or inositol to synthesize phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI)11.
We applied metabolomics to measure low molecular weight molecules in the Finnish prospective population-based Metabolic Syndrome In Men (METSIM) study to obtain a comprehensive understanding of metabolites associated with rs780094 of GCKR12. A special focus in our study is on the associations of this genetic variant with different lipids because they play an important role in liver disease, especially in NAFLD.
Results
Principal component analysis and correlations
We performed the principal component analysis (PCA) for the lipids. As shown in Supplementary Table S1, 33 metabolites resulted in 7 components explaining 77.2% of total variance. The first principal component (PC) having the largest loading for oleoyl-linoleoyl-glycerol (18:1/18:2)2 and other glycerolipids explained 38.5% of total variance. The second PCs had the highest loading for 1-stearoyl-2-linoleoyl-GPE (18:0/18:2) and explained 12.6%, and the third PC having the highest loading for 1-myristoyl-2-palmitoyl-GPC (14:0/16:0) explained 8.0% of total variance. The second and third PCs had high loadings for glycerophospholipids. These three PCs explained 59.1% of total variance.
The heat map shows the correlations between the lipids (Supplementary Fig. S1). Glycerolipids (the lower left corner of Supplementary Fig. S1) were loaded mainly on the first PC (PC1). Glycerophospholipids were loaded on both PC2 and PC3, and had high intercorrelations. Sphingolipids were mainly loaded on PC4 (Supplementary Table S1) and had no correlation or negative correlations with all other lipids demonstrating the heterogeneity among lipid species. The strongest correlation (r = 0.840) was observed between the two isomers of oleoyl-arachidonoyl-glycerol (18:1/20:4)1 and oleoyl-arachidonoyl-glycerol (18:1/20:4)2. Lipids and carbohydrates were also significantly correlated, 1-stearoyl-2-oleoyl-GPE (18:0/18:1) and pyruvate had a correlation of 0.358, as well as lipids and amino acids, 1-myristoyl-2-arachidonoyl-GPC (14:0/20:4) and alanine had a correlation of 0.287.
Association of the T allele of GCKR rs780094 with amino acids, carbohydrates, and other metabolites
After adjustment for batch effect, age and fasting glucose GCKR rs780094-T showed a significant positive association (P < 5.8 × 10−5) with alanine (P = 7.9 × 10−10), and downstream metabolites of the tryptophan pathway metabolites, indolelactate and N-acetyltryptophan (P = 2.1 × 10−6, and P = 5.2 × 10−7, respectively), and novel negative associations with serine (P = 2.1 × 10−6), threonine (P = 4.8 × 10−11), and 3-aminoisobutyrate (P = 3.5 × 10−10) which is a downstream metabolite of the valine pathway Table 1. We also found significant associations of GCKR rs780094-T with lactate, pyruvate and mannose, as previously published. Novel negative associations were also found with gamma-glutamylthreonine (P = 4.9 × 10−6), taurocholenate sulfate (P = 8.1 × 10−6), and a novel positive association with retinol (P = 4.8 × 10−6) (Table 1). Nominally significant associations (P < 0.05) of GCKR rs780094-T with the metabolites are shown in Supplementary Table S2, and non-significant associations of GCKR rs780094-T with metabolites in Supplementary Table S3.
Table 1.
Associations of GCKR rs780094-T with amino acids, carbohydrates and other metabolites.
| Pathway | beta | P value | P* value | Novel |
|---|---|---|---|---|
| Amino acids | ||||
| Alanine | 0.080 | 1.3E-08 | 7.9E10-10 | No |
| Serine | −0.065 | 3.5E-06 | 2.1E10-6 | Yes |
| Threonine | −0.094 | 1.8E-11 | 4.8E10-11 | Yes |
| Tryptophan pathway: | ||||
| Indolelactate | 0.066 | 2.1E-06 | 2.1E10-6 | Yes |
| N-acetyltryptophan | 0.066 | 2.1E-06 | 5.2E10-7 | Yes |
| Valine pathway: | ||||
| 3-aminoisobutyrate | −0.082 | 4.1E-09 | 3.5E10-10 | Yes |
| Carbohydrates | ||||
| Lactate | 0.066 | 2.6E-06 | 3.5E10-7 | No |
| Mannose | −0.325 | 1.8E-125 | 8.5E10-128 | No |
| Pyruvate | 0.083 | 2.9E-09 | 1.4E10-10 | No |
| Other metabolites | ||||
| Gamma-glutamylthreonine | −0.065 | 3.3E-06 | 4.9E10-6 | Yes |
| Taurocholenate sulfate | −0.063 | 6.7E-06 | 8.1E10-6 | Yes |
| Retinol (Vitamin A) | 0.063 | 6.6E-06 | 4.8E10-7 | Yes |
| 3-hydroxybutyrate | −0.057 | 4.7E-05 | 1.4E10-5 | No |
Beta and P values were obtained from linear regression and adjusted for batch effect. Only metabolites that were associated significantly (P < 5.8 × 10−5) are shown. Beta and P values were obtained from linear regression, and were adjusted for batch effect, P* values were adjusted for batch effect, age and fasting glucose.
Association of rs780094 of GCKR with lipids
Glycerolipids
We found 12 novel significant positive associations of GCKR rs780094-T with GLs after the adjustment for batch effect, age and fasting glucose. Nine of them were 1,2-DAGs and three monoacylglycerides Table 2, Fig. 1.
Table 2.
Association of GCKR rs780094-T with lipids.
| Pathway | Beta | P value | P* value | Sub class | Direct parent | Novel |
|---|---|---|---|---|---|---|
| Glycerolipids | ||||||
| Triacylglycerides* | 0.107 | 2.1E-14 | 1.6E10-16 | TAG | TAG | No |
| Palmitoleoyl-linoleoyl-glycerol (16:1/18:2) (1)** | 0.109 | 3.8E-09 | 6.9E10-10 | DAG | 1,2-DAG | Yes |
| Myristoyl-linoleoyl-glycerol (14:0/18:2) (1)** | 0.120 | 7.4E-11 | 2.2E10-11 | DAG | 1,2-DAG | Yes |
| Palmitoyl-linoleoyl-glycerol (16:0/18:2) (2)** | 0.057 | 4.9E-05 | 2.4E10-5 | DAG | 1,2-DAG | Yes |
| Oleoyl-linoleoyl-glycerol (18:1/18:2) (1)** | 0.065 | 3.4E-06 | 1.3E10-6 | DAG | 1,2-DAG | Yes |
| Oleoyl-linoleoyl-glycerol (18:1/18:2) (2)** | 0.061 | 1.1E-05 | 4.9E10-6 | DAG | 1,2-DAG | Yes |
| DAG (12:0/18:1, 14:0/16:1, 16:0/14:1) (2)** | 0.091 | 9.0E-07 | 3.0E10-7 | DAG | 1,2-DAG | Yes |
| Oleoyl-arachidonoyl-glycerol (18:1/20:4) (1)** | 0.095 | 3.1E-07 | 8.0E10-8 | DAG | 1,2-DAG | Yes |
| Oleoyl-arachidonoyl-glycerol (18:1/20:4) (2)** | 0.087 | 2.4E-06 | 9.5E10-7 | DAG | 1,2-DAG | Yes |
| Oleoyl-oleoyl-glycerol (18:1/18:1) (2)** | 0.087 | 2.7E-06 | 7.7E10-7 | DAG | 1,2-DAG | Yes |
| 1-palmitoleoylglycerol (16:1) | 0.082 | 1.6E-07 | 4.4E10-8 | MAG | 1-MAG | Yes |
| 1-oleoylglycerol (18:1) | 0.057 | 5.0E-05 | 3.2E10-5 | MAG | 1-MAG | Yes |
| 1-myristoylglycerol (14:0) | 0.073 | 2.0E-07 | 9.8E10-8 | MAG | 1-MAG | Yes |
| Glycerophospholipids | ||||||
| 1-stearoyl-2-arachidonoyl-GPI (18:0/20:4) | 0.058 | 3.5E-05 | 1.2E10-5 | GPI | PI | Yes |
| 1-palmitoyl-2-oleoyl-GPE (16:0/18:1) | 0.085 | 1.3E-09 | 4.0E10-10 | GPE | PE | Yes |
| 1-palmitoyl-2-docosahexaenoyl-GPE (16:0/22:6) | 0.087 | 2.1E-08 | 1.2E10-8 | GPE | PE | Yes |
| 1-stearoyl-2-docosahexaenoyl-GPE (18:0/22:6) | 0.098 | 3.1E-10 | 6.1E10-11 | GPE | PE | Yes |
| 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | 0.063 | 6.6E-06 | 7.7E10-6 | GPE | PE | Yes |
| 1-stearoyl-2-oleoyl-GPE (18:0/18:1) | 0.075 | 9.0E-08 | 1.7E10-8 | GPE | PE | Yes |
| 1-stearoyl-2-linoleoyl-GPE (18:0/18:2) | 0.07 | 5.0E-07 | 2.2E10-6 | GPE | PE | Yes |
| 1-stearoyl-2-arachidonoyl-GPE (18:0/20:4) | 0.064 | 4.2E-06 | 1.2E10-6 | GPE | PE | Yes |
| 1-oleoyl-2-docosahexaenoyl-GPE (18:1/22:6) | 0.082 | 1.0E-05 | 6.3E10-6 | GPE | PE | Yes |
| 1-oleoyl-2-arachidonoyl-GPE (18:1/20:4) | 0.078 | 2.7E-05 | 3.8E10-5 | GPE | PE | Yes |
| 1-stearoyl-GPE (18:0) | 0.077 | 3.6E-08 | 3.0E10-8 | GPE | Lyso-PEth | Yes |
| 1-palmitoyl-GPE (16:0) | 0.068 | 1.2E-06 | 2.7E10-6 | GPE | Lyso-PEth | Yes |
| 1-myristoyl-2-arachidonoyl-GPC (14:0/20:4) | 0.092 | 3.0E-09 | 2.9E10-10 | GPC | PC | Yes |
| 1-myristoyl-2-palmitoyl-GPC (14:0/16:0) | 0.065 | 2.9E-05 | 2.1E10-5 | GPC | PC | Yes |
| 1-palmitoyl-2-palmitoleoyl-GPC (16:0/16:1) | 0.064 | 5.1E-06 | 9.8E10-7 | GPC | PC | Yes |
| 1-palmitoleoyl-GPC (16:1) | 0.065 | 3.5E-06 | 1.1E10-6 | GPC | Lyso-PCho | Yes |
| 1-(1-enyl-palmitoyl)-2-palmitoleoyl-GPC (P-16:0/16:1) | -0.070 | 4.8 E-07 | 1.6E10-7 | GPC | PlCho | Yes |
| 1-(1-enyl-palmitoyl)-2-linoleoyl-GPC (P-16:0/18:2) | -0.069 | 9.7 E-07 | 1.6E10-7 | GPC | PlCho | Yes |
| 1-(1-enyl-palmitoyl)-2-oleoyl-GPC (P-16:0/18:1) | -0.063 | 5.9E-06 | 1.1E10-6 | GPC | PlCho | Yes |
| Sphingolipids | ||||||
| lactosyl-N-nervonoyl-sphingosine (d18:1/24:1) | -0.081 | 1.1E-05 | 2.7E10-6 | GSL | Lactosylceramide | Yes |
| lactosyl-N-palmitoyl-sphingosine (d18:1/16:0) | -0.061 | 1.3E-05 | 2.8E10-6 | GSL | Lactosylceramide | Yes |
Abbreviations: DAG, Diacylglycerol; GPC, Glycerophosphocoline; GPE, Glycerophosphoethanolamine; GPI, Glycerophosphoinositol; GSL, Glycerosphingolipid; Lyso-PCho, Lysophosphatidylcholine; Lyso-PEth, Lysophosphatidylethanolamine; MAG, Monoacylglycerol; PC, Phosphatidylcholine; PE, Phosphatidylethanolamine; PI, Phosphatidylinositol; PlCho, Plasmalogen-Choline; TAG, Triacylglycerol.
*TAG was measured using an enzymatic method.
Beta and P-values were obtained from linear regression.
P-value*: P value adjusted for batch effect, age and fasting glucose.
**The numbers (1) and (2) in parentheses refer to different stereoisomers of the metabolites.
Abbreviations: DAG, Diacylglycerol; GPC, Glycerophosphocoline; GPE, Glycerophosphoethanolamine; GPI, Glycerophosphoinositol; GSL, Glycerosphingolipid; Lyso-PCho, Lysophosphatidylcholine; Lyso-PEth, Lysophosphatidylethanolamine; MAG, Monoacylglycerol; PC, Phosphatidylcholine; PE, Phosphatidylethanolamine; PI, Phosphatidylinositol; PlCho, Plasmalogen-Choline; TAG, Triacylglycerol.
*TAG was measured using an enzymatic method. **The numbers (1) and (2) in parentheses refer to different stereoisomers of the metabolites. Only metabolites that were associated significantly (P < 5.8 × 10−5) with GCKR rs780094-T are shown. Beta and P values were obtained from linear regression, and were adjusted for batch effect, P* values were adjusted for batch effect, age and fasting glucose.
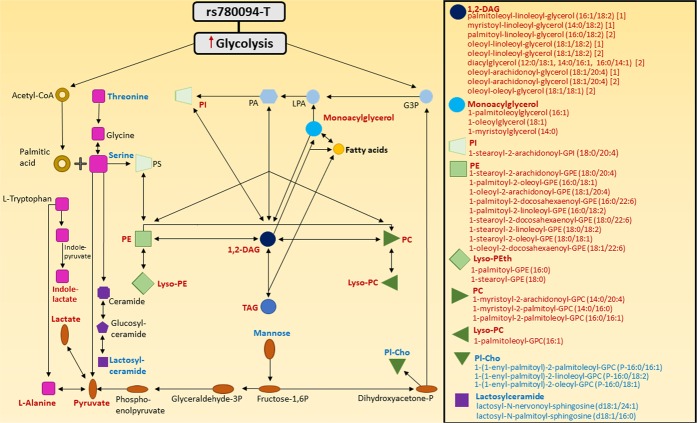
Figure 1.

Increased levels of metabolites in the lipid pathways in carriers of GCKR rs780094-T are marked by red frames, decreased metabolite levels by blue frames, and other metabolites by brown frames. Increased glycolysis stimulates the formation of glycerol-3-phosphate (G3P) and acetyl coenzyme A (acetyl-CoA). G3P conjugates with fatty acyl coenzyme A (FA-CoA) to generate LPA (lysophosphatidic acid) and phosphatidic acid (PA), which is the precursor for all glycerolipids (GLs) and glycerophospholipids (GPLs). GLs are composed of mono- (MAG), di- (DAG), and trisubstituted glycerols (TAG) that are hydrolyzed to free fatty acids (FFAs). GPLs produce phosphatidic acid (PA) and DAG. PA can also be formed via phosphorylation of DAG, and dephosphorylated to generate 1,2-DAG or bound with choline, ethanolamine or inositol to synthetize phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI). Increased glycolysis and low serine level enhance de novo lipogenesis and the FFA/GL cycling. Acetyl-CoA initiates the de novo lipogenesis pathway by generating saturated fatty acids, such as palmitic acid and monounsaturated fatty acid, oleic acid. The excess of palmitic acid is directed to the FFA/GL cycling by unsaturated fatty acids, such as oleate, and leads to increasing levels of DAG, TAG and GPLs, resulting in fat accumulation in the liver. Low availability of serine leads to an increase in palmitic acid levels, and a decrease in ceramide and lactosylceramide levels. LPC, Lysophosphatidylcholine; LPE, Lysophosphatidylethanolamine.
Glycerophospholipids
We found 19 novel statistically significant associations of GCKR rs780094-T with GPLs after the adjustment for batch effect, age and fasting glucose (16 positive associations, and 3 inverse associations with plasmalogen-cholines).
Sphingolipids
Two lactosylceramides, lactosyl-N-nervonoyl-sphingosine (d18:1/24:1) and lactosyl-N-palmitoyl-sphingosine (d18:1/16:0) were significantly and negatively associated with GCKR rs780094-T.
Palmitic acid
GCKR rs780094-T was nominally associated with palmitic acid (P = 0.033).
We performed further adjustments for all metabolites significantly associated with GCKR rs780094-T (Supplementary Tables S4–S5). First, we adjusted P values for bath effect, age, and total triglycerides (Supplementary Tables S4–S5, P*), and finally for bath effect, age, total triglycerides, fasting glucose and for all other metabolites (N = 46) showing significant associations with GCKR rs780094-T of (Supplementary Tables S4–S5, P**). Since GCKR rs780094-T was significantly associated with total triglycerides and other lipids it was not surprising that the P values were substantially less significant or non-significant after adding total triglycerides as an independent variable in the model. Similarly, P values further weakened when the adjustment was done for all metabolites significantly associated with GCKR rs780094 (Supplementary Tables S4–S5).
Correlations of lipids with insulin sensitivity, body mass index and ALT
We applied linear regression analysis (adjusted for the bath effect and age) to investigate if the metabolites significantly associated with GCKR rs780094-T (Tables 1–2) are also significantly associated with insulin sensitivity, BMI and ALT (Supplementary Tables S6–S8). Amino acids or their downstream metabolites (except for serine and 3-aminoisobutyrate), carbohydrates and other metabolites mentioned in Table 1 (except for 3-hydroxubuturate) were associated with an increase in insulin resistance (a decrease in Matsuda ISI), and a majority of these metabolites were also associated with increases in BMI and ALT (Supplementary Tables S6 and S7). All GLs and GPLs, which were positively associated with GCKR rs780094-T (Table 2), were associated with an increase in insulin resistance, BMI and ALT, except for 1-stearoyl-GPE (18:0) and 1-palmitoyl-GPE (16:0) which are lysophoshatidylethanolamines. Plasmalogen-cholines 1-(1-enyl-palmitoyl)-2-palmitoleoyl-GPC (P-16:0/16:1), 1-(1-enyl-pamitoyl)-2-linoleoyl-GPC (P-16:0/18:2), 1-(1-enyl-pamitoyl)-2-oleoyl-GPC (P-16:0/18:1)], and sphingolipids lactosyl-N-nervonoyl-spingosine (d18:1/24:1), lactosyl-N-palmitoyl-spingosine (d18:1/16:0) increased insulin sensitivity and decreased BMI and ALT. We performed linear regression analysis also separately for GCKR rs780094-T and the GCKR rs780094-CC genotype (Supplementary Tables S8 and S9). Associations remained quite similar in these groups.
Discussion
We and other investigators have previously shown that GCKR rs780094 is associated with several metabolites and diseases2–7. In the present study, we performed an extensive metabolomics analysis to obtain a comprehensive view of the associations of GCKR rs780094-T with several metabolites belonging to different metabolic pathways. We found novel associations of GCKR rs780094-T with amino acids and their downstream metabolites (threonine, indolelactate, N-acetyltryptophan), and especially with lipids.
Previous studies have reported associations of GCKR rs780094-T with several amino acids7. We additionally found novel negative associations of GCKR rs780094-T with serine and threonine, and positive associations with the metabolites indolelactate and N-acetyltryptophan in the tryptophan pathway. Indole, the most abundant metabolite of tryptophan, is produced by bacteria species and modulates the secretion of glucagon-like peptidi-1, and thus insulin secretion13. N-acetyltryptophan is also an indicator of the gut microbial metabolism14. GCKR rs780094-T was inversely associated with 3-aminoisobutyrate. This metabolite is generated by the catabolism of valine, and contributes to exercise-induced protection from metabolic diseases in humans15.
We found multiple new positive associations of GCKR rs780094-T with GLs. Most of these associations were with 1,2-DAG which can be acetylated to form TAG or redirected to generate phospholipids10. TAGs can be hydrolyzed to generate DAGs and FFAs, and DAGs to monoacylglyceride (MAG) and FFAs. The cells can store TAG in lipid droplets which prevents lipotoxicity16,17.
We also found several new positive associations of GCKR rs780094-T with GPLs which included PEs (N = 9), PCs (N = 3), lysophospatidylethanolamine (N = 2), PI (N = 1), and lysophophatidylcholine (N = 1). Given the fact that the degradation of these GPLs results in FFAs, this results in increased de novo lipid synthesis and fat accumulation in the liver. We found negative associations of GCKR rs780094-T with three plasmalogen-cholines and two lactosylceramides. Plasmalogens have preventive effects against fat accumulation in the liver since they activate PPARα which increases FA oxidation18. Impaired FA oxidation leads to increased FFA levels and synthesis of TAGs in the liver. The negative association of GCKR rs780094-T with lactosylceramides (d18:1/24:1) and (d18:1/16:0) suggests that the ceramide pathway may contribute indirectly to fat accumulation in the liver, since downregulation of the ceramide pathway results in an increase in the GL/FFA cycling.
Glycolysis stimulates the formation of acetyl-CoA and synthesis of saturated FAs (Fig. 1), and consequently hepatic TAG accumulation either via de novo lipid synthesis or by interrupting β-oxidation19. Palmitic acid, a saturated fatty acid, is the first product of de novo lipid synthesis, promoting inflammation and endoplasmic reticulum stress20. GCKR rs780094-T has been previously shown to be associated with high levels of palmitic acid21. We also found a similar nominally significant positive association of GCKR rs780094-T with palmitic acid.
Ceramides are synthesized from the condensation of serine with palmitic acid and are the precursors for the synthesis of lactosylceramides (Fig. 1). The role of lactosylceramides in the development of insulin resistance has not been well characterized. Lematre et al. measured 15 ceramide and sphingomyelin species in fasting baseline samples from a prospective cohort of 2086 American Indians22. Their study showed that high levels of lactosylceramide 16 associated with lower fasting insulin, HOMA-IR, and HOMA-B compared to other ceramide species in cross-sectional analyses22. This could be explained by the fact that lactosylceramides are ganglioside precursors23 which enhance insulin sensitivity24. Another study performed in a rodent model showed that lactosylceramide (d18:1/24:1) had a negative correlation with HOMA-IR, although this association was not statistically significant25. We found a positive correlation of the lactosylceramides d18:1/24:1 and d18:1/16:0 with insulin sensitivity, and therefore our results agree with previous studies22,25 suggesting that lactosylceramides may increase insulin sensitivity. We found low levels of serine and lactosylceramides in the carriers of GCKR rs780094-T, which could result in increased level of palmitic acid26 and the glycerolipids/FFA cycling11, and accumulation of TAGs in the liver.
We found that GCKR rs780094-T was inversely associated with gamma-glutamylthreonine which is a dipeptide composed of gamma-glutamate and threonine. A recent study reported that dietary sucrose was positively associated with gamma-glutamylthreonine27. We also found a significant positive association of GCKR rs780094-T with retinol (vitamin A). Retinol stimulates the secretion of RBP4 protein by hepatocytes28, and stimulates de novo lipogenesis29, thus promoting fat accumulation into the liver.
Figure 2 summarizes the associations of GCKR rs780094-T with the metabolites belonging to amino acid, carbohydrate and lipid pathways in our study. GCKR rs780094-T was associated with the metabolites from all these pathways. Serine is a key metabolite connecting amino acid, lipid and carbohydrate pathway. In our study GCKR rs780094-T was inversely associated with serine but positively associated with multiple GLs and GPLs in agreement with the stimulation of de novo lipogenesis as previously reported26. Serine has also effects on carbohydrate metabolism because it can be deaminated to pyruvate and further to lactate. GCKR rs780094-T was inversely associated also with threonine which is expected because this amino acid can be converted to serine.
Figure 2.
Summary of the associations of rs780094-T of GCKR with metabolites in lipid, carbohydrate and amino acid pathways. Increased metabolite levels are marked by red color, decreased metabolite levels by blue color, and other metabolites by black color. A variant rs780094-T of GCKR is associated with increased glycolysis and glycogen synthesis, and de novo lipid synthesis. Serine is a key metabolite connecting amino acid, lipid and carbohydrate pathways. Low availability of serine and increased palmitic acid increase lipid levels. Serine can be deaminated to pyruvate and further to lactate or alanine. Mannose generates fructose-1,6P which can through multiple reactions generate pyruvate. Dihydroxyacetone-P is the precursor for Pl-Cho and G3P, showing that mannose is a connector between carbohydrate and lipid pathways. Tryptophan can be converted via several steps to alanine and also to indole-lactate. Threonine can be metabolized to generate glycine and serine. Acetyl-CoA, Acetyl coenzyme A; DAG, Diacylglycerol; G3P, Glycerol-3-phosphate; GPC, Glycerophosphocholine; GPE, Glycerophosphoethanolamine; GPI, Glycerophosphoinositol; LPA, Lysophosphatidic acid; Lyso-PC, Lysophosphatidylcholine; Lyso-PE, Lysophosphatidylethanolamine; PA, Phosphatidic acid; PC, Phosphatidylcholine; PE, Phosphatidylethanolamine; PI, Phosphatidylinositol; Pl-Cho, Choline plasmalogen; PS, Phosphatidylserine; TAG, Triacylglycerol.
Our study may have clinical significance for NAFLD/NASH. Variants of the genes that encode proteins in the lipogenesis pathway have been associated with NAFLD and NASH, including a missense variant rs738409 in the patatin like phospholipase 3 (PNPLA3)30 and rs58542926 in the transmembrane 6 superfamily member 2 (TM6SF2) genes31. GCKR rs780094, which is in linkage disequilibrium with rs126036, is also involved in the NAFLD/NASH pathogenesis, since it activates de novo lipogenesis, TAG and cholesterol synthesis32. Increased de novo lipid synthesis together with impaired β-oxidation leads to fat accumulation in the liver. We found multiple positive associations of GCKR rs780094-T with GLs and GLPs, and these lipids were also associated with insulin resistance, obesity and elevation of ALT concentrations. Several of them, e.g. palmitoyl-linoleoyl-glycerol (16:0/18:2), oleoyl-linoleoyl-glycerol (18:1/18:2), oleoyl-oleoyl-glycerol (18:1/18:1), 1-palmitoyl-2-docosahexaenoyl-GPE (16:0/22:6), and 1-stearoyl-2-docosahexaenoyl-GPE (18:0/22:6) have been previously associated in plasma and liver biopsies with NAFLD or NASH33. In another study total hepatic lipid content was markedly increased in NAFLD and NASH (P < 0.001), driven mainly by increased TAG and DAG34. Interestingly, a recent report demonstrated that a rare loss-of-function variant Arg227Ter of GCKR was associated with a rapidly progressive form of nonalcoholic steatohepatitis giving further evidence on the role of GCKR in liver diseases35.
The strength of our study is a large size of our population-based study and detailed metabolite analyses. The limitations of our study are that our study was cross-sectional, and only middle-aged and elderly Finnish men were included in the study. Therefore, we do not know if the results are valid for women, all age groups and other ethnic and racial groups. In conclusion, our study adds novel information about the pleiotropy of the GCKR gene, and suggests a role of this gene in the regulation of lipids. These results may add to our understanding how GCKR rs780094-T is associated with the risk of liver diseases, especially NAFLD.
Materials and Methods
Subjects
The METSIM study comprises 10,197 Finnish men randomly selected from the population register of Kuopio, Eastern Finland, aged from 45 to 73 years, and examined in 2005–2010. The study design has been described previously12,36. Our study includes 5,181 participants of the METSIM study without diabetes at baseline (age 57 ± 7 years, body mass index 26.5 ± 3.5 kg/m2, mean ± standard deviation). Their fasting plasma glucose was 5.6 ± 0.4 mmol/l and 2-hour glucose 5.9 ± 1.6 mmol/l. This subset of the METSIM study had similar clinical and laboratory characteristics as the entire METSIM population, and therefore it is a representative of the entire METSIM cohort. At baseline, 40% men had normal glucose tolerance in an oral glucose tolerance test (OGTT) (N = 2,092), 46.5% had isolated impaired fasting glucose (IIFG, N = 2,430), 4.1% had isolated impaired glucose tolerance (IIGT, N = 213), and 8.5% had IFG + IGT (N = 446), according to American Diabetes Association criteria37. The study was approved by the Ethics Committee of the University of Kuopio and Kuopio University Hospital. All study participants gave written informed consent.
Methods
All laboratory methods, including metabolomics analysis, were performed in accordance with the relevant guidelines and regulations.
Clinical and laboratory measurements
Height was measured without shoes to the nearest 0.5 cm. Weight was measured in light clothing with a calibrated digital scale (Seca 877, Hamburg, Germany). Body mass index (BMI) was calculated as weight (kg) divided by height (m) squared. Laboratory studies after 12 h fasting included the following measurements, plasma glucose and insulin, lipids, lipoproteins, and mass spectrometry metabolomics (Metabolon, Durham, NC). An OGTT was performed to evaluate glucose tolerance (75 g of glucose). Clinical and laboratory measurement methods have been previously published36. Briefly, plasma glucose was measured by enzymatic hexokinase photometric assay (Konelab Systems Reagents, Thermo Fischer Scientific, Vantaa, Finland). Insulin was determined by immunoassay (ADVIA Centaur Insulin IRI, no 02230141, Siemens Medical Solutions Diagnostics, Tarrytown, NY, USA). Serum alanine aminotransferase (ALT) was measured by an enzymatic photometric test (Konelab Reagent System, Thermo Fisher Scientific, Vantaa, Finland).
Metabolomics analysis
Metabolites were measured by using Metabolon Inc.’s untargeted Discovery HD4 platform based on Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectroscopy (UPLC-MS/MS) (Metabolon, Morrisville, NC, USA). Samples stored at −80 C prior analysis were prepared using the automated MicroLab STAR® system from Hamilton Company. Several recovery standards were added prior to the first step in the extraction process for quality control (QC) purposes. A pooled matrix sample generated by taking a small volume of each experimental sample served as a technical replicate throughout the data set. Extracted water samples served as process blanks, and QT standards that were carefully chosen not to interfere with the measurement of endogenous compounds were spiked into every analyzed sample, allowed instrument performance monitoring and aided chromatographic alignment. Overall process variability was determined by calculating the median relative standard deviation for all endogenous metabolites present in 100% of the pooled matrix samples. Data normalization step was performed to correct variation resulting from instrument inter-day tuning differences in studies spanning multiple days. Experimental samples were randomized across the platform run with QC samples spaced evenly. Raw data was extracted, peak-identified and QC processed using Metabolon’s hardware and software, and peaks quantified using area-under-the-curve. Compounds were identified by comparison to library entries of purified standards or recurrent unknown entities. Library matches for each compound were checked for each sample and corrected if necessary. Each metabolite was rescaled to set the median equal to 1. After standardization of the metabolite missing values were replaced with the minimum value for each metabolite. The average percent of imputed metabolites among all metabolites was 5%.
The determination of metabolites was performed in three batches. Batch one included 999 samples with 717 metabolites identified, batch two 1,231 samples with 778 metabolites, and batch three 3,000 samples with 843 metabolites identified. All metabolites having >50% missing values were omitted in statistical analyses. A total of 857 unique metabolites were included in current statistical analysis. The sub-classification of the lipids was based on the Human Metabolome Database (http://www.hmdb.ca).
Calculations
The Matsuda insulin sensitivity index (ISI) was calculated as previously described36,38. The selection of Matsuda ISI as a marker of insulin sensitivity was based on our previous validation study36.
Genotyping
We genotyped rs780094 using specific TaqMan assays (ThermoFisher) in a 7500 Real-Time PCR System (Applied Biosystems) as previously described4. GCKR variants rs780094 and rs1260326 are in high linkage disequilibrium (0.91 in the METSIM study), and therefore the results are shown only for rs780094. The frequency of the minor T allele of rs780094 of the GCKR gene (GCKR rs780094-T) was 38.4% in our study (N = 5,181).
Statistical analysis
All statistical analyses were performed using IBM SPSS Statistics 25. We performed association analyses between GCKR rs780094-T and metabolites using linear regression analysis adjusted for batch effect, and confounding risk factors. We give the results as standardized beta coefficients and P values with the metabolite as a dependent variable. We used Pearson correlation analysis to analyze the inter-correlations of metabolites of interest, and principal component analyses with Varimax rotation to evaluate the number of components formed from metabolites included in statistical analyses. All variables were log-transformed to correct for their skewed distribution. Pearson correlation plot was generated using ggplot2 package based on R. P < 5.8 × 10−5 was considered as statistically significant given 857 metabolites measured, and P < 0.05 as nominally significant.
Supplementary information
Acknowledgements
The research leading to these results has received support from the Innovative Medicines Initiative Joint Undertaking under EMIF grant agreement no. 115372 (M.L.). The METSIM study was supported by grants from Academy of Finland (321428), Sigrid Juselius Foundation, Finnish Foundation for Cardiovascular Research, Kuopio University Hospital, and Centre of Excellence of Cardiovascular and Metabolic Diseases supported by the Academy of Finland (M.L.).
Author Contributions
L.F.S. conceived the study, performed statistical analyses, wrote and revised the manuscript. J.V. performed statistical analyses, and revised the manuscript. T.K. researched metabolomics data and revised the manuscript. M.L. conceived the study, wrote, reviewed the manuscript and supervised the entire study and is the guarantor of the study.
Data Availability
All datasets generated during the current study can be found within the manuscript or the Supplementary Information. Other datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-46750-3.
References
- 1.Flannick J, Florez JC. Type 2 diabetes genetic data sharing to advance complex disease research. Nature Reviews Genetics. 2016;17:535–549. doi: 10.1038/nrg.2016.56. [DOI] [PubMed] [Google Scholar]
- 2.Stančáková A, et al. Effects of 34 risk loci for type 2 diabetes or hyperglycemia on lipoprotein subclasses and their composition in 6,580 nondiabetic Finnish men. Diabetes. 2011;60:1608–1616. doi: 10.2337/db10-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suhre K, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stancáková A, et al. Hyperglycemia and a common variant of GCKR are associated with the levels of eight amino acids in 9,369 Finnish men. Diabetes. 2012;61:1895–902. doi: 10.2337/db11-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahendran Y, et al. Association of ketone body levels with hyperglycemia and type 2 diabetes in 9,398 Finnish men. Diabetes. 2013;62:3618–3626. doi: 10.2337/db12-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan HL, et al. Association of glucokinase regulatory gene polymorphisms with risk and severity of non-alcoholic fatty liver disease: an interaction study with adiponutrin gene. J Gastroenterol. 2014;49:1056–1064. doi: 10.1007/s00535-013-0850-x. [DOI] [PubMed] [Google Scholar]
- 7.Brouwers MCGJ, Chantal J, Bast A, Stehouwer CDA, Schape NC. Modulation of glucokinase regulatory protein: a double-edged sword? Trends Mol Med. 2015;21:583–594. doi: 10.1016/j.molmed.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Zelent B, et al. Analysis of the co-operative interaction between the allosterically regulated proteins GK and GKRP using tryptophan fluorescence. Biochem. J. 2014;459:551–564. doi: 10.1042/BJ20131363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agius L, Peak M, Newgard CB, Gomez-Foix AM, Guinovart JJ. Evidence for a role of glucose-induced translocation of glucokinase in the control of hepatic glycogen synthesis. J Biol Chem. 1996;271:30479–30486. doi: 10.1074/jbc.271.48.30479. [DOI] [PubMed] [Google Scholar]
- 10.Prentki M, Madiraju SR. Glycerolipid metabolism and signaling in health and disease. Endocrine Rev. 2008;29:647–676. doi: 10.1210/er.2008-0007. [DOI] [PubMed] [Google Scholar]
- 11.Hermansson M, Hokynar K, Somerharju P. Review: Mechanisms of glycerophospholipid homeostasis in mammalian cells. Prog Lipid Res. 2011;50:240–257. doi: 10.1016/j.plipres.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Laakso M, et al. The Metabolic Syndrome in Men study: a resource for studies of metabolic and cardiovascular diseases. J Lipid Res. 2017;58:481–493. doi: 10.1194/jlr.O072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chimerel C, et al. Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep. 2014;9:1202–1208. doi: 10.1016/j.celrep.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavlova T, et al. Urinary intermediates of tryptophan as indicators of the gut microbial metabolism. Anal Chim Acta. 2017;22(987):72–80. doi: 10.1016/j.aca.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 15.Roberts LD, et al. β-aminoisobutyric acid induces browning of white fat and hepatic β-oxidation and Is inversely correlated with cardiometabolic risk factors. Cell Metab. 2014;19:96–108. doi: 10.1016/j.cmet.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Listenberger LL, et al. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci USA. 2003;100:3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dugail I, Hajduch EA. new look at adipocyte lipid droplets: towards a role in the sensing of triacylglycerol stores? Cell Mol Life Sci. 2007;64:2452–2458. doi: 10.1007/s00018-007-7277-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang JE, et al. Protective role of endogenous plasmalogens against hepatic steatosis and steatohepatitis. Hepatology. 2017;66:416–431. doi: 10.1002/hep.29039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rui L. Energy metabolism in the liver. Compr Physiol. 2014;4:177–97. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei Y, Wang D, Topczewski F, Pagliassotti MJ. Saturated fatty acids induce endoplasmic reticulum stress and apoptosis independently of ceramide in liver cells. Am J Physiol Endocrinol Metab. 2006;291:275–281. doi: 10.1152/ajpendo.00644.2005. [DOI] [PubMed] [Google Scholar]
- 21.Wu JHY, et al. Genome-wide association study identifies novel loci associated with concentrations of four plasma phospholipid fatty acids in the de novo lipogenesis pathway: results from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. Circ Cardiovasc Genet. 2013;6:171–183. doi: 10.1161/CIRCGENETICS.112.964619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemaitre RN, et al. Circulating sphingolipids, insulin, HOMA-IR, and HOMA-B: The Strong Heart Family Study. Diabetes. 2018;67:1663–1672. doi: 10.2337/db17-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu RK, Tsai YT, Ariga T, Yanagisawa M. Structures, biosynthesis, and functions of gangliosides: an overview. J Oleo Sci. 2011;60:537–44. doi: 10.5650/jos.60.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chavez JA, et al. Ceramides and glucosylceramides are independent antagonists of insulin signaling. J Biol Chem. 2014;289:723–734. doi: 10.1074/jbc.M113.522847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wigger L, et al. Plasma dihydroceramides are diabetes susceptibility biomarker candidates in mice and humans. Cell Rep. 2017;18:2269–2279. doi: 10.1016/j.celrep.2017.02.019. [DOI] [PubMed] [Google Scholar]
- 26.Gao X, et al. Serine availability influences mitochondrial dynamics and function through lipid metabolism. Cell Rep. 2018;22:3507–3520. doi: 10.1016/j.celrep.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Y, Yu B, Alexander D, Steffen LM, Boerwinkle E. Human metabolome associates with dietary intake habits among African Americans in the atherosclerosis risk in communities study. Am J Epidemiol. 2014;179:1424–1433. doi: 10.1093/aje/kwu073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ronne H, et al. Ligand-dependent regulation of intracellular protein transport: Effect of vitamin A on the secretion of the retinol-binding protein. J Cell Biol. 1983;96:907–910. doi: 10.1083/jcb.96.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia M, et al. Retinol binding protein 4 stimulates hepatic SREBP-1 and increases lipogenesis through PGC-1beta-dependent pathway. Hepatology. 2013;58:564–575. doi: 10.1002/hep.26227. [DOI] [PubMed] [Google Scholar]
- 30.Santoro N, et al. A common variant in the patatin-like phospholipase 3 gene (PNPLA3) is associated with fatty liver disease in obese children and adolescents. Hepatology. 2010;52:1281–1290. doi: 10.1002/hep.23832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macaluso FS, Maida M, Petta S. Genetic background in nonalcoholic fatty liver disease: A comprehensive review. World J Gastroenterol. 2015;21:11088–11111. doi: 10.3748/wjg.v21.i39.11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rees MG, et al. Cellular characterization of the GCKR P446L variant associated with type 2 diabetes risk. Diabetologia. 2012;55:114–122. doi: 10.1007/s00125-011-2348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorden DL, et al. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J Lipid Res. 2015;56:722–736. doi: 10.1194/jlr.P056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puri P, et al. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 2007;46:1081–1090. doi: 10.1002/hep.21763. [DOI] [PubMed] [Google Scholar]
- 35.Pirola CJ, et al. A rare nonsense mutation in the glucokinase regulator gene is associated with a rapidly progressive clinical form of nonalcoholic steatohepatitis. Hepatol Commun. 2018;2:1030–1036. doi: 10.1002/hep4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stancáková A, et al. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes. 2009;58:1212–1221. doi: 10.2337/db08-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2013;36:67–74. doi: 10.2337/dc13-S067. [DOI] [Google Scholar]
- 38.Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: Comparison with the euglycemic insulin clamp. Diabetes Care. 1999;22:1462–1470. doi: 10.2337/diacare.22.9.1462. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated during the current study can be found within the manuscript or the Supplementary Information. Other datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.