Abstract
Accurate estimation of carrier probabilities of cancer susceptibility gene mutations is an important part of pre-test genetic counselling. Many predictive models are available but their applicability in the Asian population is uncertain. We evaluated the performance of five BRCA mutation risk predictive models in a Chinese cohort of 647 women, who underwent germline DNA sequencing of a cancer susceptibility gene panel. Using areas under the curve (AUCs) on receiver operating characteristics (ROC) curves as performance measures, the models did comparably well as in western cohorts (BOADICEA 0.75, BRCAPRO 0.73, Penn II 0.69, Myriad 0.68). For unaffected women with family history of breast or ovarian cancer (n = 144), BOADICEA, BRCAPRO, and Tyrer-Cuzick models had excellent performance (AUC 0.93, 0.92, and 0.92, respectively). For women with both personal and family history of breast or ovarian cancer (n = 241), all models performed fairly well (BOADICEA 0.79, BRCAPRO 0.79, Penn II 0.75, Myriad 0.70). For women with personal history of breast or ovarian cancer but no family history (n = 262), most models did poorly. Between the two well-performed models, BOADICEA underestimated mutation risks while BRCAPRO overestimated mutation risks (expected/observed ratio 0.67 and 2.34, respectively). Among 424 women with personal history of breast cancer and available tumor ER/PR/HER2 data, the predictive models performed better for women with triple negative breast cancer (AUC 0.74 to 0.80) than for women with luminal or HER2 overexpressed breast cancer (AUC 0.63 to 0.69). However, incorporating ER/PR/HER2 status into the BOADICEA model calculation did not improve its predictive accuracy.
Subject terms: Breast cancer, Cancer models, Cancer genetics
Introduction
BRCA mutation carriers face 45% to 85% risk of developing breast cancer, and 10% to 46% risk of developing ovarian cancer by age 701–3. With recent advances in gene sequencing technology, detecting germline mutations in cancer susceptibility genes is becoming more widely accessible to the public. Effective risk reducing surgeries, medications, and/or intense cancer screening strategies are available to BRCA mutation carriers to manage risks of breast and ovarian cancer at an early or even pre-emptive stage. These advances introduce the possibility of substantial benefits from broad genetic screens for individuals at risk for BRCA mutation4.
However, before genetic testing, accurate estimation of the probability of carrying a germline mutation in cancer susceptibility genes is crucial. Genetic testing in a low risk individual could lead to potential harm, dissatisfaction, and misallocation of resources5,6. In fact, many professional societies recommend at-risk individuals to receive genetic counselling before undergoing genetic testing5,7–9, where the probability of carrying a germline mutation must be determined to ascertain the risks and benefits of genetic testing.
A variety of risk predictive models have been developed to determine the probability of carrying a BRCA mutation utilizing personal and family cancer history and mathematical models10–16. A threshold risk rate could be set at the discretion of the clinician or genetic counsellor to determine whether the patient should proceed with genetic testing. These predictive models have been tested and utilized extensively in European and American geographies17–20. However, only a few studies have tested their performance in Asian populations, and those that have done so exhibited mixed results in discerning risk for BRCA mutations compared to their western counterparts. Several studies reported poor model performance in Asian breast cancer cohorts, including evaluation of Manchester Scoring System14 and BOADICEA10 in a Malaysian cohort of 187 patients with BRCA mutation rate of 14.4%21, and evaluation of BRCAPRO11 and Myriad12 in a Korean cohort of 236 patients with BRCA mutation rate of 19.5%22. In contrast, an updated Manchester Scoring System that included adjustment for breast cancer receptor status and high grade serous type ovarian cancer was shown to be equally effective in BRCA mutation prediction in the Singapore cohort as in the Manchester population23. In a cohort of 212 Chinese familial breast cancer patients with BRCA mutation rate of 15.6%, BRCAPRO, Penn II16, and Myriad models showed comparable accuracy to western cohorts24. In a study of Hong Kong Chinese cohort consisted of 310 female and male breast or ovarian cancer patients with BRCA mutation rate of 13.9%25, BOADICEA appeared to be the most accurate in combined BRCA1/2 mutation prediction among the five tested models, while BRCAPRO better predicted mutations of BRCA1 alone. These two models actually performed slightly better in the Chinese cohort than in several western and Asian cohorts previously reported. BRCAPRO and Myriad models were also tested in a Korean ovarian cancer cohort of 232 with 24.6% BRCA mutation prevalence26, in which both models had acceptable performance. The aforementioned studies tested the models only in affected individuals with breast or ovarian cancer, and the cohorts were relatively small. More studies in Asian cohorts are clearly needed to validate and discern how best to use these models.
In the present study, we aimed to evaluate the mutation predictive accuracies of BOADICEA, BRCAPRO, Myriad, Penn II, and Tyrer-Cuzick models in those at risk for hereditary breast and ovarian cancer syndrome in a Chinese cohort in Taiwan. We explored model performances in various subgroups to determine how best to use the models in pre-test genetic counselling.
Results
A total of 647 female participants from 488 families were included in the study. The cohort was divided into three subgroups based on the presence or absence of personal history and family history (FH) of breast cancer (BC) or ovarian cancer (OC). The personal characteristics, cancer characteristics, and mutation frequencies for the entire cohort and for the subgroups are shown in Table 1. The mean age at study enrolment was 50.2, ranging from 16 to 96. Among them, 503 (77.7%) had a personal history of BC or OC, and 385 (59.5%) had a family history of BC or OC. The subgroup with personal history but no family history of BC or OC (BC/OC(+)FH(−)) were younger (mean age 47.8) and included more early onset cancer and triple-negative breast cancer. In the entire cohort, 48 individuals were found to be carrying a BRCA mutation (12 BRCA1, 36 BRCA2), making the carrier rate of 7.4%. The subgroup with both personal and family history of BC or OC (BC/OC(+)FH(+)) had the highest BRCA mutation carrier rate of 10.4%.
Table 1.
Participant characteristics.
| All Participants N = 647 |
BC/OC(+) FH(+) n = 241 |
BC/OC(+) FH(−) n = 262 |
BC/OC(−) FH(+) n = 144 |
|
|---|---|---|---|---|
| Age, mean (range) | 50.2 (16–96) | 53.0 (28–82) | 47.8 (18–89) | 49.8 (16–96) |
| Personal history of BC/OC | 503 (77.7%) | 241 (100%) | 262 (100%) | 0 (0%) |
| Family history of BC/OC | 385 (59.5%) | 241 (100%) | 0 (0%) | 144 (100%) |
| Mutations detected: | ||||
| BRCA1 mutation | 12 (1.9%) | 8 (3.3%) | 2 (0.8%) | 2 (1.4%) |
| BRCA2 mutation | 36 (5.6%) | 17 (7.1%) | 15 (5.7%) | 4 (2.8%) |
| Other HR gene mutation | 31 (4.8%) | 14 (5.8%) | 10 (3.8%) | 7 (4.9%) |
| In those with BC/OC: | n = 503 | n = 241 | n = 262 | n = 0 |
| Age of onset ≤40 for BC/OC | 265 (52.7%) | 90 (37.3%) | 175 (67.8%) | N/A |
| Triple-negative breast cancer | 136 (27.0%) | 50 (20.7%) | 86 (32.8%) | N/A |
| Bilateral breast cancer | 54 (10.7%) | 33 (13.7%) | 21 (8.0%) | N/A |
| Ovarian cancer | 10 (2.0%) | 5 (2.1%) | 5 (1.9%) | N/A |
All values are listed as number (% of the subgroup in each column), unless otherwise specified.
BC: breast cancer; OC: ovarian cancer; BC/OC(+): personal history of breast/ovarian cancer; FH(+): family history of breast/ovarian cancer; HR gene: homologous recombination pathway genes tested in this study other than BRCA: ATM, BARD1, BRIP1, PALB2, RAD50, RAD51C and RAD51D (detailed numbers are provided in the Supplementary Table 1S); N/A: not applicable; FH: family history, defined as two or more first-, second- or third-degree relatives on the same lineage of the family with BC or OC.
Model performance by genes
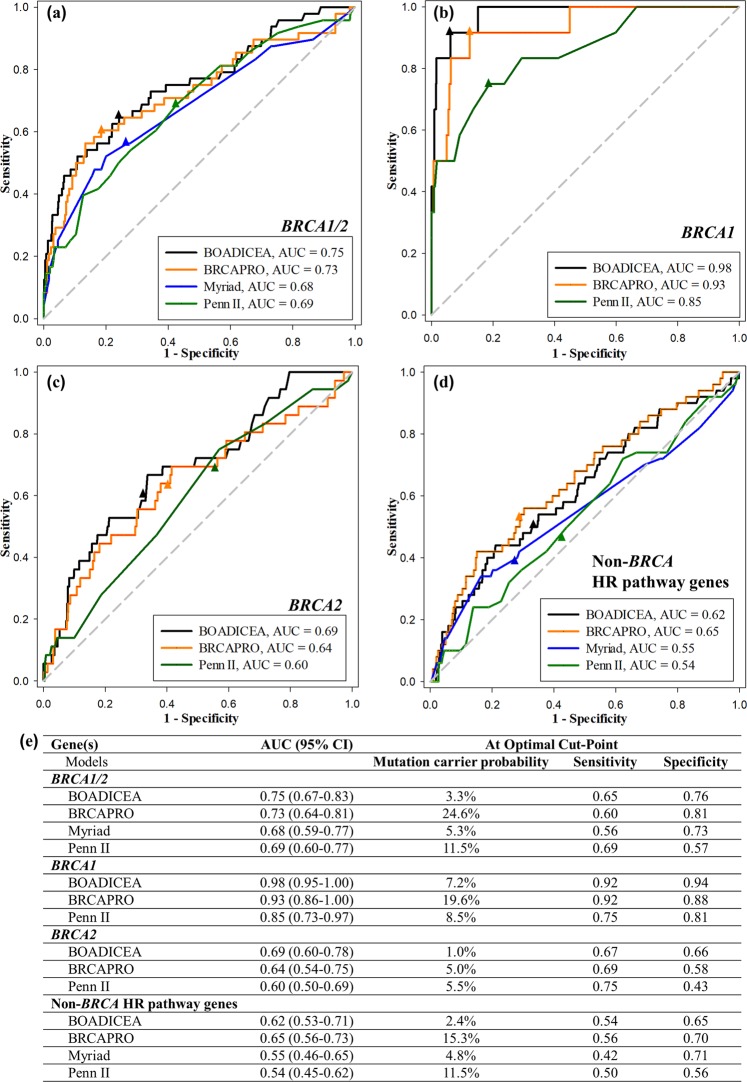
Figure 1 shows the performance of four mutation predictive models on ROC curves. The AUCs for having either a BRCA1 or a BRCA2 mutation were: 0.75 (95% CI, 0.67–0.83) for BOADICEA, 0.73 (95% CI, 0.64–0.81) for BRCAPRO, 0.68 (95% CI, 0.59–0.77) for Myriad, and 0.69 (95% CI, 0.60–0.77) for Penn II (Fig. 1a). At the optimal cut-points, defined by the closest points to the left upper corner, the values of the mutation carrier probabilities varied widely, with BOADICEA having the lowest cut-off value of 3.3%, BRCAPRO having the highest cut-off value of 24.6%, and Myriad and Penn II having the middle values of 5.3% and 11.5%, respectively. The sensitivities at the optimal cut-points were between 0.56 and 0.69, while the specificities were between 0.57 and 0.81 (Fig. 1e).
Figure 1.
Performance of four BRCA mutation risk predictive models using ROC curves; the respective AUC for each model is shown at the right lower corner of the curves and in panel (e); the optimal cut-points (closest point to the left upper corner) are shown as triangles on each curve. (a) BRCA1/2: probability of BRCA1 or BRCA2 mutation prediction using the BOADICEA, BRCAPRO, Myriad, and Penn II models; (b) BRCA1 only: probability of BRCA1 mutation prediction using the BOADICEA, BRCAPRO, and Penn II models; (c) BRCA2 only: probability of BRCA2 mutation prediction using the BOADICEA, BRCAPRO, and Penn II models; (d) Non-BRCA HR pathway genes: probability of HR pathway gene other than BRCA1/2 (ATM, BARD1, BRIP1, PALB2, RAD50, RAD51C, RAD51D) mutation prediction using the BOADICEA, BRCAPRO, Myriad, and Penn II models; (e) AUC with 95% confidence interval (CI) for each model and each gene set, as well as mutation carrier probability, sensitivity, and specificity at the optimal cut-point for each curve are listed.
Three of the models also predict the BRCA1 and BRCA2 mutation carrier probabilities separately. The models performed very well for BRCA1 and worse for BRCA2. Figure 1b shows the AUCs for BRCA1-only predictions: 0.98 (95% CI, 0.95–1.00) for BOADICEA, 0.93 (95% CI, 0.86–1.00) for BRCAPRO, and 0.85 (95% CI, 0.73–0.97) for Penn II. The optimal cut-point values for BRCA1 mutation carrier probability were: 7.2% for BOADICEA, 19.6% for BRCAPRO, and 8.5% for Penn II. Figure 1c shows the ROC curves for BRCA2-only prediction, and the AUCs were: 0.69 (95% CI, 0.60–0.78) for BOADICEA, 0.64 (95% CI, 0.54–0.75) for BRCAPRO, and 0.60 (95% CI, 0.50–0.69) for Penn II. The optimal cut-point values for BRCA2 mutation carrier probability were: 1.0% for BOADICEA, 5.0% for BRCAPRO, and 5.5% for Penn II. Table S2 shows the numbers and proportions of BRCA mutation carriers in each predicted range category of the mutation carrier probability. BOADICEA gave good correlation between actual mutation rates and predicted mutation probability ranges. For the other models, the mutation rates had an upward trend with increasing ranges of predicted probabilities, but the actual rate values did not always fit the predicted probability ranges.
When the models were used to predict non-BRCA homologous recombination (HR) pathway gene mutation probabilities, the accuracies were generally poor (Fig. 1d). BRCAPRO was the only model that performed similarly to that of BRCA2, with AUC of 0.65 (95% CI, 0.56–0.73). The other models performed less well than that of BRCA genes, with AUC of 0.62 (95% CI, 0.53–0.71) for BOADICEA, 0.55 (95% CI, 0.46–0.65) for Myriad, and 0.54 (95% CI, 0.45–0.62) for Penn II.
Model performance by clinical subgroups
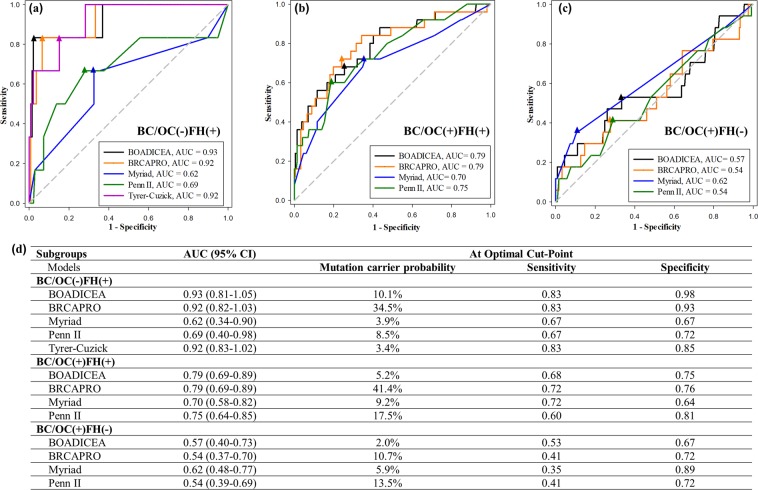
To find the population group where the models are the most applicable, we did subgroup analyses based on personal and familial BC/OC status. In the subgroup that had no personal history of BC/OC but had family history of BC/OC (designated BC/OC(−)FH(+) in Fig. 2a), three models had superb performance: AUC 0.93 (95% CI, 0.81–1.05) for BOADICEA, 0.92 (95% CI, 0.82–1.03) for BRCAPRO, and 0.92 (95% CI, 0.83–1.02) for the Tyrer-Cuzick model. In the BC/OC(+)FH(+) subgroup, the models performed fairly well with AUCs between 0.70 and 0.79 (Fig. 2b). In the BC/OC(+)FH(−) subgroup (Fig. 2c), the models had poor accuracy, except Myriad, which had comparable AUC (0.62) as those with family history (BC/OC(−)FH(+) 0.62 or BC/OC(+)FH(+) 0.70). The optimal cut-off values for mutation carrier probability were much higher for BRCAPRO than for all other models (Fig. 1d).
Figure 2.
Performance of five BRCA mutation risk predictive models in the three subgroups using ROC curves; the respective area under the curve (AUC) for each model is shown at the right lower corner of the curves and in panel (d); the optimal cut-points (closest point to the left upper corner) are shown as triangles on each curve. (a) BC/OC(−)FH(+): in unaffected women with no personal history but with family history of breast or ovarian cancer, the probability of BRCA1 or BRCA2 mutation prediction using the BOADICEA, BRCAPRO, Myriad, Penn II and Tyrer-Cuzick models; (b) BC/OC(+)FH(+): in women with both personal history and family history of breast or ovarian cancer, the probability of BRCA1 or BRCA2 mutation prediction using the BOADICEA, BRCAPRO, Myriad, and Penn II models; (c) BC/OC(+)FH(−): in women with personal history but no family history of breast or ovarian cancer, the probability of BRCA1 or BRCA2 mutation prediction using the BOADICEA, BRCAPRO, Myriad, and Penn II models; (d) AUC with 95% confidence interval (CI) for each subgroup and each model, as well as mutation carrier probability, sensitivity, and specificity at the optimal cut-point for each curve are listed.
The observed and expected number of BRCA mutation carriers for the entire cohort and the subgroups are shown in Table 2. Examining the expected/observed (E/O) ratio, we found that the BOADICEA model gave a fairly accurate estimation of mutation rate (E/O 0.91) in the BC/OC(+)FH(+) subgroup but an underestimation in the other two subgroups. The Myriad prevalence table gave a good estimation of mutation rates in all subgroups (E/O 0.79 to 0.86), probably because the Myriad table was based on prevalence rates and was constructed by personal and family history of BC/OC, similar to our subgroup division. The BRCAPRO and Penn II models gave an approximately 2-fold overestimation in all subgroups.
Table 2.
Comparison of observed and expected (model predicted) mutation proportions.
| Subgroups | N | Obs | BOADICEA | BRCAPRO | Myriad | Penn II | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AC | Exp | E/O | AC | Exp | E/O | AC | Exp | E/O | AC | Exp | E/O | |||
| All | 647 | 48 | 5.0% | 32.2 | 0.67 | 17.4% | 112.5 | 2.34 | 6.1% | 39.6 | 0.83 | 12.7% | 82.3 | 1.71 |
| BC/OC(+)FH(+) | 241 | 25 | 9.4% | 22.7 | 0.91 | 29.3% | 70.6 | 2.83 | 8.8% | 21.2 | 0.85 | 16.0% | 38.6 | 1.54 |
| BC/OC(+)FH(−) | 262 | 17 | 2.3% | 6.0 | 0.35 | 9.8% | 25.7 | 1.51 | 5.1% | 13.4 | 0.79 | 12.4% | 32.5 | 1.91 |
| BC/OC(−)FH(+) | 144 | 6 | 2.5% | 3.6 | 0.60 | 11.3% | 16.3 | 2.71 | 3.6% | 5.2 | 0.86 | 7.8% | 11.2 | 1.87 |
Obs: observed number of mutation carriers; AC: predicted average carrier rate; Exp: expected number of mutation carriers = AC × N.
Model performance by breast cancer pathology subtypes
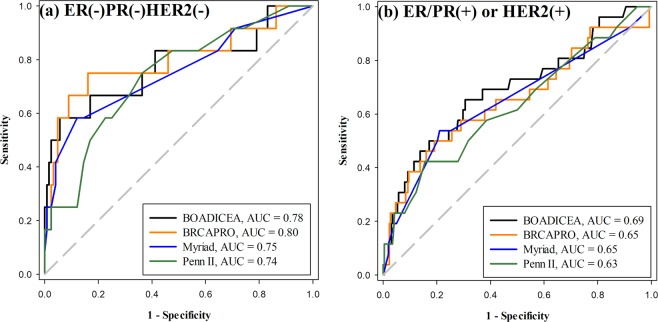
In our cohort, 424 women with personal history of breast cancer had estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) expression status of the breast tumor available. We divided these patients into three pathology subtypes based on the receptor status: triple negative (ER(−)PR(−)HER2(−)), luminal (ER or PR(+)HER2(−)), and HER2 overexpressed (HER2(+)) subtypes (Table 3). Figure 3 shows that the BOADICEA, BRCAPRO, Myriad, and Penn II models had better predictive accuracies in the triple negative breast cancer group (Fig. 3a, AUC 0.78, 0.80, 0.75, and 0.74, respectively) than in the luminal or HER2-overexpressed breast cancer group (Fig. 3b, AUC 0.69, 0.65, 0.65, and 0.63, respectively).
Table 3.
Prediction accuracy of the BOADICEA model in women with breast cancer and available ER/PR/HER2 data, with and without incorporating receptor status.
| Group | No. | BRCA1/2 mutation rate | BOADICEA - AUC (95% CI) | |
|---|---|---|---|---|
| w/o receptor status | w/ receptor status | |||
| All | 424 | 38 (9.0%) | 0.71 (0.61–0.81) | 0.70 (0.59–0.80) |
| By ER/PR/HER2 status | ||||
| ER(−)PR(−)HER2(−) | 136 | 12 (8.8%) | 0.78 (0.60–0.95) | 0.77 (0.58–0.95) |
| ER/PR(+)HER2(−) | 209 | 22 (10.5%) | 0.69 (0.56–0.82) | 0.69 (0.55–0.82) |
| HER2(+) | 79 | 4 (5.1%) | 0.69 (0.36–1.00) | 0.60 (0.23–0.97) |
| By family history | ||||
| BC(+)FH(+) | 192 | 22 (11.5%) | 0.78 (0.66–0.89) | 0.76 (0.64–0.88) |
| BC(+)FH(−) | 232 | 16 (6.9%) | 0.60 (0.44–0.77) | 0.59 (0.43–0.76) |
ER: estrogen receptor; PR: progesterone receptor; HER2: human epidermal growth factor receptor 2; ER/PR(+): ER(1+, 2+, or 3+) or PR(1+, 2+, or 3+); ER(−)PR(−): ER(−) and PR(−); HER2(+): HER2 overexpressed; HER2(−): HER2 not overexpressed. BC: breast cancer; BC(+): personal history of breast cancer; FH(+): family history of breast or ovarian cancer;
Figure 3.
Performance of four BRCA mutation risk predictive models for women with breast cancer of different pathology subtypes (n = 424); the respective area under the ROC curve (AUC) for each model is shown at the right lower corner of the curves. (a) ER(−)PR(−)HER2(−) (triple negative) breast cancer, the probability of BRCA1 or BRCA2 mutation prediction using the BOADICEA, BRCAPRO, Myriad, and Penn II models; (b) ER/PR(+) (luminal type) or HER2(+) (HER2 overexpressed) breast cancer, the probability of BRCA1 or BRCA2 mutation prediction using the BOADICEA, BRCAPRO, Myriad, and Penn II models.
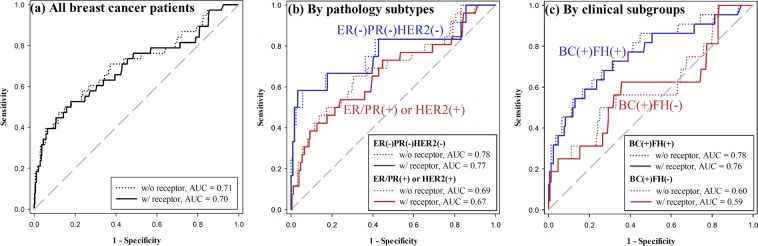
The BOADICEA model allows inclusion of the ER/PR/HER2 data in the calculation for BRCA mutation prediction. We compared the predictive accuracy with and without including the receptor status into the model, as shown in Table 3 and Fig. 4. The BOADICEA model performance did not improve with the additional receptor information for the group as a whole (AUC 0.71 to 0.70, Fig. 4a), or for any of the pathology or clinical subgroups (Fig. 4b,c).
Figure 4.
Performance of the BOADICEA model for women with breast cancer and ER/PR/HER2 status available (n = 424); the respective area under the ROC curve (AUC) for each subgroup is shown at the right lower corner of the curves. (a) For the entire group, the probability of BRCA1 or BRCA2 mutation prediction using BOADICEA without (dotted line) and with (solid line) incorporating the ER/PR/HER2 receptor status; (b) Comparison of pathology subtypes: triple negative ER(−)PR(−)HER2(−) (blue lines) and luminal ER/PR(+) or HER2(+) (red lines), the probability of BRCA1 or BRCA2 mutation prediction using BOADICEA without (dotted lines) and with (solid lines) incorporating the ER/PR/HER2 receptor status; (c) Comparison of clinical subgroups: BC(+)FH(+) (blue lines) and BC(+)FH(−) (red lines), the probability of BRCA1 or BRCA2 mutation prediction using BOADICEA without (dotted lines) and with (solid lines) incorporating the ER/PR/HER2 receptor status.
Setting the positive test threshold
The performance measures using sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) for each predictive model are shown in Table 4, with the positive test threshold for carrying a BRCA1 or BRCA2 mutation set at 10% or 20% carrier probability. For the entire study cohort, at the same threshold level, BRCAPRO had a relatively high sensitivity while BOADICEA had the highest specificity and PPV. Moving the threshold from 10% to 20%, the performance of BOADICEA and BRCAPRO were affected only slightly, while the performance measures of Penn II were significantly affected with a large drop in sensitivity (0.82 to 0.25) and a large rise in specificity (0.39 to 0.91). The positive predictive values (PPVs) were generally low due to the low mutation prevalence in the study cohort, although using the 20% threshold, BOADICEA could give a PPV of 0.46. The negative predictive values (NPVs) of all four models were high (range 0.94–0.96). In the BC/OC(-)FH(+) subgroup at a positive test threshold of 10%, Tyrer-Cuzick, BOADICEA, and BRCAPRO models gave good sensitivities (0.67, 0.83, and 0.83, respectively) and specificities (0.96, 0.98, and 0.70, respectively). Moving the threshold to 20% markedly lowered the sensitivity to 0.33 for Tyrer-Cuzick and BOADICEA without much gain in specificity. BRCAPRO’s performance did not differ much between the 10% and 20% thresholds.
Table 4.
Performance measures for each model at the 10% or 20% threshold for BRCA1/2 mutation carrier probability.
| Carrier number/rate | Test parameters at set threshold | |||||
|---|---|---|---|---|---|---|
| Entire cohort | ||||||
| 10% threshold | <10% | ≥10% | Sensitivity | Specificity | PPV | NPV |
| BOADICEA | 26/583 | 22/64 | 0.46 | 0.93 | 0.34 | 0.96 |
| BRCAPRO | 14/370 | 34/277 | 0.71 | 0.59 | 0.12 | 0.96 |
| Myriad | 25/526 | 23/121 | 0.48 | 0.84 | 0.19 | 0.95 |
| Penn II | 9/239 | 41/408 | 0.82 | 0.39 | 0.10 | 0.96 |
| 20% threshold | <20% | ≥20% | Sensitivity | Specificity | PPV | NPV |
| BOADICEA | 32/612 | 16/35 | 0.33 | 0.97 | 0.46 | 0.95 |
| BRCAPRO | 19/474 | 29/173 | 0.60 | 0.76 | 0.17 | 0.96 |
| Myriad | 37/612 | 11/35 | 0.23 | 0.96 | 0.31 | 0.94 |
| Penn II | 36/584 | 12/63 | 0.25 | 0.91 | 0.19 | 0.94 |
| BC/OC(−)FH(+) subgroup | ||||||
| 10% threshold | <10% | ≥10% | Sensitivity | Specificity | PPV | NPV |
| Tyrer-Cuzick | 2/134 | 4/10 | 0.67 | 0.96 | 0.40 | 0.99 |
| BOADICEA | 1/136 | 5/8 | 0.83 | 0.98 | 0.62 | 0.99 |
| BRCAPRO | 1/97 | 5/47 | 0.83 | 0.70 | 0.11 | 0.99 |
| Myriad | 6/144 | 0/0 | 0.00 | 1.00 | NaN | 0.96 |
| Penn II | 3/28 | 3/116 | 0.50 | 0.18 | 0.03 | 0.89 |
| 20% threshold | <20% | ≥20% | Sensitivity | Specificity | PPV | NPV |
| Tyrer-Cuzick | 4/141 | 2/3 | 0.33 | 0.99 | 0.67 | 0.97 |
| BOADICEA | 4/141 | 2/3 | 0.33 | 0.99 | 0.67 | 0.97 |
| BRCAPRO | 1/112 | 5/32 | 0.83 | 0.80 | 0.16 | 0.99 |
| Myriad | 6/144 | 0/0 | 0.00 | 1.00 | NaN | 0.96 |
| Penn II | 6/142 | 0/2 | 0.00 | 0.99 | 0.00 | 0.96 |
| BC/OC(+)FH(+) subgroup | ||||||
| 10% threshold | <10% | ≥10% | Sensitivity | Specificity | PPV | NPV |
| BOADICEA | 11/196 | 14/45 | 0.56 | 0.86 | 0.31 | 0.94 |
| BRCAPRO | 3/94 | 22/147 | 0.88 | 0.42 | 0.15 | 0.97 |
| Myriad | 7/145 | 18/96 | 0.72 | 0.64 | 0.19 | 0.95 |
| Penn II | 2/59 | 23/182 | 0.92 | 0.26 | 0.13 | 0.97 |
| 20% threshold | <20% | ≥20% | Sensitivity | Specificity | PPV | NPV |
| BOADICEA | 13/212 | 12/29 | 0.48 | 0.92 | 0.41 | 0.94 |
| BRCAPRO | 4/134 | 21/107 | 0.84 | 0.60 | 0.20 | 0.97 |
| Myriad | 16/208 | 9/33 | 0.36 | 0.89 | 0.27 | 0.92 |
| Penn II | 16/202 | 9/39 | 0.36 | 0.86 | 0.23 | 0.92 |
| BC/OC(+)FH(−) subgroup | ||||||
| 10% threshold | <10% | ≥10% | Sensitivity | Specificity | PPV | NPV |
| BOADICEA | 14/253 | 3/9 | 0.18 | 0.98 | 0.33 | 0.94 |
| BRCAPRO | 10/180 | 7/82 | 0.41 | 0.69 | 0.09 | 0.94 |
| Myriad | 12/237 | 5/25 | 0.29 | 0.92 | 0.20 | 0.95 |
| Penn II | 4/64 | 13/198 | 0.76 | 0.24 | 0.07 | 0.94 |
| 20% threshold | <20% | ≥20% | Sensitivity | Specificity | PPV | NPV |
| BOADICEA | 15/259 | 2/3 | 0.12 | 1.00 | 0.67 | 0.94 |
| BRCAPRO | 14/228 | 3/34 | 0.18 | 0.87 | 0.09 | 0.94 |
| Myriad | 15/260 | 2/2 | 0.12 | 1.00 | 1.00 | 0.94 |
| Penn II | 14/240 | 3/22 | 0.18 | 0.92 | 0.14 | 0.94 |
PPV: positive predictive value; NPV: negative predictive value; NaN: not a number
Discussion
Our study tested five widely-used BRCA mutation risk predictive models in a large Chinese cohort in Taiwan, which included breast or ovarian cancer patients with or without family history of breast or ovarian cancer, as well as unaffected women with family history of breast or ovarian cancer. Participants were enrolled to the study with comprehensive personal and pedigree data collection, and the same experimental and data analytical protocols for genetic testing were used for all participants. Our data thus allowed consistent evaluation of model performance not only in the cohort as a whole, but also in different subgroups of women with or without personal or family history of cancer. Using AUCs in the ROC curves for combined BRCA1/2 predictions, we showed that BOADICEA and BRCAPRO models performed equally well in this Chinese cohort (AUCs 0.75 and 0.73) as previous studies in western cohorts (AUCs ranging from 0.71 to 0.77), while Myriad and Penn II models performed less well (AUCs 0.68 and 0.69) than those in western cohorts (AUCs ranging from 0.71–0.79)16,27–30.
In pre-test genetic counselling, it is important to know which model(s) are best used for which type of patients. We showed that BOADICEA and BRCAPRO were particularly well suited for unaffected women with family history (BC/OC(-)FH(+) subgroup), achieving AUCs of 0.93 and 0.92 respectively. These two models also performed fairly well (AUCs 0.79 and 0.79) for women with both personal and family history of breast or ovarian cancer (BC/OC(+)FH(+) subgroup), but they were close to unhelpful (AUCs 0.57 and 0.54) for those without family history. The Tyrer-Cuzick model can only be applied to unaffected women with family history and it worked very well for this subgroup and achieved AUC of 0.92. The Penn II and Myriad models worked the best for women with both personal and family history (AUC 0.75 and 0.70), and less well for unaffected women with family history (AUC 0.69 and 0.62). For women with breast or ovarian cancer but no family history, Myriad performed relatively well (AUC 0.62) compared to the other models.
Among women with breast cancer and known ER/PR/HER2 receptor status, we found that all models performed better for those with triple negative cancer than for those with luminal or HER2 overexpressed cancer, although none of the models actually used the receptor data in the prediction. In fact, incorporating the receptor status in the BOADICEA model had almost no effect on its predictive accuracy for the whole group or any of the subgroups.
All the models that predict separate BRCA1 and BRCA2 probabilities performed much better for BRCA1 than for BRCA2: AUC of 0.98 vs 0.69 for BOADICEA, 0.93 vs 0.64 for BRCAPRO, and 0.85 vs 0.54 for Penn II. Similar difference was observed in previous studies but not as profound as our results16,21,25,29. This difference could possibly explain the poorer model performance in some Asian cohorts since BRCA2 mutations seemed to be found more often than BRCA1 mutations in Asians while it is the opposite in Whites31–33. Most models performed even poorer for non-BRCA HR pathway genes probably because these genes have lower penetrance in phenotype than BRCA1/2. However, BRCAPRO model gave similar AUC for these HR genes (0.65) to that of BRCA2 (0.64).
A mutation risk threshold of 10% is often used to recommend genetic testing. The threshold could be set lower or higher depending on resources available to the individual or to the healthcare system. The availability of new cancer treatment options targeted for BRCA-mutated tumors, such as PARP inhibitors, could lower the threshold for testing. Moreover, the threshold should also depend on the model used to determine the risk. Our study showed that at the optimal points on the ROC curves, the cut-off carrier probability values were much higher for BRCAPRO (24.6%) than for BOADICEA (3.3%) (Fig. 1), consistent with the results shown in Table 2 that BRCAPRO seemed to overestimate mutation risk (E/O 2.34) while BOADICEA seemed to underestimate risk (E/O 0.67). Thus, setting a universal risk threshold for genetic testing may not be appropriate.
It is clear that most models rely heavily on family history of cancer. The models that performed better overall in the cohort (e.g. BOADICEA, BRCAPRO, Tyrer-Cuzick) utilized detailed pedigree data rather than the categorical yes/no or age cut-off clinical variables in the other models, while the models (e.g. Myriad) that incorporated more personal cancer information performed better for affected women without family history of breast or ovarian cancer. However, family histories are often limited due to many factors, including inaccurate or unavailable information, small families, scarcity of females in a family, premature death due to war or natural causes, migration or separation within a family. In modern societies, extended family history will probably become more and more limited. With more genetic testing results available, new models may be developed using personal history, and clinical and genetic information of nuclear families.
There are several limitations in our study. First, despite the relatively large Chinese cohort, our sample size was still limited. A larger pool of at risk individuals with genetic data available would make the model assessment more accurate, and new models could possibly be developed. Second, our cohort was consisted of a quite uniform Chinese population. The results may not be able to extend to other Asian ethnic groups. Third, the cohort included only women and a very small group of ovarian cancer patients, and therefore the results may not be applicable to men or women with ovarian cancer.
In summary, the five mutation predictive models performed generally well in this Chinese cohort as compared with western cohorts. The predictions were the most accurate for unaffected women with family history of breast or ovarian cancer using the BOADICEA, BRCAPRO and Tyrer-Cuzick models. The predictions were also fairly accurate for women with both personal and family history of breast or ovarian cancer, as well as for women with triple negative breast cancer. For breast or ovarian cancer patients with no family history, the predictions were quite unreliable. Between the two better-performed models, BOADICEA seemed to underestimate mutation risk while BRCAPRO seemed to overestimate mutation risk, thus we recommend setting higher risk threshold for genetic testing when using BRCAPRO (e.g. 20%) and lower risk threshold when using BOADICEA (e.g. 5%).
Methods
Study cohort and data collection
The study was conducted in accordance with the Declaration of Helsinki, and the study protocol was approved by the Institutional Review Board Committee at Koo Foundation Sun Yat-Sen Cancer Center (case No. 20141222A). Written informed consent was obtained from each study participant. Eligible individuals were enrolled between July 2015 and April 2017 at Koo Foundation Sun Yat-Sen Cancer Center (KF-SYSCC) to participate in germline testing of a panel of cancer susceptibility genes. Participants had to fulfil at least one of the following eligibility criteria: family history of breast or ovarian cancer at any age (2 or more individuals on the same lineage of the family), personal history of breast cancer or ovarian cancer with age of diagnosis less than or equal to 40, bilateral breast cancer, triple negative breast cancer, or both breast and ovarian cancer in the same individual. None of the participants had known mutation status in any cancer susceptibility genes prior to enrolment. Through participant surveys, detailed personal and family history regarding all cancers were collected, and pedigrees were extended to third-degree relatives as much as possible. The data of each pedigree was manually checked and formulated into a relational database for analysis. For the analyses in this study, male probands were excluded. ER, PR, and HER2 immunohistochemical (IHC) stains were available for a majority of invasive breast tumors in this cohort. ER(+) or PR(+) were defined as 1+, 2+ or 3+ on IHC stain. HER2(+) was defined as HER2 overexpression (3+ on IHC stain, or positive on dual in situ hybridization or fluorescence in situ hybridization).
Genetic testing for BRCA1/2 and other cancer susceptibility genes
Exonal and exon-flanking regions of twenty cancer susceptibility genes were sequenced on a next generation sequencing platform and variants were identified using standard protocols, details of which have been published previously34. We classified only protein-truncating variants including nonsense, frameshift, and splice-site mutations as pathogenic mutations. In addition to BRCA1 and BRCA2, seven genes including ATM, BARD1, BRIP1, PALB2, RAD50, RAD51C and RAD51D were denoted as homologous recombination pathway genes for predictive model analyses.
Calculation of germline mutation carrier probabilities
Relevant proband and pedigree information were formatted and stored in a local database, and input data for running the models were generated by in-house scripts (Perl, PHP, R script and shell script) in an automated fashion. Five mutation predictive models, BOADICEA, BRCAPRO, Myriad, Penn II, and Tyrer-Cuzick, were used for estimation of carrier probability of BRCA gene mutations. The prediction results were filtered and stored in the local database for statistical analyses.
The BOADICEA and BRCAPRO models compute the individual BRCA mutation carrier probability based on individual information on the proband and each of her relatives, including current age or age of death, incidence of breast, ovarian and other cancers, age at diagnosis and relationship to the proband. For BOADICEA, the predicted probability of carrying either a BRCA1 or a BRCA2 mutation was generated using the BOADICEA web application v3, (https://pluto.srl.cam.ac.uk/cgi-bin/bd3/v3/bd.cgi). BOADICEA allows input of the breast tumor ER/PR/HER2 (receptor) data. For comparison among the models, receptor status was not included in the calculation. Separate analyses comparing the predictive accuracies of BOADICEA with and without inclusion of the receptor status were performed. For BRCAPRO, the BayesMendel R package version 2.1–3 (available at https://projects.iq.harvard.edu/bayesmendel/bayesmendel-r-package) was used. The inputs for the Penn II and the Myriad model use a summary of personal and family cancer history. The Penn II predictions were carried out through a web interface (available at https://pennmodel2.pmacs.upenn.edu/penn2/). The Myriad model considers only the combined probability of carrying a mutation in either BRCA1 or BRCA2, by using a mapping table downloaded from the Myriad Genetics website (https://myriadgenetics.eu/healthcare-professional-treating-diseases/hereditary-cancer-testing/hereditary-breast-and-ovarian-cancer-hboc-syndrom/prevalence-tables/). The Tyrer-Cuzick model is only applicable to individuals without personal history of breast or ovarian cancer but have family history of these cancers. The International Breast Cancer Intervention Study (IBIS) breast cancer risk evaluation tool (v8) was used to calculate the mutation carrier probabilities.
Statistical analysis
Characteristics of the cohort were summarized using descriptive statistics stratified by subgroups. To determine the performance of the predictive models, receiver operating characteristics (ROC) curves were constructed, and the areas under the curve (AUCs) and 95% confidence interval (CI) were calculated. For each model, mutation carrier probability value, sensitivity, and specificity at the optimal cut-off point, which is the closest value to the left upper corner, were recorded. Subgroup analyses were done based on personal history and family history status. The applicable models were compared within the subgroups. Goodness of fit was assessed by comparing the expected/observed ratio (E/O) of the predicted probability to the actual frequency of the mutations, and calibration evaluated how well the model performed in each subgroup. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated for each risk model at the 10% and 20% thresholds for mutation carrier probability. All ROC curves were plotted using SigmaPlot (Systat Software, Inc.) version 12.0.
Supplementary information
Acknowledgements
The authors wish to thank the National Center for Genome Medicine at Academia Sinica (Taipei, Taiwan) for their technical and bioinformatics support in the next generation sequencing of the germline gene panel. This work was supported by the health and welfare surcharge of tobacco products in Taiwan (Ministry of Health and Welfare) [MOHW107-TDU-B-212-114021], the Ministry of Science and Technology [MOST104-0210-01-09-02, MOST105-0210-01-13-01, MOST106-0210-01-15-02], and by the Taiwan Protein Project [MOST105-0210-01-12-01, MOST106-0210-01-15-04].
Author Contributions
Y.A.W. conceptualized and designed the study, and was a major contributor in data interpretation and manuscript writing. F.H.H. coordinated data input and pedigree construction, and did a significant amount of work on statistical analysis, data interpretation, and manuscript writing. J.W.J. and H.P.P. contributed to pedigree construction, data analyses, and manuscript writing. L.L.H. and C.F.H. contributed to data acquisition and clinical data analysis. H.P.P., J.W.J. and A.S.Y. set up the bioinformatics pipeline to identify and annotate the genomic variants. M.M.Y. contributed to data interpretation and manuscript writing. A.S.Y. acquired funding and made substantial contributions to study design, data interpretation and manuscript writing. All authors read, contributed to, and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fei-Hung Hung and Yong Alison Wang contributed equally.
Contributor Information
Yong Alison Wang, Email: wang@kfsyscc.org.
An-Suei Yang, Email: yangas@gate.sinica.edu.tw.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-46707-6.
References
- 1.Ford D, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. American journal of human genetics. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoniou A, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. American journal of human genetics. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25:1329–1333. doi: 10.1200/jco.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gill J, Obley AJ, Prasad V. Direct-to-Consumer Genetic Testing: The Implications of the US FDA’s First Marketing Authorization for BRCA Mutation Testing. JAMA: the journal of the American Medical Association. 2018;319:2377–2378. doi: 10.1001/jama.2018.5330. [DOI] [PubMed] [Google Scholar]
- 5.Moyer VA. Risk assessment, genetic counseling, and genetic testing for BRCA-related cancer in women: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2014;160:271–281. doi: 10.7326/m13-2747. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong J, et al. Utilization and Outcomes of BRCA Genetic Testing and Counseling in a National Commercially Insured Population: The ABOUT Study. JAMA oncology. 2015;1:1251–1260. doi: 10.1001/jamaoncol.2015.3048. [DOI] [PubMed] [Google Scholar]
- 7.Robson ME, et al. American Society of Clinical Oncology Policy Statement Update: Genetic and Genomic Testing for Cancer Susceptibility. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2015;33:3660–3667. doi: 10.1200/JCO.2015.63.0996. [DOI] [PubMed] [Google Scholar]
- 8.ACOG bulletin 182 Hereditary Breast and Ovarian Cancer Syndrome 2017.pdf. [DOI] [PubMed]
- 9.Berliner JL, Fay AM, Cummings SA, Burnett B, Tillmanns T. NSGC practice guideline: risk assessment and genetic counseling for hereditary breast and ovarian cancer. Journal of genetic counseling. 2013;22:155–163. doi: 10.1007/s10897-012-9547-1. [DOI] [PubMed] [Google Scholar]
- 10.Lee AJ, et al. BOADICEA breast cancer risk prediction model: updates to cancer incidences, tumour pathology and web interface. British journal of cancer. 2014;110:535–545. doi: 10.1038/bjc.2013.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parmigiani G, Berry D, Aguilar O. Determining carrier probabilities for breast cancer-susceptibility genes BRCA1 and BRCA2. American journal of human genetics. 1998;62:145–158. doi: 10.1086/301670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank TS, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2002;20:1480–1490. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 13.Couch FJ, et al. BRCA1 mutations in women attending clinics that evaluate the risk of breast cancer. The New England journal of medicine. 1997;336:1409–1415. doi: 10.1056/nejm199705153362002. [DOI] [PubMed] [Google Scholar]
- 14.Evans DG, et al. A new scoring system for the chances of identifying a BRCA1/2 mutation outperforms existing models including BRCAPRO. J Med Genet. 2004;41:474–480. doi: 10.1136/jmg.2003.017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyrer J, Duffy SW, Cuzick J. A breast cancer prediction model incorporating familial and personal risk factors. Statistics in medicine. 2004;23:1111–1130. doi: 10.1002/sim.1668. [DOI] [PubMed] [Google Scholar]
- 16.Lindor NM, et al. Predicting BRCA1 and BRCA2 gene mutation carriers: comparison of PENN II model to previous study. Familial cancer. 2010;9:495–502. doi: 10.1007/s10689-010-9348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cintolo-Gonzalez JA, et al. Breast cancer risk models: a comprehensive overview of existing models, validation, and clinical applications. Breast cancer research and treatment. 2017;164:263–284. doi: 10.1007/s10549-017-4247-z. [DOI] [PubMed] [Google Scholar]
- 18.Arnold AG, et al. Assessment of individuals with BRCA1 and BRCA2 large rearrangements in high-risk breast and ovarian cancer families. Breast cancer research and treatment. 2014;145:625–634. doi: 10.1007/s10549-014-2987-6. [DOI] [PubMed] [Google Scholar]
- 19.Culver J, Lowstuter K, Bowling L. Assessing breast cancer risk and BRCA1/2 carrier probability. Breast Dis. 2006;27:5–20. doi: 10.3233/BD-2007-27102. [DOI] [PubMed] [Google Scholar]
- 20.Antoniou AC, et al. BRCA1 and BRCA2 mutation predictions using the BOADICEA and BRCAPRO models and penetrance estimation in high-risk French-Canadian families. Breast cancer research: BCR. 2006;8:R3. doi: 10.1186/bcr1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thirthagiri E, et al. Evaluation of BRCA1 and BRCA2 mutations and risk-prediction models in a typical Asian country (Malaysia) with a relatively low incidence of breast cancer. Breast cancer research: BCR. 2008;10:R59. doi: 10.1186/bcr2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang E, et al. Accuracy of BRCA1/2 mutation prediction models in Korean breast cancer patients. Breast cancer research and treatment. 2012;134:1189–1197. doi: 10.1007/s10549-012-2022-8. [DOI] [PubMed] [Google Scholar]
- 23.Chew W, et al. Evaluation of the relative effectiveness of the 2017 updated Manchester scoring system for predicting BRCA1/2 mutations in a Southeast Asian country. J Med Genet. 2018;55:344–350. doi: 10.1136/jmedgenet-2017-105073. [DOI] [PubMed] [Google Scholar]
- 24.Rao NY, et al. Evaluating the performance of models for predicting the BRCA germline mutations in Han Chinese familial breast cancer patients. Breast cancer research and treatment. 2009;116:563–570. doi: 10.1007/s10549-008-0181-4. [DOI] [PubMed] [Google Scholar]
- 25.Kwong A, et al. Accuracy of BRCA1/2 mutation prediction models for different ethnicities and genders: experience in a southern Chinese cohort. World journal of surgery. 2012;36:702–713. doi: 10.1007/s00268-011-1406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eoh KJ, et al. BRCA1 and BRCA2 mutation predictions using the BRCAPRO and Myriad models in Korean ovarian cancer patients. Gynecol Oncol. 2017;145:137–141. doi: 10.1016/j.ygyno.2017.01.026. [DOI] [PubMed] [Google Scholar]
- 27.Antoniou AC, et al. Predicting the likelihood of carrying a BRCA1 or BRCA2 mutation: validation of BOADICEA, BRCAPRO, IBIS, Myriad and the Manchester scoring system using data from UK genetics clinics. J Med Genet. 2008;45:425–431. doi: 10.1136/jmg.2007.056556. [DOI] [PubMed] [Google Scholar]
- 28.Euhus DM, et al. Pretest prediction of BRCA1 or BRCA2 mutation by risk counselors and the computer model BRCAPRO. Journal of the National Cancer Institute. 2002;94:844–851. doi: 10.1093/jnci/94.11.844. [DOI] [PubMed] [Google Scholar]
- 29.Kurian AW, et al. Performance of BRCA1/2 mutation prediction models in Asian Americans. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26:4752–4758. doi: 10.1200/jco.2008.16.8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marroni F, et al. Evaluation of widely used models for predicting BRCA1 and BRCA2 mutations. J Med Genet. 2004;41:278–285. doi: 10.1136/jmg.2003.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, et al. Prevalence and characterization of BRCA1 and BRCA2 germline mutations in Chinese women with familial breast cancer. Breast cancer research and treatment. 2012;132:421–428. doi: 10.1007/s10549-011-1596-x. [DOI] [PubMed] [Google Scholar]
- 32.de Bruin MA, et al. Breast cancer risk factors differ between Asian and white women with BRCA1/2 mutations. Familial cancer. 2012;11:429–439. doi: 10.1007/s10689-012-9531-9. [DOI] [PubMed] [Google Scholar]
- 33.Kim H, Choi DH. Distribution of BRCA1 and BRCA2 Mutations in Asian Patients with Breast Cancer. Journal of breast cancer. 2013;16:357–365. doi: 10.4048/jbc.2013.16.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang YA, et al. Germline breast cancer susceptibility gene mutations and breast cancer outcomes. BMC cancer. 2018;18:315. doi: 10.1186/s12885-018-4229-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.