Abstract
Aim: The study aimed to identify the underlying differentially expressed genes (DEGs) and mechanism of macrophage-enriched rupture atherosclerotic plaque using bioinformatics methods.
Methods: GSE41571, which includes six stable samples and five ruptured atherosclerotic samples, was downloaded from the GEO database. After preprocessing, DEGs between ruptured and stable atherosclerotic samples were identified using LIMMA. Gene Ontology biological process (GO_BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of DEGs were performed using the Database for Annotation, Visualization, and Integration Discovery (DAVID) online tool. Based on the STRING database, protein-protein interactions (PPIs) network among DEGs were constructed. Regulatory relationships between miRNAs/transcriptional factors (TFs) and target genes were predicted using Enrichr, and regulatory networks were visualized using Cytoscape.
Results: A total of 268 DEGs (64 up-regulated and 204 down-regulated DEGs) were identified between ruptured and stable samples. In the PPI network, collagen type III alpha 1 chain (COL3A1), collagen type I alpha 2 chain (COL1A2), and asporin (ASPN) were more than 15 interaction degrees. In the miRNA-target network, miR21 was highlighted with highest degrees and ASPN could be targeted by miR21. Functional enrichment analysis showed that COL3A1 and COL1A2 were significantly enriched in extracellular matrix organization and cell adhesion GO_BP terms. Pre-platelet basic protein (PPBP) was the most significantly up-regulated gene in ruptured atherosclerotic samples and enriched in immune response and inflammatory response GO_BP terms.
Conclusions: Down-regulated COL3A1, COL1A2 and ASPN, and up-regulated PPBP might perform critical promotional roles in atherosclerotic plaque rupture. Furthermore, miR21 might be potential target to prevent atherosclerotic rupture.
Keywords: Atherosclerosis, Ruptured atherosclerotic plaque, Macrophages, Differentially Expressed Genes, Transcription factors, microRNA
Introduction
Atherosclerosis, the most common vascular disease, is the main cause of death of 17.5 million people annually1). Atherosclerosis (AS) is a chronic inflammatory disease characterized by medium- and large-sized atherosclerotic plaques on the vascular walls, which consists of two types: stable and ruptured plaques. Erosion and rupture of atherosclerotic plaques lead to atherosclerotic embolization, which contributes to the majority of acute coronary syndromes2). Until now, differences in cellular composition between ruptured and stable plaques have been well established3), and the immune system has been identified to play a basic role in the initial and development of atherosclerosis, as well as the mechanism of unstable plaques, which are prone to rupture4). During plaque rupture, however, the specific molecular mechanisms of macrophage involved in the rupture of atherosclerotic plaques are complex and not fully characterized.
Multiple gene expression studies have been performed between whole plaques and normal tissue. For example, visfatin is reported to function as an inflammatory mediator, which mediates destabilization of atherosclerotic plaques5). The formation of AS plaques can be inhibited by PRDM16 via enhancing the function of periaortic brown adipose tissue6). It has also been reported that the balance between inflammation and fibrosis plays a key role in the transition of atherosclerotic plaques from stable to rupture-prone7). A recent review study summarized that inflammation also promotes plaque calcification to regulate the progression and regression of atherosclerosis8). To further reveal the mechanism of macrophage-enriched plaque rupture, a transcriptome dataset numbered GSE41571 was deposited by Lee et al., and it has revealed that FABP4 and leptin were significantly elevated in rupture atherosclerotic plaques, indicating that down-regulated adipocytokine/PPAR signaling might be a promising therapeutic potential for AS3). However, the potential regulatory mechanisms of transcription factors (TFs) and miRNAs involved in the progression of rupture have not been revealed.
In the current study, the dataset of GSE41571 was re-analyzed using bioinformatics methods, including differentially expressed genes (DEGs) screening, functional enrichment analysis, protein-protein interaction, and the regulatory relationship prediction between transcriptional factors (TF)/microRNAs (miRNAs) and DEGs. With these analyses, we hope to identify new insights for the understanding of macrophage-enriched ruptures in atherosclerotic plaques.
Materials and Methods
Data Resources
The mRNA expression profiles of GSE415713) (human), which was assayed on the platform of GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array chip, were obtained from Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41571). A total of 11 macrophage-rich regions of carotid AS samples, including six stable samples and five ruptured samples, were isolated from patients undergoing surgery of the carotid artery stenosis at the Regional Neurosurgical Center, Newcastle-upon-Tyne. The Local Research Ethics Committee authorized construction of this dataset, and informed consents of all patients were obtained.
Data Preprocessing
The raw data of CEL profiles were downloaded and read using affy in R package9) (Version: 1.50.0, http://www.bioconductor.org/packages/release/bioc/html/affy.html), and subsequently processed using the robust multi-array average method10, 11), including background correction, normalization, and expression calculation. Furthermore, probes were annotated using the annotation profile provided by the platform, and unmatched probes were discarded. For multiple probes matched to one gene symbol, the mean values of probes were calculated as the final expression of the gene.
Identification of DEGs
After processed, DEGs between groups were screened out using the Linear Models for Microarray (LIMMA; Version: 3.30.3) affy in R package12). q-value between gene expressions was determined using the T test and adjusted using Benjamin Hochberg (BH) method. Those genes with the cut-off criteria of q-value < 0.05 and |log2 fold change (FC)| > 1 were considered as DEGs.
Functional and Pathway Enrichment Analysis
The Gene Ontology (GO) term13) and Kyoto Encyclopedia of Genes and Genomes (KEGG)14) pathway enrichment analyses of DEGs were performed using the online tool of the DAVID, version: 6.8)15) with the thresholds of count ≥ 2 and p-value < 0.05.
To observe the function of DEGs visually, the ClueGO plugin (Version 2.2.6, http://apps.cytoscape.org/apps/ClueGO)16) of Cytoscape software17) was used to visualize GO biological processes (BPs) and KEGG pathways enriched for up- and down-regulated DEGs under the threshold of p-value < 0.05.
PPI Network Analysis
Based on the Search tool for the retrieval of interacting genes/proteins (STRING)18) online database (http://string-db.org/), PPIs among DEGs-encoded proteins were predicted with the threshold of PPI score (medium confidence) ≥ 0.4. Then, PPIs were visualized using Cytoscape software (version: 3.2.0, http://cytoscape.org/). The degree centrality, a network topology index, was used to analyze the scores of nodes in the network. The nodes with higher degree played an important role in PPI network, and might be key nodes. Nodes represent proteins encoded by DEGs, and degree is considered as the number of protein interaction pairs of a certain protein.
Subnet Module Analysis
Proteins produced by genes in the same module tend to have the same or similar function, and they act as a module to function in a same biological role. Using the plugin of MCODE (Multi-Contrast Delayed Enhancement)19) in Cytoscape, the most significantly clustered modules in the network were analyzed. The score of each module was calculated using the MCODE algorithm. The higher the score of the module presented, the greater the number of close interactions and enrichments it possessed. Furthermore, GO and KEGG functional enrichments of modules with score ≥ 3 and node ≥ 5 were analyzed using DAVID.
Prediction of miRNA-Target Regulatory Networks
Regulatory relationships between miRNA and targeted-DEGs were predicted using Enrichr (http://amp.pharm.mssm.edu/Enrichr/)20) and adjusted by BH21) followed by the super geometric test. With the threshold of q-value < 0.05, miRNA-target regulatory network was visualized using the Cytoscape software.
Prediction of TF-Target Regulatory Networks
Based on TRANSFAC and JASPAR database, regulatory relationships between TFs and targeted-DEGs were predicted using Enrichr, and the TF-target regulatory networks were visualized by Cytoscape.
Results
Identification of DEGs
A total of 268 DEGs were identified in ruptured samples compared to stable samples, including 64 upregulated genes and 204 down-regulated genes (Supplementary Table 1). COL1A2 and PPBP were the most significantly down-regulated and up-regulated genes in ruptured atherosclerotic samples, respectively. The Hierarchical clustering heat map of DEGs showed that these identified DEGs were well distinguished from the different kinds of samples (Fig. 1).
Supplementary Table 1. The list of all differentially expressed genes.
| logFC | AveExpr | t | P.Value | adj.P.Val | B | |
|---|---|---|---|---|---|---|
| PIK3R6 | 1.2 | 6.319 | 8.201 | 3.85E-06 | 0.011196886 | 4.626 |
| CEMIP | −1.25669178 | 7.687 | −8.95594463 | 1.59E-06 | 0.011196886 | 5.387 |
| CTNNAL1 | −1.50635456 | 7.069 | −8.76434724 | 1.98E-06 | 0.011196886 | 5.2 |
| ROBO1 | −2.70565648 | 8.496 | −8.84771249 | 1.79E-06 | 0.011196886 | 5.282 |
| ITGBL1 | −3.36215802 | 7.681 | −8.27010501 | 3.54E-06 | 0.011196886 | 4.699 |
| METRNL | 1.002 | 9.818 | 8.071 | 4.51E-06 | 0.011665271 | 4.487 |
| CD38 | 1.565 | 6.27 | 7.222 | 1.33E-05 | 0.014239233 | 3.526 |
| MPP1 | 1.133 | 10.398 | 7.139 | 1.49E-05 | 0.014239233 | 3.426 |
| TMA16 | −1.37306826 | 6.025 | −7.14752281 | 1.47E-05 | 0.014239233 | 3.436 |
| LNX1 | −1.43840671 | 5.596 | −7.69369652 | 7.22E-06 | 0.014239233 | 4.073 |
| NT5E | −1.45802181 | 6.667 | −7.10333002 | 1.56E-05 | 0.014239233 | 3.383 |
| PMEPA1 | −1.62651784 | 7.781 | −6.9731982 | 1.86E-05 | 0.014239233 | 3.224 |
| CRTAC1 | −1.76339168 | 7.001 | −7.42291778 | 1.02E-05 | 0.014239233 | 3.763 |
| SULF1 | −1.79939429 | 10.654 | −7.25296984 | 1.28E-05 | 0.014239233 | 3.563 |
| HECW2 | −1.89456819 | 5.37 | −7.158743 | 1.45E-05 | 0.014239233 | 3.45 |
| GLIS2 | −2.1429111 | 7.666 | −7.34832308 | 1.13E-05 | 0.014239233 | 3.675 |
| FAP | −2.28958226 | 8.494 | −7.04661664 | 1.68E-05 | 0.014239233 | 3.314 |
| LTBP2 | −2.56032838 | 9.309 | −7.53162444 | 8.88E-06 | 0.014239233 | 3.888 |
| ASPN | −3.12774161 | 5.907 | −6.95861939 | 1.90E-05 | 0.014239233 | 3.206 |
| FNDC1 | −3.6200187 | 7.689 | −6.99541145 | 1.80E-05 | 0.014239233 | 3.251 |
| COL21A1 | −4.4469972 | 7.136 | −7.14767824 | 1.47E-05 | 0.014239233 | 3.437 |
| SLC7A8 | 1.015 | 6.912 | 6.849 | 2.20E-05 | 0.014542207 | 3.07 |
| SKI | −1.01671528 | 5.422 | −6.84693447 | 2.21E-05 | 0.014542207 | 3.067 |
| SGCB | −1.18421525 | 6.196 | −6.81390243 | 2.31E-05 | 0.014542207 | 3.026 |
| LOC100132891 | −1.82346745 | 6.17 | −6.82874716 | 2.26E-05 | 0.014542207 | 3.045 |
| FZD7 | −2.02231618 | 6.228 | −6.83837974 | 2.23E-05 | 0.014542207 | 3.057 |
| MRAP2 | −1.7051916 | 5.87 | −6.78409852 | 2.41E-05 | 0.014749787 | 2.988 |
| ZFPM2 | −2.70450016 | 5.713 | −6.72406374 | 2.62E-05 | 0.014749787 | 2.912 |
| PLSCR4 | −3.02986415 | 7.889 | −6.71207272 | 2.66E-05 | 0.014749787 | 2.897 |
| KDELC1 | −1.07781756 | 5.894 | −6.66357421 | 2.85E-05 | 0.014873881 | 2.835 |
| EBF1 | −1.93542721 | 5.188 | −6.6531068 | 2.89E-05 | 0.014873881 | 2.822 |
| EFCAB7 | −1.26119539 | 5.316 | −6.55903778 | 3.30E-05 | 0.015351409 | 2.701 |
| LRRC17 | −1.52743734 | 6.263 | −6.5605111 | 3.29E-05 | 0.015351409 | 2.703 |
| CTHRC1 | −2.28289422 | 11.407 | −6.52568507 | 3.46E-05 | 0.015776567 | 2.658 |
| FKBP7 | −1.08592784 | 5.062 | −6.40842257 | 4.08E-05 | 0.016113508 | 2.504 |
| LBH | −1.76630203 | 7.774 | −6.47488815 | 3.71E-05 | 0.016113508 | 2.591 |
| CRNDE | −1.81466313 | 6.371 | −6.43567139 | 3.93E-05 | 0.016113508 | 2.54 |
| RSPO3 | −2.25938915 | 6.74 | −6.41389175 | 4.05E-05 | 0.016113508 | 2.511 |
| UCP2 | 1.046 | 10.197 | 6.356 | 4.40E-05 | 0.016270683 | 2.434 |
| AFAP1 | −1.21925021 | 5.084 | −6.36304591 | 4.36E-05 | 0.016270683 | 2.444 |
| FRMD6 | −2.16411717 | 7.524 | −6.37293311 | 4.30E-05 | 0.016270683 | 2.457 |
| NTN4 | −3.27525684 | 6.969 | −6.29606061 | 4.80E-05 | 0.017460006 | 2.355 |
| NCF4 | 1.354 | 8.239 | 6.285 | 4.88E-05 | 0.017468685 | 2.34 |
| HEPH | −1.2555782 | 6.127 | −6.26631153 | 5.01E-05 | 0.017676079 | 2.315 |
| IGHM | 1.254 | 6.413 | 6.147 | 5.97E-05 | 0.017810879 | 2.153 |
| TPBG | −1.14822882 | 6.065 | −6.15880954 | 5.86E-05 | 0.017810879 | 2.17 |
| STK38L | −1.26875147 | 9.209 | −6.15941448 | 5.86E-05 | 0.017810879 | 2.17 |
| ART4 | −1.72536951 | 5.02 | −6.16575484 | 5.80E-05 | 0.017810879 | 2.179 |
| KCNS3 | −1.91768639 | 6.684 | −6.19561586 | 5.55E-05 | 0.017810879 | 2.22 |
| NAP1L3 | −2.51867182 | 5.597 | −6.19671351 | 5.54E-05 | 0.017810879 | 2.221 |
| HMCN1 | −3.85610413 | 7.39 | −6.06690228 | 6.71E-05 | 0.018098086 | 2.044 |
| PTPRK | −1.50735965 | 5.155 | −6.03902761 | 6.99E-05 | 0.018288638 | 2.006 |
| FBN1 | −2.24618566 | 8.156 | −6.03920907 | 6.99E-05 | 0.018288638 | 2.006 |
| LOC100507165 | −1.26563477 | 6.349 | −5.99392548 | 7.48E-05 | 0.018713617 | 1.943 |
| LUM | −1.89877639 | 9.986 | −6.00525254 | 7.35E-05 | 0.018713617 | 1.959 |
| PRSS23 | −1.40485461 | 7.92 | −5.96238674 | 7.83E-05 | 0.01915319 | 1.899 |
| CTSLP8 | 1.546 | 6.208 | 5.91 | 8.48E-05 | 0.019311481 | 1.826 |
| TRIM59 | −1.06753541 | 4.988 | −5.90024542 | 8.60E-05 | 0.019311481 | 1.813 |
| THBS2 | −2.42982068 | 10.65 | −5.90527251 | 8.53E-05 | 0.019311481 | 1.82 |
| FMO2 | −3.06556932 | 7.134 | −5.89211025 | 8.70E-05 | 0.019311481 | 1.801 |
| EGFLAM | −1.15809758 | 6.61 | −5.84266913 | 9.38E-05 | 0.020211564 | 1.732 |
| SGIP1 | −2.38144236 | 5.731 | −5.81487254 | 9.78E-05 | 0.020508377 | 1.692 |
| LAIR1 | 1.123 | 9.363 | 5.804 | 9.93E-05 | 0.020557983 | 1.678 |
| ISLR | −2.49728223 | 7.143 | −5.8014734 | 9.98E-05 | 0.020557983 | 1.674 |
| FRY | −1.05072072 | 5.327 | −5.74316552 | 0.000109014 | 0.02098725 | 1.591 |
| ACTA2 | −2.91276358 | 5.63 | −5.74280218 | 0.000109074 | 0.02098725 | 1.59 |
| STON1 | −1.1288534 | 5.259 | −5.70844297 | 0.00011494 | 0.021749286 | 1.541 |
| RBMS3 | −1.20418414 | 4.754 | −5.70335452 | 0.000115837 | 0.021749286 | 1.534 |
| SNX7 | −1.01351647 | 8.664 | −5.66744557 | 0.00012238 | 0.021750042 | 1.482 |
| TRPC1 | −1.48467559 | 5.479 | −5.68361084 | 0.000119387 | 0.021750042 | 1.505 |
| LOC102725271 | −2.08021698 | 6.195 | −5.6792456 | 0.000120188 | 0.021750042 | 1.499 |
| FZD6 | −2.52508467 | 6.587 | −5.66876178 | 0.000122134 | 0.021750042 | 1.484 |
| TMEM200A | −2.65799647 | 7.649 | −5.68436322 | 0.00011925 | 0.021750042 | 1.506 |
| LTBP1 | −2.66774268 | 9.446 | −5.63414494 | 0.000128799 | 0.021887182 | 1.434 |
| HCK | 1.027 | 10.062 | 5.586 | 0.00013864 | 0.022335508 | 1.365 |
| WISP1 | −1.09763938 | 5.262 | −5.58419252 | 0.000139105 | 0.022335508 | 1.362 |
| GPX8 | −2.09217016 | 7.453 | −5.58734641 | 0.000138429 | 0.022335508 | 1.366 |
| ECM2 | −2.26273265 | 6.217 | −5.56641706 | 0.00014298 | 0.022645381 | 1.336 |
| ZNF532 | −1.17796726 | 7.645 | −5.54560073 | 0.000147664 | 0.022803536 | 1.306 |
| KCNT2 | −3.25602404 | 6.031 | −5.54458613 | 0.000147897 | 0.022803536 | 1.304 |
| FSTL1 | −1.2961765 | 8.781 | −5.53364296 | 0.000150428 | 0.02299253 | 1.288 |
| POSTN | −2.00535336 | 7.828 | −5.5290006 | 0.000151516 | 0.02299253 | 1.281 |
| LCP1 | 1.126 | 9.603 | 5.458 | 0.000169281 | 0.023576697 | 1.177 |
| ASAP2 | −1.0195605 | 8.974 | −5.42846544 | 0.000177234 | 0.023576697 | 1.134 |
| LOC100996668 | −1.03219502 | 5.509 | −5.43837456 | 0.000174505 | 0.023576697 | 1.148 |
| LOXL1-AS1 | −1.29412144 | 4.801 | −5.46806046 | 0.000166593 | 0.023576697 | 1.192 |
| PPIC | −1.33677848 | 8.976 | −5.42561755 | 0.000178026 | 0.023576697 | 1.129 |
| EXT1 | −1.34917431 | 6.22 | −5.49825972 | 0.000158932 | 0.023576697 | 1.236 |
| GATA6 | −1.35281251 | 6.764 | −5.47930665 | 0.000163695 | 0.023576697 | 1.208 |
| F2R | −1.59271311 | 6.752 | −5.49532218 | 0.000159661 | 0.023576697 | 1.232 |
| DKK3 | −1.86007416 | 6.333 | −5.45341254 | 0.000170448 | 0.023576697 | 1.17 |
| MEIS2 | −1.94522503 | 6.595 | −5.48118849 | 0.000163216 | 0.023576697 | 1.211 |
| LAMB1 | −2.36891793 | 7.601 | −5.41921172 | 0.000179823 | 0.023576697 | 1.12 |
| MIR100HG | −3.0403574 | 6.304 | −5.47572941 | 0.000164611 | 0.023576697 | 1.203 |
| OXCT1 | −1.04892338 | 6.861 | −5.40242719 | 0.000184621 | 0.023747807 | 1.095 |
| FAM114A1 | −1.19667178 | 8.182 | −5.37032694 | 0.00019418 | 0.023982939 | 1.047 |
| ZNF415 | −1.64499095 | 5.195 | −5.36622657 | 0.000195438 | 0.023982939 | 1.041 |
| LEP | 2.268 | 6.42 | 5.34 | 0.00020371 | 0.024447275 | 1.002 |
| LIFR | −1.04740797 | 4.788 | −5.34280558 | 0.000202791 | 0.024447275 | 1.006 |
| LIMCH1 | −1.44928729 | 5.869 | −5.34224492 | 0.000202971 | 0.024447275 | 1.006 |
| GLA | 1.142 | 11.256 | 5.328 | 0.000207585 | 0.024471802 | 0.984289808 |
| SPOCK1 | −2.00207978 | 7.99 | −5.30897156 | 0.000213935 | 0.024471802 | 0.955807089 |
| CAV2 | −1.71618358 | 6.47 | −5.2679935 | 0.000228303 | 0.025240194 | 0.894323182 |
| CNN3 | −2.40155517 | 6.511 | −5.27633466 | 0.000225298 | 0.025240194 | 0.906860152 |
| FABP4 | 2.335 | 5.645 | 5.246 | 0.000236293 | 0.025499036 | 0.861768833 |
| FCGR3A | 2.28 | 5.843 | 5.253 | 0.00023395 | 0.025499036 | 0.871203283 |
| DHRS9 | 1.974 | 8.98 | 5.246 | 0.000236569 | 0.025499036 | 0.860667129 |
| EVA1C | −1.73126087 | 6.248 | −5.24772741 | 0.000235783 | 0.025499036 | 0.863816319 |
| S100A9 | 1.814 | 9.17 | 5.231 | 0.000242257 | 0.02599181 | 0.83817444 |
| TSPAN13 | 1.364 | 10.132 | 5.224 | 0.000245051 | 0.026121045 | 0.827316374 |
| FIBIN | −2.18712548 | 6.245 | −5.21228387 | 0.000249493 | 0.026283656 | 0.810304916 |
| TMEM263 | −1.09452026 | 9.279 | −5.17428412 | 0.00026513 | 0.027145577 | 0.752711892 |
| XG | −1.14122656 | 5.25 | −5.1770576 | 0.000263954 | 0.027145577 | 0.756923192 |
| ANO1 | −1.5220958 | 5.903 | −5.17976835 | 0.000262811 | 0.027145577 | 0.761038048 |
| PLS3 | −2.61058898 | 10.003 | −5.16190948 | 0.000270443 | 0.027196827 | 0.733907168 |
| IGLJ3 | 1.328 | 6.847 | 5.138 | 0.000280903 | 0.027589235 | 0.697929449 |
| COL8A1 | −1.59954307 | 5.177 | −5.10344528 | 0.000297106 | 0.028177964 | 0.644736003 |
| PODXL | −2.5532906 | 8.65 | −5.10308659 | 0.000297278 | 0.028177964 | 0.644187253 |
| CPE | −2.41341692 | 6.49 | −5.08342649 | 0.000306862 | 0.028692184 | 0.614078827 |
| MGP | −2.7960331 | 7.892 | −5.06861727 | 0.000314295 | 0.029120038 | 0.591359135 |
| PPBP | 2.977 | 4.934 | 5.047 | 0.000325516 | 0.029568025 | 0.558054325 |
| LINC00847 | 1.701 | 6.722 | 5 | 0.000351428 | 0.029568025 | 0.485294566 |
| CKAP2 | 1.444 | 6.347 | 5.046 | 0.000326105 | 0.029568025 | 0.556335592 |
| NINJ1 | 1.227 | 9.489 | 5.017 | 0.00034164 | 0.029568025 | 0.512134064 |
| ZNF385A | 1.002 | 7.307 | 5.015 | 0.000342894 | 0.029568025 | 0.50865254 |
| TCF4 | −1.11012014 | 8.861 | −5.03410741 | 0.000332364 | 0.029568025 | 0.538282068 |
| PRELP | −1.83415382 | 8.551 | −5.00604766 | 0.000347861 | 0.029568025 | 0.494988219 |
| C1R | −1.86165559 | 9.136 | −5.04079479 | 0.000328779 | 0.029568025 | 0.548582002 |
| GLT8D2 | −1.93529812 | 6.747 | −4.99897494 | 0.000351886 | 0.029568025 | 0.484056257 |
| TMEM47 | −2.65208148 | 7.739 | −5.01707979 | 0.000341679 | 0.029568025 | 0.512024529 |
| NEXN | −2.73532684 | 6.243 | −5.03303355 | 0.000332943 | 0.029568025 | 0.53662745 |
| AK021804 | −3.30885199 | 7.223 | −5.03233933 | 0.000333318 | 0.029568025 | 0.535557691 |
| CRIM1 | −1.84112926 | 8.548 | −4.97768255 | 0.000364302 | 0.030183899 | 0.451098671 |
| VCAM1 | −1.74604498 | 11.444 | −4.97321252 | 0.000366966 | 0.030189802 | 0.444170766 |
| TSPYL4 | −1.16572958 | 8.755 | −4.95426541 | 0.000378491 | 0.030398027 | 0.41477112 |
| FAT1 | −2.46760386 | 8.673 | −4.95239161 | 0.000379651 | 0.030398027 | 0.411860598 |
| DNM3OS | −2.26298358 | 5.665 | −4.94189258 | 0.000386222 | 0.030514411 | 0.395542649 |
| SLC37A3 | −1.00679181 | 7.362 | −4.90755169 | 0.000408561 | 0.031189135 | 0.342050316 |
| GNAI1 | −1.02458618 | 4.511 | −4.90353054 | 0.000411265 | 0.031189135 | 0.335774788 |
| STXBP1 | −1.06233296 | 7.563 | −4.90389178 | 0.000411021 | 0.031189135 | 0.336338656 |
| NHS | −1.18994264 | 5.633 | −4.90527988 | 0.000410086 | 0.031189135 | 0.338505158 |
| GRN | 1.017 | 12.203 | 4.901 | 0.000412639 | 0.031191784 | 0.332599348 |
| TNFAIP6 | −2.22807951 | 7.13 | −4.86846811 | 0.000435649 | 0.031943596 | 0.280950449 |
| ZBTB20 | −1.00515779 | 6.98 | −4.84421694 | 0.000453401 | 0.03248024 | 0.242921225 |
| SUGCT | −1.88509353 | 7.086 | −4.84428645 | 0.000453349 | 0.03248024 | 0.243030364 |
| CDH11 | −1.95194678 | 7.012 | −4.84519063 | 0.000452674 | 0.03248024 | 0.244449829 |
| KIAA1377 | −1.11080072 | 4.659 | −4.82196057 | 0.000470362 | 0.032662868 | 0.20794168 |
| CCND1 | −1.15263195 | 8.102 | −4.8216234 | 0.000470624 | 0.032662868 | 0.207411186 |
| CDH2 | −1.2169333 | 7.663 | −4.8223331 | 0.000470072 | 0.032662868 | 0.208527793 |
| NOX4 | −1.82320944 | 4.658 | −4.82715307 | 0.000466346 | 0.032662868 | 0.216109247 |
| HMOX1 | 2.226 | 11.122 | 4.813 | 0.000477479 | 0.032773348 | 0.193633653 |
| IL10RA | 1.003 | 10.987 | 4.812 | 0.000478063 | 0.032773348 | 0.192469301 |
| BICC1 | −1.10999716 | 4.796 | −4.79730384 | 0.000489933 | 0.032967077 | 0.169102285 |
| AX747132 | −1.48765545 | 5.276 | −4.78862768 | 0.000497021 | 0.03306183 | 0.15541382 |
| SFRP2 | −3.62509444 | 8.232 | −4.78137924 | 0.000503025 | 0.033365917 | 0.143969241 |
| ARRB2 | 1.21 | 8.361 | 4.759 | 0.000521784 | 0.033462477 | 0.109069249 |
| SPATA18 | −1.01167048 | 6.085 | −4.7517964 | 0.000528332 | 0.033462477 | 0.097179376 |
| PRTFDC1 | −1.20002167 | 5.839 | −4.75113323 | 0.000528915 | 0.033462477 | 0.096128966 |
| HEG1 | −1.29863636 | 9.213 | −4.75486385 | 0.000525647 | 0.033462477 | 0.10203709 |
| PCDHGA4 | −1.61801494 | 4.893 | −4.76503003 | 0.00051685 | 0.033462477 | 0.11812655 |
| COL1A2 | −2.23065822 | 10.887 | −4.75182652 | 0.000528306 | 0.033462477 | 0.097227086 |
| MYH10 | −2.4727541 | 7.291 | −4.75829436 | 0.000522661 | 0.033462477 | 0.107468095 |
| SLC48A1 | 1.098 | 7.003 | 4.744 | 0.000535212 | 0.033625891 | 0.084843842 |
| PTPRG | −1.06299347 | 4.626 | −4.72336071 | 0.00055392 | 0.034040944 | 0.052081228 |
| PDGFA | −1.20814415 | 7.245 | −4.7231203 | 0.000554141 | 0.034040944 | 0.051699445 |
| STEAP1 | −1.59703261 | 7.015 | −4.71504336 | 0.000561647 | 0.034411224 | 0.03886755 |
| TPM1 | −1.28919304 | 7.279 | −4.71171411 | 0.000564772 | 0.03441254 | 0.033575558 |
| NEU1 | 1.14 | 8.864 | 4.701 | 0.000575252 | 0.034515567 | 0.016035637 |
| CX3CR1 | −1.15611971 | 6.078 | −4.69985497 | 0.000576051 | 0.034515567 | 0.014711631 |
| RP4-614O4 | −1.30864069 | 5.283 | −4.69918825 | 0.000576693 | 0.034515567 | 0.01365048 |
| GUCY1A3 | −2.45288651 | 6.228 | −4.69733727 | 0.000578476 | 0.034533555 | 0.010704154 |
| DPT | −2.10253648 | 6.383 | −4.68924907 | 0.000586339 | 0.034865462 | −0.002176312 |
| COL5A2 | −2.51114645 | 9.242 | −4.68842114 | 0.00058715 | 0.034865462 | −0.003495332 |
| MZB1 | 2.34 | 6.786 | 4.665 | 0.00061014 | 0.035032694 | −0.040144013 |
| ATP6V0B | 1.249 | 10.161 | 4.67 | 0.000605709 | 0.035032694 | −0.033189336 |
| FERMT2 | −1.32252556 | 6.945 | −4.66808806 | 0.000607445 | 0.035032694 | −0.035920518 |
| YAP1 | −1.72909066 | 6.431 | −4.66877616 | 0.000606747 | 0.035032694 | −0.034822218 |
| EML1 | −1.99655211 | 5.432 | −4.66075832 | 0.000614941 | 0.035032694 | −0.047624057 |
| CFH | −2.1218776 | 10.83 | −4.67777279 | 0.000597689 | 0.035032694 | −0.020468742 |
| 2018/3/2 | −1.049441 | 6.229 | −4.64018199 | 0.000636504 | 0.035582405 | −0.080520385 |
| IGFBP7 | −1.17251974 | 8.893 | −4.61773173 | 0.000660938 | 0.036207263 | −0.116482484 |
| DZIP1 | −1.21446462 | 5.069 | −4.61329159 | 0.000665885 | 0.036207263 | −0.123603551 |
| IQCJ-SCHIP1 | −1.89678603 | 7.193 | −4.59387208 | 0.000687985 | 0.036631536 | −0.154781644 |
| TAF1A | −1.29817562 | 5.537 | −4.57505232 | 0.000710136 | 0.037323023 | −0.185048194 |
| MXRA5 | −2.08246518 | 9.942 | −4.5760163 | 0.000708983 | 0.037323023 | −0.183496667 |
| MYO1B | −1.06000364 | 5.771 | −4.56172834 | 0.000726268 | 0.037659193 | −0.206506663 |
| SLCO2B1 | 1.319 | 8.457 | 4.533 | 0.000762207 | 0.038274727 | −0.252652647 |
| GSTT2 | −1.10875678 | 6.235 | −4.53840847 | 0.000755434 | 0.038274727 | −0.244123997 |
| AEBP1 | −2.35590453 | 10.096 | −4.52961448 | 0.000766748 | 0.038274727 | −0.25832946 |
| FCGRT | 1.177 | 10.833 | 4.52 | 0.000778957 | 0.038504641 | −0.273426586 |
| FAT4 | −2.56303105 | 6.048 | −4.51762232 | 0.000782464 | 0.038596008 | −0.27771853 |
| INHBA | −1.86991676 | 6.789 | −4.51503663 | 0.000785896 | 0.038683361 | −0.281901738 |
| IGKV1-17 | 2.579 | 7.067 | 4.507 | 0.00079641 | 0.038894789 | −0.294602761 |
| CYP27A1 | 1.326 | 11.108 | 4.498 | 0.000808921 | 0.039131153 | −0.30950161 |
| LOC100506119 | 1.01 | 7.294 | 4.488 | 0.000822133 | 0.039303695 | −0.324987906 |
| VEGFC | −1.83703081 | 7.396 | −4.48856265 | 0.000821959 | 0.039303695 | −0.324785545 |
| GAA | 1.167 | 9.206 | 4.473 | 0.000843911 | 0.039994585 | −0.349981918 |
| NFIB | −1.32832364 | 5.712 | −4.45439285 | 0.00087108 | 0.040260819 | −0.38027823 |
| SERTAD4 | −1.21609373 | 4.817 | −4.44402046 | 0.00088659 | 0.040401598 | −0.397154852 |
| PLTP | 1.311 | 10.929 | 4.432 | 0.000904488 | 0.040436634 | −0.416267419 |
| CTSA | 1.163 | 10.809 | 4.441 | 0.000890772 | 0.040436634 | −0.401654419 |
| SLC16A4 | −1.39699797 | 4.694 | −4.42089439 | 0.00092222 | 0.040517383 | −0.434835104 |
| FOXC1 | −2.12951548 | 8.291 | −4.42836534 | 0.000910549 | 0.040517383 | −0.422654485 |
| IGKV1OR2-108 | 1.868 | 7.811 | 4.397 | 0.000960932 | 0.040756283 | −0.47416657 |
| TJP1 | −1.08938082 | 6.622 | −4.40604957 | 0.000945876 | 0.040756283 | −0.459060347 |
| ZNRF3 | −1.12957808 | 5.084 | −4.39928713 | 0.000956861 | 0.040756283 | −0.470105748 |
| MPDZ | −1.54103226 | 6.522 | −4.40894973 | 0.000941205 | 0.040756283 | −0.45432526 |
| NET1 | −1.60153131 | 7.662 | −4.39586793 | 0.000962467 | 0.040756283 | −0.475692803 |
| SNAI2 | −2.33153198 | 6.709 | −4.39421282 | 0.000965192 | 0.040756283 | −0.478397851 |
| TANC1 | −1.04522496 | 5.558 | −4.38674571 | 0.000977589 | 0.040935653 | −0.490606346 |
| IGKC | 1.285 | 6.009 | 4.382 | 0.000985827 | 0.040965943 | −0.498633937 |
| ACOT13 | 1.03 | 7.829 | 4.375 | 0.000997539 | 0.040965943 | −0.509933389 |
| SPARC | −1.20354323 | 11.219 | −4.3753052 | 0.000996905 | 0.040965943 | −0.509325556 |
| GSTA4 | −1.29851562 | 6.86 | −4.36613168 | 0.001012682 | 0.040965943 | −0.524347965 |
| ZBTB8A | −1.37631649 | 7.052 | −4.37997542 | 0.000988972 | 0.040965943 | −0.50168195 |
| FST | −1.76779273 | 6.563 | −4.36991914 | 0.001006137 | 0.040965943 | −0.51814434 |
| TWIST1 | −2.11250906 | 7.358 | −4.36900418 | 0.001007714 | 0.040965943 | −0.519642824 |
| AOC3 | −2.28787858 | 7.915 | −4.3647634 | 0.001015057 | 0.040965943 | −0.52658959 |
| ATL1 | −1.00687259 | 6.814 | −4.3604407 | 0.0010226 | 0.041059108 | −0.533672997 |
| CRISPLD1 | −3.53397277 | 5.399 | −4.35427774 | 0.001033455 | 0.041200168 | −0.543776176 |
| SSPN | −1.17667481 | 5.806 | −4.35112305 | 0.001039058 | 0.041292889 | −0.548949711 |
| PPP1R15A | 1.035 | 7.244 | 4.335 | 0.001068043 | 0.041862246 | −0.575277661 |
| MGLL | 1.074 | 10.068 | 4.323 | 0.001090989 | 0.042231298 | −0.595619862 |
| CFHR2 | −1.05882399 | 6.425 | −4.3235495 | 0.001089399 | 0.042231298 | −0.594224097 |
| AMOTL2 | −2.27119759 | 7.063 | −4.3168058 | 0.001102093 | 0.04241147 | −0.605311898 |
| TNFRSF17 | 2.009 | 4.822 | 4.298 | 0.001138401 | 0.043046977 | −0.636334631 |
| SLC24A3 | −1.37012793 | 6.638 | −4.29952451 | 0.001135333 | 0.043046977 | −0.633751967 |
| NDC80 | −1.4974775 | 7.142 | −4.30164087 | 0.001131207 | 0.043046977 | −0.630266987 |
| AP1B1 | 1.11 | 8.429 | 4.28 | 0.001174262 | 0.043593147 | −0.666022436 |
| H2BFS | 1.222 | 5.121 | 4.271 | 0.001191769 | 0.04384424 | −0.68018847 |
| MAP4K4 | −1.04172561 | 7.215 | −4.27171029 | 0.001191044 | 0.04384424 | −0.679606089 |
| PLD3 | 1.916 | 10.825 | 4.264 | 0.001208022 | 0.043913909 | −0.693155632 |
| ENPP1 | −1.14642047 | 4.826 | −4.26575893 | 0.00120333 | 0.043913909 | −0.689430104 |
| ITGA10 | −1.85620797 | 7.214 | −4.26030907 | 0.001214696 | 0.043913909 | −0.69843019 |
| IGKV1-37 | 1.831 | 7.021 | 4.236 | 0.001267234 | 0.044290054 | −0.738968381 |
| GPC6 | −1.1298533 | 6.394 | −4.23566454 | 0.001267508 | 0.044290054 | −0.739175243 |
| C9orf3 | −1.47756582 | 6.871 | −4.24153164 | 0.001254722 | 0.044290054 | −0.729468257 |
| GXYLT2 | −1.69386575 | 5.555 | −4.24355961 | 0.001250334 | 0.044290054 | −0.726114027 |
| VMO1 | 1.666 | 7.962 | 4.229 | 0.001281973 | 0.044349043 | −0.750040061 |
| MGARP | −1.55504243 | 5.353 | −4.19551 | 0.001358735 | 0.045692391 | −0.805723339 |
| GLUL | 1.145 | 11.202 | 4.182 | 0.001390136 | 0.046038611 | −0.827601836 |
| CD27 | 1.21 | 7.39 | 4.179 | 0.001399117 | 0.046139169 | −0.833769133 |
| MYLK | −1.14019221 | 6.013 | −4.17280728 | 0.001413311 | 0.046410017 | −0.843434981 |
| LOC728061 | −2.05163732 | 5.984 | −4.16582764 | 0.001430544 | 0.04677797 | −0.855041301 |
| COL3A1 | −2.07311618 | 10.754 | −4.16094377 | 0.001442732 | 0.046974735 | −0.86316603 |
| SYNM | −2.20201792 | 7.297 | −4.13903732 | 0.001498743 | 0.047474464 | −0.899643872 |
| IL4I1 | 1.077 | 7.981 | 4.13 | 0.001523648 | 0.047937254 | −0.915428286 |
| POR | 1.289 | 7.363 | 4.114 | 0.001564286 | 0.048233071 | −0.940638903 |
| LURAP1L | −1.87326982 | 5.808 | −4.11317906 | 0.001567777 | 0.048233071 | −0.942774118 |
| PEPD | 1.135 | 8.808 | 4.105 | 0.00159094 | 0.048355035 | −0.956822001 |
| SYNC | −1.48850046 | 5.161 | −4.10469247 | 0.001591146 | 0.048355035 | −0.956946087 |
| OMD | −2.78152739 | 5.449 | −4.10474212 | 0.001591009 | 0.048355035 | −0.956863151 |
| PROCR | −1.00314734 | 7.544 | −4.09518 | 0.001617771 | 0.048689939 | −0.972840972 |
| CD180 | 1.345 | 7.32 | 4.083 | 0.001653047 | 0.048855534 | −0.993502199 |
| PPFIBP1 | −1.13010206 | 5.456 | −4.07920891 | 0.001663519 | 0.048855534 | −0.999550951 |
| PRRX1 | −1.6549019 | 6.099 | −4.07511267 | 0.001675467 | 0.048855534 | −1.0064061 |
| CFH | −2.33548068 | 9.12 | −4.07508164 | 0.001675558 | 0.048855534 | −1.00645803 |
| BGN | −1.62394918 | 8.095 | −4.06341277 | 0.001710086 | 0.04915337 | −1.02599645 |
| COL15A1 | −1.89798428 | 9.581 | −4.06441292 | 0.001707098 | 0.04915337 | −1.02432119 |
| IGLL3P | 2.172 | 9.334 | 4.06 | 0.001721112 | 0.049322321 | −1.03215249 |
| CYAT1 | 1.22 | 7.027 | 4.059 | 0.001723207 | 0.049322321 | −1.03331779 |
| ZNF404 | −1.06352944 | 4.472 | −4.05407513 | 0.001738248 | 0.049353524 | −1.04164231 |
| SFRP4 | −2.93801794 | 6.929 | −4.05424937 | 0.001737718 | 0.049353524 | −1.04135026 |
| IGLC1 | 1.596 | 7.517 | 4.046 | 0.001763359 | 0.049473158 | −1.05538118 |
| HLA-DMA | 1.306 | 11.378 | 4.045 | 0.001765836 | 0.049473158 | −1.05672574 |
| CEP290 | −1.0804849 | 6.391 | −4.04268175 | 0.001773262 | 0.049621504 | −1.0607457 |
| CXCL3 | 1.317 | 7.891 | 4.039 | 0.001784045 | 0.049684365 | −1.06655264 |
| MS4A6E | 1.668 | 4.81 | 4.037 | 0.001791087 | 0.049820889 | −1.07032628 |
| ACKR4 | −1.52268155 | 4.42 | −4.02988751 | 0.001813455 | 0.050044606 | −1.0822148 |
| EFEMP1 | −1.54844572 | 8.354 | −4.02769537 | 0.001820435 | 0.050157839 | −1.08589504 |
| LHFP | −1.81885212 | 6.486 | −4.02237604 | 0.00183749 | 0.050516661 | −1.09482749 |
| ADM | 1.107 | 11.237 | 4.02 | 0.001845287 | 0.050543483 | −1.09888318 |
| SEMA5A | −1.41324129 | 6.863 | −4.01790982 | 0.001851938 | 0.050606592 | −1.10232973 |
| C3orf80 | −1.72303094 | 4.81 | −4.01381707 | 0.001865281 | 0.050838319 | −1.10920649 |
| ZNF667-AS1 | −1.74621215 | 6.251 | −3.99983639 | 0.001911615 | 0.051114758 | −1.13271066 |
| PCDH10 | −2.08087587 | 5.158 | −4.00221063 | 0.001903663 | 0.051114758 | −1.12871765 |
| CD52 | 1.832 | 10.092 | 3.997 | 0.001922413 | 0.051145657 | −1.13810607 |
| TCN2 | 1.51 | 9.023 | 3.996 | 0.001924853 | 0.051145657 | −1.13932131 |
| PILRA | 1.096 | 8.679 | 3.99 | 0.001943711 | 0.051191763 | −1.14866009 |
| RP11-182L21 | −1.17416448 | 6.41 | −3.98819442 | 0.001951108 | 0.051191763 | −1.15229877 |
| METTL7B | 2.348 | 7.441 | 3.974 | 0.00199995 | 0.05152927 | −1.17598256 |
| HIST1H2BK | 1.241 | 8.407 | 3.972 | 0.002006245 | 0.051555629 | −1.17899306 |
| CALD1 | −1.15502391 | 6.474 | −3.97143398 | 0.002009454 | 0.051581145 | −1.18052373 |
| MIR143HG | −1.7863461 | 6.767 | −3.96581334 | 0.002029422 | 0.051859691 | −1.18999551 |
| PTN | −1.23570748 | 5.634 | −3.93877431 | 0.002128393 | 0.052998117 | −1.23560607 |
| GJA1 | −1.84211055 | 9.798 | −3.93940584 | 0.002126025 | 0.052998117 | −1.23453994 |
| SLITRK4 | −1.09949375 | 4.732 | −3.92826729 | 0.002168187 | 0.053248877 | −1.25334957 |
| ZNF680 | −1.15108211 | 6.536 | −3.92920137 | 0.002164618 | 0.053248877 | −1.25177171 |
| RP11-305O6 | −1.5668101 | 5.155 | −3.92047009 | 0.002198215 | 0.053590398 | −1.26652398 |
| PEAR1 | −1.45350304 | 6.879 | −3.91007867 | 0.002238903 | 0.053945632 | −1.28409097 |
| PCOLCE | −1.46136398 | 8.89 | −3.90848164 | 0.002245225 | 0.053945632 | −1.28679173 |
| SLIT2 | −1.73248051 | 7.539 | −3.91111181 | 0.002234823 | 0.053945632 | −1.28234394 |
| MID1 | −1.21316819 | 6.613 | −3.90211625 | 0.002270606 | 0.054219745 | −1.29755876 |
| OGN | −3.53385479 | 5.648 | −3.90268222 | 0.002268337 | 0.054219745 | −1.29660127 |
| NKG7 | 1.198 | 5.988 | 3.9 | 0.002277495 | 0.054328521 | −1.30046024 |
| PHOSPHO2 | −1.27042123 | 5.769 | −3.89654933 | 0.002293047 | 0.054587647 | −1.30697841 |
| CD72 | 1.917 | 7.487 | 3.881 | 0.002356347 | 0.055377521 | −1.33305954 |
| TMEM133 | −1.46004818 | 4.868 | −3.88095243 | 0.002357148 | 0.055377521 | −1.3333853 |
| SYTL2 | −1.07422641 | 5.833 | −3.87363677 | 0.00238785 | 0.055649564 | −1.34577929 |
| FAIM3 | 1.669 | 7.098 | 3.868 | 0.002412198 | 0.055904268 | −1.35549554 |
| IGLC1 | 1.244 | 5.63 | 3.865 | 0.002424514 | 0.055963427 | −1.36037311 |
| COL16A1 | −1.47126191 | 7.34 | −3.85744434 | 0.002457281 | 0.056102143 | −1.37322983 |
| PRORSD1P | −1.25635926 | 5.119 | −3.8419094 | 0.002525854 | 0.056873241 | −1.39958844 |
| CSPG4 | −1.70433251 | 6.435 | −3.83657651 | 0.002549847 | 0.05703302 | −1.40864197 |
| C1QA | 1.726 | 10.971 | 3.824 | 0.002607739 | 0.057988982 | −1.43014086 |
| PCDH18 | −1.20010692 | 5.687 | −3.82245976 | 0.0026145 | 0.057988982 | −1.43262 |
| SLC2A10 | −1.82473554 | 6.629 | −3.81256837 | 0.002660806 | 0.058145044 | −1.44943155 |
| LOC101929787 | −1.52065348 | 6.235 | −3.79932411 | 0.002724138 | 0.058493484 | −1.47195507 |
| ACP2 | 1.293 | 9.064 | 3.798 | 0.002728784 | 0.058500499 | −1.47358656 |
| AC128677 | 1.314 | 7.689 | 3.796 | 0.002740529 | 0.058599953 | −1.47769888 |
| CDH13 | −1.59536168 | 7.516 | −3.7881186 | 0.002778933 | 0.058710644 | −1.49102324 |
| TM4SF1 | −1.13482268 | 6.372 | −3.78568638 | 0.002790976 | 0.05882372 | −1.49516351 |
| SETBP1 | −2.04001615 | 5.492 | −3.78550728 | 0.002791865 | 0.05882372 | −1.49546841 |
| AKR1B1 | 1.001 | 11.004 | 3.773 | 0.002855397 | 0.059250757 | −1.51701084 |
| CD14 | 1.06 | 12.276 | 3.77 | 0.002869783 | 0.059352832 | −1.52182209 |
| GM2A | 1.026 | 8.328 | 3.763 | 0.00290519 | 0.059646061 | −1.53356152 |
| CHN1 | −1.43327294 | 7.218 | −3.75742871 | 0.002934899 | 0.060044226 | −1.54330163 |
| TGFBR3 | −1.94883598 | 7.354 | −3.75376994 | 0.002954086 | 0.060277845 | −1.54953931 |
| PKIG | −1.33129041 | 7.111 | −3.75201347 | 0.002963342 | 0.060360921 | −1.55253423 |
| P2RX4 | 1.357 | 8.316 | 3.749 | 0.002977745 | 0.060495517 | −1.55717564 |
| LY86 | 1.05 | 10.429 | 3.741 | 0.003019884 | 0.060768315 | −1.57062683 |
| IGKC | 1.131 | 6.415 | 3.737 | 0.003044466 | 0.060946901 | −1.57838682 |
| LINC00702 | −1.44549954 | 5.255 | −3.71950703 | 0.003140146 | 0.061821447 | −1.60800455 |
| FCER1G | 1.055 | 10.016 | 3.71 | 0.003193331 | 0.061904366 | −1.62407893 |
| PLA2G7 | 1.197 | 11.348 | 3.709 | 0.003202244 | 0.061936817 | −1.62674627 |
| LINC00657 | −1.36968121 | 8.481 | −3.69566712 | 0.003276684 | 0.062315323 | −1.64873798 |
| COL5A1 | −1.56308726 | 8.873 | −3.69034404 | 0.003307996 | 0.062351143 | −1.65783893 |
| SDS | 1.479 | 7.287 | 3.683 | 0.003350515 | 0.062655973 | −1.67005981 |
| TREM2 | 1.627 | 9.53 | 3.681 | 0.003364255 | 0.062761682 | −1.67397568 |
| RRN3P2 | 1.172 | 5.753 | 3.668 | 0.003442043 | 0.063601298 | −1.69584691 |
| COL6A3 | −1.78321343 | 11.797 | −3.6438087 | 0.003595174 | 0.064386793 | −1.73748812 |
| HBA1 | 2.739 | 8.398 | 3.641 | 0.003611872 | 0.064536915 | −1.74192076 |
| PTPN13 | −1.03095065 | 4.503 | −3.64078073 | 0.003614725 | 0.064538358 | −1.74267599 |
| HAMP | 1.271 | 7.182 | 3.626 | 0.003709523 | 0.065529521 | −1.76743745 |
| AHNAK2 | −1.16667494 | 7.662 | −3.60878191 | 0.003828129 | 0.066019364 | −1.79753723 |
| CHSY3 | −1.22204212 | 6.676 | −3.60638586 | 0.00384462 | 0.066019364 | −1.80164787 |
| NR2F2 | −1.52328366 | 5.648 | −3.60405828 | 0.003860709 | 0.066019364 | −1.80564141 |
| NOV | −1.91939829 | 7.518 | −3.61275996 | 0.00380091 | 0.066019364 | −1.79071333 |
| ADH1B | −2.23275369 | 5.263 | −3.60611297 | 0.003846503 | 0.066019364 | −1.80211607 |
| KIAA1462 | −1.23390712 | 5.005 | −3.59957708 | 0.00389188 | 0.066123504 | −1.81333093 |
| CD200 | −1.23990656 | 6.373 | −3.59120956 | 0.003950777 | 0.066308294 | −1.82769256 |
| C11orf96 | −1.8970491 | 9.023 | −3.59326057 | 0.003936256 | 0.066308294 | −1.82417191 |
| NDP | −1.52712861 | 4.978 | −3.58699312 | 0.0039808 | 0.066576401 | −1.8349311 |
| CCL23 | 1.185 | 4.999 | 3.578 | 0.004043445 | 0.066907955 | −1.84985935 |
| LOC100506585 | 1 | 6.671 | 3.573 | 0.00408413 | 0.067199089 | −1.8594304 |
| LOC100506558 | −1.09670933 | 5.314 | −3.57323447 | 0.004080407 | 0.067199089 | −1.85855858 |
| FMOD | −1.50776967 | 8.355 | −3.57284018 | 0.004083299 | 0.067199089 | −1.85923586 |
| PCSK5 | −1.28009798 | 6.504 | −3.56713186 | 0.004125401 | 0.067544018 | −1.86904207 |
| UACA | −1.05013878 | 6.654 | −3.55904856 | 0.004185783 | 0.068006558 | −1.88293145 |
| IGLV1-44 | 1.525 | 6.707 | 3.553 | 0.004232359 | 0.068367945 | −1.89350824 |
| IGK | 2.096 | 8.425 | 3.548 | 0.004269896 | 0.068623023 | −1.90194756 |
| USP1 | −1.14547526 | 7.686 | −3.53669661 | 0.004357506 | 0.068950885 | −1.92135753 |
| AK025288 | −1.98571356 | 8.033 | −3.5350646 | 0.004370323 | 0.068982952 | −1.92416424 |
| GZMB | 1.385 | 6.613 | 3.498 | 0.004675599 | 0.070549122 | −1.9886731 |
| CTSD | 1.568 | 11.244 | 3.494 | 0.004708177 | 0.070735539 | −1.99530536 |
| LRRC15 | −1.12921205 | 5.814 | −3.48577685 | 0.004776139 | 0.071144001 | −2.00899357 |
| SLC9B2 | −1.35623012 | 5.458 | −3.48354936 | 0.004795364 | 0.071167995 | −2.01283012 |
| ADAM1A | −1.43593993 | 6.533 | −3.47742566 | 0.004848625 | 0.071537196 | −2.02337853 |
| SLC22A18 | 1.314 | 8.866 | 3.476 | 0.00485765 | 0.071624955 | −2.02515427 |
| TPRA1 | 1.382 | 7.212 | 3.467 | 0.004939522 | 0.071966184 | −2.04111418 |
| PFN2 | −1.92215403 | 7.835 | −3.45452005 | 0.005053263 | 0.073210991 | −2.06284955 |
| MT1M | −1.20995452 | 5.25 | −3.45398354 | 0.00505816 | 0.073236371 | −2.06377435 |
| PLAGL1 | −1.17697226 | 8.294 | −3.4374629 | 0.005211362 | 0.074103921 | −2.09225691 |
| VGLL3 | −1.86051455 | 5.492 | −3.43570056 | 0.005227981 | 0.074103921 | −2.09529592 |
| TGFB1I1 | −1.91203016 | 6.9 | −3.43770237 | 0.005209107 | 0.074103921 | −2.09184396 |
| PBX1 | −1.01705906 | 5.355 | −3.43106813 | 0.005271926 | 0.074208576 | −2.1032848 |
| FPR1 | 1.084 | 6.106 | 3.429 | 0.005290933 | 0.074385801 | −2.10671942 |
| DDIT4 | 1.49 | 9.718 | 3.425 | 0.005334084 | 0.074579661 | −2.1144709 |
| VCAN | −1.5582745 | 9.014 | −3.42256774 | 0.005353546 | 0.074582358 | −2.11794629 |
| TLCD2 | −1.01274834 | 5.575 | −3.41837151 | 0.005394312 | 0.074845274 | −2.12518497 |
| COL24A1 | −1.20468207 | 5.625 | −3.41642392 | 0.00541334 | 0.074938028 | −2.12854486 |
| FAM171A1 | −1.23232038 | 6.785 | −3.40813696 | 0.005495071 | 0.075042307 | −2.14284264 |
| PDGFD | −1.65623364 | 5.872 | −3.40000426 | 0.005576505 | 0.075615545 | −2.15687664 |
| LOC101928916 | −1.24868866 | 9.412 | −3.38908463 | 0.005687782 | 0.075974146 | −2.17572328 |
| abParts | 1.055 | 6.89 | 3.378 | 0.005800963 | 0.076912316 | −2.19451488 |
| TNFRSF11B | −2.32614996 | 8.959 | −3.37306976 | 0.005855088 | 0.077249334 | −2.20337088 |
| CCRL2 | 1.069 | 9.279 | 3.365 | 0.005937457 | 0.077894866 | −2.21669122 |
| CCDC113 | −1.17122181 | 4.579 | −3.36082978 | 0.005986329 | 0.07824588 | −2.22450672 |
| OLFML1 | −1.85943197 | 6.329 | −3.35750026 | 0.006022545 | 0.078508896 | −2.23025681 |
| ZNF302 | −1.01985971 | 7.658 | −3.35151039 | 0.00608826 | 0.078859711 | −2.24060208 |
| RUNDC3B | −1.47075415 | 4.742 | −3.34993023 | 0.006105717 | 0.078867572 | −2.24333138 |
| IGLL5 | 2.546 | 6.618 | 3.341 | 0.006207258 | 0.079187602 | −2.25905202 |
| SAMSN1 | 1.212 | 6.716 | 3.3 | 0.006686271 | 0.082084201 | −2.32987484 |
| RGS5 | −1.47176028 | 5.433 | −3.29976294 | 0.006687231 | 0.082084201 | −2.33001146 |
| CCL18 | 1.7 | 12.378 | 3.297 | 0.006715839 | 0.082111048 | −2.33407705 |
| PXDN | −1.61084531 | 8.374 | −3.29718734 | 0.006718561 | 0.082111048 | −2.334463 |
| PLK2 | −1.72107139 | 6.446 | −3.29524778 | 0.006742253 | 0.082290914 | −2.3378153 |
| FCRL5 | 1.034 | 4.663 | 3.289 | 0.00681584 | 0.08239289 | −2.34815202 |
| C12orf75 | −1.56631522 | 6.387 | −3.29035853 | 0.006802353 | 0.08239289 | −2.34626606 |
| DHRS3 | 1.17 | 8.695 | 3.287 | 0.006843462 | 0.082497646 | −2.35200311 |
| BACE2 | −1.13729452 | 8.3 | −3.2866389 | 0.00684844 | 0.082497646 | −2.35269546 |
| DCN | −1.27797183 | 6.941 | −3.27884117 | 0.006946086 | 0.082871272 | −2.36617454 |
| PCDHB16 | −2.0403688 | 4.744 | −3.28141758 | 0.006913668 | 0.082871272 | −2.36172088 |
| FPR3 | 1.108 | 8.998 | 3.275 | 0.006992625 | 0.082958974 | −2.37253155 |
| FDX1 | 1.044 | 9.311 | 3.276 | 0.006982964 | 0.082958974 | −2.37121541 |
| FBXL7 | −1.10298254 | 6.347 | −3.27704144 | 0.006968823 | 0.082958974 | −2.36928565 |
| RP11-379H18 | −1.26161514 | 5.742 | −3.27700657 | 0.006969264 | 0.082958974 | −2.36934592 |
| SDSL | 1.193 | 7.509 | 3.272 | 0.007038201 | 0.083119231 | −2.37871581 |
| HACE1 | −1.01249208 | 7.259 | −3.2716711 | 0.007037119 | 0.083119231 | −2.37856938 |
| HLA-DMB | 1 | 11.958 | 3.26 | 0.007185634 | 0.083941761 | −2.39844726 |
| EPHX1 | 1.281 | 6.325 | 3.242 | 0.007430938 | 0.084942411 | −2.43038762 |
| FNBP1L | −1.17944856 | 7.479 | −3.2415762 | 0.007432599 | 0.084942411 | −2.43060032 |
| TNC | −1.37578741 | 7.69 | −3.23325442 | 0.007545881 | 0.085104671 | −2.44498911 |
| AQP1 | −1.49341568 | 7.025 | −3.22534816 | 0.007655127 | 0.085609579 | −2.45865982 |
| RP11-124L9 | −1.12643933 | 5.719 | −3.22277474 | 0.007691029 | 0.085757925 | −2.46310956 |
| CXCL1 | 1.137 | 6.775 | 3.22 | 0.007727722 | 0.085890838 | −2.46763557 |
| SLC15A3 | 1.214 | 9.406 | 3.212 | 0.007839458 | 0.086081652 | −2.48128533 |
| HS3ST2 | 1.206 | 11.404 | 3.213 | 0.007834345 | 0.086081652 | −2.480665 |
| ANGPT1 | −1.16253508 | 4.798 | −3.19883173 | 0.008033349 | 0.08653032 | −2.5045106 |
| HBB | 2.301 | 8.438 | 3.188 | 0.008200276 | 0.087217375 | −2.52405622 |
| LOC101928620 | 1.214 | 4.851 | 3.184 | 0.008254768 | 0.08769952 | −2.53034954 |
| LRRC49 | −1.14867086 | 5.705 | −3.17829006 | 0.008339302 | 0.088217178 | −2.54002987 |
| LAMA2 | −1.0481289 | 5.445 | −3.1670848 | 0.008511135 | 0.088660508 | −2.55940451 |
| ZNF423 | −1.04457418 | 4.446 | −3.16092116 | 0.008607175 | 0.089221833 | −2.57006149 |
| ZNF23 | −1.11669749 | 7.011 | −3.14224086 | 0.008904983 | 0.090230763 | −2.60235781 |
| LINC01279 | −2.02681924 | 8.138 | −3.14175538 | 0.00891286 | 0.090230763 | −2.6031971 |
| CTB-167B5 | −2.31293162 | 5.307 | −3.14420574 | 0.008873174 | 0.090230763 | −2.5989609 |
| FOLR2 | 1.192 | 7.817 | 3.138 | 0.008980412 | 0.090353121 | −2.61036421 |
| RAPGEF4 | −1.03668555 | 4.378 | −3.13513916 | 0.009020911 | 0.090543007 | −2.6146349 |
| LOC100127972 | −1.01216172 | 5.313 | −3.10834255 | 0.009472216 | 0.092466307 | −2.66095306 |
| COMP | −2.66774652 | 7.887 | −3.10283885 | 0.009567691 | 0.092947345 | −2.67046473 |
| RP11-690I21 | −1.02869854 | 6.001 | −3.10092157 | 0.009601178 | 0.09298905 | −2.67377809 |
| SBDS | −1.11130408 | 6.895 | −3.09837781 | 0.009645789 | 0.093140037 | −2.678174 |
| IL13RA2 | −1.84607884 | 5.783 | −3.08858504 | 0.009819488 | 0.093888012 | −2.69509576 |
| PLA2G15 | 1.04 | 9.387 | 3.086 | 0.009873995 | 0.094254348 | −2.70034338 |
| COL4A1 | −1.59461938 | 9.74 | −3.08497929 | 0.009884235 | 0.094301373 | −2.70132592 |
| MICU3 | −1.65121416 | 4.776 | −3.07900138 | 0.009992527 | 0.094778654 | −2.71165409 |
| MRGPRF | −1.32811473 | 6.587 | −3.07627303 | 0.010042348 | 0.09500344 | −2.71636764 |
| SHISA3 | −2.89115811 | 7.35 | −3.07619179 | 0.010043835 | 0.09500344 | −2.71650798 |
| SCG5 | −1.79294998 | 6.654 | −3.07435902 | 0.010077448 | 0.095134323 | −2.71967421 |
| ESF1 | −1.01991963 | 7.362 | −3.07194815 | 0.010121835 | 0.09529178 | −2.723839 |
| C1QB | 1.594 | 11.646 | 3.061 | 0.010327621 | 0.096081559 | −2.74290853 |
| PPAP2A | −1.18883309 | 8.397 | −3.0473476 | 0.010586162 | 0.09748556 | −2.76632741 |
| LOXL1 | −1.45575401 | 7.401 | −3.04001176 | 0.010728726 | 0.09786511 | −2.77899386 |
| EFNB2 | −1.7614713 | 6.572 | −3.03366254 | 0.010853673 | 0.098363256 | −2.78995536 |
| MFAP5 | −1.99102483 | 7.296 | −3.01954597 | 0.011136735 | 0.099381171 | −2.81432163 |
| PRPF39 | −1.08112002 | 7.339 | −3.01372934 | 0.011255515 | 0.099836985 | −2.82435944 |
| HLA-DRA | 1.05 | 12.472 | 3.008 | 0.011374911 | 0.100236116 | −2.83434154 |
| PALD1 | −1.13409746 | 6.356 | −3.00434742 | 0.011449785 | 0.100236116 | −2.84054718 |
| LIMS3-LOC440895 | −1.00361209 | 4.943 | −3.00361724 | 0.011465045 | 0.100236269 | −2.8418069 |
| SRGAP2C | −1.12202496 | 8.51 | −2.99612123 | 0.01162289 | 0.100589895 | −2.85473787 |
| LILRB5 | 1.058 | 7.406 | 2.992 | 0.011711091 | 0.100834918 | −2.861886 |
| GUCY1B3 | −1.09481873 | 6.043 | −2.98824718 | 0.011791042 | 0.101074761 | −2.86831842 |
| MAP1B | −1.26295569 | 7.014 | −2.98678584 | 0.011822517 | 0.10115834 | −2.87083853 |
| SLC14A1 | −2.08714663 | 5.023 | −2.97554416 | 0.012067475 | 0.102481695 | −2.8902218 |
| CAV1 | −1.54266666 | 9.273 | −2.97162638 | 0.012154033 | 0.102795272 | −2.89697559 |
| CAP2 | −1.3285113 | 6.031 | −2.97096325 | 0.012168746 | 0.10280373 | −2.89811867 |
| MCOLN1 | 1.463 | 8.13 | 2.969 | 0.01221883 | 0.102923357 | −2.90199944 |
| LUC7L3 | −1.05672069 | 8.768 | −2.96672843 | 0.012263123 | 0.103136633 | −2.90541804 |
| ZNF185 | 1.021 | 8.026 | 2.962 | 0.012375046 | 0.103527067 | −2.91400079 |
| GEM | −1.31254579 | 9.126 | −2.94762008 | 0.012698168 | 0.104648883 | −2.9383431 |
| G6PD | 1.013 | 7.245 | 2.944 | 0.01278875 | 0.104955796 | −2.94505403 |
| SMOC2 | −1.32742429 | 6.086 | −2.9416023 | 0.012838349 | 0.105122304 | −2.94870821 |
| UHRF1 | −1.19022326 | 6.062 | −2.93775939 | 0.012928677 | 0.105337267 | −2.95532626 |
| SMC3 | −1.25436837 | 6.196 | −2.93208304 | 0.013063263 | 0.105677166 | −2.96510026 |
| ABHD17A | 1.049 | 8.667 | 2.93 | 0.013104232 | 0.105861458 | −2.96805519 |
| ANKRD36BP2 | 1.259 | 4.886 | 2.911 | 0.01358772 | 0.107455605 | −3.00223375 |
| GOLGA8A | −1.33502678 | 7.411 | −2.90660944 | 0.013684694 | 0.107892663 | −3.00893968 |
| CCDC80 | −1.47135906 | 7.984 | −2.9019688 | 0.013801041 | 0.10811435 | −3.01692182 |
| GPR124 | −1.04704431 | 6.619 | −2.90075678 | 0.013831591 | 0.108178391 | −3.01900633 |
| GPRASP1 | −1.21408354 | 6.161 | −2.90013968 | 0.013847171 | 0.10821844 | −3.02006763 |
| CSRP2 | −1.0528362 | 7.237 | −2.88943576 | 0.014120222 | 0.108979328 | −3.03847238 |
| EDNRA | −1.11557607 | 5.042 | −2.87552836 | 0.01448303 | 0.110117829 | −3.06237382 |
| AJ420595 | −1.93827636 | 6.9 | −2.87065154 | 0.014612442 | 0.110360104 | −3.07075202 |
| PCED1B | 1.082 | 7.195 | 2.87 | 0.014631696 | 0.110385766 | −3.07199205 |
| FLJ45482 | −1.15599542 | 5.962 | −2.86957912 | 0.014641055 | 0.110385766 | −3.07259417 |
| TRIB2 | −1.33020834 | 6.242 | −2.86777486 | 0.014689318 | 0.110606956 | −3.07569325 |
| SUSD5 | −1.7300781 | 4.347 | −2.86337332 | 0.014807725 | 0.110957286 | −3.08325256 |
| CD300LF | 1.196 | 8.828 | 2.858 | 0.014956045 | 0.111461795 | −3.09263505 |
| RCAN2 | −2.3402395 | 7.039 | −2.85563237 | 0.015018278 | 0.111639705 | −3.09654358 |
| COX7A1 | −1.12472813 | 7.883 | −2.85246805 | 0.015105203 | 0.111993538 | −3.10197535 |
| ID4 | −1.25669749 | 5.511 | −2.84854904 | 0.015213555 | 0.112270462 | −3.10870155 |
| BD495725 | −1.14536337 | 6.277 | −2.84711125 | 0.015253501 | 0.112454719 | −3.11116894 |
| CYR61 | −1.64729732 | 9.399 | −2.8461505 | 0.015280251 | 0.11249641 | −3.11281758 |
| MMP19 | 1.16 | 8.042 | 2.844 | 0.015336492 | 0.112556839 | −3.11627422 |
| GCHFR | 1.108 | 8.674 | 2.845 | 0.015325977 | 0.112556839 | −3.11562898 |
| ACP5 | 1.066 | 11.566 | 2.842 | 0.015406513 | 0.112789619 | −3.12055977 |
| MME | 1.103 | 6.469 | 2.838 | 0.015503553 | 0.113203814 | −3.12646624 |
| L3MBTL3 | −1.16706759 | 6.383 | −2.83821982 | 0.015502856 | 0.113203814 | −3.1264239 |
| PSENEN | 1.4 | 7.63 | 2.835 | 0.015600401 | 0.113510457 | −3.13232348 |
| ADORA3 | 1.506 | 8.249 | 2.807 | 0.01639745 | 0.115441619 | −3.17916393 |
| FHOD3 | −1.36684403 | 4.999 | −2.80469636 | 0.016480005 | 0.115626385 | −3.18388196 |
| SYNPO2 | −1.04598587 | 5.51 | −2.80004775 | 0.01662025 | 0.116096583 | −3.19184194 |
| IGLJ3 | 2.196 | 7.381 | 2.789 | 0.016943387 | 0.116649653 | −3.2099243 |
| INTU | −1.06120671 | 5.354 | −2.78933493 | 0.016947975 | 0.116649653 | −3.21017851 |
| TTC14 | −1.02237882 | 7.064 | −2.78734066 | 0.017009688 | 0.116668823 | −3.21359085 |
| SGCE | −1.54727914 | 5.697 | −2.78702188 | 0.017019573 | 0.116668823 | −3.21413627 |
| ALDH1A1 | 1.366 | 9.167 | 2.775 | 0.01740769 | 0.11764985 | −3.23529852 |
| CWC27 | −1.03998318 | 7.371 | −2.77446547 | 0.017413512 | 0.11764985 | −3.23561225 |
| PCDHB10 | −1.2483677 | 4.317 | −2.77218427 | 0.017486046 | 0.11771822 | −3.23951233 |
| COL4A2 | −1.23543218 | 8.964 | −2.75287084 | 0.018112264 | 0.1194927 | −3.2725113 |
| JAM2 | −1.57791951 | 5.778 | −2.75027808 | 0.018198006 | 0.119618316 | −3.27693846 |
| MS4A4A | 1.212 | 10.225 | 2.74 | 0.018540224 | 0.120631263 | −3.29439839 |
| TNFSF4 | −1.3513573 | 5.39 | −2.73276259 | 0.018787856 | 0.121184544 | −3.30682809 |
| GHR | −1.27758998 | 4.535 | −2.71595095 | 0.019371777 | 0.122391782 | −3.33548585 |
| GRAMD3 | −1.12266645 | 5.579 | −2.70354869 | 0.019814009 | 0.123078962 | −3.35660713 |
| TMEM178A | −1.10816793 | 4.327 | −2.68917108 | 0.020339164 | 0.124880621 | −3.38107034 |
| LRRC32 | −1.29488862 | 7.886 | −2.68879414 | 0.020353115 | 0.124930459 | −3.38171138 |
| BRE-AS1 | 1.21 | 5.413 | 2.68 | 0.020664605 | 0.125584268 | −3.39590751 |
| CRYL1 | 1.018 | 9.3 | 2.673 | 0.020957562 | 0.126506082 | −3.40906024 |
| LOC101930415 | −1.13138302 | 7.538 | −2.6697484 | 0.021070457 | 0.126805168 | −3.41407856 |
| ITGA1 | −1.43419542 | 5.197 | −2.66572768 | 0.021225051 | 0.127197331 | −3.42090588 |
| FHL2 | −1.36563838 | 8.325 | −2.65435513 | 0.021668391 | 0.128432655 | −3.44020578 |
| LOC100134445 | −1.11977348 | 8.329 | −2.64334506 | 0.022106281 | 0.129903695 | −3.45887473 |
| CYB561A3 | 1.151 | 9.832 | 2.642 | 0.02215034 | 0.129947681 | −3.46073208 |
| SNORD50A | −1.04346702 | 6.105 | −2.63943635 | 0.022263823 | 0.13022485 | −3.46549862 |
| ZNF367 | −1.03406552 | 5.621 | −2.63477152 | 0.022453286 | 0.130646243 | −3.4734012 |
| SCG2 | −2.46514311 | 5.894 | −2.62786288 | 0.022736797 | 0.131191602 | −3.48509962 |
| PLN | −1.77310428 | 4.955 | −2.62374641 | 0.022907396 | 0.131588944 | −3.49206697 |
| LINC00622 | −1.29760971 | 4.816 | −2.62204858 | 0.022978124 | 0.131800122 | −3.49493996 |
| CCDC3 | −1.14629604 | 8.171 | −2.61901746 | 0.02310493 | 0.131984439 | −3.50006811 |
| BDP1 | −1.02156787 | 6.396 | −2.60396943 | 0.023744702 | 0.133564668 | −3.52550781 |
| ATP6V0D2 | 1.299 | 7.947 | 2.603 | 0.023769522 | 0.133639702 | −3.52648047 |
| CDC7 | −1.2299785 | 5.748 | −2.59773007 | 0.024015042 | 0.134500408 | −3.53604642 |
| LOC100507311 | −1.1014119 | 4.723 | −2.59494021 | 0.024136896 | 0.134638546 | −3.54075683 |
| C2orf40 | −2.00780375 | 5.604 | −2.58958986 | 0.024372284 | 0.134949872 | −3.54978718 |
| EFEMP2 | −1.01495407 | 8.312 | −2.57717436 | 0.024927207 | 0.13607391 | −3.57072584 |
| SGCG | −1.23332282 | 5.997 | −2.57103843 | 0.02520601 | 0.136733928 | −3.58106551 |
| EFHD1 | −1.39488703 | 6.468 | −2.57032983 | 0.025238403 | 0.136745109 | −3.58225919 |
| KMO | 1.092 | 8.409 | 2.57 | 0.02527123 | 0.136797207 | −3.58346726 |
| IGFBP6 | −1.09339489 | 8.386 | −2.563997 | 0.025529721 | 0.137399954 | −3.59292392 |
| HSPA2 | −1.16363559 | 6.492 | −2.5622374 | 0.025611248 | 0.137645677 | −3.59588606 |
| INHBB | 1.075 | 5.453 | 2.559 | 0.025760526 | 0.137945413 | −3.60128476 |
| GYPC | 1.031 | 8.17 | 2.558 | 0.025791692 | 0.13801015 | −3.60240783 |
| RGS4 | −1.0175785 | 6.26 | −2.55787658 | 0.025814394 | 0.138036455 | −3.60322504 |
| CADPS2 | −1.19152589 | 6.48 | −2.55757742 | 0.025828388 | 0.13807957 | −3.6037284 |
| RARRES1 | 1.253 | 9.202 | 2.55 | 0.026188027 | 0.139012687 | −3.61656913 |
| AQP9 | 1.41 | 9.605 | 2.545 | 0.026432749 | 0.139674824 | −3.62520339 |
| LOC389834 | −1.2184531 | 7.174 | −2.54446388 | 0.026449147 | 0.139698017 | −3.62577899 |
| TMEM45A | −1.20152293 | 8.765 | −2.53745321 | 0.026786972 | 0.14073182 | −3.63755621 |
| GPR183 | 1.037 | 9.969 | 2.533 | 0.026986459 | 0.141112074 | −3.64443905 |
| LINC00619 | −1.0745705 | 3.743 | −2.52008766 | 0.027642021 | 0.14272822 | −3.6666939 |
| SPRY4 | −1.15340389 | 7.363 | −2.51484388 | 0.027905412 | 0.143207217 | −3.67548254 |
| FAM20A | 1.074 | 7.481 | 2.514 | 0.027932064 | 0.143209492 | −3.67636706 |
| PKIB | −1.69038403 | 6.538 | −2.50121819 | 0.028601315 | 0.145050097 | −3.69829742 |
| PPP1R14A | −1.94935478 | 6.21 | −2.49821224 | 0.028757099 | 0.145327282 | −3.70332625 |
| DNASE2B | 1.139 | 6.444 | 2.486 | 0.029411775 | 0.146772313 | −3.72415661 |
| VMP1 | 1.562 | 9.523 | 2.483 | 0.029580001 | 0.147133627 | −3.7294319 |
| IGJ | 3.104 | 8.864 | 2.482 | 0.029625818 | 0.147193406 | −3.73086328 |
| PLOD2 | −1.16827633 | 8.908 | −2.47499807 | 0.029988334 | 0.14802969 | −3.74210872 |
| LOC101927069 | −1.60316052 | 5.125 | −2.47180935 | 0.030161414 | 0.148104476 | −3.74742831 |
| SPARCL1 | −1.88778385 | 9.426 | −2.47061628 | 0.030226422 | 0.148241776 | −3.74941817 |
| GPX3 | 1.22 | 10.365 | 2.463 | 0.030651314 | 0.149356193 | −3.76231603 |
| PRICKLE2 | −1.08501938 | 4.945 | −2.45977406 | 0.030823469 | 0.149757159 | −3.76748936 |
| THBS1 | −1.00450902 | 6.406 | −2.45649882 | 0.031006068 | 0.150173348 | −3.77294402 |
| PPP1R3C | −1.10842436 | 5.881 | −2.45049465 | 0.031343543 | 0.15083513 | −3.78293825 |
| CTGF | −1.68043237 | 10.245 | −2.44316364 | 0.031760431 | 0.151849734 | −3.79513177 |
| PLGLB1 | −1.04046038 | 5.02 | −2.44033293 | 0.031922839 | 0.152238333 | −3.79983726 |
| ZNF521 | −1.25923381 | 4.897 | −2.43804712 | 0.032054572 | 0.152398313 | −3.80363583 |
| PTPRZ1 | −1.04325 | 4.2 | −2.42396632 | 0.032877759 | 0.154295501 | −3.82701284 |
| GPM6B | −1.15925929 | 4.968 | −2.41770644 | 0.033250266 | 0.155136811 | −3.8373929 |
| AK021977 | −1.05674346 | 5.415 | −2.40910828 | 0.033768584 | 0.156085006 | −3.85163739 |
| OLFML2A | −1.13887278 | 6.219 | −2.39552362 | 0.034603473 | 0.157597428 | −3.87411218 |
| MANSC1 | −1.20061235 | 5.162 | −2.37742265 | 0.035746982 | 0.159649267 | −3.90399891 |
| CX3CL1 | −1.20643257 | 6.702 | −2.37411628 | 0.035959764 | 0.160110005 | −3.90945057 |
| MEOX1 | −1.3648004 | 5.619 | −2.37259553 | 0.036058041 | 0.160155144 | −3.91195724 |
| FIGN | −1.16179673 | 4.34 | −2.37020584 | 0.036212997 | 0.160409247 | −3.91589518 |
| F3 | −1.2706323 | 7.831 | −2.36731755 | 0.036401141 | 0.160690719 | −3.9206531 |
| AC012065 | −1.36960739 | 7.269 | −2.36455566 | 0.036581934 | 0.161096543 | −3.92520112 |
| HSP90B1 | −1.0931333 | 6.992 | −2.35529786 | 0.037194283 | 0.162507108 | −3.94043371 |
| FAM46C | 1.128 | 6.681 | 2.331 | 0.038840666 | 0.164775579 | −3.98013075 |
| DYSF | 1.147 | 8.407 | 2.325 | 0.039302486 | 0.16534884 | −3.99095213 |
| AKR1C3 | −1.37012153 | 8.401 | −2.3184279 | 0.039732384 | 0.166435652 | −4.00090724 |
| LOC100506076 | −1.03285843 | 6.345 | −2.30883743 | 0.040419389 | 0.167696452 | −4.01658575 |
| AK5 | −1.13886628 | 5.649 | −2.30470529 | 0.040718893 | 0.167998808 | −4.02333424 |
| BC017398 | −1.18816092 | 4.847 | −2.29570776 | 0.041378431 | 0.169190839 | −4.03801454 |
| SRGAP2B | −1.49183273 | 7.338 | −2.28478755 | 0.042192668 | 0.170586274 | −4.05580544 |
| LOC100130872 | 1.199 | 7.965 | 2.283 | 0.042330561 | 0.170715418 | −4.0587829 |
| PDGFRB | −1.21137854 | 7.133 | −2.26745861 | 0.043516326 | 0.172980554 | −4.08397666 |
| CRYAB | −1.00937569 | 7.311 | −2.26452563 | 0.043744255 | 0.173371994 | −4.08873726 |
| IBSP | −1.27075954 | 8.743 | −2.23484703 | 0.046115718 | 0.176965926 | −4.13678496 |
| JPX | −1.01960017 | 4.715 | −2.22531267 | 0.04690324 | 0.178402425 | −4.15217142 |
| IFIT2 | −1.27238246 | 7.817 | −2.19997112 | 0.04905913 | 0.181687766 | −4.19294792 |
Fig. 1.
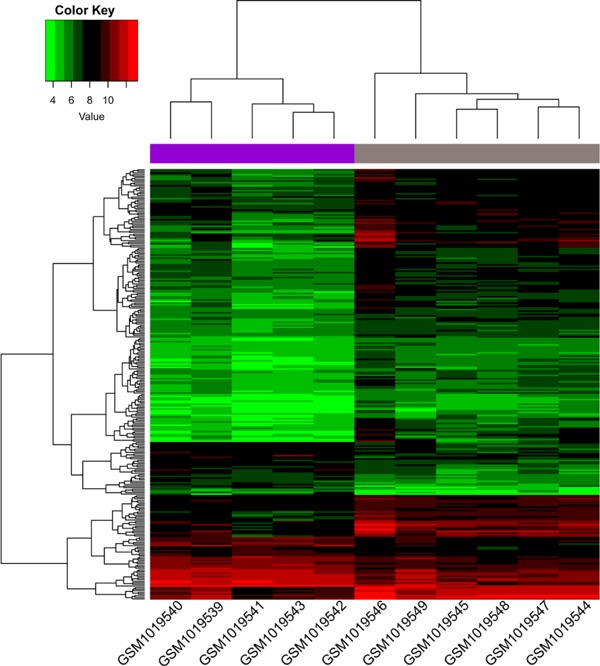
Clustered heat map of DEGs. The abscissa represents different samples, and the ordinate represents different genes. The red boxes indicate up-regulated genes, and the green boxes indicate down-regulated genes.
DEGs: Differentially Expressed Genes.
Functional Enrichment Analysis
GO functional and KEGG pathway analyses revealed that the up-regulated DEGs were significantly enriched in GO BP terms of the ‘immune response’ (p-value = 4.82E-05), ‘inflammatory response’ (p-value = 0.001192) in GO BP, and ‘Lysosome’ pathway (pvalue = 1.08E-04) and ‘Cytokine-cytokine receptor interaction pathways’ (Table 1). The down-regulated DEGs identified in ruptured atherosclerotic plaque were enriched in GO_BP terms of ‘extracellular matrix organization’ (p-value = 1.44E-08) and ‘cell adhesion’ (p-value = 3.32E-15), and the KEGG pathways of ‘focal adhesion’ (p-value = 7.82E-06) and ‘ECM-receptor interaction’ (Table 2).
Table 1. The top five results of functional enrichment analyses of up-regulated DEGs ranked by P-value.
| Term | Count | Gene | P-value |
|---|---|---|---|
| GO-BPs | |||
| GO:0006955∼immune response | 9 | PPBP, IGKV1-17, CXCL3, NCF4, FCGR3A, IGKC, HLA-DMA, IGLC1, CD27 | 4.82E-05 |
| GO:0050853∼B cell receptor signaling pathway | 4 | CD38, IGKC, IGHM, IGLC1 | 6.57E-04 |
| GO:0038096∼Fc-gamma receptor signaling pathway involved in phagocytosis | 5 | IGKV1-17, HCK, FCGR3A, IGKC, IGLC1 | 6.86E-04 |
| GO:0006954∼inflammatory response | 7 | PPBP, CXCL3, HCK, S100A9, MGLL, CD27, CD180 | 0.001192 |
| GO:0050776∼regulation of immune response | 5 | LAIR1, IGKV1-17, FCGR3A, IGKC, IGLC1 | 0.002385 |
| KEGG pathways | |||
| hsa04142:Lysosome | 6 | AP1B1, GLA, GAA, NEU1, CTSA, ATP6V0B | 1.08E-04 |
| hsa04060:Cytokine-cytokine receptor interaction | 5 | LEP, PPBP, IL10RA, TNFRSF17, CD27 | 0.013065 |
| hsa04145:Phagosome | 4 | NCF4, FCGR3A, HLA-DMA, ATP6V0B | 0.023255 |
| hsa03320:PPAR signaling pathway | 3 | CYP27A1, FABP4, PLTP | 0.02977 |
| hsa05140:Leishmaniasis | 3 | NCF4, FCGR3A, HLA-DMA | 0.033129 |
GO, Gene Ontology; BP, biological process; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes.
Table 2. The top five results of functional enrichment analyses of down-regulated DEGs ranked by P-value.
| Term | Count | Gene | P-value |
|---|---|---|---|
| GO-BPs | |||
| GO:0007155∼cell adhesion | 30 | CTNNAL1, IGFBP7, ITGA10, SPOCK1, POSTN, CDH2, ITGBL1, ISLR, VCAM1, WISP1, ROBO1, FAT1, FAP, COL8A1, LAMB1, THBS2, DPT, PTPRK, MPDZ, PODXL, COL15A1, PPFIBP1, SSPN, TPBG, TNFAIP6, OMD, CX3CR1, MYH10, CDH11, AOC3 | 3.32E-15 |
| GO:0030198∼extracellular matrix organization | 15 | PDGFA, LUM, COL3A1, FBN1, ITGA10, POSTN, SPARC, ECM2, COL5A2, VCAM1, EGFLAM, BGN, COL1A2, LAMB1, COL8A1 | 1.44E-08 |
| GO:0030199∼collagen fibril organization | 7 | SFRP2, LUM, COL3A1, COL1A2, FOXC1, COL5A2, DPT | 2.61E-06 |
| GO:0001501∼skeletal system development | 10 | AEBP1, COL3A1, FBN1, COL1A2, POSTN, FOXC1, EXT1, COL5A2, PRELP, CDH11 | 1.17E-05 |
| GO:0001503∼ossification | 8 | MGP, LRRC17, FOXC1, SPARC, EXT1, COL5A2, CDH11, TWIST1 | 1.77E-05 |
| KEGG pathways | |||
| hsa04510:Focal adhesion | 11 | VEGFC, CAV2, CCND1, PDGFA, COL3A1, COL1A2, ITGA10, LAMB1, THBS2, COL5A2, MYLK | 7.82E-06 |
| hsa04611:Platelet activation | 7 | GNAI1, COL3A1, COL1A2, GUCY1A3, COL5A2, MYLK, F2R | 7.86E-04 |
| hsa04512:ECM-receptor interaction | 6 | COL3A1, COL1A2, ITGA10, LAMB1, THBS2, COL5A2 | 8.27E-04 |
| hsa04151:PI3K-Akt signaling pathway | 10 | VEGFC, CCND1, PDGFA, COL3A1, COL1A2, ITGA10, LAMB1, THBS2, COL5A2, F2R | 0.002384 |
| hsa04974:Protein digestion and absorption | 5 | COL21A1, COL3A1, COL1A2, COL15A1, COL5A2 | 0.00656 |
GO, Gene Ontology; BP, biological process; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes.
PPI Network Analysis
A total of 154 nodes and 322 interactional pairs were included in the PPI network. Among them, only eight genes with degrees ≥ 15 in the PPI network were found, including type III alpha 1 chain (COL3A1), collagen type I alpha 2 chain (COL1A2), LUM, ACTA2, SPARC, ASPN, GNAI1 and COL5A2. Further, three functional subnet modules (mcode 1, mcode 2, and mcode 3) were selected from the PPI network. Specifically, mcode 1 consisted of 1 up-regulated node (LCP1) and 6 down-regulated nodes (Fig. 2A), mcode 2 was made up of five down-regulated nodes (e.g. COL3A1, COL1A2 and COL5A2) (Fig. 2B), and mcode 3 consisted of twelve down-regulated nodes (e.g. ASPN) (Fig. 2C). In total, 5 nodes (e.g. COL3A1, COL1A2, COL5A2) of mcode 2 were mainly enriched in the “ECM-receptor interaction” (hsa04512) pathway, and nodes in mcode 3 was only enriched in the “Protein digestion and absorption” (hsa04974). Whereas, no pathway related to the nodes in mcode 1 was enriched.
Fig. 2.
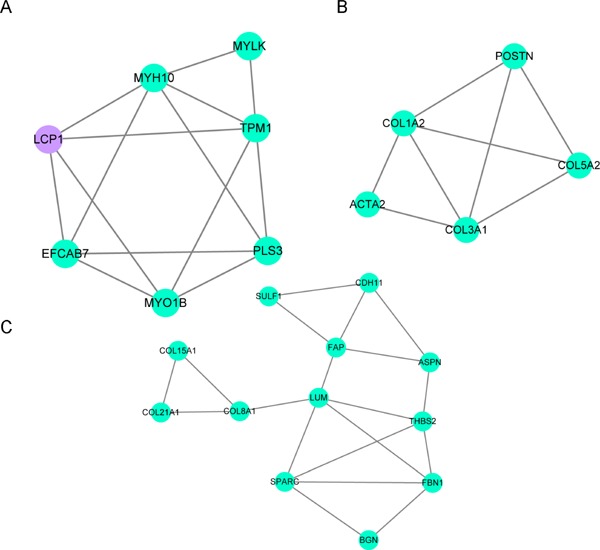
Results of subnet module analysis of the PPI network. A: mcode 1; B: mcode 2; C: mcode 3. The green circles represent down-regulated DEG-encoded proteins and the purple circles represent upregulated DEG-encoded proteins.
PPI: Protein-Protein Interaction; DEG, differentially expressed gene.
Analysis of miRNA-Target and TF-Target Regulating Networks
In the miRNA-target regulating network, 114 nodes were detected, including 100 DEGs (e.g. COL3A1, ASPN) and 14 miRNAs (e.g. miR21), and miR21 was highlighted with highest degrees. Among these nodes involved in the PPI network, 310 regulatory relationships were identified between miRNAs and down-regulated DEGs (e.g. miR21-ASPN), but no regulatory relationship was observed between miRNA and the up-regulated DEGs (Fig. 3). One TF (TEA Domain Transcription Factor 2; TEAD2) was predicted to target 25 down-regulated DEGs (e.g. CCND1, BGN and COL8A1), and one TF (Spi-1 Proto-Oncogene; SFPI1) was predicted to regulate 10 up-regulated DEGs (e.g., PPBP) (Fig. 4).
Fig. 3.
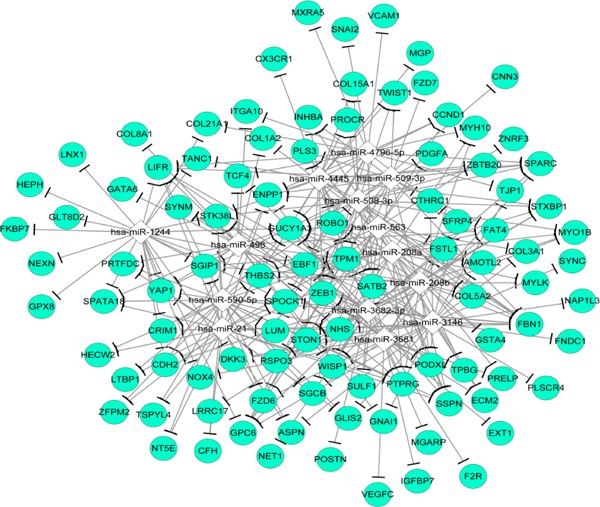
MiR-target DEGs regulatory network. The diamond represents miRNA, and the green circle represents down-regulated DEGs.
DEGs: Differentially Expressed Genes
Fig. 4.
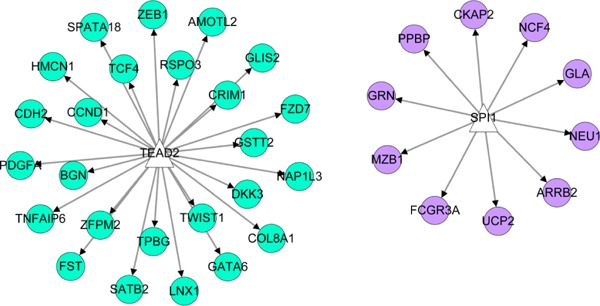
TF-target DEGs regulatory network. The triangle represents TF, and the green circle and purple circle represent down-regulated and up-regulated DEGs, respectively.
TF: Transcription Factor; DEGs: Differentially Expressed Genes
Discussion
In the present study, 268 DEGs were identified in ruptured samples compared with stable samples. Specifically, COL3A1 and COL1A2 were important downregulated genes, and PPBP was the most significant up-regulated gene in ruptured atherosclerotic samples. With a high degree in the PPI network, ASPN could be targeted by miR-21. Both COL3A1 and COL1A2 were enriched in the extracellular matrix organization and cell adhesion biological processes. PPBP was mostly enriched in immune and inflammatory response-related biological processes.
According to previous reports, balance between macrophage and fibrosis performs a critical role in the stability of atherosclerotic plaques22, 23). Ma et al. have demonstrated that deletion of matrix metalloproteinase (MMP)-28 in the infarct region induces a reduction in the expression of COL1A1, COL3A1, COL5A1, and MMP-9, which are closely associated with the progression of fibrosis24). Among these genes, COL3A1 encodes the pre-α1 chain of type III collagen, which has been identified to be related to the thickened intima of atherosclerotic lesions25). Mutation of the COL3A1 gene is known to cause Ehlers-Danlos syndrome type IV (including rupture and stroke), which is related to early-onset occurrence of arterial ruptures26). Variants of COL3A1 are risk factors for coronary and the recurrence of coronary events and artery disease in an Irish population27). The decrease in transcriptional activity of COL3A1 results in reduced stability, rupture, and re-occlusion of small vessel plaques by reducing the production of type III collagen28). COL1A2 is the encoding gene of the pro-alpha 2 chain of type I collagen, which is a collagen to form fibril. Type I collagen is the main component of the vessel wall and plays a key role in disease progression29). The content of collagen in atherosclerotic plaques and structural integrity of the fibrous cap are signs of plaque stability30). An unstable plaque that is prone to rupture is characterized by less collagen expression, a thinner fibrous cap, and a larger necrotic core30). In the current study, COL3A1 and COL1A2 were identified to be down-regulated in the macrophage-enriched rupture samples and interacted with each other. In coronary atherosclerosis, macrophages are reported to have cross talk between smooth muscle cells (SMCs), inhibiting the synthesis of MMPs and collagen proteins. This might be the potential explanation for the down-regulations of collagen related genes in rupture samples, but the in-depth mechanism for this crosstalk remains unclear. Improvement of plaque extracellular matrix (ECM) can enhance plaque morphology and stabilize lesions, indicating that change of ECM may be a potential new approach to improve atherosclerotic lesions31–33). In the current study, COL3A1 and COL1A2 were obviously enriched in the extracellular matrix organization and collagen fibril organization biological processes and focal adhesion and ECM-receptor interaction pathways. These findings indicated that down-regulation of COL3A1 and COL1A2 might promote rupture via altering ECM components.
In the present study, among the 14 predicted miRNAs, miR21 had the most targeted-DEGs in the miRNA-target regulatory network. miR21 is implicated to play an important role in progression of various diseases including cancer, osteoarthritis, cardiovascular diseases and inflammation34). Liang et al have highlighted that the proatherosclerotic genes COX2, VCAM1, ICAM1 and MCP1 are regulated by miRNA-21 in macrophages of atherosclerotic plaques, indicating the importance of miRNA-21 in the development of atherosclerotic plaques35). It also reports that miR-21 may promote inflammation underlying atherogenesis in VSMCs and macrophages36). In our study, ASPN was significantly down-regulated in macrophage-enriched rupture atherosclerotic plaques samples and predicted to be a target of miRNA-21. Asporin (ASPN) as a member of the small leucine-rich proteoglycan family, is a major non-collagen components of the ECM, and it has a possible role in calcification of collagen in atherosclerotic plaques37), speculating that the low expression level of ASPN may reduce the calcification of collagen in atherosclerotic plaques. Interestingly, it has been demonstrated that unstable (known as noncalcified) plaques undergo thinning of the fibrous cap prior to rupture, possibly as a result of macrophages releasing proteolytic matrix-degrading enzymes which may degrade the fibrous cap38). What' more, another excellent study have supported that level of miR-21 in non-calcified coronary plaque patients' macrophages was higher than that in the coronary calcified groups, uncovering that miR-21 overexpression can enhance the instability of coronary atherosclerotic plaque in macrophages39). In consistent with our predicted regulatory pair, wang et al have suggested that ASPN may be regulated by miR-21 in the renal cortex of hypertensive patients40). Thus, it was speculated that miR-21 might play a role in calcification of collagen in macrophages of atherosclerotic plaques via regulating ASPN.
Pre-platelet basic protein (PPBP), also known as chemokine (Cxc Motif ) Ligand 7 (CXCL7), is a platelet-derived growth factor that belongs to the CXC chemokine family. Chemokines play an important role on atherosclerosis development through attracting circulating inflammatory cells and stem/progenitor cells41). PPBP in connection with CTAP-IIII and CXCL4 regulate neutrophil adhesion and transendothelial migration42). As reported, monocyte-derived CXCL7 can promote macrophages chemotaxis into the tumor microenvironment43). CXCL4, PPBP structurally related gene, may affect the differentiation of monocytes and induces a specific M4 macrophage phenotype, which is associate with plaque instability44). In addition, CXCL1, an important paralog of PPBP, is up-regulated in carotid atherosclerotic plaques45). Similarly, our result revealed that PPBP was significantly up-regulated in ruptured atherosclerotic plaque samples, and among all up-regulated DEGs, its express change was greatest. Moreover, PPBP was significantly enriched in the immune and inflammatory response-related biological processes, which might be owing to the increased accumulation of macrophages. Therefore, it is deduced that accumulated macrophages might promote the expression of PPBP, increasing the destabilization of the atherosclerotic plaque.
However, some limitations should also be strengthened in this study. First, all the findings contained in this study were obtained using in silico methods, and the results experimental verifications were still deficient owing to insufficient tissue samples. Second, only small samples were included in this dataset, which might affect the values of these findings, or some other information might be ignored during these processes. Thus, a further analysis, combined experimental confirmation, and a larger sample size are required in future investigations.
Conclusions
In conclusion, findings in the current study demonstrated that the rupture of atherosclerotic plaques might be the result of imbalance between macrophages and fibrosis. Crosstalk between macrophages and SMCs may be involved in this process. Specifically, downregulated COL3A1, COL1A2 and ASPN, and up-regulated PPBP performed important promotional effects for the rupture of atherosclerotic plaques. Moreover, miR-21 was the potential up-stream regulators for ASPN and could serve as underlying targets for AS rupture prevention. These findings provide a theoretical basis for the future study of specific mechanisms of atherosclerotic rupture.
Acknowledgments
Not applicable.
List of Abbreviations
atherosclerosis (AS); differentially expressed genes (DEGs); Gene Ontology biological process (GO_BP); Kyoto Encyclopedia of Genes and Genomes (KEGG); protein-protein interactions (PPIs); transcriptional factors (TFs); collagen type III alpha 1 chain (COL3A1); collagen type I alpha 2 chain (COL1A2); Asporin (ASPN); Pre-platelet basic protein (PPBP); TEA Domain Transcription Factor 2 (TEAD2); Spi-1 Proto-Oncogene (SFPI1); microRNAs (miRNAs); Gene Expression Omnibus (GEO); Benjamin Hochberg (BH); fold change (FC); Gene Ontology (GO); biological processes (BPs); Search tool for the retrieval of interacting genes/proteins (STRING); Multi-Contrast Delayed Enhancement (MCODE); matrix metalloproteinase (MMP); extracellular matrix (ECM); smooth muscle cells (SMCs); lipoprotein receptor-related protein 6 (LRP6)
Declarations
Ethics Approval and Consent to Participate
Construction of this dataset was authorized by the Local Research Ethics Committee.
Consent for Publication
Informed consents of all the patients were obtained.
Availability of Data and Materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare that they have no competing interests.
Funding
Not applicable.
Authors' Contributions
Conception and design of the research and drafting the manuscript: Hao Wang; acquisition of data, analysis and interpretation of data and statistical analysis: Dongyuan Liu; drafting the manuscript and revision of manuscript for important intellectual content: Hongbing Zhang.
References
- 1). Stefanadis C, Antoniou CK, Tsiachris D, Pietri P: Coronary Atherosclerotic Vulnerable Plaque: Current Perspectives. J Am Heart Assoc, 2017; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2). Virmani R, Kolodgie FD, Burke AP, Farb A, Schwartz SM: Lessons from sudden coronary death: a comprehensive morphological classification scheme for atherosclerotic lesions. Arterioscler Thromb Vasc Biol, 2000; 20: 1262-1275 [DOI] [PubMed] [Google Scholar]
- 3). Lee K, Santibanez-Koref M, Polvikoski T, Birchall D, Mendelow AD, Keavney B: Increased expression of fatty acid binding protein 4 and leptin in resident macrophages characterises atherosclerotic plaque rupture. Atherosclerosis, 2013; 226: 74-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4). Cochain C, Zernecke A: Macrophages in vascular inflammation and atherosclerosis. Pflugers Arch, 2017; 469: 485-499 [DOI] [PubMed] [Google Scholar]
- 5). Dahl TB, Yndestad A, Skjelland M, E Ø, Dahl A, Michelsen A, Damås JK, Tunheim SH, Ueland T, Smith C: Increased expression of visfatin in macrophages of human unstable carotid and coronary atherosclerosis: possible role in inflammation and plaque destabilization. Circulation, 2007; 115: 972. [DOI] [PubMed] [Google Scholar]
- 6). Cong X, He Z, Cui H, Yang J, Huang Z, Li N, Chen W, Wang Q, Ren Y, Liang C: PRDM16 inhibits atherosclerosis plaque forming through enhancing the function of periaortic brown adipose tissue. 2016
- 7). Lutgens E, Gijbels M, Smook M, Heeringa P, Gotwals P, Koteliansky VE, Daemen MJ: Transforming growth factor-beta mediates balance between inflammation and fibrosis during plaque progression. Arterioscler Thromb Vasc Biol, 2002; 22: 975. [DOI] [PubMed] [Google Scholar]
- 8). Shioi A, Ikari Y: Plaque Calcification During Atherosclerosis Progression and Regression. J Atheroscler Thromb, 2018; 25: 294-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9). Gautier L, Cope L, Bolstad BM, Irizarry RA: affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics, 2004; 20: 307-315 [DOI] [PubMed] [Google Scholar]
- 10). Bolstad BM, Irizarry RA, Astrand M, Speed TP: A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics, 2003; 19: 185-193 [DOI] [PubMed] [Google Scholar]
- 11). Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP: Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 2003; 4: 249-264 [DOI] [PubMed] [Google Scholar]
- 12). Smyth GK, Limma: linear models for microarray data, In, Bioinformatics and computational biology solutions using R and Bioconductor, Springer, 2005: 397-420 [Google Scholar]
- 13). Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT: Gene Ontology: tool for the unification of biology. Nat Genet, 2000; 25: 25-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14). Kanehisa M, Goto S: KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res, 2000; 28: 27-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15). Da Wei Huang BTS, Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc, 2008; 4: 44-57 [DOI] [PubMed] [Google Scholar]
- 16). Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J: ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics, 2009; 25: 1091-1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17). Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T: Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res, 2003; 13: 2498-2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18). Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP: STRING v10: protein — protein interaction networks, integrated over the tree of life. Nucleic Acids Res, 2014: gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19). Bandettini WP, Kellman P, Mancini C, Booker OJ, Vasu S, Leung SW, Wilson JR, Shanbhag SM, Chen MY, Arai AE: MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson, 2012; 14: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20). Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A: Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics, 2013; 14: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21). Benjamini Y, Hochberg Y: Controlling The False Discovery Rate - A Practical And Powerful Approach To Multiple Testing. J R Stat Soc, 1995; 57: 289-300 [Google Scholar]
- 22). Chinettigbaguidi G, Colin S, Staels B: Macrophage subsets in atherosclerosis. Nat Rev Cardiol, 2015; 12: 10-17 [DOI] [PubMed] [Google Scholar]
- 23). Lutgens E, Lutgens SP, Faber BC, Heeneman S, Gijbels MM, de Winther MP, Frederik P, Van dMI, Daugherty A, Sijbers AM: Disruption of the cathepsin K gene reduces atherosclerosis progression and induces plaque fibrosis but accelerates macrophage foam cell formation. Circulation, 2006; 113: 98. [DOI] [PubMed] [Google Scholar]
- 24). Ma Y, Halade GV, Zhang J, Ramirez TA, Levin D, Voorhees A, Jin YF, Han HC, Manicone AM, Lindsey ML: Matrix metalloproteinase-28 deletion exacerbates cardiac dysfunction and rupture after myocardial infarction in mice by inhibiting M2 macrophage activation. Circ Res, 2013; 112: 675-688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25). Pickering JG, Ford CM, Chow LH: Evidence for rapid accumulation and persistently disordered architecture of fibrillar collagen in human coronary restenosis lesions. Am J Cardiol, 1996; 78: 633-637 [DOI] [PubMed] [Google Scholar]
- 26). Leistritz DF, Pepin MG, Schwarze U, Byers PH: COL3A1 haploinsufficiency results in a variety of Ehlers-Danlos syndrome type IV with delayed onset of complications and longer life expectancy. Genet Med, 2011; 13: 717. [DOI] [PubMed] [Google Scholar]
- 27). Muckian C, Fitzgerald A, O'Neill A, O'Byrne A, Fitzgerald DJ, Shields DC: Genetic variability in the extracellular matrix as a determinant of cardiovascular risk: association of type III collagen COL3A1 polymorphisms with coronary artery disease. Blood, 2002; 100: 1220-1223 [DOI] [PubMed] [Google Scholar]
- 28). Lv W, Lin Y, Song W, Sun K, Yu H, Zhang Y, Zhang C, Li L, Suo M, Hui R: Variants of COL3A1 are associated with the risk of stroke recurrence and prognosis in the Chinese population: a prospective study. J Mol Neurosci, 2014; 53: 196. [DOI] [PubMed] [Google Scholar]
- 29). Prockop DJ, Kivirikko KI: Collagens: molecular biology, diseases, and potentials for therapy. Annu Rev Biochem, 1995; 64: 403. [DOI] [PubMed] [Google Scholar]
- 30). Libby P, Ridker PM, Hansson GK: Progress and challenges in translating the biology of atherosclerosis. Nature, 2011; 473: 317-325 [DOI] [PubMed] [Google Scholar]
- 31). Zhang XY, Shen BR, Zhang YC, Wan XJ, Yao QP, Wu GL, Wang JY, Chen SG, Yan ZQ, Jiang ZL: Induction of Thoracic Aortic Remodeling by Endothelial-Specific Deletion of MicroRNA-21 in Mice. PLoS One, 2013; 8: e59002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32). Melo SF, Fernandes T, Baraúna VG, Matos KC, Santos AA, Tucci PJ, Oliveira EM: Expression of MicroRNA-29 and Collagen in Cardiac Muscle after Swimming Training in Myocardial-Infarcted Rats. Cell Physiol Biochem, 2014; 33: 657. [DOI] [PubMed] [Google Scholar]
- 33). Maegdefessel L, Azuma J, Toh R, Merk DR, Deng A, Chin JT, Raaz U, Schoelmerich AM, Raiesdana A, Leeper NJ: Inhibition of microRNA-29b reduces murine abdominal aortic aneurysm development. J Clin Invest, 2012; 122: 497-506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34). Kumarswamy R, Volkmann I, Thum T: Regulation and function of miRNA-21 in health and disease. RNA Biol, 2011; 8: 706-713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35). YY L, WW X, JN Z, QX L, X Z, CT Z, CY D, YH F, HH T, SJ G: miRNA-21 regulates proatherosclerotic gene expression in macrophages. South China J Cardiol, 2014 [Google Scholar]
- 36). Koroleva IA, Nazarenko MS, Kucher AN: Role of microRNA in Development of Instability of Atherosclerotic Plaques. Biochemistry, 2017; 82: 1380-1390 [DOI] [PubMed] [Google Scholar]
- 37). Kalamajski S, Oldberg A: The role of small leucinerich proteoglycans in collagen fibrillogenesis. Matrix Biol, 2010; 29: 248-253 [DOI] [PubMed] [Google Scholar]
- 38). Finn AV, Nakano M, Narula J, Kolodgie FD, Virmani R: Concept of Vulnerable/Unstable Plaque. Arterioscler Thromb Vasc Biol, 2010; 30: 1282-1292 [DOI] [PubMed] [Google Scholar]
- 39). Fan X, Gu C, Wang X, Wang F, Yang J, Cai L, Cong X, Chen X: miRNA-16/21 as a biomarker for coronary atherosclerotic unstable plaques. Cardiology, 2013; 126: 55-55 [Google Scholar]
- 40). Wang G, Wu L, Chen Z, Sun J: Identification of crucial miRNAs and the targets in renal cortex of hypertensive patients by expression profiles. Ren Fail, 2016; 39: 1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41). Duchene J, Von HP: Platelet-derived chemokines in atherosclerosis. Hämostaseologie, 2015; 35: 137-141 [DOI] [PubMed] [Google Scholar]
- 42). Schenk BI, Petersen F, Flad HD, Brandt E: Plateletderived chemokines CXC chemokine ligand (CXCL)7, connective tissue-activating peptide III, and CXCL4 differentially affect and cross-regulate neutrophil adhesion and transendothelial migration. J Immunol, 2002; 169: 2602. [DOI] [PubMed] [Google Scholar]
- 43). Pillai MM, Iwata M, Awaya N, Graf L, Torokstorb B: Monocyte-derived CXCL7 peptides in the marrow microenvironment. Blood, 2006; 107: 3520-3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44). Domschke G, Gleissner CA: CXCL4-induced macrophages in human atherosclerosis. Cytokine, 2017 [DOI] [PubMed] [Google Scholar]
- 45). Copin JC, Silva RFD, Fragasilva RA, Capettini L, Quintao S, Lenglet S, Pelli G, Galan K, Burger F, Braunersreuther V: Treatment with Evasin-3 reduces atherosclerotic vulnerability for ischemic stroke, but not brain injury in mice. J Cereb Blood Flow Metab, 2013; 33: 490-498 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


