Graphical abstract
Keywords: Holobiont, Diazotrophic plant beneficial bacteria, Azospirillum, Opportunistic human pathogens, Metagenome and transcriptome analyses, N-acyl-homoserine lactones
Highlights
-
•
History about the discovery of endophytes with the focus on Azospirillum and related diazotrophs.
-
•
Contribution of approaches to reach highest resolution of microbial diversity assessment.
-
•
Differentiation of beneficial A. brasilense and opportunistic human pathogen R. fauriae.
-
•
Osmoadaption and oxygen tolerance as major traits for endophytic bacteria.
-
•
Bacteria-plant communication with focus on bacterial N-acyl homoserine lactones.
Abstract
Analyses of the spatial localization and the functions of bacteria in host plant habitats through in situ identification by immunological and molecular genetic techniques combined with high resolving microscopic tools and 3D-image analysis contributed substantially to a better understanding of the functional interplay of the microbiota in plants. Among the molecular genetic methods, 16S-rRNA genes were of central importance to reconstruct the phylogeny of newly isolated bacteria and to localize them in situ. However, they usually do not allow resolution for phylogenetic affiliations below genus level. Especially, the separation of opportunistic human pathogens from plant beneficial strains, currently allocated to the same species, needs genome-based resolving techniques. Whole bacterial genome sequences allow to discriminate phylogenetically closely related strains. In addition, complete genome sequences enable strain-specific monitoring for biotechnologically relevant strains. In this mini-review we present high resolving approaches for analysis of the composition and key functions of plant microbiota, focusing on interactions of diazotrophic plant growth promoting bacteria, like Azospirillum brasilense, with non-legume host plants. Combining high resolving microscopic analyses with specific immunological detection methods and molecular genetic tools, including especially transcriptome analyses of both the bacterial and plant partners, enables new insights into key traits of beneficial bacteria-plant interactions in holobiontic systems.
Introduction and historical aspects of the discovery of endophytes with focus on Azospirillum and related diazotrophs
More than one decade ago, the hologenome theory was introduced to express the tight interaction of microbes with animals and plants as basis for a better adaptation to changing environmental conditions with implications for co-evolution and speciation [1]. Holobionts are multicellular eukaryotic organisms living together in a symbiont-like manner with different types of external and internal microorganism (e.g. endophytes), which contribute essential life traits [2], [3]. A more recent study concludes, that in order to understand speciation in the frame of the hologenome concept holobionts do not necessarily need to be viewed as units of selection, but it is sufficient to consider them as units of tight co-operation of eu- and prokaryotic organisms [4]. Looking back, it took a long time until this detailed view of omnipresent organismic interactions was established by firm evidence, because the appropriate methodological approaches had not been available. First evidences for bacterial endophytes, i.e. bacteria colonizing the interior of plants, were already published in the late 19th century. In 1887, M. L. V. Galippe reported the isolation of bacteria from the interior of different plants and postulated soil as origin of these bacteria [5]. Since he could not further prove their location and identity, these findings were heavily criticized. However, Hellriegel and Wilfarth demonstrated in 1888 the presence of endophytic bacteria within root nodules of legumes and their contribution of nitrogen for plant growth (reviewed by R.H. Burris) [6]. The general concept of the “rhizosphere” as the habitat where plant roots attract beneficial and pathogenic soil microbes by their exudates was finally coined by L. Hiltner in 1904 [7]. He found that microbes were enriched around the roots, but also recognized bacteria-like bodies within roots, which he called “bacteriorhiza” [8]. This term was coined in analogy to the term “mycorrhiza”, which had been defined in 1885 for filamentous organisms within roots by Albert Bernhard Frank, a German botanist and biologist. In 1893, Hiltner and Nobbe developed the first efficient Rhizobium-based inoculants, which they called “Nitragin”, based on their discovery of host specificities in Rhizobium-legume symbioses [9]. However, Hiltner was not successful to establish plant growth promotion by bacterial inoculation of non-leguminous plants. His quite early death in 1923 and the difficult post-world war situation in Germany contributed to slow down scientific progress in this field. For many decades no further major breakthrough on plant growth promoting bacteria was reported. Only in the 1970s new interest arose on plant beneficial bacteria after the isolation and introduction of Azospirillum spp. by Döbereiner and Day [10]. In the 1980s for example Baldani and coworkers [11], [12], and Cavalcante and Döbereiner [13] from EMBRAPA-Agrobiologia, Seropédica, RJ, Brazil, isolated and characterized new diazotrophic bacteria from roots of different important crop plants. The high engagement and dedication for their science in combination with establishing a worldwide cooperation including sharing newest results as well as newly isolated strains by Johanna Döbereiner helped enormously in developing this field of research. Most recently, a book describes Johanna Döbereineŕs life as highly engaged scientist [14]. In 1981, Walter Klingmüller, head of genetic department at the University of Bayreuth, Germany, initiated a series of six biannual workshops entitled “Azospirillum: Genetics, Physiology and Ecology” bringing together the international research community on Azospirillum and related microorganisms – the last two workshops were organized by Istvan Fendrik, Maddalena del Gallo and Jos Vanderleyden [15], [16], [17], [18], [19], [20]. While the first four workshops were focused on research about Azospirillum spp., increasing interest and research activities also on other plant-growth promoting bacteria led to a broadening of the subject in the workshops V and VI. The articles in the corresponding proceeding books not only document the groundbreaking establishment of molecular genetic tools for Azospirillum by several research groups and discoveries of new endophytic diazotrophic bacteria, but also most interesting work by different groups on outstanding physiological properties, like e.g. the cyst-formation of Azospirillum spp., and early field application trials [15], [16], [17], [18], [19], [20]. In parallel, an international symposium series “Biological Nitrogen Fixation with Non-Legumes” started in 1979; the XVIth symposium of this series was held in August 2018 in Foz de Iguacu, Brazil, attracting more than 300 participants (www.mpcp2018.com.br). Quite recently, the current status of research on endophytic diazotrophic rhizobacteria was also summarized by Reinhold et al. [21] and Kandel et al. [22].
The challenge for pioneering research on endophytic diazotrophs and other plant growth promoting bacteria was not only to understand the biochemical and genetic processes characterizing the basis for plant growth promotion, but also the ecology of the interaction with their host plants as fundament of the beneficial action. The ultimate applied goal was to use these bacteria as so-called “biofertilizer” or “biostimulants” towards the establishment of sustainable agricultural management. In the 1980s, substantial agronomic applications were still far away on the horizon, while within the last ten years several Azospirillum brasilense strains [23] and other PGPRs including biocontrol-active Gram-positive bacteria [e.g. [24]] have been applied successfully in agro-biotechnology worldwide. However, in this mini-review, the vast development of Gram-positive inoculants has not been covered. It is still a key issue to provide unequivocal evidences for the colonization and localization of the bacteria as well as their in situ activities in the rhizosphere and within the plant. Serological and molecular genetic techniques suitable for these in situ analyses have been developed over the years, but always have to be adapted for successful identification, localization and quantification of bacteria in their specific association with plants. In addition, key functions in the beneficial interaction of rhizobacteria with plants needed to be identified. Moreover, the development of culture independent approaches was necessary to overcome the bias of studying only culturable members of the plant microbiome. In this review, a number of techniques and approaches are presented from a historical to current development perspective, which allows the detailed analysis of the composition of beneficial plant microbiota – even down to the level of monitoring specific inoculant strains - and their functions leading to plant growth promotion. Furthermore, a scientific based distinction of plant beneficial from opportunistic human pathogenic bacteria is addressed.
Techniques for resolving the diversity and function of the plant microbiome at highest resolution
Serological techniques coupled with confocal laser scanning microscopy (CLSM) as identification and quantification tools
The prerequisite of creating antibodies is the availability of bacteria in pure culture, which certainly is a limitation for the application of this approach, since many plant-associated bacteria are difficult to cultivate. After developing fluorescent-labeled monoclonal antibodies against A. brasilense Sp7 which are directed against EPS-cell surface compounds [25], [26], confocal laser scanning microscopy (CLSM) was successfully used by Schloter et al. in 1993 for the first time to produce clear images of these bacteria being embedded in the rhizoplane matrix [27]. Using the confocal technique as well as silver enhancement of the antibody detection, the root colonization pattern of the plant growth promoting Rhizobium leguminosarum bv. trifolii R39 was characterized in different gramineaeous plants in 1997 by Schloter et al. [28]. In the same year, Yanni et al. [29] could also demonstrate the endophytic colonization of rice by the N2-fixing symbiont Rhizobium leguminosarum bv. trifolii strain C6 in Egyptian berseem clover (Trifolium alexandrinum) applying immunofluorescence techniques. This demonstrated for the first time an intimate colonization also of rice by Rhizobium. In the case of A. brasilense, monoclonal antibodies against the putative endophytic strain Sp245, isolated from surface disinfected wheat roots [30], [31], demonstrated a different colonization pattern of roots by the strains Sp7 and Sp245: strain Sp7 colonized wheat roots mostly at the root-surface, while strain Sp245 was able to enter the root, colonizing the apoplast tissue in wheat roots [32]. In addition, also quantitative colonization data of Sp7 and Sp245 in wheat plants could be obtained by the ELISA-technique, confirming the microscopic evidences of different colonization patterns [32]. Furthermore, in situ expression of specific enzymes (e. g. nitrogenase) in different rhizobacteria colonizing their host plant could be achieved using this technique [33].
Monoclonal or mono-specific polyclonal antibodies are also unique tools to easily enrich and cultivate a high diversity of root-associated bacteria of the same or closely related species from the root and the rhizosphere using the antibody based immuno-trapping technique [34]. For example, antibodies against whole cells of a rhizosphere isolate of Ochrobactrum anthropi were coated on microtiter plates, followed by adsorption of soil extracts. After proper washing steps, the bound bacteria were desorbed with 0.1 M KCl-solution. This resulted in a more than 100-times enrichment of this specific group of bacteria and isolates of this particular species could be easily obtained. Thus, the influence of the crop plant, management practices, and ecotoxicological effects of applied agrochemicals on the micro-diversity spectra of Ochrobactrum anthropi communities in soils and the rhizosphere could be isolated and studied [35]. Even isolates of closely related new species could be retrieved using the immuno-trapping approach [36]. The application of this immuno-enrichment technique turned out to enable access to a hidden bacterial micro-diversity and should be applied more generally. In this straightforward approach, a greater diversity of saprophytic and beneficial rhizobacteria of specific species may be achieved.
Ribosomal RNA as identification marker with limitations to separate closely related strains
The establishment of a phylogenetically based natural system of organisms for the domains Archaea, Bacteria and Eucarya by Carl R. Woese, Otto Kandler and Mark L. Wheelis in 1990 [37] was the landmark for a molecular approach to the phylogeny of Bacteria and Archaea. The 16S rRNA genes of Bacteria rapidly became the gold standard of molecular phylogenetic analysis, because the ribosomal RNA is present in all organisms and its sequence has highly conserved and variable regions. This facilitates the design of primers or oligonucleotide probes, usually 16–20 nucleotides long, with specificities to different taxonomic levels: probes complementary to conserved regions of the 16S or 23S rRNA will identify all bacteria of a high taxonomical rank, e.g. family or domain level, while for targeting bacteria on genus or in some cases - if a differentiation is possible - even species level, probes need to target highly variable regions of the rRNA specific to the taxonomic group of interest. In addition, the rRNA genes are expressed at very high levels in physiologically active cells (with copy numbers up and over 10.000), are more stable compared to mRNA due to their secondary structure and are therefore good targets for labelling the bacteria with fluorescent probes. Consequently, cells with low activity have usually low rRNA contents, resulting in low fluorescence labeling due to an insufficient number of target sites for the probes. This means on the one hand that positively labeled cells are very likely also functionally relevant for the analyzed habitat, but on the other hand also implies that this method is of limited use for targeting bacteria with low physiological activity. In addition, the cell wall penetration of applied probes has to be optimized, i.e. due to their differences in cell wall structure, Gram-negative and Gram-positive cells need to be treated with different fixation protocols to enable the phylogenetic probes to get into the cells [38]. Despite some obvious limitations of this approach, so-called “phylogenetic stains” became rather quickly a widely employed tool to identify single cells using the Fluorescence In Situ Hybridization (FISH) technique [39]. In combination with flow cytometry, FISH was successfully applied to quantify single cells [40] or to identify and localize bacterial consortia in complex natural habitats with the help of highly resolving confocal laser scanning microscopy and differentially labeled sets of oligonucleotide probes [41].
The first application of the FISH-technique coupled with CLSM-application to characterize plant microbiota was to identify and localize A. brasilense strains in the rhizosphere of wheat [42]. The inoculated A. brasilense bacteria colonizing the root surface and intercellular spaces in the epidermis had swollen cyst-like morphology harboring high ribosome content, which verified earlier evidences from light and electron-microscopic scanning [42]. The productive cooperation with the institute of Prof. Karl-Heinz Schleifer (TU München), coming from the “phylogenetic school” of Prof. Otto Kandler (LMU München), was very helpful to establish the FISH-technique for rhizosphere research. It could further be demonstrated that A. brasilense strain Sp245 could colonize wheat roots also endophytically. Some root hairs or intercellular spaces in the root cortex and even cortical cells were heavily colonized by the strain Sp245 showing high staining intensity with the rRNA-targeted oligonucleotide probes reflecting high physiological activity of the bacteria [42]. A combination of a differentially fluorescence-labeled monoclonal antibody against A. brasilense Wa3, and a species-specific oligonucleotide for A. brasilense revealed a different colonization profile of the strains Wa3 and Sp245 [43]. In the 1990s, when six Azospirillum species were known, all Azospirillum spp. could be clearly distinguished using a set of differentiating oligonucleotide probes [44]. At present, 19 different Azospirillum species are known and validly published, which makes it difficult to clearly allocate new isolates to one of these very closely related species by 16S rRNA sequences and 16S rRNA directed probes. Although the larger 23S rRNA gene and the 16S-23S rRNA intergenic regions provide higher separating power, these different species are impossible to separate with individual species-specific probes. The present solution of differentiation and even strain-specific identification is provided by the increasingly available whole genome sequences. Based on the comparison of the different available whole genome sequences within one species, strain-specific sequences could be found for e.g. A. brasilense strain FP2. Primers derived from these unique regions led to a specific and quantitative amplification of the target strain even from natural habitats like soil-grown wheat plants [45]. Thus, whole genome sequencing is becoming an ever more popular approach and currently only suffers from a lack of genome information for type and reference strains in the database.
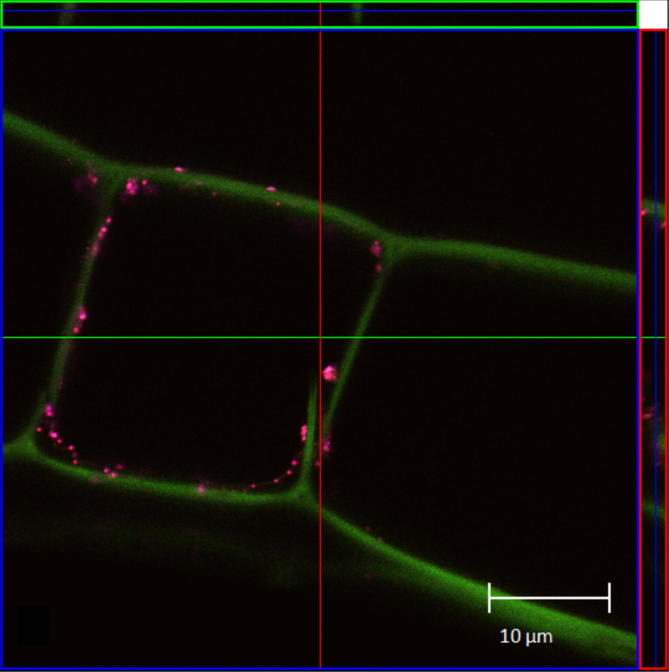
There are also severe limitations for the application of the FISH-technique to identify and localize endophytic bacteria. In many environmental samples and also in adult field grown plants, like sugarcane, multiple auto-fluorescent objects in the sizes of bacteria are present in the tissue or within cells [46] (Fig. 1). Therefore, an alternative labelling method replacing fluorescence was necessary. Schmidt et al. [47] developed a modification of the CARD-FISH-protocol using gold-particles resulting in a specific bacterial identification using scanning electron microscopy as detection method for the deposited gold-particles. Nevertheless, this technique is limited to surface scans and therefore thin sections are required for the analysis of endophytic communities.
Fig. 1.
CLSM-image with adult sugarcane (green) samples, viewing unspecific fluorescence signals in magenta ([46]).
Fluorescent protein-tagging for in situ analysis of structural and functional aspects
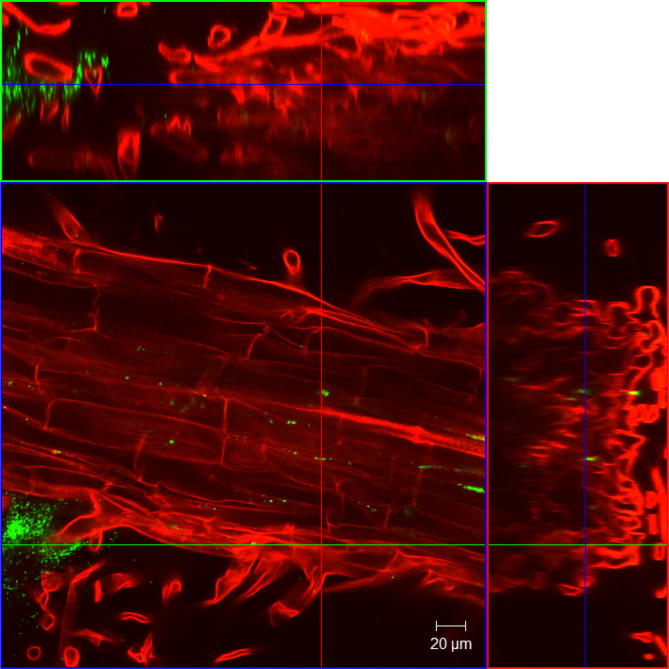
A very powerful cell labelling method is the tagging with a constitutively expressed gene coding for a fluorescing protein, like the green-fluorescent protein (GFP). The basics and variations of this approach were reviewed by Crivat and Caraska [48]. Several applications for studying rhizosphere bacteria were reviewed by Reinhold-Hurek and Hurek [49]. Fig. 2 shows fluorescence-tagged Herbaspirillum frisingense cells located within root tissue. Alternatively the tagging gene can be inserted under the control of a promotor from a gene of interest to study its expression in situ [50]. Furthermore, a GusA-kanamycin reporter gene was inserted into the nifH-genes of an A. brasilense wild type and ammonium-excreting strains to facilitate an expression analysis in barley roots [51]. Quantitative data can be retrieved even from field samples, as was demonstrated by You et al. [52]. In a GFP-tagged Herbaspirillum the expression of nifH was quantified by RT-qPCR and related to the amount of the tagged bacteria colonizing rice endophytically.
Fig. 2.
Optical sectioning through intact barley (Hordeum vulgare, red) roots (19 days old) from a monoxenic quarz sand growth system colonized by inoculated Herbaspirillum frisingense GSF30 fluorescently tagged by a constitutively expressed chromosomal gfpmut3 gene (green).
Concluding this phylogenetic and identification part, it can be stated that 16S rRNA-based phylogeny is still the prerequisite for powerful approaches of bacterial identification, including in situ localization by FISH as well as high-throughput amplicon sequencing based community analysis (discussed in the next section), but the applications are limited. Detailed resolution of diversity and functional aspects in a strain-specific resolution may also need molecular tagging approaches or advanced bioinformatic analyses based on whole genome sequence information.
Community metagenomics and functional transcriptomics of bacteria and plants
Undoubtedly, the culture-independent analysis of complex bacterial communities associated with plants would not be possible without using PCR-based amplification of different regions of the 16S rRNA gene. As prerequisite, DNA or RNA needs to be isolated from plant material and purified to remove plant substances inhibiting the PCR enzymatic reactions. While a proper quality of DNA/RNA is quite easily achievable from plant seedlings, especially, from soil free model experiments, it can be very challenging to obtain sufficiently pure DNA/RNA in enough quantity from field grown, adult plants. However, after optimization, this important initial step of microbial community analysis was achieved in several cases. For example, Fischer et al. [53] retrieved many bacterial 16S rRNA sequences from field grown sugarcane plants, which were not known from cultivation-based approaches. From their data it became obvious that a high diversity of diazotrophic bacteria colonized roots and stems and also a high diversity of nifH-genes was expressed. However, from the five inoculated strains of the EMBRAPA-inoculum (Gluconacetobacter diazotrophicus Pal5T-BR11281, Nitrospirillum amazonense Cbamc-BR11145, Herbaspirillum seropedicae HRC54-BR11335, Herbaspirillum rubrisubalbicans HCC103-BR11504, and Paraburkholderia tropica PPe8T-BR11366), only Gluconacetobacter diazotrophicus Pal5 was found to be able to colonize sugarcane roots and stems for several months [53]. A high diversity of different active Rhizobium and Bradyrhizobium species was also found in these adult, field grown sugarcane plants, based on retrieved 16S rRNA. This clear demonstration of hitherto only rarely observed diversity of Rhizobium and Bradyrhizobium strains colonizing sugarcane and other non-legume plants triggered the attempt to isolate these bacteria in scavenging experiments with broad host range legumes [54], which resulted in the successful isolation of a diversity of Bradyrhizobia. The knowledge about the high diversity of uncultured bacteria within the plant microbiota also led to isolation approaches not aiming for single bacteria through specific enrichment procedures but for whole communities in non-selective complex media. Indeed, this yielded the growth of bacterial consortia, including species which could not be isolated from the plant microbial community before. This has been exemplified for the sugarcane community yielding complex plant growth promoting consortia [55]. However, as this approach is difficult and lacks reproducibility, it seems more straightforward to isolate members of the plant microbiota using plant derived cultivation media and subsequently combining these individual pure isolates based on functional criteria (so-called “syncoms”).
The crosstalk of beneficial endophytic bacteria and their plant hosts during the interactions is of key importance to understand holobiontic interactions and to optimize the efficiency of inoculation trials. Several highlights of important ecophysiological and interactive traits for plant microbiota and their hosts in a holobiontic context could be already identified by metagenomic and especially transcriptomic studies at both the bacterial and plant side [56], [57], [58], [59]. Metagenome and transcriptome analyses on both bacterial and plant side during the interaction contribute very important functional information. However, to guarantee the reliability and reproducibility of these types of results principles for standardization have to be followed, as was learned from human microbiome research [60], [61]. Based on frequently expressed genes during the interaction of plant endophytic bacterial communities in the holobiont context, functions like e.g. osmoadaptation, phytohormone production, oxygen tolerance and quorum sensing are of particular relevance.
Discrimination of plant beneficial bacteria from closely related human pathogenic bacteria exemplified by A. brasilense and Roseomonas fauriae
The rhizosphere is a habitat, which is colonized by a phenotypically wide spectrum of bacteria: from symbionts to pathogens. This has been pointed out by Berg et al. [62] and more recently by Mendes et al. [63], who highlighted the presence of plant beneficial, plant pathogenic and human pathogenic microorganisms in the rhizosphere. Already Lorenz Hiltner had proposed that many “wanted or unwanted guests” are attracted by root derived nutrients [7]. Even within a particular rhizobacterial genus, species with plant beneficial and pathogenic phenotypes are known [64].
In recent years, isolates with almost identical 16S rRNA to A. brasilense type strain Sp7, which also have high root colonization potential [65], were retrieved from wounds and other human sources. These isolates had been originally classified as Roseomonas fauriae or R. genomospecies 6, but lately they were reallocated to the A. brasilense species [66], based on wet DNA-DNA-hybridization analysis using the re-association method according to Brenner et al. [67]. Also, the ITS region of 16S-23S rRNA genes and many household genes are almost identical (Fig. 3).
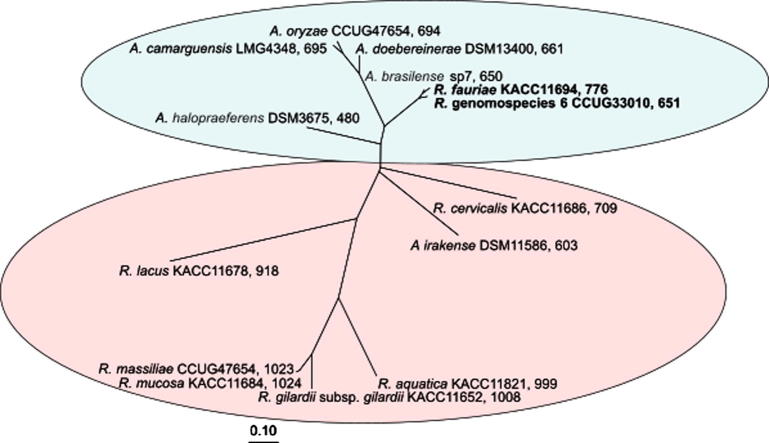
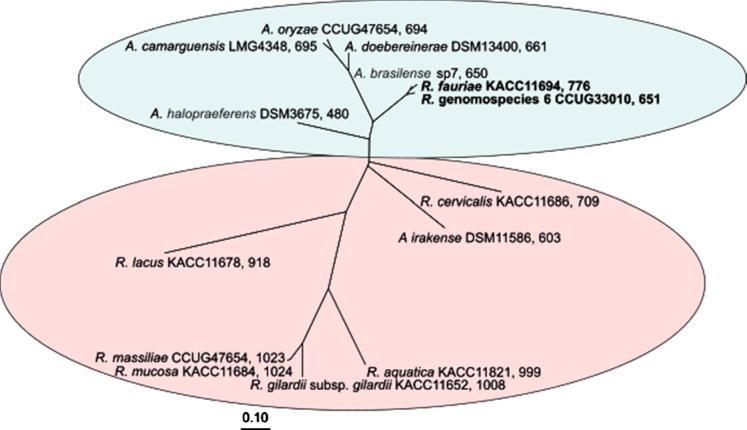
Fig. 3.
Phylogenetic tree (ITS-region of the 16S-23S rRNA genes, maximum-likelyhood method with 50% conservation filter) of Azospirillum spp. and Roseomonas spp. [68]
However, recent whole genome DNA-DNA hybridization analyses using a spectrophotometric determination of re-association kinetics [69] revealed only 61.2% and 54.4% DNA-DNA sequence identity between A. brasilense Sp7T and Roseomonas fauriae and R. genomospecies 6 (measurements of DSMZ, Braunschweig, Germany, unpublished) (Table 1). This definitely argues for a phylogenetic separation of A. brasilense from these opportunistic pathogenic Roseomonas bacteria. These results were corroborated by in silico determinations of ANI-values (Average Nucleotide Identity) based on whole genome sequences [70]. Based on a concatenated phylogenetic analysis of rpoD- and 16S rRNA gene sequences [70], it was further proposed to separate the A. brasilense strains into three closely related species: A. brasilense sensu stricto, A. formosense [71] and A. himalayense [72]. Thus, it became apparent, that there is an unresolved micro-diversity within the species of A. brasilense. In addition, the plant endophytic A. brasilense strains Sp245, Az39, and strain NH, isolated from salt-affected wheat rhizosphere from Northern Algeria [73], were all shown to have DNA-DNA-hybridization values around 50% compared to the A. brasilense Sp7T (Table 1). Therefore, further DNA-DNA hybridization studies and whole genome sequence analyses are necessary to clarify the relationship within A. brasilense and closely related species and their phylogenetic relationship to R. fauriae and R. genomospecies 6.
Table 1.
Spectrophotometric DNA-DNA hybridization analysis, according to Huss et al. [69] of A. brasilense Sp7T to several A. brasilense strains, Roseomonas fauriae, and R. genomospecies 6 (data from Deutsche Stammsammlung für Mikroorganismen and [68]).
| Azospirillum brasilense Sp7T | |
|---|---|
| Azospirillum brasilense FP2 | 96.5% |
| Azospirillum brasilense Sp245 | 54.0% |
| Azospirillum brasilense NH | 56.0% |
| Azospirillum brasilense Az39 | 48.3% |
| Azospirillum lipoferumT Sp59b | 28.7% |
| Roseomonas fauriaeT KACC1694 | 61.2% |
| Roseomonas genomospecies 6 CCUG33010 | 54.4% |
| Roseomonas mucosaT KACC11684 | 12.5% |
The application of whole genome-based comparative software tools together with the assessment of the pathogenic potential of each species [74], finally helped to clarify the difficult case of distinction between saprophytic or beneficial and pathogenic strains within the genus Burkholderia. This genus harbored a large number of species with human pathogenic or opportunistic pathogenic phenotypes as well as environmental and plant growth beneficial and symbiotic species. For a long time, there was a situation, when regulatory authorities banned every environmental release of a Burkholderia strain, including the beneficial and even symbiotic ones. Now, based on the available complete genome sequence data, conserved sequence indels (CSI) were successfully used as molecular marker for the demarcation of the Burkholderia groups [75]. Finally, there are at present three different genera within the Burkholderia cluster: (i) Burkholderia, containing the pathogens and opportunistic pathogens, (ii) Paraburkholderia, comprising the plant-associated and -beneficial species, and (iii) the Caballeronia cluster, a group of environmental species [76]. An even more complex situation is present within the species Serratia marcescens. Strains of environmental and nosocomial origins were intermixed without any handle to separate them based on a strict and efficient scientific approach. Whole genome multilocus sequence types (wgMLSTs) and core genome multilocus sequence types (cgMLSTs) were created with the PHYLIP program UPGMA algorithm creating two sectors representing strains with environmental or nosocomial origins [64]. Since there were even genomes identified, which reflected intermediary genomic situations, there is the chance to have even closer insights into steps of micro-evolution to optimize the fitness in an apparently altered habitat.
Major traits of rhizosphere bacteria for efficient root colonization
Osmoadaptation
Lack of available water is causing stress to each living organism, because all life processes and essential proteins and cellular structures are dependent in their native conformation on available water molecules. Due to their molecular structure, several small molecules, so-called osmolytes, like proline, glycine betaine, ectoin, and trehalose are able to replace water molecules to some degree [77]. During osmoadaptation, organisms activate the synthesis or uptake of these and similar substances within their cells. Since these osmolytes are functional across different organisms, microbes and higher organisms can help each other out under water stress [78]. They also enable to protect salt-sensitive enzymes and stabilize cellular structures and functions by balancing the osmotic pressure in plant cells against the outside osmotic pressure caused by salt or water deficiency. In saline soils, osmotolerance mechanisms are omnipresent. For rhizosphere bacteria, osmoadaptation has selective power also in non-saline soils, because salt is being concentrated around the roots during the continuous uptake of water by the plant, resulting in an accumulation of ions in the rhizosphere. In addition, during daytime, the transpiration stream causes water deficiency in the rhizoplane, which may only be replenished during night time by slow diffusion of water from root-distant soil habitats. This water dynamics and the increasing salt-pollution of soils made osmo-adaptation and osmo-tolerance important traits in rhizosphere bacteria [79]. Moreover, the salt-tolerant IAA-producing rhizobacterium A. brasilense NH isolated from salt-affect rhizosphere soil of wheat in northern Algeria, can replenish specific phytohormones, like indole acetic acid (IAA, i.e. auxin), which are not sufficiently produced by salt-stressed root tissues [80]. In salt-affected soils, the 1-aminocyclopropane-1-carboxylate (ACC)-deaminase activity of rhizobacteria is of particular relevance, because due to this enzymatic activity, elevated levels of ethylene are reduced in roots, which would inhibit plant activities drastically [81], [82]. It is remarkable that the occurrence of the ACC-deaminase gene is rather frequent in plant-associated bacteria from saline habitats and there are indications of horizontal gene transfer of this beneficial trait [83].
Among Azospirillum spp. different levels of osmotolerance can be found [84]. A. halopraeferens has the highest salt-tolerance and it could be shown that it is able to synthesize glycine betaine or take up and transform choline into betaine [85], while A. brasilense is only able to take up betaine glycine [86]. Trehalose is not significantly used as osmolyte by A. brasilense. However, when transformed with a plasmid harboring a trehalose biosynthesis gene-fusion from Saccharomyces cerevisiae, A. brasilense Cd accumulates trehalose under water stress and is able to grow up to 0.5 M NaCl. Furthermore, maize plants inoculated with this engineered bacterium were able to withstand drought stress and increase its biomass and grain yield [87]. The ability of salt-tolerant A. brasilense and A. halopraeferens strains to utilize proline and other amino acids as C-source for growth was only rather limited [88]. A. brasilense strains with increased NaCl-tolerance could be isolated which proved to be spontaneously resistant to the toxic proline antimetabolite dehydroproline under mild salt stress conditions [89]. Another relevant stress adaptation in Azospirillum is the cyst formation, which occurs when cells are challenged with nutrient deprivation or desiccation. In Azospirillum this regularly occurs, when cells are inoculated to roots as was shown in several independent techniques [42]. The induction of cyst formation can also be triggered by the application of fructose and nitrate as C- and N-sources in laboratory media. Malinich and Bauer [90] recently compared the metabolic and replicative gene expression by transcriptome analysis in vegetative and cyst states of A. brasilense.
Phytohormones and other growth enhancers
Besides IAA and derived substances with auxin activity, also nitrogen oxide (NO) is often found as plant growth regulating compound in rhizosphere bacteria. In the case of A. brasilense, which is a most successful and widely used PGPR, it is documented that besides IAA also NO has a pronounced effect on the stimulation of root growth [91].
It has been shown in inoculation experiments of mutants, which produced only very low levels of NO, that root morphology was almost not changed in contrast to the inoculation with the NO-producing A. brasilense Sp245 wild type [92]. Similarly, IAA-deficient mutants lost the activity of root growth stimulation. The level of IAA-production could be increased in mutants of A. brasilense SpCd, resistant to the antimetabolite 5-fluor-tryptophan [93]. Inoculation of maize plants in an axenic system with the IAA-overproducing mutant FT326 showed root growth stimulation only at low inoculation densities and very low nitrate levels compared to the wild type inoculation [94]. In a similar way, mutants which show ammonium excretion could be selected from A. brasilense Sp7 by Machado et al. [95] using the antimetabolite ethylenediamine for ammonium assimilation. Using the ammonium-excreting mutant HM053 as inoculant for maize or wheat, nitrogen fixation and N-assimilation in inoculated plants were changed compared to the wild type inoculation [96], [97].
Thus, the application of mutations resulting in drastically reduced or increased functions or the production of certain effector molecules are of central importance in the assessment of functional relevance of interaction traits. A detailed collection of physiological properties of Azospirillum spp. by Hartmann and Zimmer can be found in Yaacov Okon's book on Azospirillum/plant associations [98].
Oxygen tolerance
Induction of reactive oxygen species is a key element of defense reaction of plants. Thus, bacteria which approach plants need to be equipped with defense measures against these toxic oxygen species. In the case of the plant endophytic diazotroph Gluconacetobacter diazotrophicus Pal5, mutants devoid of catalase and superoxide dismutase were unable to colonize rice roots and to establish an endophytic life style [99]. Another oxygen defense mechanism uses O2-diffusion protection by gum production. Consequently, mutants of Pal5 in gum-production lacked endophytic colonization too [100]. In the case of the interaction of the diazotrophic Burkholderia australis Q208 with sugarcane, a downregulation of reactive oxygen production of plants could be demonstrated by RNAseq during colonization by B. australis Q208 [59]. On the bacterial side, LPS- and flagella-production, which are well-known elicitors for pathogen-associated molecular patterns, were reduced in strain Q208 during the root colonization process. Since also strain Q208 harbors the QS-related genes for N-acyl-homoserine production [59], which are usually activated during biofilm production and root colonization, it is quite possible that they are involved in regulatory processes in the physiological changes occurring during root colonization and the interaction with plants (see below).
Bacteria-plant communication with focus on N-acyl homoserine lactones
Bacterial quorum sensing signals are involved in many important ecological functions, like biofilm formation, induction of antibiotic production and virulence. In Gram-negative bacteria N-acyl-homoserine lactones (AHL) were often found regulating these processes through an activation of the luxI/luxR-type regulatory circuit [101]. It has been shown using AHL-biosensor constructs that the production of AHL-molecules was heavily induced during the colonization of root surfaces by bacteria harboring the luxI/luxR-type auto-inducing system [102], [103]. The auto-induction of AHL-synthesis can be activated already in micro-colonies at the root surface due the spatial accumulation of the AHL-compounds [103]. However, the excreted quorum sensing molecules are not only sensed by neighboring rhizosphere bacteria, but also by the plant hosts [104]. This trans-kingdom signaling induces different responses in the plants, depending on the type of AHLs (diffusible, water-soluble AHLs with short C-side chains or lipophilic, water-insoluble AHLs with C-side chains from 12 to 14 C-units). Water-soluble AHLs are taken up actively into the plant shoots inducing gene expression of antioxidative and xenobiotic degradation genes in roots and shoots as well as phytohormonal changes in the whole plant [105], [106], [107]. Also NO-accumulation and membrane hyperpolarization accompanied by increased K+ uptake are early events after AHL application to barley roots [108]. In contrast, water-insoluble AHLs prime the induction of systemic resistance response in the plant hosts [109] and finally confer increased resistance towards biotrophic and hemi-biotrophic pathogens in wheat and Arabidopsis [110], [111]. The central involvement of QS-regulation in endophytic colonization of rhizobacteria could also be demonstrated, when mutants devoid of luxI or luxR homologous genes were tested for endophytic colonization. For example, a negative mutant for AHL synthesis of the beneficial root endophyte Acidovorax radicis N35 had reduced endophytic colonization abilities. In contrast to the wild type, the AHL synthesis mutant caused induction of the flavonoid biosynthesis genes, which are known to be part of the plant defense response [112]. Thus, the AHL-lacking mutant may not be recognized by the plant as beneficial bacterium. Furthermore, an AHL receptor mutant of Gluconacetobacter diazotrophicus Pal5 was also no longer able to colonize the plant host endophytically, since the QS-coordination was not functioning (Hofmann A and Baldani JI, unpublished results). Thus, QS-signaling in bacteria-plant interactions may not only act through direct interaction with the plant, but also by establishing and coordinating an adapted gene expression of traits like biofilm formation, necessary for endophytic colonization. A. brasilense strain Ab-V5 (originally derived from A. brasilense Sp7T), applied in large scale for about 10 years in Brazilian agriculture, was recently shown to respond to N-acyl-homoserine lactones (especially 3-oxo-C8-HSL). It showed increased biofilm and exopolysaccharide formation as well as cell motility, because it harbours a luxR, but no luxI homologous gene [113]. Interestingly, while luxI homologues are missing in A. brasilense, they are present in most of the A. lipoferum strains [114]. Transconjugants of Ab-V5 carrying a plasmid with the N-acyl-homoserine lactonase gene abolished the PGPR effect of the wild type. The functionality of so-called luxR-solos reflect the release of AHL-mimic compounds by the plant host [115] or by the accompanying plant microbiome. As one important mechanism of stimulation of plant performance, AHLs induce priming effects, which are specific plant responses in the crosstalk of root-colonizing bacteria with their plant hosts leading to an alert state towards the attack of plant pathogens. It has been shown that a wide variety of molecules can induce priming, besides AHLs also including antibiotically active compounds, like lipopeptides of pseudomonads and bacilli, as well as certain volatile compounds [116], [117]. The effects of priming are not visible in the absence of pathogens, but in the situation of pathogen attack, the defense responses are rapid and enhanced.
Conclusions and further perspectives
Thanks to the great methodological progress in the last two decades, there are now quite some “eye-opening insights” into many structural and functional details of the plant associated microbiome and key interactions between the plant microbiome and the host plant in the holobiont context [118]. However, the complexity of interactions is overwhelming and thus the collection and careful interpretation of further metagenome and transcriptome data needs to be intensified for a deeper understanding. This should be supported by isolation approaches of novel bacteria leading to defined inoculation experiments and testing of functional hypothesis with mutant studies. In addition, the improvement of their environmental fitness and key interaction traits with the plant host (phytohormone production, ammonium excretion) by spontaneous selection or chemical mutagenesis of already established inoculation strains should be considered, since in some cases these appeared quite feasible. The final goal is to implement the knowledge about plant microbiome/host interactions under field conditions into practical applications. Ideally, this would mean to utilize synergistic effects in “synthetic” holobionts, where a specifically tailored set of beneficial microbes is introduced to plants which have been improved by selection, breeding or genetic modification in supporting the beneficial plant microbiome in a most productive manner. Within the “Plant Phytobiome” concept [119] aiming to integrate biological, soil, climate and agricultural management, a deeper understanding of key interaction and communication processes of the plant and its microbiome within the holobiontic context is urgently needed.
Acknowledgments
Conflict of interest
The authors have declared no conflict of interest.
Compliance with Ethical Requirements
This article does not contain any studies with human or animal subjects.
Acknowledgements
We greatly appreciate the intramural funding of the focus area “Molecular signalling in the rhizosphere” for more than 10 years between several institutes of the Department of Environmental Sciences by the Helmholtz Zentrum München, German Research Center for Environmental Health. The excellent expertise and engagement of Gudrun Kirchhof, Marion Stoffels, and Michael Schmid, leaders of the group “Molecular microbial ecology” within the Research Unit “Microbe-Plant Interactions” is greatly acknowledged.
Biographies

Anton Hartmann studied biochemistry at the University Tübingen, and got the doctoral degree in 1980. He was postdoc at University of Wisconsin, Madison, USA, from 1983-1985, and was habilitated at University Bayreuth. He finally joined the Helmholtz Zentrum München (HMGU) in 1989, and was teaching at Ludwig-Maximilians-University München. In his research unit at HMGU, fluorescence-labelled rRNA-directed probes together with laser scanning microscopy were applied in the rhizosphere and new diazotrophic bacteria were identified with molecular phylogenetic techniques. Structural and functional aspects of the plant microbiome, especially nitrogen fixation and the interkingdom communication based on quorum sensing signaling compounds were studied.

Doreen Fischer studied Biology in Regensburg and Oldenburg (Germany, 2000-6). She accomplished her PhD at the Helmholtz Zentrum München in the working group of Microbe-Plant Interactions under supervision of Prof. Dr. Anton Hartmann, focusing on diazotrophic bacteria associated with sugarcane. 2010-15 she joined the Institute of Soil Ecology and the Research Unit Terrestrial Ecogenetics, later the Research Unit Environmental Genomics at the Helmholtz Zentrum München as a Postdoctoral researcher, investigating soil-microbe and plant-microbe interactions, biocontrol, microbial ecology and ecosystem services. 2015-17 she joined EMBRAPA Agrobiologia (Brasil) as senior scientist in the group of Veronica Massena Reis. After a stay at the University of Kassel in 2017-18 where she was doing bioinformatics in microbial ecology, she came back to the Research Unit Comparative Microbiome Analyses at the HMGU in Munich in 2019 focusing on food microbiome.

Linda Kinzel completed her diploma thesis about Comparative and phenotypic characterization of Roseomonas spp. and Azospirillum spp. with focus on bacterial taxonomic classification at the LMU and the GSF in Munich (Germany) in 2008. In 2008-14 she did her PhD with focus on molecular radiation biology and radiation oncology at the LMU in Munich. In 2014 she worked as postdoc in the same field and changed her occupation afterwards towards sales specialist and Medical Science Liaison Manager Oncology.

Soumitra Paul Chowdhury completed his Master of Science (M.Sc.) in Botany from the University of Calcutta, Kolkata, India in 1999, with specialization in Plant Physiology, Biochemistry and Molecular Biology. He received his PhD degree in Biotechnology from the Banaras Hindu University, Varanasi, India in 2006. In 2007, he joined as a Postdoctoral research fellow at the Max Planck Institute for Terrestrial Microbiology, Marburg, Germany. From April 2010, Soumitra joined the group Molecular Microbiology in the research unit Microbe-Plant Interactions at the Helmholtz Zentrum München as a Postdoctoral researcher. From February 2017 he is a part of the newly founded Institute of Network Biology at the Helmholtz Zentrum München, where he is a researcher at the working group Molecular Microbial Ecology.

Andreas Hofmann studied Biology at the Technical University of Munich (1999 – 2006). He accomplished his PHD at the Helmholtz Zentrum München in the department of Microbe-Plant-Interactions under the supervision of Prof. Dr. Anton Hartmann, focusing on the transfer of human pathogenic bacteria in the course of organic vegetable production (2007 – 2011). In the years 2012 – 2014 he joined the Institute of Soil Ecology of the Technical University of Munich and the department Environmental Genomics of the Helmholtz Zentrum München focusing on soil microbiology and ecology. After a stay at the EMBRAPA Agrobiologia, Seropédica, Brazil, focusing on the microbe-plant interactions of G. diazotrophicus and rice in the working group of Dr. Ivo Baldani (2015 – 2017), he joined the University of Kassel, Section of Organic Plant Breeding and Agrobiodiversity focusing on plant genetics (2017 - 2018).

José Ivo Baldani is employed of the Brazilian Enterprise on Agricultural Research, Embrapa Agrobiology, Seropédica, RJ, Brazil. He studied Agronomy and MSc. in Soil Sciences at the Federal Rural University – UFRRJ, Seropédica, RJ and holds a PhD in Soil Sciences from the Texas A&M University, Texas, USA. He has worked since 1976 on Biological Nitrogen Fixation with Gramineous plants and for many years had the privilege to work and share the knowledge with Dr. Johanna Döbereiner, the pioneer on BNF with Grasses and isolation of many associative and endophytic diazotrophic bacteria. Along the years, he has been involved in the isolation and identification of new diazotrophic genera and species, particularly Herbaspirillum with the species H. seropedicae, H. rubrisubalbicans as well as the Nitrospirillum amazonense. Strains belonging to these species are now being used as inoculants in sugarcane, maize and rice crops. More recently, his group has applied molecular approaches (genomic, transcriptomic and proteomic) to understand the plant- bacteria interaction of mainly those related to sugarcane inoculant strains. In addition, he has dedicated part of his research on the biocontrol area involving the use of endophytic bacteria with activity against phytopathogens (bacteria and fungi) and pest insects of economic importance to Brazilian crops.

Michael Rothballer studied Biology (Diploma) at the Technical University of Munich (1994-2000). He accomplished his PhD at GSF-Research Center for Environment and Health in Neuherberg in the working group Plant-Microbe Interaction of the Institute of Soil Ecology under supervision of Prof. Dr. Anton Hartmann. 2004-6 he was a scientific employee at the GSF-Research Center in the Department of Rhizosphere Biology in the Institute of Soil Ecology and from 2006 he was deputy group leader of the working group Molecular Microbial Ecology in the research unit Microbe-Plant Interactions at the Helmholtz Zentrum München. From June 2016 until January 2017 he was acting as a head of the Research Unit Microbe-Plant Interactions. Finally, in February 2017 he became part of the newly founded Institute of Network Biology at the Helmholtz Zentrum München under Prof. Pascal Falter-Braun, where he leads the working group Molecular Microbial Ecology.
Footnotes
Peer review under responsibility of Cairo University.
Contributor Information
Anton Hartmann, Email: ahartmanndr@gmail.com.
Michael Rothballer, Email: rothballer@helmholtz-muenchen.de.
References
- 1.Zilber-Rosenberg I., Rosenberg E. Role of micro-organisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev. 2008;32:723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 2.Mc Fall-Ngai M., Hadfield M.G., Bosch T.C.G., Carey H.V., Donazet-Loso T., Douglas A.E. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–3232. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moran N.A., Sloan D.B. The hologenome concept: helpful or hollow? PLOS Biol. 2015;13:e1002311. doi: 10.1371/journal.pbio.1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stencel A., Wloch-Salamon D.M. Some theoretical insights into the hologenome theory of evolution and the role of microbes in speciation. Theory Biosci. 2018;137:197–206. doi: 10.1007/s12064-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galippe V. Note sur la presence de micro-organismes dans les tissus végétaux. C R Hebd Sci Mem Soc Biol. 1887;39:410–416. [Google Scholar]
- 6.Burris R.H. Biological nitrogen fixation: a scientific perspective. Plant Soil. 1988;108:7–14. [Google Scholar]
- 7.Hiltner L. Über neuere Erfahrungen und Probleme auf dem Gebiete der Bodenbakteriologie unter besonderer Berücksichtigung der Brache. Arb Dtsch Landwirtsch Gesellschaft. 1904;98:59–78. [Google Scholar]
- 8.Hartmann A., Rothballer M., Schmid M., Hiltner Lorenz. A pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil. 2008;312:7–14. [Google Scholar]
- 9.Nobbe F., Hiltner L. Impfet den Boden! Sächsische landwirtschaftl. Zeitschrift. 1893;16:1–5. [Google Scholar]
- 10.Döbereiner J., Day J.M. Associative symbiosis in tropical grasses: characterization of microorganisms and dinitrogen fixing sites. In: Newton W.E., Nyman C.J.N., editors. Proc. 1st Int Symp Nitrogen Fixation. Washington State Univ. Press; Washington; Pullman: 1976. pp. 518–538. [Google Scholar]
- 11.Baldani J.I., Baldani V.L.D., Seldin L., Döbereiner J. Characterization of Herbaspirillum seropedicae gen. nov, spec. nov., a root-associated nitrogen-fixing bacterium. Int J Syst Bacteriol. 1986;36:86–93. [Google Scholar]
- 12.Baldani V.L.D., Baldani J.I., Doebereiner J. Effects of Azospirillum inoculation on root infection and nitrogen incorporation in wheat. Can J Microbiol. 1983;29:924–929. [Google Scholar]
- 13.Cavalcante V.A., Doebereiner J. A new acid-tolerant niotrogen fixing bacterium associated with sugarcane. Plant Soil. 1988;108:23–31. [Google Scholar]
- 14.Michahelles K. Hanne: Johanna Döbereiner, uma vida dedicada à ciencia. 2018.
- 15.Klingmüller W. (Ed.) Azospirillum I: Geneticvs, Physiology, Ecology. EXS42: Experientia Supplementum 42, Birkhäuser Verlag, Basel, Boston, Stuttgart 1982; pp 149, ISBN 3-7643-1330-7.
- 16.Klingmüller W. (Ed.) Azospirillum II: Genetics, Physiology, Ecology. EXS 48: Experientia Supplementum 48 Birkhäuser Verlag, Basel Boston, Stuttgart 1983; pp 194, ISBN 3-7643-1576-8.
- 17.Klingmüller W. (Ed.) Azospirillum III: Genetics, Physiology, Ecology. Springer-Verlag Berlin, Heidelberg, New York, Tokyo 1985; pp 263, ISBN 3-540-15914-2.
- 18.Klingmüller W. (Ed.) Azospirillum IV: Genetics, Physiology, Ecology. Springer-Verlag Berlin, Heidelberg, New York, Tokyo 1987, ISBN 3-642-73072-8.
- 19.Del Gallo M, Fendrik I. (Eds.) Azospirillum and Related Microorganisms V: Genetics, Physiology, Ecology. Symbiosis 1992;13: pp 315.
- 20.Fendrik I, del Gallo M, Vanderleyden J, de Zamaroczy M. (Eds.) Azospirillum and Related Microorganisms VI: Genetics, Physiology, Ecology. NATO ASI Series G: Ecological Sciences 1995;3: pp 570, Springer-Verlag Berlin, Heidelberg, New York, pp 570, ISBN 3-540-60107-4.
- 21.Reinhold-Hurek B., Buenger W., Burbano C.S., Sabale M., Hurek T. Roots shaping their microbiome: global hot spots for microbial activity. Annu Rev Phytopathol. 2015;53:403–424. doi: 10.1146/annurev-phyto-082712-102342. [DOI] [PubMed] [Google Scholar]
- 22.Kandel S.L., Joubert P.M., Doty S.L. Bacterial endophytes: colonization and distribution within plants. Microorganisms. 2017;5:77. doi: 10.3390/microorganisms5040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia M.M., Pereira L.C., Braccini A.L., Angelotti P., Suzukwa A.K., Marteli D.C.V. Effects of Azospirillum brasilense on growth and yield components of maize grown at nitrogen limitaing conditions. Revista de Ciencia Agrárias. 2017;40:353–362. [Google Scholar]
- 24.Berendsen R.L., Pieterse C.M.J., Bakker P.A.H.M. The rhizospere microbiome and plant health. Cell. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Schloter M., Bode W., Hartmann A. Characterization of monoclonal antibodies against cell surface structures of Azospirillum brasilense Sp7. Symbiosis. 1992;13:37–45. [Google Scholar]
- 26.Schloter M., Moens S., Croos C., Reidel G., Esquenet M., De Mot R. Characterization of cell surface components of Azospirillum brasilense Sp7 as antigenic determinants for strain specific monoclonal antibodies. Microbiol. 1994;140:823–828. [Google Scholar]
- 27.Schloter M., Borlinghaus R., Bode W., Hartmann A. Direct identification and localization of Azospirillum in the rhizosphere of wheat using fluorescence-labelled monoclonal antibodies and confocal scanning laser microscopy. J Microscopy. 1993;171:173–177. [Google Scholar]
- 28.Schloter M., Wiehe W., Assmus B., Steindl H., Becke H., Höflich G. Root colonization of different plants by plant growth promoting Rhizobium leguminosarum bv. trifolii R39 studied with monospecific polyclonal antisera. Appl Environ Microbiol. 1997;63:2038–2046. doi: 10.1128/aem.63.5.2038-2046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanni Y.G., Dazzo F.B., Squartini A. Assessment of the natural endophytic association between Rhizobium and wheat and its ability to increase wheat production in the Nile delta. Plant Soil. 1997;407:367–383. [Google Scholar]
- 30.Baldani J.I., Carnoa L., Baldani V.L.D., Goi S.R., Doebereiner J. Recent advances in BNF with non-legume plants. Soil Biol Biochem. 1992;29:911–922. [Google Scholar]
- 31.Schloter M., Hartmann A. Production and characterization of strain-specific monoclonal antibodies against outer membrane components of Azospirillum brasilense Sp245. Hybridoma. 1996;15:225–232. doi: 10.1089/hyb.1996.15.225. [DOI] [PubMed] [Google Scholar]
- 32.Schloter M., Hartmann A. Endophytic and surface colonization of wheat roots (Triticum aestivum) by different Azospirillum brasilense strains studied with strain-specific monoclonal antibodies. Symbiosis. 1998;25:159–179. [Google Scholar]
- 33.Hartmann A., James E.K., de Brujin F., Schwab S., Rothballer M., Schmid M. In situ localization and strain-specific quantification of Azospirillum and other diazotrophic plant growth-promoting rhizobacteria (PGPR) using antibodies and molecular probes. In: Cassan F.D., Okon Y., Creus C., editors. In: Handbook for Azospirillum. Technical Issues and Protocols. Springer International Publishing; Switzerland: 2015. pp. 45–64. [Google Scholar]
- 34.Schloter M., Assmus B., Hartmann A. The use of immunological methods to detect and identify bacteria in the environment. Biotech Adv. 1995;13:75–90. doi: 10.1016/0734-9750(94)00023-6. [DOI] [PubMed] [Google Scholar]
- 35.Schloter M., Zelles L., Hartmann A., Munch J.C. New quality of assessment of microbial diversity in arable soils using molecular and biochemical methods. J Plant Nutr Soil Sci. 1998;161:425–431. [Google Scholar]
- 36.Lebuhn M., Achouak W., Schloter M., Berge O., Meier H., Hartmann A. Taxonomic characterization of Ochrobactrum sp. isolates from soil samples and wheat roots, and description of Ochrobactrum tritici sp. nov. and Ochrobactrum grignonense sp. nov. Int J Syst Evol Microbiol. 2000;50:2207–2223. doi: 10.1099/00207713-50-6-2207. [DOI] [PubMed] [Google Scholar]
- 37.Woese C.R., Kandeler O., Wheelis M.L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria and Eucarya. Proc Natl Acad Sci USA. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeLong E.F., Wickham G.S., Pace N.R. Phylogenetic stains: ribosomal RNA based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 39.Schmid M., Rothballer M., Hartmann A. Analysis of microbial communities in soil microhabitats using fluorescence in situ hybridization. In: van Elsas J.D., Jansson J.K., Trevors J.T., editors. In: Modern Soil Microbiology. 2nd ed. CRC-Press, Boca Raton; Florida, USA: 2007. pp. 317–355. [Google Scholar]
- 40.Wallner G., Amann R., Beisker W. Optimizing fluorescence in situ hybridization with rRNA-targeted oligonucleotides. Cytometry. 1993;14(2):136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 41.Amann R.I., Ludwig W., Schleifer K.H. Phylogenetic identification and in situ detection of individual cells. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Assmus B., Hutzler P., Kirchhof G., Amann R.I., Lawrence J.R., Hartmann A. In situ localization of A. brasilense in the rhizosphere of wheat using fluorescently labeled, rRNA-targeted oligonucleotide probes and scanning confocal laser microscopy. Appl Env Microbiol. 1995;61:1013–1019. doi: 10.1128/aem.61.3.1013-1019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Assmus B., Schloter M., Kirchhof G., Hutzler P., Hartmann A. Improved in situ tracking of rhizosphere bacteria using dual staining with fluorescence-labeled antibodies and rRNA-targeted oligonucleotides. Microb Ecol. 1997;33:32–40. doi: 10.1007/s002489900005. [DOI] [PubMed] [Google Scholar]
- 44.Stoffels M., Castellanos T., Hartmann A. Design and application of new 16S rRNA-targeted oligonucleotide probes for the Azospirillum-Skermanella-Rhodocista-cluster. Syst Appl Microbiol. 2001;24:83–97. doi: 10.1078/0723-2020-00011. [DOI] [PubMed] [Google Scholar]
- 45.Stets M.I., Alqueres S.M.C., Souza E.M., Pedrosa F.O., Schmid M., Hartmann A. Quantification of Azospirillum brasilense FP2 bacteria in wheat roots by strain-specific quantitative PCR. Appl Environ Microbiol. 2015;81:6700–6709. doi: 10.1128/AEM.01351-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doreen Fischer, Doktorarbeit/PhD thesis, Ludwig-Maximilians-Universität München, 2010. title: Molekulare Analyse diazotropher Bakterien in Zuckerrohr (Saccharum officinarum).
- 47.Schmidt H., Eickhorst T., Mussmann T. GOLD-FISH: a new approach for the in situ detection of single microbial cells combining fluorescence and scanning electron microscopy. Syst Appl Microbiol. 2012;35:518–525. doi: 10.1016/j.syapm.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 48.Crivat G., Taraska J.W. Imaging proteins inside cells with fluorescent tags. Trends Biotechnol. 2012;30(1):8–16. doi: 10.1016/j.tibtech.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reinhold-Hurek B., Hurek T. Living inside plants: bacterial endophytes. Curr Opin Plant Biol. 2011;14:435–443. doi: 10.1016/j.pbi.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Rothballer M., Schmid M., Fekete A., Hartmann A. Comparative in situ analysis of ipdC-gfpmut3 promotor fusions of Azospirillum brasilense strains Sp7 and Sp245. Environ Microbiol. 2005;7:1839–1846. doi: 10.1111/j.1462-2920.2005.00848.x. [DOI] [PubMed] [Google Scholar]
- 51.Santos A.R.S., Etto R.M., Furmam R.W., de Freitas D.L., Santos K.F.D.N., Souza E.M. Labeled Azospirillum brasilense wild type and ammonium-excreting strains in association with barley roots. Plant Physiol Biochem. 2017;118:422–426. doi: 10.1016/j.plaphy.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 52.You M., Nishiguchi T., Saito A., Isawa T., Mitsui H., Minamisawa K. Expression of the nifH gene of a Herbaspirillum endophyte in wild rice species: daily rhythm during the light-dark cycle. Appl Env Microbiol. 2005;71:8183–8190. doi: 10.1128/AEM.71.12.8183-8190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fischer D., Pfitzner B., Schmid M., Simoes-Araújo Reis V, Pereira W. Molecular characterization of the diazotrophic bacterial community in non-inoculated and inoculated field-grown sugarcane (Saccharum sp.) Plant Soil. 2012;356:83–99. [Google Scholar]
- 54.Rouws L.F.M., Leite J., de Matos G.F., Zilli J.E., Coelho M.R., Xavier G.R. Endophytic Bradyrhizobium spp. isolates from sugarcane obtained through different culture strategies. Environ Microbiol Rep. 2014;6:354–363. doi: 10.1111/1758-2229.12122. [DOI] [PubMed] [Google Scholar]
- 55.Armanhi JSL, de Souza RSC, de Brito Damasceno N, de Araújo LM, Imperial J, Arruda P. A community-based culture collection for targeting novel plant growth-promoting bacteria from the sugarcane microbiome. Front Plant Sci 2018; 8: 2191. [DOI] [PMC free article] [PubMed]
- 56.Sessitsch A., Hardoim P., Döring J., Weilharter A., Krause A., Woyke T. Functional characterization of an endophyte community colonizing rice roots as revealed by metagenome analysis. Mol Plant Microbe Interact. 2012;25:28–36. doi: 10.1094/MPMI-08-11-0204. [DOI] [PubMed] [Google Scholar]
- 57.Drogue B., Sanguin H., Borland S., Prigent-Combaret C., Wisniewski-Dyé F. Genome wide profiling of Azospirillum lipoferum 4B gene expression during interaction with rice roots. FEMS Microbiol Ecol. 2014;87:543–555. doi: 10.1111/1574-6941.12244. [DOI] [PubMed] [Google Scholar]
- 58.Camilios-Neto D., Bonato P., Wassem R., Tadra-Sfeir M.Z., Brusamarello-Santos L.C.C., Valdameri G. Dual RNA-seq transcriptional analysis of wheat roots colonized by Azospirillum brasilense reveals up-regulation of nutrient acquisition and cell cycle genes. BMC Geno. 2014;15:378. doi: 10.1186/1471-2164-15-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paungfoo-Lonhienne C., Lonhienne T.G.A., Yeoh Y.K., Donose B.C., Webb E.I., Parsons J. Crosstalk between sugarcane and a plant-growth promoting Burkholderia species. Sci Rep. 2016;6:37389. doi: 10.1038/srep37389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Urseil L.K., Metcaf J.L., Parfrey L.W., Knight R. Defining the human microbiome. Nutr Rev. 2012;70(Suppl 1):S38–S44. doi: 10.1111/j.1753-4887.2012.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sczyrba A., Hofmann P., Belmann P., Koslicki D., Janssen S. Critical assessment of metagenome interpretation – a benchmark of metagenomic software. Nat Methods. 2017;14:1063–1071. doi: 10.1038/nmeth.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berg G., Eberl L., Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Env Microbiol. 2005;7:1673–1685. doi: 10.1111/j.1462-2920.2005.00891.x. [DOI] [PubMed] [Google Scholar]
- 63.Mendes R., Garbeva P., Raaijmakers J. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 64.Abreo E., Altier N. Pangenome of Serratia marcescens strains from nosocomial and environmental origins reveals different populations and the link between them. Sci Rep. 2019;9:46. doi: 10.1038/s41598-018-37118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cohen M.F., Han X.Y., Mazzola M. Molecular and physiological comparison of Azospirillum spp. isolated from Rhizoctonia solani mycelia, wheat rhizosphere and human skin wounds. Can J Microbiol. 2004;50:291–297. doi: 10.1139/w04-007. [DOI] [PubMed] [Google Scholar]
- 66.Helsel L.O., Hollis D.G., Steigerwalt A.G., Levett P.N. Reclassification of Roseomonas fauriae Rihs et al. 1998 as a later heterotypic synoym of Azospirillum brasilense Tarrand et al. 1979 . Int J Syst Evol Microbiol. 2006;56:2753–2755. doi: 10.1099/ijs.0.64549-0. [DOI] [PubMed] [Google Scholar]
- 67.Brenner D.J., McWhorter A.C., Knutson J.K., Steigerwalt A.G. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J Clin Microbiol. 1982;15:1133–1140. doi: 10.1128/jcm.15.6.1133-1140.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Linda Kinzel. Vergleichende phylogenetische und phänotypische Charakterisierung von Roseomonas spp. und Azospirillum spp. zur taxonomischen Einordnung von Roseomonas fauriae und Roseomonas genomospecies 6, Lehrstuhl für Mikrobiologie LMU, Munich, September 2008. Diploma thesis.
- 69.Huss V.A.R., Festl H., Schleifer K.H. Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol. 1983;4:184–192. doi: 10.1016/S0723-2020(83)80048-4. [DOI] [PubMed] [Google Scholar]
- 70.Maroniche G.A., Garcia J.E., Salcedo F. Creus CM. Molecular identification of Azospirillum spp.: limitations of 16S rRNA and qualities of rpoD as genetic markers. Microbiol Res. 2017;195:1–10. doi: 10.1016/j.micres.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 71.Lin S.Y., Shen F.T., Young L.S., Zhu Z.L., Chen W.M., Young C.C. Azospirillum formosense sp. nov., a diazotroph from agricultural soil. Int J Syst Evol Micro biol. 2012;62:1185–1190. doi: 10.1099/ijs.0.030585-0. [DOI] [PubMed] [Google Scholar]
- 72.Tyagi S., Singh D.K. Azospirillum himalayense sp. nov., a nifH-bacterium isolated from Himalayan valley soil. Ann. Microbiol. 2014;64:259–266. [Google Scholar]
- 73.Nabti E., Sahnoune M., Adjrad S., van Dommelen A., Ghoul M., Schmid M. A halophilic and osmotolerant Azospirillum brasilense strain from Algerian soil restores wheat growth under saline conditions. Eng Life Sci. 2007;7:354–360. [Google Scholar]
- 74.Angus A.A., Agapakis C.M., Fong S., Yerrapragada S., Estrada-de los Santos P. Plant associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLoS ONE. 2014;9:e83779. doi: 10.1371/journal.pone.0083779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sawana A., Adeolu M., Gupta R.S. Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet. 2014;5:429. doi: 10.3389/fgene.2014.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dobritsa A.P., Samadpour M. Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int J Syst Evol Microbiol. 2016;66:2836–2846. doi: 10.1099/ijsem.0.001065. [DOI] [PubMed] [Google Scholar]
- 77.Wood J.P. Bacterial responses to osmotic challenges. J Gen Physiol. 2015;145(5):381. doi: 10.1085/jgp.201411296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nabti E., Schmid M., Hartmann A. Application of halotolerant bacteria to restore plant growth under salt stress. In: Dinesh K.M., Meenu S., editors. Halophiles. Springer International Publishing; New York: 2015. pp. 235–259. [Google Scholar]
- 79.Miller K.J., Wood J.M. Osmoadaptation by rhizosphere bacteria. Annu Rev Microbiol. 1996;50:101–136. doi: 10.1146/annurev.micro.50.1.101. [DOI] [PubMed] [Google Scholar]
- 80.Nabti E., Sahnoune M., Ghoul M., Fischer D., Hofmann A., Rothballer M. Restoration of growth of durum wheat (Triticum aestivum var. waha) under saline conditions due to inoculation with the rhizosphere bacterium Azospirillum brasilense NH and extracts of the marine alga Ulva lactuca. J Plant Growth Regul. 2010;29:6–22. [Google Scholar]
- 81.Glick B.R. Bacteria with ACC-deaminase can promote plant growth and help to feed the world. Microbiol Res. 2014;169:30–39. doi: 10.1016/j.micres.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 82.Nascimento F.X., Rossi M.J., Soares C.R.F.S., Mc Conkey B.J., Glick B.R. New insights into 1-aminocyclopropane-1-carbocylate (ACC) deaminase phylogeny, evolution and ecological significance. PLoS ONE. 2014;9(6):e99168. doi: 10.1371/journal.pone.0099168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jha B., Gontia I., Hartmann A. The roots of the halophyte Salicornia brachiata are source of new halotolerant diazotrophic bacteria with plant growth promoting potential. Plant Soil. 2012;356:265–277. [Google Scholar]
- 84.Hartmann A. Ecophysiological effects on growth and nitrogen fixation in Azospirillum spp. Plant Soil. 1988;110:225–238. [Google Scholar]
- 85.Hartmann A., Prabhu S.R., Galinski E.A. Osmotolerance of diazotrophic rhizosphere bacteria. Plant Soil. 1991;137:105–109. [Google Scholar]
- 86.Riou N., Le Rudulier D. Osmoregulation in Azospirillum brasilense: glycine betaine transport enhances growth and nitrogen fixation under salt stress. J Gen Microbiol. 1990;136:1455–1461. doi: 10.1099/00221287-136-8-1455. [DOI] [PubMed] [Google Scholar]
- 87.Rodriguez-Salazar J., Suarez R., Caballero-Mellado J., Iturriaga G. Trehalose accumulation in Azospirillum brasilense improves drought tolerance and biomass in maize plants. FEMS Microbiol Lett. 2009;296:52–59. doi: 10.1111/j.1574-6968.2009.01614.x. [DOI] [PubMed] [Google Scholar]
- 88.Hartmann A., Fu H., Burris R.H. Influence of amino acids on nitrogen fixation activity and growth in Azospirillum spp. Appl Environ Microbiol. 1988;54:87–93. doi: 10.1128/aem.54.1.87-93.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hartmann A., Gündisch C., Bode W. Azospirillum mutants improved in iron acquisition and osmotolerance as tools for the investigation of environmental fitness traits. Symbiosis. 1992;13:271–279. [Google Scholar]
- 90.Malinich E.A., Bauer C.E. Transcriptome analysis of Azospirillum brasilense vegetative and cyst states reveals large-scale alterations in metabolic and replicative gene expression. Microb Genom. 2018;4:200. doi: 10.1099/mgen.0.000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fibach-Paldi S., Burdman S., Okon Y. Key physiological properties contributing to rhizosphere adaptation and PGP-abilities of Azospirillum brasilense. FEMS Microbiol Lett. 2012;326:99–108. doi: 10.1111/j.1574-6968.2011.02407.x. [DOI] [PubMed] [Google Scholar]
- 92.Molina-Favero C., Creus C.M., Simontacchi M., Puntarulo S., Lamattina L. Aerobic nitrite oxide production of Azospirillum brasilense Sp245 and its influence on root architecture in tomato. Mol Plant Microbe Interact. 2006;21:1001–1009. doi: 10.1094/MPMI-21-7-1001. [DOI] [PubMed] [Google Scholar]
- 93.Hartmann A., Singh M., Klingmüller W. Isolation and characterization of Azospirillum mutants excreting high amounts of indole acetic acid. Can J Microbiol. 1983;29:916–923. [Google Scholar]
- 94.Hartmann A, Fußeder A, Klingmüller W. Mutants of Azospirillum affected in nitrogen fixation and auxin production. In: Azospirillum II: Genetics, Physiology, Ecology. (Ed.: W. Klingmüller) Birkhäuser Verlag, Basel, Boston, Stuttgart; Experientia Suppl 1983;48:78-88.
- 95.Machado H.B., Funayama S., Rigo L.U., Pedrosa F.O. Excretion of ammonium by Azospirillum brasilense mutants resistant to ethylenediamine. Can J Microbial. 1991;37:549–553. [Google Scholar]
- 96.Pankievicz V.C.S., Amaral F.P., Santos K.F.D.N., Agtuca B., Xu Y., Schueller M.J. Robust biological nitrogen fixation in a model grass-bacterial association. Plant J. 2015;81:907–919. doi: 10.1111/tpj.12777. [DOI] [PubMed] [Google Scholar]
- 97.Santos K.F.D.N., Moure V.R., Hauer V., Santos A.R.S., Donatti L., Galvao C.W. Wheat colonization by an Azospirillum brasilense ammonium-excreting strain reveals upregulation of nitrogenase and superior plant growth promotion. Plant Soil. 2017;415:245–255. [Google Scholar]
- 98.Hartmann A, Zimmer W. Physiology of Azospirillum. In: Azospirillum/Plant Associations. (Okon Y. ,Ed.) CRC Press, Boca Raton, USA. 1994, pp. 15–39.
- 99.Alqueres S., Menses C., Rouws L.M., Rothballer M., Baldani I., Schmid M. The bacterial superoxide dismutase and glutathione reductase are crucial for endophytic colonization of rice roots by Gluconacetobacter diazotrophicus strain PAL5. Mol Plant Microbe Interact. 2013;26:937–945. doi: 10.1094/MPMI-12-12-0286-R. [DOI] [PubMed] [Google Scholar]
- 100.Meneses C.H., Rouws L.F., Simoes-Araujo J.L., Vidal M.S., Baldani J.I. Exopolysaccharide production is required for biofilm formation and plant colonization by the nitrogen-fixing endophyte Gluconacetobacter diazotrophicus PAL5. Mol Plant Microbe Interact. 2011;24:1448–1458. doi: 10.1094/MPMI-05-11-0127. [DOI] [PubMed] [Google Scholar]
- 101.Hense B.A., Kuttler C., Müller J., Rothballer M., Hartmann A., Kreft J.U. Does efficiency sensing unify diffusion and quorum sensing? Nature Rev Microbiol. 2007;5:230–239. doi: 10.1038/nrmicro1600. [DOI] [PubMed] [Google Scholar]
- 102.Gantner S., Schmid M., Duerr C., Schuhegger R., Steidle A., Hutzler P. In situ spatial scale of calling distances and population density-dependent N-acylhomoserine lactone mediated communication by rhizobacteria colonized on plant roots. FEMS Microbiol Ecol. 2006;56:188–194. doi: 10.1111/j.1574-6941.2005.00037.x. [DOI] [PubMed] [Google Scholar]
- 103.Dazzo F.B., Yanni Y., Jones A., Elsadany A. CMEIAS bioimage informatics that define the landscape ecology of immature microbial biofilms developed on plant rhizoplane surfaces. AIMS Bioeng. 2015;2:469–486. [Google Scholar]
- 104.Hartmann A., Schikora A. Quorum sensing of bacteria and trans-kingdom interactions of N-acylhomoserine lactones with eucaryotes. J Chem Ecol. 2012;38:704–713. doi: 10.1007/s10886-012-0141-7. [DOI] [PubMed] [Google Scholar]
- 105.von Rad U., Klein I., Dobrev P.I., Kottova J., Zazimalova E., Fekete A. The response of Arabidopsis thaliana to N-hexanoyl-DL-homoserine lactone, a bacterial quorum sensing molecule produced in the rhizosphere. Planta. 2008;229:73–85. doi: 10.1007/s00425-008-0811-4. [DOI] [PubMed] [Google Scholar]
- 106.Sieper T., Forczek S., Matucha M., Krämer P., Hartmann A., Schröder P. N-acylhomoserine lactone uptake and systemic transport in barley rest upon active parts of the plant. New Phytol. 2014;201:545–555. doi: 10.1111/nph.12519. [DOI] [PubMed] [Google Scholar]
- 107.Götz-Rösch C., Riedel T., Schmitt-Kopplin P., Hartmann A., Schröder P. Influence of bacterial N-acylhomoserine lactones on growth parameters, pigments, antioxidative capacities and the xenobiotic phase II detoxification enzymes in barley and yam bean. Front Plant Sci. 2015;6:205. doi: 10.3389/fpls.2015.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rankl S., Gunsé B., Sieper T., Schmid C., Poschenrieder C., Schroeder P. Microbial N-acyl-homoserine lactones (AHLs) are effectors of root morphological changes in barley. Plant Sci. 2016;253:130–140. doi: 10.1016/j.plantsci.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 109.Schenk S.T., Hernández-Reyes C., Samans B., Stein E., Neumann C., Schikora M. N-acyl-homoserine lactone primes plants for cell wall reinforcement and induces resistance to bacterial pathogens via the salicylic acid/oxylipin pathway. Plant Cell. 2014;26:2708–2723. doi: 10.1105/tpc.114.126763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schikora A., Schenk S.T., Hartmann A. Beneficial effects of bacteria-plant communication based on quorum sensing molecules of the N-acyl-homoserine lactone group. Plant Mol Biol. 2016;90:605–612. doi: 10.1007/s11103-016-0457-8. [DOI] [PubMed] [Google Scholar]
- 111.Schikora A., Schenk S.T., Stein E., Molitor A., Zuccaro A., Kogel K.H. N-acyl- homoserine lactone confers resistance towards biotrophic and hemibiotrophic pathogens via altered activation of AtMPK6. Plant Physiol. 2011;157:1407–1418. doi: 10.1104/pp.111.180604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Han S., Li D., Trost E., Mayer K.F., Vlot A.C., Heller W. Systemic responses of barley to the 3-hydroxy-decanoyl-homoserine lactone producing plant beneficial endophyte Acidovorax radicis N35. Front Plant Sci. 2016;7:1868. doi: 10.3389/fpls.2016.01868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fukami J., Abrantes J.L.F., del Cerro P., Nogueira M.A., Ollero F.J., Megias M. Revealing strategies of quorum sensing in Azospirillum brasilense strains Ab-V5 and Ab-V6. Arch Microbiol. 2018;200:47–56. doi: 10.1007/s00203-017-1422-x. [DOI] [PubMed] [Google Scholar]
- 114.Vial L., Cuny C., Gluchoff-Fiasson K., Comte G., Oger P.M., Faure D. N-acyl-homoserine lactone-mediated quorum sensing in Azospirillum: an exception rather than a rule. FEMS Microbiol Ecol. 2006;58:155–168. doi: 10.1111/j.1574-6941.2006.00153.x. [DOI] [PubMed] [Google Scholar]
- 115.Patel H.K., Suárez-Moreno Z.R., Degrassi G., Subramoni S., González J.F., Venturi V. Bacterial LuxR solos have evolved to respond to different molecules including signals from plants. Front Plant Sci. 2013;4:447. doi: 10.3389/fpls.2013.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chowdhury S.P., Hartmann A., Gao X., Borriss R. Biocontrol mechanism of root-associated Bacillus amyloliquefaciens FZB42 – a review. Front Microbiol. 2015;6:780. doi: 10.3389/fmicb.2015.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sharifi R., Ryu C.M. Revisiting bacterial volatile-mediated plant growth promotion: lessons from the past and objectives for the future. Ann Botany. 2018;122:349–358. doi: 10.1093/aob/mcy108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sánchez-Canizares C., Jorrin B., Poole P.S., Tkacz A. Understanding the holobiont: the interdependence of plants and their microbiome. Curr Opin Microbiol. 2017;38:188–196. doi: 10.1016/j.mib.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 119.Leach J.E., Triplett L.R., Argueso C.T., Trivedi P. Communication in the Phytobiome. Cell. 2017;169:587–596. doi: 10.1016/j.cell.2017.04.025. [DOI] [PubMed] [Google Scholar]