Abstract
Cadmium is a highly toxic heavy metal for biological systems. Cupriavidus metallidurans CH34 is a model strain to study heavy metal resistance and bioremediation as it is able to deal with high heavy metal concentrations. Biofilm formation by bacteria is mediated by the second messenger bis-(3′–5′)-cyclic dimeric guanosine monophosphate (c-di-GMP). The aim of this study was to characterize the response of C. metallidurans CH34 planktonic and biofilm cells to cadmium including their c-di-GMP regulatory pathway. Inhibition of the initiation of biofilm formation and EPS production by C. metallidurans CH34 correlates with increased concentration of cadmium. Planktonic and biofilm cells showed similar tolerance to cadmium. During exposure to cadmium an acute decrease of c-di-GMP levels in planktonic and biofilm cells was observed. Transcription analysis by RT-qPCR showed that cadmium exposure to planktonic and biofilm cells induced the expression of the urf2 gene and the mercuric reductase encoding merA gene, which belong to the Tn501/Tn21 mer operon. After exposure to cadmium, the cadA gene involved in cadmium resistance was equally upregulated in both lifestyles. Bioinformatic analysis and complementation assays indicated that the protein encoded by the urf2 gene is a functional phosphodiesterase (PDE) involved in the c-di-GMP metabolism. We propose to rename the urf2 gene as mrp gene for metal regulated PDE. An increase of the second messenger c-di-GMP content by the heterologous expression of the constitutively active diguanylate cyclase PleD correlated with an increase in biofilm formation and cadmium susceptibility. These results indicate that the response to cadmium in C. metallidurans CH34 inhibits the initiation of biofilm lifestyle and involves a decrease in c-di-GMP levels and a novel metal regulated PDE.
Keywords: c-di-GMP, Cupriavidus metallidurans, cadmium, phosphodiesterase, biofilm, urf2 gene, mer gene, PleD
Introduction
Cadmium is a highly toxic heavy metal for biological systems. Cadmium induces indirectly oxidative stress, disturbs manganese acquisition, and depletes the cellular reduced thiol pool (Vallee and Ulmer, 1972; Nies, 1999; Begg et al., 2015). This heavy metal is also carcinogenic (World Health Organization [WHO], 2011). Global cadmium production has risen >1,000-fold in the last century due to its incorporation into diverse manufacture applications (Begg et al., 2015). The Word Health Organization included cadmium in the top 10 of the “chemicals of major public health concern” (World Health Organization [WHO], 2011).
The remediation of heavy metals in polluted waters and soils is a major challenge for sustainable development (Orellana et al., 2018). Microorganisms are used to immobilize and/or transform heavy metals into less toxic forms (Rojas et al., 2011; Mosa et al., 2016; Orellana et al., 2018). Heavy metal bioremediation is commonly carried out in fixed bed bioreactors wherein the formation of attached multicellular communities – i.e., biofilm – is crucial. Biofilm cells produce extracellular polymeric substances (EPS) such as polysaccharides, lipids, proteins, and extracellular DNA, providing different functional groups that may bind heavy metal ions via passive biosorption processes that immobilize heavy metals and protect cells (Harrison et al., 2007). Biofilms are also involved in bioreduction processes through electrically conductive components (Harrison et al., 2004b; Chen et al., 2018; Espinoza-Tofalos et al., 2018). Biofilms have been described as an advantageous lifestyle promoted by stress, toxic compounds, or heavy metals, which allow the selection of resistant cells (Harrison et al., 2007). However, in contrast to the bacterial response to antibiotics, planktonic and biofilm cells are in most cases almost equally susceptible to heavy metal oxyanions and cations (Harrison et al., 2004b; Gugala et al., 2017). Consequently, the minimum biofilm-eradication concentration (MBEC) is often not higher than the minimum bactericidal concentration (MBC) observed under planktonic growth conditions. Specific bacterial biofilms are even more susceptible to some heavy metal oxyanions and cations than planktonic cells (Harrison et al., 2004a). Metal tolerance in bacterial biofilms is dependent on the bacterial strain, the metal, and the culture medium (Harrison et al., 2004a, 2005; Gugala et al., 2017).
Biofilm formation in response to environmental conditions is mediated by the second messenger bis-(3′–5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) (Jenal et al., 2017). Intracellular pools of c-di-GMP seem to be dynamically regulated by diguanylate cyclases (DGCs) and c-di-GMP-specific phosphodiesterases (PDEs) that catalyze the formation and the degradation of c-di-GMP, respectively. DGC activity depends on the characteristic amino acid motif GGDEF (Wassmann et al., 2007), whereas PDE activity is mediated by the two amino acid motifs EAL and HD-GYP (Tchigvintsev et al., 2010; Lovering et al., 2011). In addition to the catalytic motifs, DGCs and PDEs contain accessory motifs, which are responsible for the regulation of their activity under different conditions. Most DGCs and PDEs act at local manner and do not impact the cellular c-di-GMP levels (Sarenko et al., 2017; Dahlstrom et al., 2018). At high concentrations, c-di-GMP interacts with a broad spectrum of c-di-GMP binding effectors including riboswitches and a wide diversity of proteins. The effectors include GGDEF/EAL domain proteins (Duerig et al., 2009; Jain et al., 2017) and PilZ domain proteins (Habazettl et al., 2011). The c-di-GMP binding with specific effectors may trigger a reduction of cell motility and an increase of EPS matrix synthesis, which promote biofilm formation (Duerig et al., 2009; Römling et al., 2013; Hengge et al., 2016; Jain et al., 2017). The c-di-GMP signaling system is more complex, since flagella play roles in biofilm formation and matrix synthesis during stationary phase in liquid culture (Hengge et al., 2016). However, the link between the c-di-GMP signaling pathway, bacterial lifestyle, and heavy metal tolerance has not yet been explored.
Cupriavidus metallidurans CH34 is a model heavy metal-resistant strain that can tolerate millimolar concentrations of heavy metal concentrations. Strain CH34 has been used for bioremediation, recovery, and reduction of heavy metals (Mergeay et al., 1985; Collard et al., 1994; Janssen et al., 2010). C. metallidurans CH34 harbors numerous heavy metal resistance determinants involved in diverse mechanisms, including complexation, efflux systems, reduction, and reductive precipitation, which enable cell detoxification and survival (Nies, 2016; Maes et al., 2017; Montero-Silva et al., 2018). A high number of CH34 gene products confer tolerance to high concentrations of heavy metals, including the CzcCBA, CzcD, CzcP, and CadA proteins that are involved in cadmium resistance (Nies, 2016). Nevertheless, molecular mechanisms driving the bacterial lifestyle in the presence of heavy metals remain unexplored. The aim of this study was to characterize the response to cadmium of C. metallidurans CH34 planktonic and biofilm cells including their c-di-GMP regulatory pathway.
Materials and Methods
Organisms, Plasmids, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. C. metallidurans strains were cultured in low phosphate Tris-buffered mineral salts (LPTMS) broth [Tris 6.06 g L-1, NaCl 4.68 g L-1, NH4Cl 1.07 g L-1, KCl 1.49 g L-1, Na2SO4⋅10H2O 0.98 g L-1, MgCl2⋅6H2O 0.2 g L-1, CaCl2⋅2H20 0.03 g L-1, Na2HPO4 0.085 g L-1, Fe(III)(NH4) citrate 0.005 g L-1, succinate 0.3%, and 1 mL L-1 of SL7 trace solution of Biebl and Pfennig] (Mergeay et al., 1985). The culture medium was adjusted to pH 7.0. Escherichia coli and Pseudomonas aeruginosa strains were cultured in LB broth (yeast extract 5 g L-1, peptone 10 g L-1, NaCl 5 g L-1). In this study, plasmids pJB3Tc19 (Blatny et al., 1997) and pJBpleD∗ (Pérez-Mendoza et al., 2014) were used. The PleD∗ protein is a DGC from Caulobacter crescentus that contains four point mutations and retains its activity independently of phosphorylation input by a cognate kinase (Paul et al., 2004).
Table 1.
Plasmids and strains used in this study.
| Plasmids/strains | Description | References |
|---|---|---|
| pJB3Tc19 | Apr, Tcr; expression vector | Blatny et al., 1997 |
| pJBurf2.2 | Apr, Tcr; pJBTc19 derivate bearing a 1423 bp XbaI/EcoRI fragment containing RMET_RS31035 | This study |
| pJBpleD∗ | Apr, Tcr; pJB3Tc19 derivate bearing a 1423 bp XbaI/EcoRI fragment containing pleD∗ | Pérez-Mendoza et al., 2014 |
| C. metallidurans CH34 | Heavy metal multiresistant strain | Mergeay et al., 1985 |
| CH34 pJB3Tc19 | CH34 strain transformed with an empty vector | This study |
| CH34 pJBurf2.2 | CH34 strain transformed with pJBurf2.2 construct | This study |
| CH34 pJBpleD∗ | CH34 strain transformed with pJBpleD∗ construct | This study |
| P. aeruginosa PAO1 | Opportunistic pathogen | Rao et al., 2008 |
| ΔrocR pJB3Tc19 | rocR null mutant PAO1 strain transformed with pJB3Tc19 | This study |
| ΔrocR pJBurf2.2 | rocR null mutant PAO1 strain transformed with pJBurf2.2 | This study |
Biofilm Formation Assay
Cupriavidus metallidurans CH34 cell attachment in the absence or presence of cadmium was assessed by an adapted protocol developed to monitor initial stages of biofilm formation (O’Toole and Kolter, 1998; Merritt et al., 2007). Briefly, a stationary culture in LPTMS broth at 30°C was centrifuged and the cells were suspended in fresh LPTMS broth to a turbidity at 600 nm of 1.0. The culture was distributed by placing 200 μL per well into a 96-well microtiter plate. A concentrated cadmium solution was added to reach different concentrations. After 15 h of incubation at 30°C in static condition, planktonic cells were removed. Wells were washed with sterile distilled water and the biofilm adhered to the bottom was stained with crystal violet (200 μL per well, 0.1% solution for 15 min). Plate was air-dried for 16 h and the stained biomass was suspended in 200 μL of acetic acid 30%. Absorbance of this suspension was measured either at 550 or 595 nm. This value was normalized by the turbidity600nm of the planktonic fraction removed previously. Viability of each planktonic cell fraction was quantified by serial dilution and plating in triplicate on tryptic soy agar (pancreatic digest of casein 15 g L-1, peptic digest of soybean meal 5 g L-1, sodium chloride 5 g L-1, and agar 15 g L-1). Colony forming units (CFUs) were counted on plates.
Qualitative Assay for Extracellular Polymeric Substances
Levels of EPS synthesis by C. metallidurans CH34 macrocolonies were assessed in salt-free LB agar in presence of Congo Red 50 μg mL-1 and Coomassie blue 10 μg mL-1 (Romero et al., 2010). Plates were incubated for 5 days at room temperature and visualized with an Olympus MVX10 lens. The red staining intensity of the macrocolony correlates to its EPS content.
Analysis of Biofilm and Planktonic Lifestyles of C. metallidurans CH34
For comparisons between biofilm and planktonic lifestyles, two aliquots of an LPTMS broth culture (108 cells mL-1) were taken in parallel. For biofilm growth, 1 mL was vacuum filtered through a 0.2-μm polycarbonate filter (Whatman) and the filter was placed on solid LPTMS (0.8% agar) with the cell filtrate facing upward. The second 1 mL aliquot was inoculated into 9 mL of fresh LPTMS broth for planktonic growth. The cultures were incubated at 30°C. The biofilm was incubated without shaking, whereas the planktonic culture was incubated at 150 rpm. This procedure synchronized cultures of both lifestyles starting from the same growth phase and biomass. After 13 h at 30°C, cultures were exposed to 2 mM cadmium (Supplementary Figure S1), either by translating the filter to a new LPTMS agar with cadmium (biofilm cells) or by directly adding a cadmium concentrated solution (planktonic cells). After 45 min incubation in presence of cadmium, cells were processed for c-di-GMP determination and qRT-PCR.
Extraction and Quantification of c-di-GMP
After 45 min incubation in presence or absence of cadmium, 9 of the 10 mL of planktonic culture were collected, leaving 1 mL for total protein content determination. To discard sample processing bias, biofilm cells were suspended in a mixture of 10 mL of LPTMS, separating a 1 mL aliquot for total protein content determination. Cells in suspension were collected by centrifugation at 13,000 × g for 10 min at 4°C. Pellets were suspended in 300 μL of extraction solution (acetonitrile:methanol:water, 2:2:1) in 2 mL microtube vials and incubated 10 min at 95°C followed by cooling on ice. Cell lysates were centrifuged at 13,000 × g for 10 min at 4°C and supernatants were transferred into new microtube vials. Extractions were repeated twice omitting the heat incubation step. Combined supernatants of three extraction steps (about 700 μL) were stored at -20°C overnight. Remaining cellular debris was removed by centrifugation at 13,000 × g for 10 min at 4°C. Finally, supernatants were evaporated to dryness in Speed-Vac at room temperature. Nucleotide extraction from planktonic cells of P. aeruginosa strains followed the same procedure. Quantification of c-di-GMP was achieved by temperature-controlled liquid chromatography (using a C18 RP column) coupled to tandem mass spectrometer (LC–MS/MS) equipped with an electro spray ionization (ESI) source (positive ionization mode) at BIOLOG Life-Sciences Institute (Bremen, Germany) based on the method reported previously (Spangler et al., 2010).
Susceptibility Tests in Calgary Biofilm Device
Cupriavidus metallidurans cultures in the Calgary Biofilm Device (96-well format with lid containing pegs that insert into each well) were performed as reported (Harrison et al., 2010). Briefly, colonies grown on LPTMS agar during 48 h incubation at 30°C were suspended in NaCl 0.9% to reach a turbidity equivalent to McFarland standard 1.0 (Thermo R20421). The suspension was diluted (15-fold) in LPTMS broth, loaded into the Calgary Biofilm Device (Innovotech) and incubated for 24 h at 150 rpm and 30°C. Different cadmium concentrations were titrated across the microtiter plates. After incubation, the peg lids were removed thus separating the biofilms adhered to the lid pegs from the planktonic fraction in the well. Cadmium susceptibility was determined by CFU counting in tryptic soy agar after 24 h of cadmium challenge incubations. MBC and MBEC were defined as the point where no growth was observed on the plate (Workentine et al., 2008).
Determination of Minimal Inhibitory Concentration (MIC)
Minimal inhibitory concentration was defined as the cadmium concentration where no increase of turbidity at 600 nm was observed in microtiter plate cultures after 96 h incubation at 150 rpm and 30°C (Monsieurs et al., 2011).
Functional Prediction of Genes Encoding DGCs, PDEs, and PilZ Domain Effectors in the C. metallidurans CH34 Genome
A list of genes coding for DGCs and PDEs in the C. metallidurans CH34 genome (Supplementary Table S1) was retrieved from the NCBI website http://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html, according to NCBI annotations. GGDEF, EAL, and HD-GYP domain proteins coding genes were analyzed. The catalytic functionality of each protein was evaluated by analyzing the conservation of the active amino acid residues in domain sequence motifs residues Asp327, Lys332, Asn335, Asp344, Asp370, Lys442, and Arg446 for GGDEF; residues Glu523, Arg527, Glu546, Asn584, Glu616, Asp646, Asp647, Glu703, and Gln723 for EAL; and residues His183, Asp184, His212, His237, Glu(His)238, Asn(Glu)265, and Asn(Arg)269 for HD-GYP, using as reference functional domains of PleD (GGDEF) from Caulobacter crescentus (Wassmann et al., 2007), TBD_1265 (EAL) from Thiobacillus denitrificans (Tchigvintsev et al., 2010), and Bd1817 (HD-GYP) from Bdellovibrio bacteriovorus (Lovering et al., 2011), respectively. Motif conservation in GGDEF, EAL, and HD-GYP domains was determined by multiple sequence alignment using Clustal W (Thompson et al., 1994).
The c-di-GMP binding capability of the PilZ domains of C. metallidurans CH34 was predicted based on the conservation of the “c-di-GMP switch” domain (RxxxR and D/NzSxxG), which undergoes the conformational change that wraps around the c-di-GMP molecule (Duerig et al., 2009). The PilZ domain of PA4608 protein from P. aeruginosa was used as the active form model (Römling et al., 2013), whereas the PilZ domain of protein XC1028 from Xanthomonas campestris was used as the inactive form reference (Li et al., 2009). The search of c-di-GMP effectors related to Burkholderia cenocepacia Bcam1349 (Fazli et al., 2014) and P. aeruginosa PelD in the C. metallidurans CH34 genome was assessed by local alignment tools and pairwise sequence alignment. Motif conservation in PilZ domains was determined by global multiple sequence alignment using the MAFFT software that identifies rapidly homologous regions by the fast Fourier transform (Katoh et al., 2002). Motif conservation was visualized using Jalview software1 reported by Waterhouse et al. (2009). Orthology inference was achieved through sequence similarity searching using BlastP algorithm (Altschul et al., 1997) and confirmed by conservation of domains architectures obtained from NCBI’s CDART tool (Geer et al., 2002).
RNA Extraction and Reverse Transcription
After incubation of cells in presence or absence of 2 mM cadmium, 10 mL planktonic culture was mixed with 2 mL stop solution [water-saturated phenol 5% (pH < 7.0), ethanol 95%] (Bernstein et al., 2002; Sung et al., 2003). To discard sample processing bias, biofilm cells were suspended in a mixture of 10 mL LPTMS and 2 mL stop solution. For both samples, cells were collected by centrifugation at 1,920 × g for 10 min at 4°C and suspended in 700 μL of lysis buffer (sodium acetate 20 mM, EDTA 1 mM, SDS 0.5%, β-mercaptoethanol 1%, pH 5.5) and incubated 5 min at room temperature. Pre-heated acidic phenol (700 μL) was added, mixed by inversion, and incubated 10 min at room temperature. Mixtures were centrifuged at 16,000 × g for 5 min at 4°C. The aqueous phase was mixed with 1 volume of acid phenol:chloroform:isoamyl alcohol (25:24:1) and incubated for 5 min at room temperature. The mixture was centrifuged at 16,000 × g for 5 min at 4°C and the aqueous phase was mixed with 1 volume of chloroform, incubated 5 min at room temperature, and centrifuged at 16,000 × g for 5 min at 4°C. The resulting aqueous phase was mixed with 1 volume of isopropanol and 0.1 volume of sodium acetate 3 M (pH 5.2). The mixture was incubated overnight at -20°C. RNA was precipitated through centrifugation at 16,000 × g for 30 min at 4°C and washed twice with ethanol 70% (v/v). RNA samples were air dried, suspended in nuclease-free water, and treated with TURBO DNAse (AM1907) following manufacturer’s instructions. Absence of genomic DNA was controlled by PCR of the cadA gene with the same primers used for qPCR (Supplementary Table S2). Reverse transcription reactions were carried out using 500 ng of RNA with the ImProm II Reverse Transcription System (Promega A3800) with random primers, following manufacturer’s instructions.
Quantitative Polymerase Chain Reaction of cDNA
Quantitative PCR was carried out with the Fast SYBRGreen Master Mix (Applied Biosystems, 4385612) in a qPCR Step One Machine (Applied Biosystems, 271003314). Primers (Supplementary Table S2) were used at 0.5 μM each. PCR conditions included a hot start step at 95°C for 20 s, followed by 40 cycles of two steps amplification (95°C for 3 s, 60°C for 30 s). For each condition, three independent experiments each with at least two technical replicates were conducted. Bacteria incubated in absence of cadmium were used as control. Ct values were normalized to expression of housekeeping rpoZ and gyrB genes as described (Hellemans et al., 2007). Fold changes to the control condition were assessed by one sample t-test. Fold changes outside of [+1, -1] range and with P-values below 0.05 were reported as significant.
Construction of Recombinant C. metallidurans and P. aeruginosa Strains
Cloning procedures were performed according to standard protocols provided by the manufacturers. Primer sequences for cloning are shown in Supplementary Table S2. To characterize the function of the urf2 gene product, the urf2.2 gene (RMET_RS31035) was overexpressed in C. metallidurans and P. aeruginosa. The urf2.2 gene was specifically amplified based on its 3′-end sequence that is absent in the urf2.1 gene. The urf2.2 gene was amplified from CH34 genomic DNA using an EcoRI restriction site flanked forward primer and a HindIII site flanked reverse primer. This amplicon was cloned into pGEM-T vector (Promega A1360) using T4 ligase. The recombinant vector was introduced into E. coli DH5α by electroporation. The urf2.2 insert recovered from recombinant plasmid by EcoRI/HindIII digestion was ligated into the EcoRI/HindIII digested pJBTc19 vector. Thereafter, the recombinant plasmid pJBurf2.2 was introduced into E. coli SL17.1 λ pir by electroporation. The transfer of plasmid pJBurf2.2 into C. metallidurans CH34 was achieved by conjugation, where cell suspensions of donor and recipient strains were concentrated through vacuum filtration over a 0.2-μm polycarbonate filter and incubated in nutrient agar (beef extract 1 g L-1, yeast extract 2 g L-1, peptone 5 g L-1, and NaCl 5 g L-1) for 4 days at 30°C. Cells grown on the filter were spread on LPTMS agar supplemented with ampicillin 100 μg mL-1, tetracycline 15 μg mL-1, and kanamycin 1 mg mL-1. P. aeruginosa was transformed by electroporation and selection on LB agar supplemented with ampicillin 100 μg mL-1 and tetracycline 15 μg mL-1. Transformations were confirmed by double EcoRI/HindIII digestion of plasmid extraction product.
Results
Cadmium Exerts Inhibition of Biofilm Formation Through Decay in c-di-GMP Levels
Cell attachment assays showed that cadmium exerts a dose-dependent inhibition of early stages of biofilm formation in C. metallidurans CH34 (Figure 1A). Cell viability (as shown by CFU/mL) was not affected by the cadmium concentration range used, thus discarding a bias due to cell death (Figure 1A). At increasing cadmium concentration, C. metallidurans macrocolonies on plates showed decreasing levels of EPS that was observed by a reduction in Congo Red staining intensity (Figure 1B). A colorless extracellular matrix was observed at 8 mM cadmium. To assess the role of the c-di-GMP pathway in these phenotypes, nucleotide extracts were obtained from planktonic and biofilm cells after incubation in absence or presence of 2 mM cadmium during 45 min. The cadmium concentration 2 mM was chosen, since that concentration corresponded to the half of the MIC of cadmium for C. metallidurans CH34 in LPTMS broth (Table 2). In absence of cadmium, planktonic cells showed twofold higher c-di-GMP levels (∼0.8 picomol/mg protein) than biofilm cells (∼0.4 picomol/mg protein) (Figure 1C). Despite that generally higher c-di-GMP levels are expected in biofilm than in planktonic cells, it has been reported that c-di-GMP levels are dynamically modified by DGCs and PDEs in bacteria. For example, in planktonic E. coli cells a twofold increase of c-di-GMP levels (from 0.7 to 1.4 picomol/mg protein) in early stationary phase compared to late exponential phase has been observed (Sarenko et al., 2017). Dynamic c-di-GMP levels in biofilm cells have also been reported (Römling et al., 2013). In our study, control planktonic C. metallidurans cells were at post exponential phase, whereas biofilm cells were in early stage of biofilm formation. Cadmium decreased c-di-GMP levels in cells of both planktonic and biofilm lifestyles (Figure 1C). However, cadmium exerted a stronger c-di-GMP level decrease in planktonic cells, triggering an eightfold drop of c-di-GMP concentrations compared to control cells.
Figure 1.
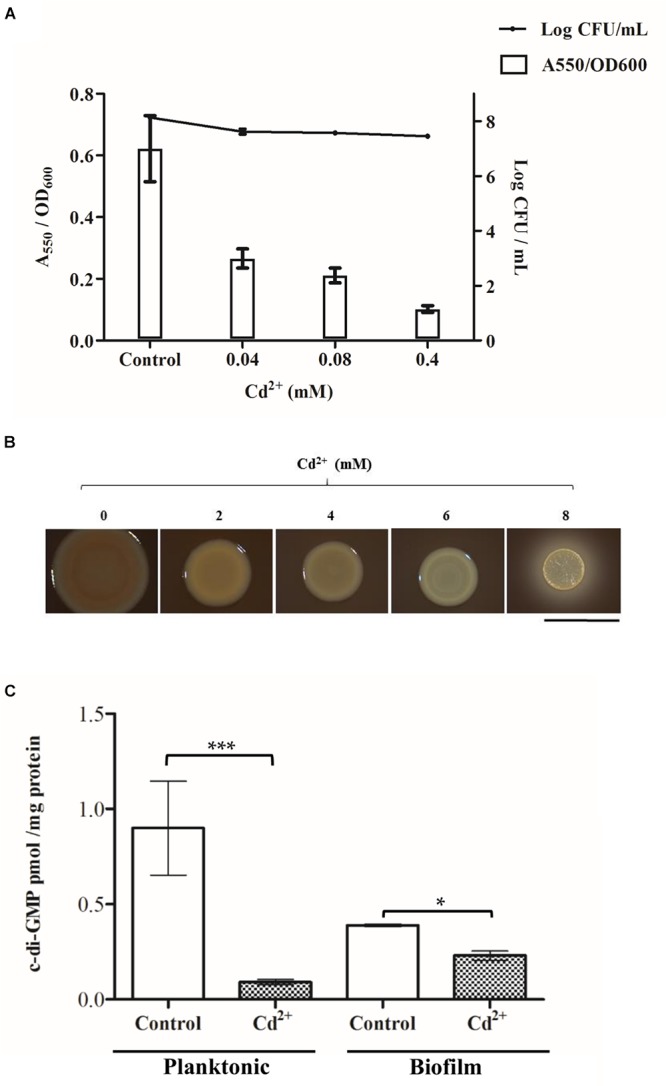
Effect of cadmium on C. metallidurans CH34 biofilm formation and c-di-GMP levels. (A) Cell attachment and cell viability after 15 h of incubation in LPTMS broth at 30°C. The means of three independent experiments and standard deviations are shown. (B) Macrocolonies grown on Congo Red LB agar plates at different cadmium concentrations. Images were taken after 5 days of incubation at room temperature. The red staining intensity of the macrocolony indicates the presence of EPS. The bar represents 1 cm. (C) c-di-GMP levels after 2 mM cadmium exposure during 45 min in LPTMS broth (planktonic) or on agar (biofilm). The c-di-GMP concentration was normalized with respect to total protein content in each sample. The means of three independent experiments and standard deviations are shown. Significant differences assessed by t-test: ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Table 2.
Cadmium susceptibility parameters in C. metallidurans strains.
| Strain | MIC (mM) | MBC (mM) (planktonic) | MBEC (mM) (biofilm) |
|---|---|---|---|
| C. metallidurans CH34 | 4 | 8 | 8 |
| CH34 pJB3Tc19 | 4 | 8 | 8 |
| CH34 pJBurf2.2 | 4 | 8 | 8 |
| CH34 pJBpleD∗ | 2 | 2 | 2 |
The susceptibility to cadmium of C. metallidurans CH34 planktonic and biofilm cells was determined. For microtiter determination of antimicrobial susceptibility, the Calgary Biofilm Device was used, which allows batch culture of biofilms on peg lids (Harrison et al., 2010). Microbial biocidal endpoints were determined quantitatively using viable cell counting (CFU) following a MBEC assay (Table 2; Workentine et al., 2008; Harrison et al., 2010). Planktonic and biofilm cells showed similar tolerance levels to cadmium (Table 2).
Searching and Selection of Potential Functional DGCs and PDEs of C. metallidurans CH34
Due to the potential role of c-di-GMP in the cadmium response (Figure 1C), genes encoding DGC and PDE domain-containing proteins were searched in the C. metallidurans CH34 genome. The functionality of proteins with GGDEF, EAL, and HD-GYP domains was predicted through analysis of the conservation of amino acid sequence motifs involved in the catalytic activity (Supplementary Figures S2–S4; Wassmann et al., 2007; Tchigvintsev et al., 2010; Lovering et al., 2011). The CH34 genome encodes 18 different GGDEF domain proteins, 10 EAL domain proteins, 12 GGDEF/EAL hybrid proteins, and 2 HD-GYP domain proteins. The functionality of each protein was predicted based on conservation of the active-site amino acids (with a maximum of one substitution), as defined by structures obtained by crystallography (Supplementary Tables S3–S5). Seventeen of 18 predicted GGDEF domain proteins showed conservation of at least six of seven amino acids that compose the active site of the GGDEF domain and, therefore, were predicted to possess DGC enzymatic activity. Nine of 10 annotated EAL domain proteins were predicted to have PDE enzymatic activity, due the conservation of at least 9 of the 10 amino acids involved in the EAL catalytic motif. In contrast, the RMET_RS19345 gene product was predicted to be non-active.
Among the 12 hybrid proteins, 9 GGDEF domains were predicted to be active, whereas RMET_RS19120, RMET_RS24490, and RMET_RS29670 gene products were predicted to be non-active (Supplementary Table S5). In addition, 10 EAL domains of hybrid proteins were predicted to be active, whereas RMET_RS24490 and RMET_RS29670 gene products were predicted to be non-active. Finally, the two loci encoding HD-GYP domain PDEs (RMET_RS09120 and RMET_RS20080) were predicted to be non-active and were not further analyzed (Supplementary Figure S5).
In summary, 36 genes coding for proteins of the c-di-GMP metabolism of C. metallidurans CH34 were predicted to be active.
An accessory domain analysis was performed with the proteins predicted to possess catalytically active c-di-GMP domains (Figure 2 and Supplementary Table S6). The cytoplasmic HAMP domain, important for receptor signal(s) transduction (Matamouros et al., 2015), and the extracellular CACHE sensor domains (Upadhyay et al., 2016) were encoded exclusively in DGCs. The ionic strength sensitive domain, cystathionine beta synthase (CBS), (Biemans-Oldehinkel et al., 2006) was located exclusively in three hybrid proteins, including an uncommon localization between EAL and GGDEF domains in the RMET_RS24295 gene product. An uncharacterized DUF3330 domain was observed in carboxy termini of the EAL domain PDE proteins encoded by the RMET_RS30310 and RMET_RS31035 genes (Figure 2).
Figure 2.
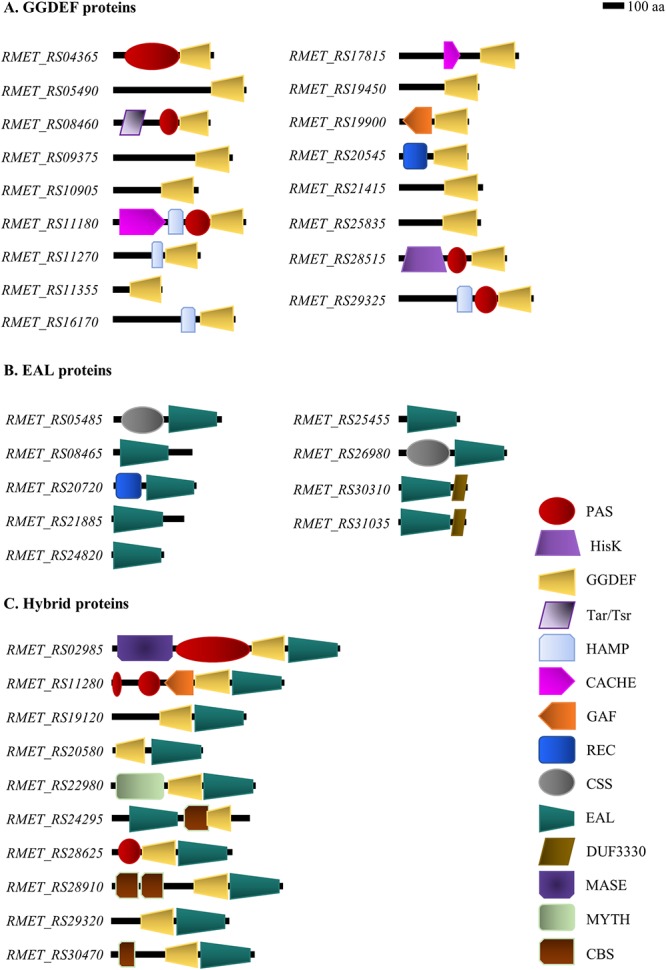
Domain-level depiction of c-di-GMP metabolism proteins from C. metallidurans CH34. Proteins with GGDEF/EAL domains that are catalytically active according to bioinformatic analyses are shown. (A) Diguanylate cyclases. (B) Phosphodiesterases. (C) Hybrid GGDEF/EAL proteins.
In DGC proteins, a Tar/Tsr chemoreceptor domain (Tajima et al., 2011) was observed in the RMET_RS08460 gene product, a nucleotide binding GAF domain (Heikaus et al., 2009) in the RMET_RS19900 gene product, and a histidine kinase domain HisK in the RMET_RS28515 gene product. The oxidative stress sensitive domain CSS was identified in the EAL domain RMET_RS05485 and RMET_RS26980 gene products (Huang et al., 2013). On the other hand, an integral membrane domain associated to gaseous ligand sensing (MHYT) (Galperin et al., 2005) was identified in the hybrid protein RMET_RS22980, and a membrane-anchoring domain MASE1 (Nikolskaya et al., 2003) in the hybrid protein RMET_RS02985.
Other accessory domains were transversally distributed between different proteins of the c-di-GMP metabolism. For example, the phosphorylation receiver domains (REC) were identified in DGC RMET_RS20545 and in PDE RMET_RS20720 gene products. The gaseous ligand binding domain PAS (Henry and Crosson, 2011) was observed in five DGCs (RMET_RS04365, RMET_RS08460, RMET_RS11180, RMET_RS28515, and RMET_RS29325) and three hybrid proteins (RMET_RS02985, RMET_RS11280, and RMET_RS28625). The domain organization analyses revealed six stand-alone GGDEF, three stand-alone EAL, and three stand-alone GGDEF/EAL domains (Figure 2 and Supplementary Table S6).
A gene orthology analysis based on comparison with the genomes of E. coli and P. aeruginosa model bacteria was performed. Based on sequence similarity and architectural domain profiles, nine CH34 orthologous DGC genes were identified in E. coli and P. aeruginosa (Supplementary Table S6). Three CH34 DGCs (RMET_RS09375, RMET_RS11270, and RMET_RS19900) showed a high percentage of identical residues with YedQ (27%) (Sanchez-Torres et al., 2011), TpbB (55%) (Xu et al., 2015), and YeaP (41%) (Ryjenkov et al., 2005), respectively. All aforementioned gene products are enzymes that promote EPS production and surface adherence toward robust biofilms. The analyses of the PDEs revealed that the CH34 RMET_RS05485 and RMET_RS26980 gene products possess high percentage of identical residues (33–34%) with the PDE YjcC from E. coli. It has been reported that YjcC is involved in the oxidative stress response and motility in K. pneumoniae (Huang et al., 2013). Similarly, the RMET_RS20720 gene product has 30% of identical residues with RocR, which is involved in biofilm formation and virulence gene expression in P. aeruginosa (Rao et al., 2008). Finally, the hybrid RMET_RS22980 gene product has 65% of identical residues with the bifunctional enzyme MucR, which is involved in biofilm dispersion in P. aeruginosa (Li et al., 2013).
The genomic context of CH34 c-di-GMP pathway genes and genes related to heavy metal resistance, motility, and biofilm formation was analyzed. Five c-di-GMP pathway genes were located in plasmids associated to heavy metal resistance. The RMET_RS30470 gene that encodes a bifunctional DGC/PDE was located in a genomic island cop-sil-nre-ncc that is tagged as not functional (Monsieurs et al., 2011), which codes for the heavy metal resistance machinery in pMOL30. In addition, the RMET_RS29670 gene that codes a predicted non-active protein is located in plasmid pMOL30, but outside of the heavy metal resistance genomic islands described in this replicon (Monsieurs et al., 2011). The PDE encoding RMET_RS30310 gene is located in the pMOL30 Tn4380 mer operon between the merE gene and a gene encoding a recombinase, overlapping their ORFs in 94 bases (Supplementary Figure S5). An identical context shows the PDE encoding RMET_RS31035 gene in the pMOL28 Tn4378 mer operon. Both EAL domain PDEs showed 95% sequence identity and possess the DUF3330 domain at the carboxy terminus. Both amino acidic sequences differ mainly at the carboxy terminus but conserve the domain architecture. The RMET_RS30310 and RMET_RS31035 genes are annotated as urf2 (for unknown related function) genes.
Some c-di-GMP pathway genes identified are associated with biofilm formation. The RMET_RS11355 gene is located downstream to an operon gene organization that is associated with cellulose synthesis (Supplementary Figure S5). The RMET_RS20545 gene is located downstream to a gene cluster associated to chemotaxis response, and the RMET_RS25455 gene is located upstream to a fimbrial synthesis gene cluster (Supplementary Figure S5).
Search and Analyses of c-di-GMP Effectors of C. metallidurans CH34
Cupriavidus metallidurans CH34 possesses four genes (RMET_RS05745, RMET_RS08705, RMET_RS09140, and RMET_RS11325) encoding PilZ domain proteins. The PilZ domain protein encoded by the RMET_RS09140 gene was discarded as c-di-GMP effector due to the absence of a c-di-GMP binding motif (Supplementary Figure S6). The hybrid GGDEF/EAL domain protein encoded by the RMET_RS29670 gene, previously predicted to be catalytically non-active, was included as potential effector of a c-di-GMP pathway in C. metallidurans CH34 (Supplementary Table S5). Local and pairwise sequence alignments determined the CH34 genes encoding orthologous proteins of the c-di-GMP effectors Bcam1349 (RMET_RS18290), a transcription factor that regulates biofilm formation in B. cenocepacia (Fazli et al., 2014; Supplementary Figure S7) and PelD (RMET_RS RS21490), a degenerate DGC that regulates cationic exopolysaccharide synthesis in P. aeruginosa (Supplementary Figure S8).
Effects of Cadmium on the Expression of Genes Coding for PDEs, DGCs, and Heavy Metal Resistance Proteins
A subset of 19 genes coding DGCs and PDEs were selected based on three criteria: (i) genes coding for accessory domains associated to extracellular signals sensing, (ii) genomic context associated to biofilm, motility, or heavy metal resistance, and (iii) orthology with E. coli or P. aeruginosa genes that encode enzymes of the c-di-GMP metabolism. Thus, the transcription levels of these 19 genes (eight DGCs, five PDEs, and six hybrids) were quantified after 45 min exposure to 2 mM cadmium in both biofilm and planktonic cells (Figure 3A). In addition, six transcripts of c-di-GMP effectors were also quantified: three transcripts encoding PilZ domain proteins that conserve the “c-di-GMP switch” binding motif (RMET_RS05745, RMET_RS08705, and RMET_RS11325), one transcript encoding a degenerate hybrid GGDEF/EAL domain protein (RMET_RS29670), one transcript encoding for a PelD-like c-di-GMP effector (RMET_RS21490), and one transcript (RMET_RS18290) that encodes a protein with a high percentage of identical residues (53%) to Bcam1349 Figure 3B).
Figure 3.
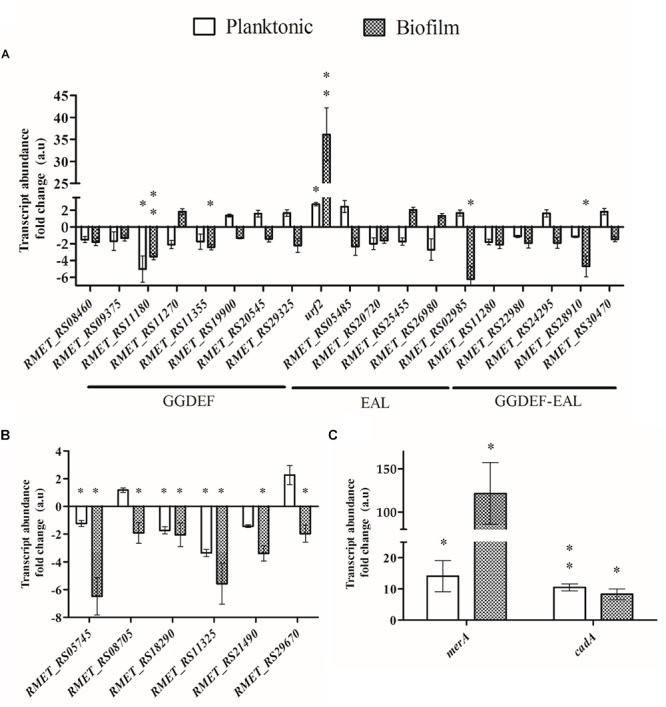
Effect of cadmium on the transcription of c-di-GMP metabolic and heavy metal resistance genes in planktonic and biofilm C. metallidurans CH34 cells. The transcription was measured after exposure to cadmium (2 mM) for 45 min. (A) Genes involved in c-di-GMP metabolism. The term urf2 represents both urf2.1 and urf2.2 genes. (B) Genes encoding c-di-GMP effectors. (C) Mercury and cadmium resistance genes. Each bar represents the mean of values of three independent experiments, each analyzed in duplicate, and standard deviations. Significant differences in expression to the control condition in absence of cadmium, assessed by t-test, are shown in asterisk codes: ∗P < 0.05, ∗∗P < 0.01.
In both lifestyles, most of the selected transcripts (DGCs, PDEs, and hybrids) showed a non-significant abundance fold change in response to cadmium (Figure 3A). In planktonic cells, the expression of two genes changed significantly in response to cadmium. The urf2 genes expression was upregulated (approximately threefold). Primers amplified the two urf2 genes transcripts, hence this value is the sum of both urf2.1 and urf2.2 transcripts. The RMET_RS11180 gene that encodes a DGC that possesses HAMP and CACHE extracellular signal sensing domains was strongly downregulated (approximately fivefold). In biofilm cells, the transcription of urf2 and RMET_RS11180 genes also changed significantly in response to cadmium. The urf2 gene transcription increased very strongly (∼36-fold) in biofilm cells (Figure 3A). The RMET_RS11180 gene transcription was also significantly downregulated (approximately threefold). In addition, the transcription of two hybrids proteins in biofilm cells was significantly downregulated by cadmium: RMET_RS02985 (approximately sixfold) that encodes an integral membrane protein with a MASE1 domain, and RMET_RS28910 (approximately fourfold) that encodes a protein with two CBS domains.
Almost all effector transcripts assessed were significantly downregulated after cadmium exposure, in higher magnitude in biofilm cells (Figure 3B). Only the RMET_RS08705 and RMET_RS29670 transcripts were non-significantly regulated in planktonic conditions.
In addition, the transcription of two heavy metal resistance genes was analyzed. The transcription of the cadA gene that encodes a Cd2+/ATPase protein transporter involved in cadmium resistance was monitored. As expected, after exposure to cadmium the cadA gene was strongly upregulated in both lifestyles at similar levels (∼10-fold) (Figure 3C). The effect of cadmium on the expression of the merA genes that encodes a mercuric reductase and belongs to the mer operons Tn4378 and Tn4380, which also harbor the urf2 genes, was also analyzed. The primers used amplify both merA transcripts. Interestingly, cadmium induced the transcription of the merA genes in both lifestyles (Figure 3C), suggesting an induction of mercury resistance by cadmium. The upregulation by cadmium of the merA genes was much stronger in biofilms (121-fold) than in planktonic cells (14-fold).
The urf2.2 Gene Encodes a Functional PDE
The urf2.2 gene is a component of the Tn501 mer operon. Although Tn501 and its derivatives were described decades ago (Brown et al., 1986), the Urf2 function remains unknown (Ei Mouali et al., 2017). Here, we are able for the first time to suggest a function. A complementation assay was developed in the PDE RocR null mutant strain P. aeruginosa PAO1 ΔrocR to evaluate the effects of the protein encoded by the urf2.2 gene on biofilm formation and c-di-GMP levels. RocR is a PDE response regulator in the RocS1S2AR two-component signaling system, which controls bacterial biofilm formation in P. aeruginosa (Kuchma et al., 2005; Kulasekara et al., 2005). The complementation by the urf2.2 gene restored biofilm formation and c-di-GMP levels (Figure 4) at levels comparable to the wild-type PAO1 strain, indicating that the urf2.2 gene encodes a functional PDE. Based on these data we propose to rename this specific gene as mrp2 gene for metal regulated PDE and its copy on plasmid pMOL28 as mrp1 gene.
Figure 4.
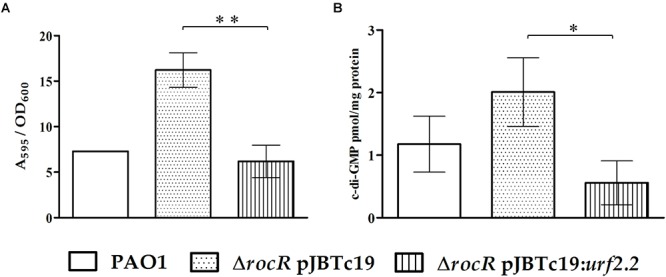
Complementation effects in P. aeruginosa PAO1 ΔrocR with pJBurf2.2. (A) Biofilm formation at 6 h of incubation in 96-well plates without cadmium. (B) The c-di-GMP content in planktonic cells at 6 h of incubation in Erlenmeyer flasks. The means of values of three independent experiments and standard deviations are shown. Significant differences assessed by t-test: ∗P < 0.05, ∗∗P < 0.01.
Effects of PDEs on Biofilm Formation, EPS Content, and c-di-GMP Levels
The relevance of c-di-GMP levels in C. metallidurans CH34 for biofilm formation was also assessed through the heterologous expression of the constitutively active DGC PleD∗ (Pérez-Mendoza et al., 2014) and the overexpression of the urf2.2 gene. Interestingly, the heterologous expression of the PleD∗ DGC increased biofilm formation in absence of cadmium (Figure 5A). In addition, cells expressing PleD∗ DGC increased their EPS content (Figure 5B) and c-di-GMP levels (up to >10-fold) either in absence or presence of cadmium (Figure 5C). Interestingly, in presence of cadmium c-di-GMP levels were not decreased in cells that express PleD∗ DGC. In contrast, the overexpression of the urf2.2 gene has no significant effects on biofilm formation (Figure 5A) and EPS content in macrocolonies (Figure 5B). However, the overexpression of the urf2.2 gene led to a slight decrease in c-di-GMP levels in the control planktonic cells in absence of cadmium (Figure 5C). After exposure to cadmium, c-di-GMP levels in cells that overexpress the urf2.2 gene decreased as observed with control cells.
Figure 5.
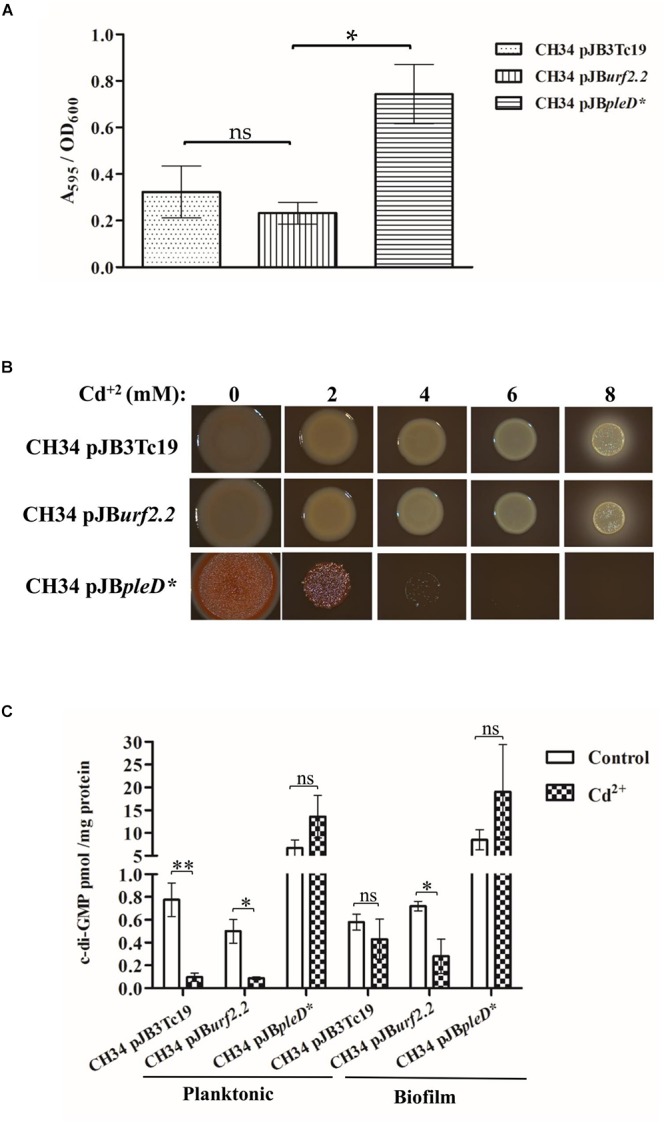
Effect of urf2.2 overexpression and pleD∗ heterologous expression on C. metallidurans CH34 biofilm formation and c-di-GMP metabolism. (A) Biofilm formation in the absence of cadmium. The means of three independent experiments and standard deviations are shown. Significant differences assessed by t-test: ∗P < 0.05; ns, not significant. (B) Macrocolonies morphotype in the presence of cadmium. The red staining of the macrocolony indicates its EPS content. (C) Response in c-di-GMP levels after cadmium exposure. The means of three independent experiments and standard deviations are shown. Significant differences assessed by t-test: ∗P < 0.05, ∗∗P < 0.01; ns, non-significant.
The effects of these alterations on cadmium susceptibility were further studied through the determination of MBC and MBEC. Interestingly, the heterologous expression of PleD∗ DGC resulted in significant detrimental effects on cadmium susceptibility (Table 2) in both bacterial lifestyles. Conversely, the overexpression of the urf2.2 gene has no effect on cadmium susceptibility (Table 2).
Discussion
This study shows that cadmium inhibits the initiation of biofilm formation in the heavy metal-resistant extremophile C. metallidurans CH34, which correlates with a drop in the c-di-GMP levels (Figure 1). The upregulation by cadmium of the PDE encoded by the urf2 genes (Figure 3A) suggests that this enzyme could be involved in the decrease in c-di-GMP concentration. In addition, the downregulation of a DGC (RMET_RS11180) and two hybrid proteins (RMET_RS02985 and RMET_RS28910) may be involved in the second messenger decrease and should be further analyzed in future studies. We propose to rename the two urf2 genes as mrp genes based on this study and the location of the two copies (RMET_RS30310/mrp1 and RMET_RS31035/mrp2) within the two mer operons of the pMOL28 and pMOL30 plasmids. These mer operons are part of transposons belonging to the Tn501/Tn21 family, the most widespread transposon families in terrestrial and marine environments (Brown et al., 1986; Liebert et al., 1999).
Similar to what we have observed here, a stressor reducing biofilm levels has been reported by Agulló et al. (2017) for the strain Paraburkholderia xenovorans LB400. This organism is a model aromatic compound-degrader, yet the toxic aromatic p-cymene reduced biofilm formation. This reduced biofilm formation in LB400 cells grown on p-cymene correlated with a decrease of the DGC protein levels that probably reduced c-di-GMP levels (Agulló et al., 2017). It has been also reported that cadmium and nickel cations inhibit biofilm formation by Burkholderia multivorans (Vega et al., 2014).
In our study, of the 42 proteins potentially involved in c-di-GMP metabolism that are encoded in the C. metallidurans CH34 genome, 36 proteins with high conservation of amino acid determinants (with a maximum of one substitution) for the catalytic activity were predicted as active, whereas 6 proteins were predicted to be non-active. The domain organization revealed a high proportion of stand-alone GGDEF (nine) and EAL (six) domains. This is in stark contrast with the 0.03 and 20% representation of these kinds of architectures within the Pfam database (Ei Mouali et al., 2017). The extremophile strain CH34 is especially well adapted to harsh niches in which heavy metals are present (Mergeay et al., 1985); therefore, the presence of unknown accessory domains in the c-di-GMP signaling pathway proteins should not be excluded.
Until this study, the role of the urf2 gene from the mercury resistance operon was unknown (Ei Mouali et al., 2017). This urf2 gene was the first gene encoding a protein with an EAL domain to be reported in Tn501 (Brown et al., 1986) and Tn21 transposon families (Liebert et al., 1999). It has been previously described that urf2.1 (orf2.1) and urf2.2 (orf2.2) genes are upregulated by mercury, cadmium, zinc, and lead (Monchy et al., 2007; Monsieurs et al., 2011). These heavy metals clustered into the same transcriptomic profile, i.e., regulate the same set of genes, based on microarrays data (Monsieurs et al., 2011). Our study establishes for the first time the catalytic functionality of the urf2 (mrp) gene product. Its ability to functionally replace RocR in P. aeruginosa PAO1 ΔrocR in complementation assays provides the insight. RocR is a PDE response regulator in the RocS1S2AR (or SadARS) two-component signaling system in P. aeruginosa (Kuchma et al., 2005). This system controls bacterial biofilm formation by regulating the transcription of the cup fimbrial gene cluster by a poorly understood mechanism (Kulasekara et al., 2005). Structural and mechanistic studies revealed that RocR is a constitutively inhibited tetrameric PDE, which is activated after phosphorylation, a modification that exposes the catalytic EAL domains (Chen et al., 2012). This mechanism explains the absence of biofilm-related phenotype effects either by overexpression or transposon inactivation of the rocR gene in P. aeruginosa (Kuchma et al., 2005). Interestingly, the results obtained in our study showed a drop in the c-di-GMP levels and decreased biofilm formation in P. aeruginosa PAO1 ΔrocR complemented with the CH34 mrp2 (urf2.2) gene (Figure 5). The discrepancy of the previous study and our work here may be explained by differences in experimental strategies used in both studies. Cells from a static culture after 8 h incubation (Kuchma et al., 2005) are in a different physiological state compared to the adhered biomass after 15 h incubation under stirred conditions (the present study). This condition generates shearing forces that promote biofilm compaction and viscosity loss in the EPS matrix (Goeres et al., 2005), which results in a biofilm that is more resistant to hydrodynamic shearing and degradation (Koechler et al., 2015). In this condition, the presence of early adherence determinants such as adhesins CupB/C could be more relevant. Adhesins expression is antagonized by RocR activity (Kulasekara et al., 2005). Thus, under the conditions used in our study, the urf2.2 gene product could replace the RocR role in biofilm formation and c-di-GMP hydrolysis.
The absence of differences on cadmium susceptibilities (Table 2) between both bacterial lifestyles along with the significant decrease of c-di-GMP concentration in planktonic cells in response to cadmium (Figure 1C) suggest that the response exceeds planktonic lifestyle promotion and might be associated with C. metallidurans survival under harsh conditions and its multilayer structure to tolerate heavy metals. In support of this idea, it has been found that specific PDEs such as YjcC in K. pneumoniae (Huang et al., 2013), CdgR in Salmonella typhimurium (Hisert et al., 2005), and YgfF in E. coli (Lacey et al., 2010) play a role in survival during oxidative stress. Heavy metals such as cadmium destroy thiol groups in proteins, triggering an oxidative stress response in bacteria (Harrison et al., 2007; Nies, 2016). It has been proposed that the synthesis of gold nanoparticles from Au (III) ions by CH34 cells is catalyzed by sulfur-rich proteins, which are oxidized (Montero-Silva et al., 2018). Cadmium probably also oxidizes sulfur-rich proteins in CH34 cells, causing oxidative stress and inducing a specific PDE that decreases c-di-GMP concentration. The heterologous expression of a constitutively active DGC (PleD∗) increased the c-di-GMP levels, biofilm formation, and cadmium susceptibility in C. metallidurans CH34 (Figure 5). As the extracellular matrix production is an energetically costly process (Karunakaran et al., 2011), a constitutive activation of this machinery could deplete ATP cellular pools, depriving the cell to achieve efficient cadmium detoxification. By overexpression of CH34 mrp gene, the role of the mrp gene product in the c-di-GMP metabolism in response to cadmium was only observed in planktonic cells with a slight decrease in c-di-GMP that correlates with a tendency to decrease biofilm formation. More pronounced changes due to the overexpression of the mrp gene from the plasmid pJBurf2.2 was not observed might be due to the high expression of the mrp genes from pMOL30 and pMOL28.
The mrp gene product domain architecture shows a C-terminal accessory domain DUF3330 (Pfam 11809), which is only present in a nitro reductase protein family. I-TASSER 3D prediction platform (Roy et al., 2010) identified five α-helix in this region beyond EAL domain toward the C-terminal region, supporting the existence of a domain in this region. Thus, the mrp gene product may be an exceptional case of domain organization in intracellular signaling proteins, combining a cytoplasmic sensor domain in its N-terminal extreme with a C-terminal output domain (Galperin, 2004). The DUF3330 domain may play a role in the c-di-GMP metabolism in C. metallidurans CH34. The mrp2 gene product is encoded in mobile genetic elements, indicating their potential distribution to other bacteria during horizontal gene transfer (Ei Mouali et al., 2017).
Notably, a strong increase (121-fold) by cadmium of the merA gene expression in C. metallidurans biofilm cells was observed. A lower induction (14-fold) by cadmium of the merA gene expression was observed in planktonic cells. Previously, we have observed the upregulation by cadmium of the expression of the merT and merP genes in C. metallidurans planktonic cells (Rojas, 2011). The mer operon is also induced by cadmium in Nitrosomonas europaea (Park and Ely, 2008). Recently, the merTPAGB operon has been associated with the cadmium resistance in C. metallidurans (Millacura et al., 2018). These studies suggest a role of the mer operon in the protection toward cadmium. The flexible nature of the transcriptional regulator MerR allows the interaction with a variety of heavy metals such as cadmium, according to in vitro transcription studies (Ralston and O’Halloran, 1990), but with lower affinity than for mercury (Wang et al., 2016). Cadmium and mercury possess similar outer shell electron configuration (d10s2) and atomic radius (ca. 150 pm). Therefore, it will be of interest to study the effects of mercury on biofilm formation and c-di-GMP metabolism in strain CH34. The higher induction of the mercury resistance merA gene in the CH34 biofilm than in planktonic cells resembles the improved mercury resistance in a Desulfovibrio desulfuricans biofilm compared to planktonic cells (Lin et al., 2013). In CH34 biofilm cells, a lower induction of the cadA gene (∼10-fold) in biofilm as well as planktonic cells was observed, indicating a differential behavior of both cad and mer operons for the sessile biofilm lifestyle.
Figure 6 illustrates the proposed signaling network activated by cadmium in C. metallidurans CH34. Cadmium cations enter the cell through metal ion transporters (MITs). In the cell, cadmium induces gene expression of the detoxification systems CadA, MerA, and RND antiporter pumps. In addition, cadmium ions upregulate the PDEs (mrp1 and mrp2 genes) encoded in the mercury resistance operons Tn4378 and Tn4380 and downregulate the DGC (RMET_RS11180). MerA proteins may act as chelator proteins of free cadmium cooperating with canonical detoxification systems. Efflux of cadmium is facilitated by CadA and RND antiporter pumps. The upregulation of PDE Mrp1/2 and downregulation of DGC RMET_ RS11180 protein decreased c-di-GMP levels. The drop of c-di-GMP levels inhibits the cell adhesion (pilus) and extracellular polysaccharide synthesis (cellulose and Pel) that may favor the planktonic lifestyle.
Figure 6.
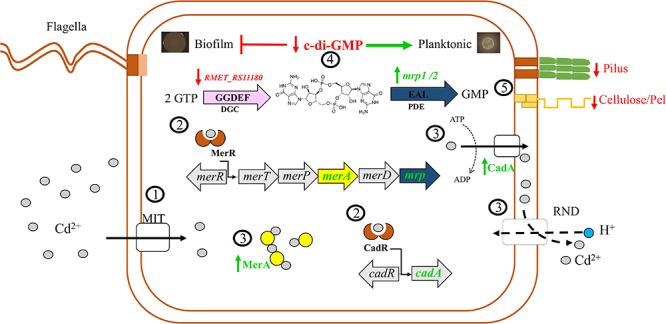
Proposed model of the C. metallidurans CH34 and its c-di-GMP signaling pathway in response to cadmium. 1, Cadmium ions (gray circles) enter the cell through metal ion transporters (MIT). 2, Cadmium induces gene expression of detoxification systems CadA, MerA, and RND antiporter pumps. Cadmium ions upregulate the phosphodiesterases (PDEs) (mrp1 and mrp2 genes) encoded in the mercury resistance operons Tn4378 and Tn4380 and downregulate the diguanylate cyclase (RMET_RS11180). 3, MerA proteins (yellow circles) may act as chelator proteins of free cadmium cooperating with canonical detoxification systems. Efflux of cadmium is facilitated by CadA and RND antiporter pumps. 4, Upregulation of Mrp1/2 and downregulation of RMET_RS11180 protein decreased c-di-GMP levels. 5, The drop of c-di-GMP levels inhibits the cell adhesion (pilus) and extracellular polysaccharide synthesis (cellulose and Pel) that may favor the planktonic lifestyle. Vertical arrows stand for upregulation (green) and downregulation (red) of molecules.
Conclusion
We studied the cadmium response on the extremophile C. metallidurans CH34, isolated from a heavy metal-polluted slurry. For this strain cadmium inhibits the initiation of biofilm formation. A decrease in c-di-GMP levels was observed in both planktonic and biofilm cells in response to cadmium. This response to cadmium suggests that the inhibition of the biofilm formation is mediated by a decrease in c-di-GMP levels. Thus, bacterial fitness under cadmium stress favors the planktonic lifestyle. However, both lifestyles showed similar tolerance toward cadmium, suggesting the adaptive advantage of planktonic cells is to provide a means to move toward a less toxic environment. Transcription analysis suggests an active role of a previously unknown catalytic functional PDE encoded by the urf2 genes of Tn4378 (pMOL28) and Tn4380 (pMOL30) mer operons from C. metallidurans CH34. Since this is the first report on the function of the urf2 gene, the gene was renamed as mrp2, for “metal regulated PDE.” In conclusion, this study indicates that cadmium inhibits the initiation of biofilm formation in C. metallidurans CH34 and that the response to cadmium involves a decrease in c-di-GMP levels and a novel metal regulated PDE.
Author Contributions
PA-G, MS, NG, and RT conceived and designed the experiments. PA-G performed the experiments. PA-G, MS, NG, RT, SF-A, and LR analyzed the data. MS, NG, and RT contributed reagents, materials, and analysis tools. PA-G, MS, NG, RT, and SF-A wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Juan Sanjuán and Dr. Daniel Pérez-Mendoza for the donation of pJB3Tc19 and pJBpleD∗ plasmids, and Natalie Gugala for support with the training and use of Calgary Biofilm Device. PA-G acknowledges a MECESUP Ph.D. fellowship (UCH 1107).
Funding. This work was supported by the FONDECYT 1160702 (NG), 1151174 (MS), and 3160424 (SF-A) (http://www.fondecyt.cl); the CONICYT Programa de Investigación Asociativa (PIA) Anillo GAMBIO ACT172128 (MS); USM (MS) (http://www.usm.cl); and the NSERC DG (RT) grants.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01499/full#supplementary-material
References
- Agulló L., Romero-Silva M. J., Domenech M., Seeger M. (2017). p-Cymene promotes its catabolism through the p-cymene and the p-cumate pathways, activates a stress response and reduces the biofilm formation in Burkholderia xenovorans LB400. PLoS One 12:e0169544. 10.1371/journal.pone.0169544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST:a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg S. L., Eijkelkamp B. A., Luo Z., Couñago R. M., Morey J. R., Maher M. J., et al. (2015). Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae. Nat. Commun. 6:6418. 10.1038/ncomms7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J. A., Khodursky A. B., Lin P.-H., Lin-Chao S., Cohen S. N. (2002). Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. U.S.A. 99 9697–9702. 10.1073/pnas.112318199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemans-Oldehinkel E., Mahmood N. A. B. N., Poolman B. (2006). A sensor for intracellular ionic strength. Proc. Natl. Acad. Sci. U.S.A. 103 10624–10629. 10.1073/pnas.0603871103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatny J. M., Brautaset T., Winther-Larsen H. C., Haugan K., Valla S. (1997). Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 63 370–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. L., Misra T. K., Winnie J. N., Schmidt A., Seiff M., Silver S. (1986). The nucleotide sequence of the mercuric resistance operons of plasmid R100 and transposon Tn501: further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Mol. Gen. Genet. 202 143–151. 10.1007/BF00330531 [DOI] [PubMed] [Google Scholar]
- Chen M., Zhou X., Liu X., Zeng R. J., Zhang F., Ye J., et al. (2018). Facilitated extracellular electron transfer of Geobacter sulfurreducens biofilm with in situ formed gold nanoparticles. Biosens. Bioelectron. 108 20–26. 10.1016/j.bios.2018.02.030 [DOI] [PubMed] [Google Scholar]
- Chen M. W., Kotaka M., Vonrhein C., Bricogne G., Rao F., Chuah M. L. C., et al. (2012). Structural insights into the regulatory mechanism of the response regulator RocR from Pseudomonas aeruginosa in cyclic di-GMP signaling. J. Bacteriol. 194 4837–4846. 10.1128/JB.00560-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard J. M., Corbisier P., Diels L., Dong Q., Jeanthon C., Mergeay M., et al. (1994). Plasmids for heavy metal resistance in Alcaligenes eutrophus CH34: mechanisms and applications. FEMS Microbiol. Rev. 14 405–414. 10.1111/j.1574-6976.1994.tb00115.x [DOI] [PubMed] [Google Scholar]
- Dahlstrom K. M., Collins A. J., Doing G., Taroni J. N., Gauvin T. J., Greene C. S., et al. (2018). A multimodal strategy used by a large c-di-GMP network. J. Bacteriol. 200 e703–e717. 10.1128/JB.00703-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerig A., Sören A., Folcher M., Nicollier M., Schwede T., Amiot N., et al. (2009). Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Gene Dev. 23 93–104. 10.1101/gad.502409.tion [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ei Mouali Y. E., Kim H., Ahmad I., Brauner A., Liu Y., Skurnik M., et al. (2017). Stand-alone EAL domain proteins form a distinct subclass of EAL proteins involved in regulation of cell motility and biofilm formation in enterobacteria. J. Bacteriol. 199:e00179-17. 10.1128/JB.00179-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Tofalos A., Daghio M., González M., Pappacchini M., Franzetti A., Seeger M. (2018). Toluene degradation by Cupriavidus metallidurans CH34 in nitrate-reducing conditions and in bioelectrochemical systems. FEMS Microbiol. Lett. 365:fny119. 10.1093/femsle/fny119/4996784 [DOI] [PubMed] [Google Scholar]
- Fazli M., Almblad H., Rybtke M. L., Givskov M., Eberl L., Tolker-Nielsen T. (2014). Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ. Microbiol. 16 1961–1981. 10.1111/1462-2920 [DOI] [PubMed] [Google Scholar]
- Galperin M. Y. (2004). Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6 552–567. 10.1111/j.1462-2920.2004.00633.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Gaidenko T. A., Mulkidjanian A. Y., Nakano M., Price C. W. (2005). MHYT, a new integral membrane sensor domain. FEMS Microbiol. Lett. 205 17–23. 10.1016/S0378-1097(01)00424-4 [DOI] [PubMed] [Google Scholar]
- Geer L. Y., Domrachev M., Lipman D. J., Bryant S. H. (2002). CDART: protein homology by domain architecture. Genome Res. 12 1619–1623. 10.1101/gr.278202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeres D. M., Loetterle L. R., Hamilton M. A., Murga R., Kirby D. W., Donlan R. M. (2005). Statistical assessment of a laboratory method for growing biofilms. Microbiology 151 757–762. 10.1099/mic.0.27709-0 [DOI] [PubMed] [Google Scholar]
- Gugala N., Lemire J. A., Turner R. J. (2017). The efficacy of different anti-microbial metals at preventing the formation of, and eradicating bacterial biofilms of pathogenic indicator strains. J. Antibiot. 70 775–780. 10.1038/ja.2017.10 [DOI] [PubMed] [Google Scholar]
- Habazettl J., Allan M. G., Jenal U., Grzesiek S. (2011). Solution structure of the PilZ domain protein PA4608 complex with cyclic di-GMP identifies charge clustering as molecular readout. J. Biol. Chem. 286 14304–14314. 10.1074/jbc.M110.209007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J. J., Ceri H., Roper N. J., Badry E. A., Sproule K. M., Turner R. J. (2005). Persister cells mediate tolerance to metal oxyanions in Escherichia coli. Microbiology 151 3181–3195. 10.1099/mic.0.27794-0 [DOI] [PubMed] [Google Scholar]
- Harrison J. J., Ceri H., Stremick C. A., Turner R. J. (2004a). Biofilm susceptibility to metal toxicity. Environ. Microbiol. 6 1220–1227. 10.1111/j.1462-2920.2004.00656.x [DOI] [PubMed] [Google Scholar]
- Harrison J. J., Ceri H., Stremick C., Turner R. J. (2004b). Differences in biofilm and planktonic cell mediated reduction of metalloid oxyanions. FEMS Microbiol. Lett. 235 357–362. 10.1016/j.femsle.2004.05.005 [DOI] [PubMed] [Google Scholar]
- Harrison J. J., Ceri H., Turner R. J. (2007). Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol. 5 928–938. 10.1038/nrmicro1774 [DOI] [PubMed] [Google Scholar]
- Harrison J. J., Stremick C. A., Turner R. J., Allan N. D., Olson M. E., Ceri H. (2010). Microtiter susceptibility testing of microbes growing on peg lids: a miniaturized biofilm model for high-throughput screening. Nat. Protoc. 5 1236–1254. 10.1038/nprot.2010.71 [DOI] [PubMed] [Google Scholar]
- Heikaus C. C., Pandit J., Klevit R. E. (2009). Cyclic nucleotide binding GAF domains from phosphodiesterases: structural and mechanistic insights. Structure 17 1551–1557. 10.1016/j.str.2009.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:R19. 10.1186/gb-2007-8-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R., Grundling A., Jenal U., Ryan R., Yildiz F. (2016). Bacterial signal transduction by cyclic di-GMP and other nucleotide second messengers. J. Bacteriol. 198 15–26. 10.1128/JB.00331-15.Editor [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry J. T., Crosson S. (2011). Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu. Rev. Microbiol. 65 261–286. 10.1146/annurev-micro-121809-151631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisert K. B., MacCoss M., Shiloh M. U., Darwin K. H., Singh S., Jones R. A., et al. (2005). A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defence and killing of macrophages: role of cyclic diGMP. Mol. Microbiol. 56 1234–1245. 10.1111/j.1365-2958.2005.04632.x [DOI] [PubMed] [Google Scholar]
- Huang C.-J., Wang Z.-C., Huang H.-Y., Huang H.-D., Peng H.-L. (2013). YjcC, a c-di-GMP phosphodiesterase protein, regulates the oxidative stress response and virulence of Klebsiella pneumoniae CG43. PLoS One 8:e66740. 10.1371/journal.pone.0066740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Sliusarenko O., Kazmierczak B. I. (2017). Interaction of the cyclic-di-GMP binding protein FimX and the type 4 pilus assembly ATPase promotes pilus assembly. PLoS Pathog. 13:e1006594. 10.1371/journal.ppat.1006594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen P. J., van Houdt R., Moors H., Monsieurs P., Morin N., Michaux A., et al. (2010). The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS One 5:e10433. 10.1371/journal.pone.0010433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U., Reinders A., Lori C. (2017). Cyclic di-GMP: second messenger extraordinaire. Nat. Rev. Microbiol. 15 271–284. 10.1038/nrmicro.2016.190 [DOI] [PubMed] [Google Scholar]
- Karunakaran E., Mukherjee J., Ramalingam B., Biggs C. A. (2011). “Biofilmology”: a multidisciplinary review of the study of microbial biofilms. App. Microbiol. Biotech. 90 1869–1881. 10.1007/s00253-011-3293-4 [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30 3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koechler S., Farasin J., Cleiss-Arnold J., Arsène-Ploetze F. (2015). Toxic metal resistance in biofilms: diversity of microbial responses and their evolution. Res. Microbiol. 166 764–773. 10.1016/j.resmic.2015.03.008 [DOI] [PubMed] [Google Scholar]
- Kuchma S. L., Connolly J. P., O’Toole G. A. (2005). A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187 1441–1454. 10.1128/jb.187.4.1441-1454.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasekara H. D., Ventre I., Kulasekara B. R., Lazdunski A., Filloux A., Lory S. (2005). A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55 368–380. 10.1111/j.1365-2958.2004.04402.x [DOI] [PubMed] [Google Scholar]
- Lacey M. M., Partridge J. D., Green J. (2010). Escherichia coli K-12 YfgF is an anaerobic cyclic di-GMP phosphodiesterase with roles in cell surface remodelling and the oxidative stress response. Microbiology 156 2873–2886. 10.1099/mic.0.037887-0 [DOI] [PubMed] [Google Scholar]
- Li T. N., Chin K. H., Liu J. H., Wang A. H.-J., Chou S. H. (2009). XC1028 from Xanthomonas campestris adopts a PilZ domain-like structure without a c-di-GMP switch. Proteins 75 282–288. 10.1002/prot.22330 [DOI] [PubMed] [Google Scholar]
- Li Y., Heine S., Entian M., Sauer K., Frankenberg-Dinkel N. (2013). NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by an MHYT domain-coupled phosphodiesterase. J. Bacteriol. 195 3531–3542. 10.1128/JB.01156-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebert C. A., Hall R. M., Summers A. O. (1999). Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63 507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T. Y., Kampalath R. A., Lin C. C., Zhang M., Chavarria K., Lacson J., et al. (2013). Investigation of mercury methylation pathways in biofilm versus planktonic cultures of Desulfovibrio desulfuricans. Environ. Sci. Technol. 47 5695–5702. 10.1021/es400079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering A. L., Capeness M. J., Lambert C., Hobley L., Sockett R. E. (2011). The structure of an unconventional HD-GYP protein from Bdellovibrio reveals the roles of conserved residues in this class of Cyclic-di-GMP phosphodiesterases. mBio 2:e00163-11. 10.1128/mBio.00163-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes S., Props R., Fitts J. P., De Smet R., Vanhaecke F., Boon N., et al. (2017). Biological recovery of platinum complexes from diluted aqueous streams by axenic cultures. PLoS One 12:e0169093. 10.1371/journal.pone.0169093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamouros S., Hager K. R., Miller S. I. (2015). HAMP domain rotation and tilting movements associated with signal transduction in the PhoQ sensor kinase. mBio 6:e00616-15. 10.1128/mBio.00616-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergeay M., Nies D., Schlegel H. G., Gerits J., Charles P., Van Gijsegem F. (1985). Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162 328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J. H., Brothers K. M., Kuchma S. L., O’Toole G. A. (2007). SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 189 8154–8164. 10.1128/JB.00585-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millacura F. A., Janssen P. J., Monsieurs P., Janssen A., Provoost A., Houdt R. V., et al. (2018). Unintentional genomic changes endow Cupriavidus metallidurans with an augmented heavy-metal resistance. Genes 9:551. 10.3390/genes9110551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monchy S., Benotmane M. A., Janssen P., Vallaeys T., Taghavi S., van der Lelie D., et al. (2007). Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. J. Bacteriol. 189 7417–7425. 10.1128/JB.00375-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsieurs P., Moors H., Van Houdt R., Janssen P. J., Janssen A., Coninx I., et al. (2011). Heavy metal resistance in Cupriavidus metallidurans CH34 is governed by an intricate transcriptional network. Biometals 24 1133–1151. 10.1007/s10534-011-9473-y [DOI] [PubMed] [Google Scholar]
- Montero-Silva F., Durán N., Seeger M. (2018). Synthesis of extracellular gold nanoparticles using Cupriavidus metallidurans CH34 cells. IET Nanobiotechnol. 12 40–46. 10.1049/iet-nbt.2017.0185 29768243 [DOI] [Google Scholar]
- Mosa K. A., Saadoun I., Kumar K., Helmy M., Dhankher O. P. (2016). Potential biotechnological strategies for the cleanup of heavy metals and metalloids. Front. Plant Sci. 7:303. 10.3389/fpls.2016.00303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies D. H. (1999). Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51 730–750. 10.1007/s002530051457 [DOI] [PubMed] [Google Scholar]
- Nies D. H. (2016). The biological chemistry of the transition metal “transportome” of Cupriavidus metallidurans. Metallomics 8 481–507. 10.1039/C5MT00320B [DOI] [PubMed] [Google Scholar]
- Nikolskaya A. N., Mulkidjanian A. Y., Beech I. B., Galperin M. Y. (2003). MASE1 and MASE2: two novel integral membrane sensory domains. J. Mol. Microbiol. Biotech. 5 11–16. 10.1159/000068720 [DOI] [PubMed] [Google Scholar]
- Orellana R., Macaya C., Bravo G., Dorochesi F., Cumsille A., Valencia R., et al. (2018). Living at the frontiers of life: extremophiles in chile and their potential for bioremediation. Front. Microbiol. 9:2309. 10.3389/fmicb.2018.02309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole G. A., Kolter R. (1998). Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, pathways convergent signalling: a genetic analysis. Mol. Microbiol. 28 449–461. 10.1046/j.1365-2958.1998.00797 [DOI] [PubMed] [Google Scholar]
- Park S., Ely R. L. (2008). Candidate stress genes of Nitrosomonas europaea for monitoring inhibition of nitrification by heavy metals. Appl. Environ. Microbiol. 74 5475–5482. 10.1128/AEM.00500-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R., Weiser S., Amiot N. C., Chan C., Schirmer T., Giese B., et al. (2004). Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 8 715–727. 10.1101/gad.289504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Mendoza D., Aragón I. M., Prada-Ramírez H. A., Romero-Jiménez L., Ramos C., Gallegos M. T., et al. (2014). Responses to elevated c-di-GMP levels in mutualistic and pathogenic plant-interacting bacteria. PLoS One 9:e91645. 10.1371/journal.pone.0091645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston D. M., O’Halloran T. V. (1990). Ultrasensitivity and heavy-metal selectivity of the allosterically modulated MerR transcription complex. Proc. Natl. Acad. Sci. U.S.A. 87 3846–3850. 10.1073/pnas.87.10.3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F., Yang Y., Qi Y., Liang Z.-X. (2008). Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 190 3622–3631. 10.1128/JB.00165-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas L. (2011). Studies of Bacterial Bioremediation in Soil Microcosms Contaminated with Mercury (II) Species, Cadmium and Copper, Chemistry. Ph.D. thesis, Universidad Técnica Federico Santa María, Valparaíso. [Google Scholar]
- Rojas L. A., Yáñez C., González M., Lobos S., Smalla K., Seeger M. (2011). Characterization of the metabolically heavy metal-resistant Cupriavidus metallidurans strain MSR33 generated for mercury remediation. PLoS One 6:e17555. 10.1371/journal.pone.0017555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D., Aguilar C., Losick R., Kolter R. (2010). Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U.S.A. 107 2230–2234. 10.1073/pnas.0910560107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U., Galperin M. Y., Gomelsky M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77 1–52. 10.1128/MMBR.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5 725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryjenkov D. A., Tarutina M., Moskvin O. V., Gomelsky M. (2005). Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 5 1792–1798. 10.1128/JB.187.5.1792-1798.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Torres V., Hu H., Wood T. K. (2011). GGDEF proteins YeaI, YedQ, and YfiN reduce early biofilm formation and swimming motility in Escherichia coli. Appl. Microbiol. Biotech. 2 651–658. 10.1007/s00253-010-3074-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarenko O., Klauck G. M., Wilke F., Pfiffer V., Richter A. M., Herbst S., et al. (2017). More than enzymes that make or break cyclic di-GMP-local signaling in the interactome of GGDEF/EAL domain proteins of Escherichia coli. mBio 8:e01639-17. 10.1128/mBio.01639-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler C., Böhm A., Jenal U., Seifert R., Kaever V. (2010). A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J. Microbiol. Meth. 81 226–231. 10.1016/j.mimet.2010.03.020 [DOI] [PubMed] [Google Scholar]
- Sung K., Khan S. A., Nawaz M. S., Khan A. A. (2003). A simple and efficient Triton X-100 boiling and chloroform extraction method of RNA isolation from Gram-positive and gram-negative bacteria. FEMS Microbiol. Lett. 229 97–101. 10.1016/S0378-1097(03)00791-2 [DOI] [PubMed] [Google Scholar]
- Tajima H., Imada K., Sakuma M., Hattori F., Nara T., Kamo N., et al. (2011). Ligand specificity determined by differentially arranged common ligand-binding residues in bacterial amino acid chemoreceptors Tsr and Tar. J. Biol. Chem. 286 42200–42210. 10.1074/jbc.M111.221887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchigvintsev A., Xu X., Singer A., Chang C., Brown G., Proudfoot M., et al. (2010). Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J. Mol. Biol. 402 524–538. 10.1016/j.jmb.2010.07.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. (1994). Clustal-W-Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay A. A., Fleetwood A. D., Adebali O., Finn R. D., Zhulin I. B. (2016). Cache domains that are homologous to, but different from PAS domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLoS Comput. Biol. 12:e1004862. 10.1371/journal.pcbi.1004862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee B. L., Ulmer D. D. (1972). Biochemical effects of mercury, cadmium, and lead. Annu. Rev. Biochem. 41 91–128. 10.1146/annurev.bi.41.070172.000515 [DOI] [PubMed] [Google Scholar]
- Vega L. M., Mathieu J., Yang Y., Pyle B. H., McLean R. J. C., Alvarez P. J. J. (2014). Nickel and cadmium ions inhibit quorum sensing and biofilm formation without affecting viability in Burkholderia multivorans. Int. Biodeter. Biodegr. 91 82–87. 10.1016/j.ibiod.2014.03.013 [DOI] [Google Scholar]
- Wang D., Huang S., Liu P., Liu X., He Y., Chen W., et al. (2016). Structural analysis of the Hg(II)-regulatory protein Tn501 MerR from Pseudomonas aeruginosa. Sci. Rep. 6:33391. 10.1038/srep33391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmann P., Chan C., Paul R., Beck A., Heerklotz H., Jenal U., et al. (2007). Structure of BeF3- -modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure 15 915–927. 10.1016/j.str.2007.06.016 [DOI] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J. (2009). Jalview Version 2- a multiple sequence alignment editor and analysis workbench. Bioinformatics 25 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workentine M. L., Harrison J. J., Stenroos P. U., Ceri H., Turner R. J. (2008). Pseudomonas fluorescens view of the periodic table. Environ. Microbiol. 10 238–250. 10.1111/j.1462-2920.2007.01448.x [DOI] [PubMed] [Google Scholar]
- World Health Organization [WHO] (2011). Safety Evaluation of Certain Food Additivies and Contaminants. Geneva: World Health Organization. [Google Scholar]
- Xu K., Li S. Y., Wen L., Kan B. A., Yuwei Xu Y., Jin J., et al. (2015). Structural and biochemical analysis of tyrosine phosphatase related to biofilm formation A (TpbA) from the opportunistic pathogen Pseudomonas aeruginosa PAO1. PLoS One 10:e0124330. 10.1371/journal.pone.0124330 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


