Abstract
Retinitis pigmentosa (RP), the most common type of inherited retinal degeneration causing blindness, initially manifests as severely impaired rod function followed by deteriorating cone function. Mutations in the rhodopsin gene (RHO) are the most common cause of autosomal dominant RP (adRP). The present study aims to identify the disease-causing mutation in a numerous, four-generation Han-Chinese family with adRP detected by whole exome sequencing and Sanger sequencing. Afflicted family members present classic adRP along with heterogeneous clinical phenotypes including differing refractive errors, cataracts, astigmatism and epiretinal membranes. A missense mutation, c.403C>T (p.R135W), in the RHO gene was identified in nine subjects and it co-segregated with family members. The mutation is predicted to be disease-causing and results in rhodopsin protein abnormalities. The present study extends the genotype–phenotype relationship between RHO gene mutations and adRP clinical findings. The results have implications for familial genetic counseling, clinical management and developing RP target gene therapy strategies.
Keywords: missense mutation, retinitis pigmentosa, rhodopsin, whole exome sequencing
Introduction
Retinitis pigmentosa (RP, OMIM 268000) is the most common type of inherited, blindness-causing retinal degeneration. Its prevalence is ∼0.020–0.033% in general populations with no apparent ethnic or geographic distinctions [1,2]. The patient initially suffered from seriously impaired rod function followed by impaired cone function. Clinical features include night blindness, visual field constriction, visual acuity impairment, fundus degeneration and late-life vision loss [1]. There are three primary Mendelian RP inheritance modes: autosomal dominant (30–40%), autosomal recessive (50–60%) and X-linked (5–15%). Mitochondrial, or digenic forms are rare but have been reported [1,3]. Most RP patients have myopic refractive errors, especially in X-linked RP [4–7]. To date, more than 79 genes have been reported as related to RP [1]. Mutations in genes including the rhodopsin gene (RHO), the pre-mRNA processing factor 31 gene (PRPF31), the RP GTPase regulator gene (RPGR), the peripherin 2 gene (PRPH2), the RP1 axonemal microtubule associated gene (RP1), the inosine monophosphate dehydrogenase 1 gene (IMPDH1), the pre-mRNA processing factor 8 gene (PRPF8) and the nuclear receptor subfamily 2 group E member 3 gene (NR2E3) can cause autosomal dominant RP (adRP), and each accounts for at least 2% of all patients [8,9]. RHO gene mutations are the most common cause of adRP and account for 16–35% of adRP cases in Western populations. The frequency is much lower (2.0–7.7%) in Chinese, Japanese, Korean and South Asian Indian patients, indicating that the frequency may be ethnicity dependent [10–12].
In the present study, whole exome sequencing (WES) and Sanger sequencing were performed to identify possible genetic causes for the presence of adRP in a four-generation Han-Chinese family. An RHO gene missense mutation, c.403C>T (p.R135W), was identified and found to co-segregate with the adRP phenotype, supporting the hypothesis that it is the genetic cause of the adRP.
Methods
Participators and clinical evaluation
A 27-person, four-generation Han-Chinese pedigree was enrolled at the Third Xiangya Hospital, Central South University, Changsha, Hunan, China (Figure 1A). Clinical data and peripheral blood samples were obtained from 14 family members, including nine affected (II:3, II:5, II:7, III:1, III:2, III:4, III:6, III:11 and IV:1) and five unaffected individuals (III:7, III:12, IV:2, IV:3 and IV:4). Blood samples were collected from 100 unrelated, ethnically matched normal controls (male/female: 50/50, age 38.5 ± 5.6 years). Informed written consent was obtained from each or their guardians. The study complied with the Declaration of Helsinki and was approved by the Ethics Committee of the Third Xiangya Hospital of Central South University in China. Participants underwent visual acuity testing by E decimal charts. Best corrected visual acuity (BCVA) was recorded as a logarithm of the minimum angle of resolution (logMAR) with cycloplegic refraction. Subjects were also examined and measured with slit-lamp biomicroscopy, fundus examination, full-field electroretinography and optical coherence tomography (OCT). A diagnosis of adRP was based on family history and typical clinical characters including presence of night blindness, reduced peripheral visual fields, typical fundus findings and abnormal OCT results [3].
Figure 1. Pedigree data of an adRP family.
(A) Pedigree of the family with adRP. N: normal; M: the RHO c.403C>T (p.R135W) mutation. Arrow indicates the proband. (B) Sequence of heterozygous c.403C>T (p.R135W) mutation. (C) Sequence of a normal control. (D) Conservation analysis of the rhodopsin p.R135 amino acid residue.
Variant genetic analysis and bioinformatics analysis
WES was performed by the Novogene Bioinformatics Institute, Beijing, China, using gDNA from the proband (III:4). After a DNA quality assessment, the captured DNA library was sequenced using a HiSeq 2000 platform to generate 150-bp paired-end reads as manufacturer protocols (Illumina Inc., San Diego, CA, U.S.A.) [13,14].
Analysis-ready BAM alignment data were obtained after duplicate removal, local alignment and base quality recalibration by Picard (http://sourceforge.net/projects/picard/), Genome Analysis Toolkit and SAMtools. All obtained variants were further screened using data from public variant databases including the Single Nucleotide Polymorphism Database (dbSNP build 137, http://www.ncbi.nlm.nih.gov/projects/SNP/snp_summary.cgi), 1000 genomes project, and the NHLBI Exome Sequencing Project (ESP) 6500 [15]. Only variants occurring in exon regions or canonical splicing sites, with a minor allele frequency of less than 0.01 were analyzed further. Sorting Intolerant from Tolerant (SIFT) and Polymorphism Phenotyping version 2 (PolyPhen-2) were used to predict functional effects of the nonsynonymous single nucleotide polymorphisms (SNPs) [16,17]. The candidate gene variant related to vision degeneration disease was prioritized for validation, as were those performed in recent studies [18–20]. Sanger sequencing was performed to confirm the potential pathogenic variant using ABI3500 sequencer (Applied Biosystems, Foster City, CA, U.S.A.) [18]. PCR amplification and Sanger sequencing primer sequences are as follows: 5′-CTCCTTAGGCAGTGGGGTCT-3′ and 5′-GGAGCTTCTTCCCTTCTGCT-3′.
Multiple sequence alignments among different species were performed using the NCBI Basic Local Alignment Search Tool (http://blast.st-va.ncbi.nlm.nih.gov/Blast.cgi). MutationTaster (http://www.mutationtaster.org/) was used to further assess amino acid alteration potential pathogenicity [20,21].
Subcellular location prediction
The rhodopsin protein consists of cytoplasmic (CP), transmembrane (TM) an extracellular (EC) domains. TransMembrane prediction using Hidden Markov Models (TMHMM) Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) was applied to predict TM helices in wild-type rhodopsin and the structural interpretation of mutated rhodopsin.
Results
Pedigree clinical characteristics
Nine family members, four males and five females, were diagnosed with RP by two independent ophthalmologists. Phenotype transmission among four generations in this pedigree supports an autosomal dominant inheritance pattern (Figure 1A). All affected patients began suffering severe night blindness at 1–2 years, exhibited characteristic clinical symptoms including reduced periphery visual fields after 30 and ultimately developed blindness in later life (II:3 at 45; II:5 at 50; and II:7 at 45). Fundus examinations presented bone spicule-like pigmentation in the inferior periphery (Figure 2A,B), retinal pigment epithelium atrophy and retinal vessel attenuation (Figure 2A-C). OCT detected severely thinned (except III:2 and IV:1) and disorganized, inner and outer, photoreceptor segments (Figure 2A,B). Non-recordable ERG results were observed in patients III:4 and IV:1 (Figure 3). Intriguingly, patients III:2 and IV:1 had serious hyperopic refractive errors. Patients II:3 and II:5 had shorter axial lengths than healthy individuals, indicating hyperopic refractive errors. Patients III:1, III:4 and III:6 have myopic refractive errors. Except for a 9-year-old boy (IV:1), all patients present with, moderate or severe, cataracts (Figure 2D). One patient (III:2) has an epiretinal membrane (EM) and six patients (III:1, III:2, III:4, III:6, III:11 and IV:1) have astigmatism. The clinical and genetic data for nine patients are summarized in Table 1.
Figure 2. Fundus, OCT and slit lamp photographs of the patients.
(A–C) Fundus photographs and OCT images for patients (III:4, III:11 and IV:1). (D) Slit lamp photographs of patients (II:3, II:5 and III:1). Bilateral eyes of the patients (II:3 and II:5) and the left eye of the patient III:1 showed cataract, and the right eye of the patient III:1 presented with glaucoma secondary to traumatism.
Figure 3. Electroretinography results for patients III:4 and IV:1.
Non-recordable ERG results were observed in patients III:4 (A) and IV:1 (B).
Table 1. Clinical and genetic data of nine patients with RHO c.403C>T (p.R135W) variant.
| Subject | II:3 | II:5 | II:7 | III:1 | III:2 | III:4 | III:6 | III:11 | IV:1 |
|---|---|---|---|---|---|---|---|---|---|
| Age (years) /Sex | 62/F | 60/F | 56/M | 39/M | 42/M | 41/F | 35/F | 32/F | 9/M |
| Onset age (years) | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 1 | 2 |
| Genotype | Heterozygote | Heterozygote | Heterozygote | Heterozygote | Heterozygote | Heterozygote | Heterozygote | Heterozygote | Heterozygote |
| BCVA (OD/OS) | LP (5 m/5 m) | LP (5 m/5 m) | NLP/NLP | NLP/0.1 | 0.3/0.3 | 0.1/0.04 | 0.2/0.2 | 0.04/0.06 | 0.5/0.4 |
| Optometry (OD) | NA | NA | NA | NA | +5.50/+1.50 × 30° | −0.75×90° | −5.00/−2.00×180° | +0.50/−3.00×180° | +3.00×105° |
| Optometry (OS) | NA | NA | NA | −9.5/−2.0 × 165° | +4.00 | −1.75×90° | −4.75/−1.50×180° | −2.75×180° | +3.25×90° |
| Axial length (mm, OD/OS) | 21.47/21.28 | 20.30/19.85 | NA | ND | ND | ND | ND | ND | ND |
| Fundus features | NA | NA | NA | Mid-peripheral pigment alterations, vessel attenuation | A pale fundus, mid-peripheral pigment alterations, vessel attenuation | A pale fundus, diffuse atrophic changes of the retinal pigment epithelium, vessel attenuation, bone spicule-like pigmentation | A blurring pale fundus, mid-peripheral pigment alterations, vessel attenuation | A pale fundus, mid-peripheral pigment alterations, vessel attenuation, bone spicule-like pigmentation | Slight mid-peripheral pigment alterations, slight vessel attenuation |
| Cataract (OD/OS) | Severe/Severe | Severe/Severe | Severe/Severe | No/Severe | Moderate/Severe | No/Moderate | Severe/Severe | Moderate/No | No/No |
| Cataract subtype (OD/OS) | TC/TC | TC/TC | TC/TC | No/PSC | CC/CC | No/PSC | PSC/PSC | PSC/No | No/No |
| OCT (CFT, μm) (OD/OS) | ND | ND | ND | ND/145 | EM (310/262) | 160/155 | 190/168 | 107/152 | 209/225 |
Abbreviations: BCVA, best corrected visual acuity; CC, cortical cataract; CFT, central foveal thickness; EM, epiretinal membrane; F, female; LP, light perception; M, male; NA, not available; ND, not detected; NLP, no light perception; OCT, optical coherence tomography; OD, right eye; OS, left eye; PSC, posterior subcapsular cataract; RHO, the rhodopsin gene; TC, total cataract.
Whole exome sequencing
Proband (III:4, Figure 1A) WES showed that ∼44.04 million reads (99.6%) mapped to the human reference genome. Average sequencing depth on the target region was 72.65. A large proportion of the region, 98.7%, was covered by the target sequence at 10× or greater. A total of 121690 SNPs were detected, including 22046 in the exon regions and 2399 in the splicing sites. Testing identified 13489 insertions/deletions, including 562 in the exon regions, and 379 in the splicing sites.
Bioinformatics analysis and RHO mutation screening
A prioritization program was applied to identify any proband pathogenic variants. Public databases, including dbSNP137, 1000 genomes project and NHLBI ESP6500 were used to filter commonly known variants. In the proband, only one heterozygous variant, c.403C>T (p.R135W), of the RHO gene exon 3 was suspected to be a pathogenic variant. No other variants in known disease-causing genes for vision degeneration-related disease were identified. Sanger sequencing confirmed the heterozygous variant present in all affected family members (II:3, II:5, II:7, III:1, III:2, III:4, III:6, III:11 and IV:1) (Figure 1B), but absent in five unaffected family members (III:7, III:12, IV:2, IV:3 and IV:4). The variant co-segregates with the disease in family members and was absent in the 100 unrelated Chinese healthy controls (Figure 1C). This suggests that the variant may be the pathogenic mutation in this family.
Arginine at position 135 (p.R135) is conserved across different species (Figure 1D). The SIFT analysis of mutation obtained a score of 0.00, which suggests a damaging mutation. PolyPhen-2 prediction showed the mutation as “probably damaging” with a score of 1.00 on HumVar (sensitivity, 0.00; specificity, 1.00). MutationTaster analysis indicated that the substitution was disease-causing with a high-confidence probability value close to 1.
Subcellular location prediction
Based on the TMHMM prediction, wild-type p.R135 locates at the second rhodopsin CP loop (amino acids from 134 to 152), but the mutated p.W135 locates at the third TM helice (amino acids from 116 to 138).
Discussion
The RHO gene, located at chromosome 3q22.1, contains five exons. It encodes a 348-amino acid rod-specific protein rhodopsin which is a typical seven TM G-protein-coupled receptor [22]. When rhodopsin absorbs the photon, the retinal chromophore (11-cis-retinal) changes to an all-trans-retinal, leads to a phototransduction cascade activation, which plays an important role in vision [11,23].
Since the first p.P23H mutation in the RHO gene was discovered in 1990 [24], more than 210 different mutations, which vary from point mutations to complex rearrangements, have been reported in different ethnic populations (HGMD Professional, http://www.hgmd.cf.ac.uk/ac/index.php).
Most RHO mutations cause adRP. However, homozygous RHO mutations p.E150K, p.W161X, p.E249X and IVS4+1G>T are reported in a few autosomal recessive RP families [22,25–30]. Heterozygous mutations (p.G90D, p.T94I, p.A295V and p.A292E) are found in patients with congenital stationary night blindness [31].
RP is exceptionally heterogeneous, genetically, allelically, phenotypically and clinically [32]. RHO mutations have been reported to be phenotypically heterogeneous in two instances: classic RP and sector RP. Classic RP is a typical RP characterized by early onset and diffuse or generalized retinal dysfunction. Sector RP is characterized by adult onset and regionalized or sectorial retinal dysfunction [11]. At least 13 RHO gene mutations associate with sector RP [11,23,33–35].
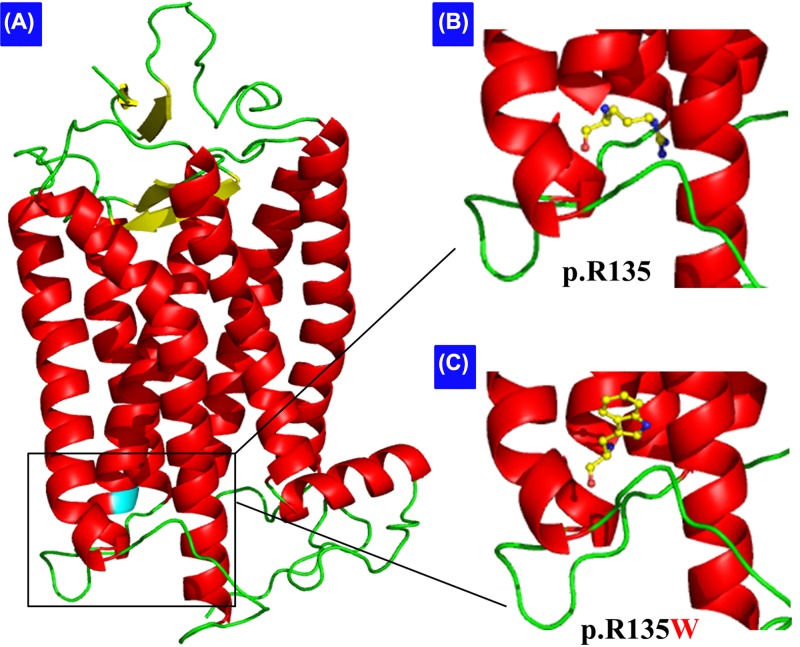
Our study found a reported RHO p.R135W mutation, which has also been found in other ethnicities, including Han-Chinese, Korean, Indian, Turkmenistan Jews, North African Jews, Caucasian, Swedish, French and others (Table 2) [2,3,10,36–40]. p.R135 locates on the border between the second CP loop and the third TM domain of rhodopsin [41]. The chain near position 135 may play a critical function in rhodopsin folding. In addition, R135 creates a salt-bridge with E134 and E247 adjacent to TM helices III and VI, respectively, forming an ionic lock which holds helices III and VI together at their CP ends while keeping the receptor as inactive state [42]. The p.R135W mutation may disrupt ionic interactions and result in partial misfolding and a reduced mutant protein ability to bind 11-cis-retinal (Figure 4) [40,42]. p.R135 seems to be a ‘hot’ mutational site with several other mutations being reported including p.R135P, p.R135L, p.R135G and p.R135W [2,3,10,36–40,43–45]. The p.R135-based mutants can be phosphorylated by rhodopsin kinase and bind arrestin, but cannot activate transducin, in the absence of 11-cis-retinal [41]. Among these mutations, p.R135W leads a more seriously and rapidly progressive RP than p.R135L, as its glycosylation state is more defective [38]. In vitro, bovine p.R135W rhodopsin mutants disrupt G protein activation, contrary to the wild-type rhodopsin [40,41].
Table 2. p.R135W mutation detected in the RHO gene in patients with RP.
| Report | Geographic and ethnic group | Detection method | Family/sporadic | Inheritance pattern | Clinical features in the patients* |
|---|---|---|---|---|---|
| Sung et al., 1991 [36] | Caucasian | DGGE | Family | AD | (1) (3) |
| Bareil et al., 1999 [37] | French | DGGE | Family | AD | (2) (3) (4) (5) |
| Iannaccone et al., 2006 [38] | Indian | Direct sequencing | Family | AD | (2) (3) (4) (5) |
| Kim et al., 2012 [2] | Korean | Direct sequencing | Family | AD | (2) (3) (4) (6) (7) |
| Wu et al., 2014 [3] | Chinese | WES | Family | AD | (1) (2) (3) (4) (6) |
| Beryozkin et al., 2016 [10] | North African Jews | Sanger sequencing | Family | AD | (2) (4) |
| Beryozkin et al., 2016 [10] | Turkmenistan Jews | Sanger sequencing | Family | AD | (2) (4) |
| Abdulridha-Aboud et al., 2016 [39] | Swedish | Genotypic microarray | Family | AD | (2) (3) (4) (5) (6) |
| Yu et al., 2016 [40] | Chinese | CNGS | Family | AD | (1) (2) (4) (5) (6) (9) (10) |
| The present study | Chinese | WES | Family | AD | (1) (2) (3) (4) (5) (6) (7) (8) (9) (10) (11) |
Clinical features in the patients: (1) night blindness; (2) reduced visual acuity; (3) reduced peripheral visual field; (4) typical fundus findings; (5) abnormal electroretinography result; (6) abnormal OCT result; (7) cataract; (8) hyperopic refractive errors; (9) myopic refractive errors; (10) astigmatism; (11) epiretinal membrane.Abbreviations: AD, autosomal dominant; CNGS, capturing next-generation sequencing; DGGE, denaturing gradient gel electrophoresis; RHO, the rhodopsin gene; WES, whole exome sequencing.
Figure 4. Cartoon representation of the model structure of the rhodopsin protein.
(A) Overall structure of the protein is displayed in three dimensions. (B,C) Amino acids at position 135, the arginine (B) and the mutated tryptophan (C), are shown as ball-and-stick models.
High clinical heterogeneity and distinct features were present in this family. All affected patients began having severe night blindness early in life, consisting with other families with RHO p.R135W mutation [3]. Nearly half, 44.4% (4/9), of the patients (II:3, II:5, III:2 and IV:1) have hyperopic refractive errors, a notably higher percentage than most reported RP patients, of whom 75–77.5% have myopia, and only 7.6% have hyperopia [4,6]. Eight of nine patients present with cataracts either unilaterally, or bilaterally, 72.2% (13/18) eyes. This is nearly twice the previously reported morbidity of ∼45% [4,5]. Early cataracts may associate with mild inflammatory reactions [46]. This observation is supported by the fact that transgenic mice with rhodopsin promoter-induced interferon-gamma (IFN-γ) expression tend to develop cataract formation [47]. Patient (III:2) has bilateral EM, and EM is present in 8.2–27.3% of the eyes of RP sufferers in recent reports [48,49]. The complex clinical manifestations observed in this family may result from many factors including genetic background, environmental effects and epigenetic modifications.
Conclusions
An RHO gene missense mutation, c.403C>T (p.R135W) was identified in a Han-Chinese family with adRP. The present study extends the genotype–phenotype relationship between the RHO gene p.R135W mutation and adRP clinical findings. These results have implications for family genetic counseling, clinical management and developing RP target gene therapy strategies.
Acknowledgments
We thank all the enrolled individuals for their participation in the present study.
Abbreviations
- adRP
autosomal dominant RP
- BCVA
best corrected visual acuity
- CP
cytoplasmic
- EM
epiretinal membrane
- ERG
electroretinography
- gDNA
genomic DNA
- IFN-γ
interferon-gamma
- IMPDH1
the inosine monophosphate dehydrogenase 1 gene
- logMAR
logarithm of the minimum angle of resolution
- NR2E3
the nuclear receptor subfamily 2 group E member 3 gene
- OCT
optical coherence tomography
- PolyPhen-2
Polymorphism Phenotyping version 2
- PRPF8
the pre-mRNA processing factor 8 gene
- PRPF31
the pre-mRNA processing factor 31 gene
- PRPH2
the peripherin 2 gene
- RHO
the rhodopsin gene
- RP
retinitis pigmentosa
- RP1
the RP1 axonemal microtubule associated gene
- RPGR
the RP GTPase regulator gene
- SIFT
Sorting Intolerant from Tolerant
- SNP
single nucleotide polymorphism
- TM
transmembrane
- TMHMM
TransMembrane prediction using Hidden Markov Model
- WES
whole exome sequencing
Author Contribution
Y.W., Y.G. and H.D. contributed to designing the study, performing experiments, researching data, discussing contents and writing the article. J.Y., H.X., L.Y. and Z.Y. contributed to providing clinical data, researching data, collecting genetic information and performing experiments. All authors reviewed the manuscript before submission.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China [grant number 2016YFC1306604]; the National Natural Science Foundation of China [grant numbers 81670216, 81800219, 81873686]; the Natural Science Foundation of Hunan Province [grant numbers 2016JJ2166, 2018JJ2556, 2019JJ50927]; Grant for the Foster Key Subject of the Third Xiangya Hospital of Central South University (Clinical Laboratory Diagnostics); and the New Xiangya Talent Project of the Third Xiangya Hospital of Central South University [grant number 20150301].
References
- 1.Zhang Q. (2016) Retinitis pigmentosa: progress and perspective. Asian Pac. J. Ophthalmol. 5, 265–271 10.1097/APO.0000000000000227 [DOI] [PubMed] [Google Scholar]
- 2.Kim C., Kim K.J., Bok J., Lee E.J., Kim D.J., Oh J.H.. et al. (2012) Microarray-based mutation detection and phenotypic characterization in Korean patients with retinitis pigmentosa. Mol. Vis. 18, 2398–2410 [PMC free article] [PubMed] [Google Scholar]
- 3.Wu J., Chen L., Tam O.S., Huang X.F., Pang C.P. and Jin Z.B. (2014) Whole exome sequencing reveals genetic predisposition in a large family with retinitis pigmentosa. Biomed Res. Int. 2014, 302487. 10.1155/2014/302487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S.H., Yu H.G., Seo J.M., Moon S.W., Moon J.W., Kim S.J.. et al. (2010) Hereditary and clinical features of retinitis pigmentosa in Koreans. J. Korean Med. Sci. 25, 918–923 10.3346/jkms.2010.25.6.918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auffarth G.U., Tetz M.R., Krastel H., Blankenagel A. and Volcker H.E. (1997) Complicated cataracts in various forms of retinitis pigmentosa. Type and incidence. Ophthalmologe 94, 642–646 10.1007/s003470050175 [DOI] [PubMed] [Google Scholar]
- 6.Sieving P.A. and Fishman G.A. (1978) Refractive errors of retinitis pigmentosa patients. Br. J. Ophthalmol. 62, 163–167 10.1136/bjo.62.3.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pruett R.C. (1983) Retinitis pigmentosa: clinical observations and correlations. Trans. Am. Ophthalmol. Soc. 81, 693–735 [PMC free article] [PubMed] [Google Scholar]
- 8.Daiger S.P., Bowne S.J. and Sullivan L.S. (2007) Perspective on genes and mutations causing retinitis pigmentosa. Arch. Ophthalmol. 125, 151–158 10.1001/archopht.125.2.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neveling K., Collin R.W., Gilissen C., van Huet R.A., Visser L., Kwint M.P.. et al. (2012) Next-generation genetic testing for retinitis pigmentosa. Hum. Mutat. 33, 963–972 10.1002/humu.22045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beryozkin A., Levy G., Blumenfeld A., Meyer S., Namburi P., Morad Y.. et al. (2016) Genetic analysis of the rhodopsin gene identifies a mosaic dominant retinitis pigmentosa mutation in a healthy individual. Invest. Ophthalmol. Vis. Sci. 57, 940–947 10.1167/iovs.15-18702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katagiri S., Hayashi T., Akahori M., Itabashi T., Nishino J., Yoshitake K.. et al. (2014) RHO mutations (p.W126L and p.A346P) in two Japanese families with autosomal dominant retinitis pigmentosa. J. Ophthalmol. 2014, 210947. 10.1155/2014/210947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim K.J., Kim C., Bok J., Kim K.S., Lee E.J., Park S.P.. et al. (2011) Spectrum of rhodopsin mutations in Korean patients with retinitis pigmentosa. Mol. Vis. 17, 844–853 [PMC free article] [PubMed] [Google Scholar]
- 13.Caburet S., Arboleda V.A., Llano E., Overbeek P.A., Barbero J.L., Oka K.. et al. (2014) Mutant cohesin in premature ovarian failure. N. Engl. J. Med. 370, 943–949 10.1056/NEJMoa1309635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi Y., Li Y., Zhang D., Zhang H., Li Y., Lu F.. et al. (2011) Exome sequencing identifies ZNF644 mutations in high myopia. PLoS Genet. 7, e1002084. 10.1371/journal.pgen.1002084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J.L., Cao L., Li X.H., Hu Z.M., Li J.D., Zhang J.G.. et al. (2011) Identification of PRRT2 as the causative gene of paroxysmal kinesigenic dyskinesias. Brain 134, 3493–3501 10.1093/brain/awr289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P.. et al. (2010) A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng P.C. and Henikoff S. (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 10.1093/nar/gkg509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan L., Wu S., Xu H., Xiao J., Yang Z., Xia H.. et al. (2015) Identification of a novel PHEX mutation in a Chinese family with X-linked hypophosphatemic rickets using exome sequencing. Biol. Chem. 396, 27–33 10.1515/hsz-2014-0187 [DOI] [PubMed] [Google Scholar]
- 19.Huang X., Deng X., Xu H., Wu S., Yuan L., Yang Z.. et al. (2015) Identification of a novel mutation in the COL2A1 gene in a Chinese family with spondyloepiphyseal dysplasia congenita. PLoS ONE 10, e0127529. 10.1371/journal.pone.0127529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y., Hu P., Xu H., Yuan J., Yuan L., Xiong W.. et al. (2016) A novel heterozygous COL4A4 missense mutation in a Chinese family with focal segmental glomerulosclerosis. J. Cell. Mol. Med. 20, 2328–2332 10.1111/jcmm.12924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng W., Chen H., Deng X., Yuan L., Yang Y., Song Z.. et al. (2016) Identification of a novel mutation in the titin gene in a Chinese family with limb-girdle muscular dystrophy 2J. Mol. Neurobiol. 53, 5097–5102 10.1007/s12035-015-9439-0 [DOI] [PubMed] [Google Scholar]
- 22.Rosenfeld P.J., Cowley G.S., McGee T.L., Sandberg M.A., Berson E.L. and Dryja T.P. (1992) A null mutation in the rhodopsin gene causes rod photoreceptor dysfunction and autosomal recessive retinitis pigmentosa. Nat. Genet. 1, 209–213 10.1038/ng0692-209 [DOI] [PubMed] [Google Scholar]
- 23.Napier M.L., Durga D., Wolsley C.J., Chamney S., Alexander S., Brennan R.. et al. (2015) Mutational analysis of the rhodopsin gene in sector retinitis pigmentosa. Ophthalmic Genet. 36, 239–243 10.3109/13816810.2014.958862 [DOI] [PubMed] [Google Scholar]
- 24.Dryja T.P., McGee T.L., Reichel E., Hahn L.B., Cowley G.S., Yandell D.W.. et al. (1990) A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature 343, 364–366 10.1038/343364a0 [DOI] [PubMed] [Google Scholar]
- 25.Van Schil K., Karlstetter M., Aslanidis A., Dannhausen K., Azam M., Qamar R.. et al. (2016) Autosomal recessive retinitis pigmentosa with homozygous rhodopsin mutation E150K and non-coding cis-regulatory variants in CRX-binding regions of SAMD7. Sci. Rep. 6, 21307. 10.1038/srep21307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saqib M.A., Nikopoulos K., Ullah E., Sher K.F., Iqbal J., Bibi R.. et al. (2015) Homozygosity mapping reveals novel and known mutations in Pakistani families with inherited retinal dystrophies. Sci. Rep. 5, 9965. 10.1038/srep09965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kartasasmita A., Fujiki K., Iskandar E., Sovani I., Fujimaki T. and Murakami A. (2011) A novel nonsense mutation in rhodopsin gene in two Indonesian families with autosomal recessive retinitis pigmentosa. Ophthalmic Genet. 32, 57–63 10.3109/13816810.2010.535892 [DOI] [PubMed] [Google Scholar]
- 28.Azam M., Khan M.I., Gal A., Hussain A., Shah S.T., Khan M.S.. et al. (2009) A homozygous p.Glu150Lys mutation in the opsin gene of two Pakistani families with autosomal recessive retinitis pigmentosa. Mol. Vis. 15, 2526–2534 [PMC free article] [PubMed] [Google Scholar]
- 29.Greenberg J., Roberts L. and Ramesar R. (2003) A rare homozygous rhodopsin splice-site mutation: the issue of when and whether to offer presymptomatic testing. Ophthalmic Genet. 24, 225–232 10.1076/opge.24.4.225.17235 [DOI] [PubMed] [Google Scholar]
- 30.Kumaramanickavel G., Maw M., Denton M.J., John S., Srikumari C.R., Orth U.. et al. (1994) Missense rhodopsin mutation in a family with recessive RP. Nat. Genet. 8, 10–11 10.1038/ng0994-10 [DOI] [PubMed] [Google Scholar]
- 31.McAlear S.D., Kraft T.W. and Gross A.K. (2010) 1 rhodopsin mutations in congenital night blindness. Adv. Exp. Med. Biol. 664, 263–272 10.1007/978-1-4419-1399-9_30 [DOI] [PubMed] [Google Scholar]
- 32.Daiger S.P., Sullivan L.S. and Bowne S.J. (2013) Genes and mutations causing retinitis pigmentosa. Clin. Genet. 84, 132–141 10.1111/cge.12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramon E., Cordomi A., Aguila M., Srinivasan S., Dong X., Moore A.T.. et al. (2014) Differential light-induced responses in sectorial inherited retinal degeneration. J. Biol. Chem. 289, 35918–35928 10.1074/jbc.M114.609958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tam B.M., Noorwez S.M., Kaushal S., Kono M. and Moritz O.L. (2014) Photoactivation-induced instability of rhodopsin mutants T4K and T17M in rod outer segments underlies retinal degeneration in X. laevis transgenic models of retinitis pigmentosa. J. Neurosci. 34, 13336–13348 10.1523/JNEUROSCI.1655-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rivera-De L.P.D., Cabral-Macias J., Matias-Florentino M., Rodriguez-Ruiz G., Robredo V. and Zenteno J.C. (2013) Rhodopsin p.N78I dominant mutation causing sectorial retinitis pigmentosa in a pedigree with intrafamilial clinical heterogeneity. Gene 519, 173–176 10.1016/j.gene.2013.01.048 [DOI] [PubMed] [Google Scholar]
- 36.Sung C.H., Davenport C.M., Hennessey J.C., Maumenee I.H., Jacobson S.G., Heckenlively J.R.. et al. (1991) Rhodopsin mutations in autosomal dominant retinitis pigmentosa. Proc. Natl. Acad. Sci. U.S.A. 88, 6481–6485 10.1073/pnas.88.15.6481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bareil C., Hamel C., Pallares-Ruiz N., Arnaud B., Demaille J. and Claustres M. (1999) Molecular analysis of the rhodopsin gene in southern France: identification of the first duplication responsible for retinitis pigmentosa, c.998999ins4. Ophthalmic Genet. 20, 173–182 10.1076/opge.20.3.173.2282 [DOI] [PubMed] [Google Scholar]
- 38.Iannaccone A., Man D., Waseem N., Jennings B.J., Ganapathiraju M., Gallaher K.. et al. (2006) Retinitis pigmentosa associated with rhodopsin mutations: Correlation between phenotypic variability and molecular effects. Vis. Res. 46, 4556–4567 10.1016/j.visres.2006.08.018 [DOI] [PubMed] [Google Scholar]
- 39.Abdulridha-Aboud W., Kjellstrom U., Andreasson S. and Ponjavic V. (2016) Characterization of macular structure and function in two Swedish families with genetically identified autosomal dominant retinitis pigmentosa. Mol. Vis. 22, 362–373 [PMC free article] [PubMed] [Google Scholar]
- 40.Yu X., Shi W., Cheng L., Wang Y., Chen D., Hu X.. et al. (2016) Identification of a rhodopsin gene mutation in a large family with autosomal dominant retinitis pigmentosa. Sci. Rep. 6, 19759. 10.1038/srep19759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi W., Sports C.D., Raman D., Shirakawa S., Osawa S. and Weiss E.R. (1998) Rhodopsin arginine-135 mutants are phosphorylated by rhodopsin kinase and bind arrestin in the absence of 11-cis-retinal. Biochemistry 37, 4869–4874 10.1021/bi9731100 [DOI] [PubMed] [Google Scholar]
- 42.Ou W.B., Yi T., Kim J.M. and Khorana H.G. (2011) The roles of transmembrane domain helix-III during rhodopsin photoactivation. PLoS ONE 6, e17398. 10.1371/journal.pone.0017398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreasson S., Ehinger B., Abrahamson M. and Fex G. (1992) A six-generation family with autosomal dominant retinitis pigmentosa and a rhodopsin gene mutation (arginine-135-leucine). Ophthalmic Paediatr. Genet. 13, 145–153 10.3109/13816819209046483 [DOI] [PubMed] [Google Scholar]
- 44.Bunge S., Wedemann H., David D., Terwilliger D.J., van den Born L.I., Aulehla-Scholz C.. et al. (1993) Molecular analysis and genetic mapping of the rhodopsin gene in families with autosomal dominant retinitis pigmentosa. Genomics 17, 230–233 10.1006/geno.1993.1309 [DOI] [PubMed] [Google Scholar]
- 45.Souied E., Soubrane G., Benlian P., Coscas G.J., Gerber S., Munnich A.. et al. (1996) Retinitis punctata albescens associated with the Arg135Trp mutation in the rhodopsin gene. Am. J. Ophthalmol. 121, 19–25 10.1016/S0002-9394(14)70530-6 [DOI] [PubMed] [Google Scholar]
- 46.Hamel C. (2006) Retinitis pigmentosa. Orphanet J. Rare. Dis. 1, 40. 10.1186/1750-1172-1-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geiger K., Howes E., Gallina M., Huang X.J., Travis G.H. and Sarvetnick N. (1994) Transgenic mice expressing IFN-gamma in the retina develop inflammation of the eye and photoreceptor loss. Invest. Ophthalmol. Vis. Sci. 35, 2667–2681 [PubMed] [Google Scholar]
- 48.Chebil A., Touati S., Maamouri R., Kort F. and El M.L. (2016) Spectral domain optical coherence tomography findings in patients with retinitis pigmentosa. Tunis. Med. 94, 265–271 [PubMed] [Google Scholar]
- 49.Triolo G., Pierro L., Parodi M.B., De Benedetto U., Gagliardi M., Manitto M.P.. et al. (2013) Spectral domain optical coherence tomography findings in patients with retinitis pigmentosa. Ophthalmic Res. 50, 160–164 10.1159/000351681 [DOI] [PubMed] [Google Scholar]