Abstract
Infiltration of leukocytes within the vessel at sites of inflammation and the subsequent generation of myeloperoxidase-derived oxidants, including hypochlorous acid, are key characteristics of atherosclerosis. Hypochlorous acid is a potent oxidant that reacts readily with most biological molecules, including DNA and RNA. This results in nucleic acid modification and the formation of different chlorinated products. These products have been used as biomarkers of inflammation, owing to their presence in elevated amounts in different inflammatory fluids and diseased tissue, including atherosclerotic lesions. However, it is not clear whether these materials are simply biomarkers, or could also play a role in the development of chronic inflammatory pathologies. In this study, we examined the reactivity of different chlorinated nucleosides with human coronary artery endothelial cells (HCAEC). Evidence was obtained for the incorporation of each chlorinated nucleoside into the cellular RNA or DNA. However, only 8-chloro-adenosine (8ClA) had a significant effect on the cell viability and metabolic activity. Exposure of HCAEC to 8ClA decreased glycolysis, and resulted in a reduction in ATP, with a corresponding increase in the chlorinated analogue, 8Cl-ATP in the nucleotide pool. 8ClA also induced sustained endoplasmic reticulum stress within the HCAEC, which resulted in activation of the unfolded protein response, the altered expression of antioxidant genes and culminated in the release of calcium into the cytosol and cell death by apoptosis. Taken together, these data provide new insight into pathways by which myeloperoxidase activity and resultant hypochlorous acid generation could promote endothelial cell damage during chronic inflammation, which could be relevant to the progression of atherosclerosis.
Keywords: Myeloperoxidase, Hypochlorous acid, Nucleoside, RNA, DNA, Inflammation
Graphical abstract
Highlights
-
•
Chlorinated nucleosides are incorporated into the RNA and DNA of endothelial cells.
-
•
8-chloro-adenosine decreases cellular metabolism and glycolysis and induces ER stress.
-
•
ER stress induced by 8-chloro-adenosine activates antioxidant responses.
-
•
ER stress induced by 8-chloro-adenosine is sustained and promotes apoptotic cell death.
Abbreviations
- 18S
18S ribosomal RNA
- 8Cl(d)A
8 chloro-(deoxy)adenosine
- 5Cl(d)C
5-chloro-(deoxy)cytidine
- 8Cl(d)G
8-chloro-(deoxy)guanosine
- 5ClU
5-chlorouracil
- Ad
adenosine
- AMPK
AMP-activated protein kinase
- ATF4
activating transcription factor 4
- ATF6
activating transcription factor 6
- BSA
bovine serum albumin
- CHOP
CCAAT-enhancer-binding protein homologous protein
- DMEM
Dulbecco's Modified Eagle's Medium
- ECAR
extracellular acidification rate
- eIF2α
endoplasmic reticulum eukaryotic translational initiation factor 2α
- ER
endoplasmic reticulum
- ETC
electron transport chain
- FCCP
carbonyl cyanide-p-trifluoromethoxyphenylhydrazone
- GADD34
growth arrest DNA damage inducible 34
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GCLC
glutamate-cysteine ligase catalytic subunit
- GPx4
glutathione peroxidase 4
- GS
glutathione synthase
- HBSS
Hank's buffered salt solution
- HCAEC
human coronary artery endothelial cells
- HO-1
heme oxygenase 1
- IRE1
inositol-requiring enzyme 1
- LDH
lactate dehydrogenase
- MPO
myeloperoxidase
- NBDG
2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)-amino]-d-glucose
- NQO1
NADPH dehydrogenase quinone
- OCR
oxygen consumption rate
- PERK
pancreatic endoplasmic reticulum eukaryotic translational initiation factor 2α (eIF2α) kinase
- RPL13
ribosomal protein L13
- sXBP1
spliced x-box binding protein 1
- TBST
Tris-buffered saline containing Tween 20
- Trx
thioredoxin
- TXNIP
thioredoxin interacting protein
- UPR
unfolded protein response
1. Introduction
Chronic inflammation and the infiltration and activation of leukocytes within the vessel wall, play a central role in the development of lesions and progression of atherosclerosis [1]. This results in the release of myeloperoxidase (MPO) and the overproduction of oxidants, particularly hypochlorous acid (HOCl), at inflammatory sites [2]. HOCl is highly reactive, and can induce the oxidation and modification of a wide range of biomolecules including proteins, lipids, RNA, and DNA [2]. This causes localised damage to the surrounding cells and tissue [3], and has been implicated in the development of numerous inflammatory pathologies, particularly atherosclerosis [4], where elevated levels of MPO are also a risk factor [5] and prognostic marker for patient outcome [6].
The reaction of HOCl with RNA and DNA forms both unstable chloramines (N–Cl species) [7] and stable chlorinated products, including 8-chloro-adenosine (8ClA), 8-chloro-guanosine (8ClG), 5-chloro-cytidine (5ClC), 8-chloro-2′-deoxyadenosine (8CldA), 8-chloro-2′-deoxyguanosine (8CldG), 5-chloro-2′-deoxycytidine (5CldC) and 5-chloro-uracil (5ClU) [[8], [9], [10], [11], [12]]. A number of these stable products have been detected in elevated amounts in the urine, plasma, inflammatory fluids or the diseased tissues of patients with different pathological conditions [9,13,14], including atherosclerosis [15]. Although chlorinated nucleosides are recognised as biomarkers, it is not well understood as to whether these materials can also play a role in promoting the progression of chronic inflammatory disease.
It is well established that the incorporation of chlorinated nucleosides into DNA is mutagenic [[16], [17], [18]], which implicates these compounds as playing a role in the development of inflammatory cancers [19,20]. Somewhat paradoxically, this ability of chlorinated nucleosides to modify cellular DNA and RNA has also been utilised therapeutically in order to kill malignant cells. For example, 5CldC has been used as a sensitizing agent, to increase the amount of damage induced during radiation therapy [21], while the toxicity of 8ClA in many cell types, including chronic lymphocytic leukemia lymphocytes [22], breast cancer [23], myeloma [24] and mantle cell lymphoma cells [25] has led to clinical trials to test its efficacy as a chemotherapeutic drug. In the malignant cell types examined, 8ClA was reported to induce apoptotic cell death by either inhibiting the transcription of anti-apoptotic proteins or through reducing the concentration of intracellular ATP [[22], [23], [24]]. In each case, 8ClA is phosphorylated by various enzymes, including adenosine kinase, to 8Cl-ATP (the ATP analogue of 8ClA) [24,26,27]. Previous studies with malignant cells are typically performed with 8ClA concentrations of 10 μM [[25], [26], [27], [28]], which is believed to be a therapeutically achievable plasma concentration based on data from previous animal studies with 8ClA [29] and clinical trials with 8-Cl-cAMP [30]. However, the levels of 8ClA formed during chronic inflammation, and whether 8ClA can influence the function of non-malignant cells, or influence cellular homeostasis by other pathways and potentially propagate the progression of disease remain unclear.
In cells, the endoplasmic reticulum (ER) is responsible for the folding, synthesis, modification and quality control of a variety of proteins. Many stresses to the ER can cause dysregulation including the activation of inflammatory pathways, the depletion of Ca2+, oxidative stress and hypoxia [31]. Prolonged ER stress leads to the activation of the unfolded protein response (UPR), which is a major pathway governing apoptotic cell death within the inflammatory environment that characterises an atherosclerotic lesion [32,33]. In particular, cell death mediated by the UPR has been implicated in endothelial damage, atherosclerotic plaque instability as well as in the clinical progression of atherosclerosis [32,34]. Furthermore, activation of the UPR has also been associated with the alteration of cellular antioxidant defence systems and other stress-related signalling pathways within cells [35].
In this study, we examined the ability of a series of model chlorinated nucleosides, including 5Cl(d)C, 8Cl(d)G, and 8Cl(d)A to incorporate into the RNA and DNA of human coronary artery endothelial cells (HCAEC) and influence cellular metabolic activity and viability. Of particular note was the sensitivity demonstrated by HCAEC in response to exposure to 8ClA. Therefore, additional studies were performed to examine the effects of this chlorinated nucleoside on stress-related signaling cascades and antioxidant responses within the cells, including ER stress and related pathways, which could play a role in propagating disease progression under chronic inflammatory conditions.
2. Materials and methods
Materials: All aqueous reagents were prepared using nanopure water (npH2O) filtered through a four-stage milli-Q system. All reagents were from Sigma-Aldrich (Castle Hill, NSW, Australia) unless otherwise noted. Chlorinated nucleosides (8ClA; 8CldA; 5ClC; 5CldC; 8ClG; 8CldG) were obtained from BioLog Life Sciences Institute (Bremen, Germany) and reconstituted in npH2O to a stock concentration of 500 μM before storage at −80 °C. Nucleosides for experimental treatments were prepared by diluting stock solutions into complete MesoEndo cell culture medium (Cell Applications) and filtered through Millex® GP 0.22 μm syringe filters to remove any contaminants prior to addition to cells.
Cell culture: HCAEC (Cell Applications, San Diego, CA, USA) from a minimum of 3 individual donors were cultured in MesoEndo cell culture medium (Cell Applications) under sterile conditions in 175 cm2 tissue culture flasks in humidified 5% CO2 at 37 °C. For experiments, cells were prepared by enzymatic dissociation using trypsin-EDTA to form a cell suspension which was normalised to 5 × 104 cells/mL. Cells between passages 3 and 6, were plated in MesoEndo cell culture medium allowed to adhere overnight prior to treatment. HCAEC were exposed to chlorinated nucleosides at varying concentrations (4, 8, 16 μM) for 2, 6, 24, 48 and 72 h. For PERK inhibition studies, HCAEC were pre-incubated with 10 μM GSK2606414 (Merck) for 1 h prior to treatment with 8ClA, with adenosine (Ad) used in analogous experiments as a non-chlorinated control for 8ClA.
Detection of chlorinated nucleosides in RNA and DNA: Following exposure of HCAEC to nucleosides, the cellular RNA and DNA were extracted using an Isolate II RNA/DNA/Protein Kit (Bioline) following manufacturer's instructions and normalised to 500 ng RNA or DNA per reaction and hydrolysed using 2 U Nuclease P1 (Sigma-Aldrich) with incubation for 2 h at 37 °C. Samples were then dephosphorylated with 10 U alkaline phosphatase (Sigma-Aldrich) for 1 h at 37 °C prior to liquid-liquid extraction with chloroform. Samples were subjected to centrifugal filtration through 10 kDa and 0.22 μm filters (Merck), before solid phase extraction using C18 cartridges (Waters). Chromatographic separation of parent and chlorinated nucleosides was achieved using a Discovery C18 column (5 μm pore size, 25 cm × 2.1 mm) (Sigma-Aldrich). A 20 μL injection of each sample was eluted at a flow rate of 0.2 mL/min using 5 mM ammonium formate in H2O (Buffer A) and 5 mM ammonium formate in 50% (v/v) acetonitrile (Buffer B). A gradient elution was performed, with an isocratic hold at 100% Buffer A for 10 min, a linear gradient to 20% Buffer B over 40 min, 50% Buffer B over 10 min and to 100% Buffer B in 5 min where buffer composition remained for 5 min before re-equilibration to 100% Buffer A over the next 5 min prior to injection of the next sample (adapted from Ref. [11]). Detection of nucleosides was achieved by UV absorbance at 260 nm (Agilent Technologies 1290 Infinity) and by positive ESI triple-quadrupole mass spectrometry (Agilent Technologies 6490). Nitrogen was used as the collision gas with source and sheath gas temperatures at 290 and 400 °C, respectively. Mass fragmentation was monitored as previously published [11,36]. The amount of chlorinated product was normalised 1:1000 to parent nucleoside.
Metabolic activity: The metabolic activity of treated cells was analysed using Prestoblue Cell Viability Reagent (Thermo Fisher Scientific) as per manufacturer's instructions. HCAEC were plated at 5 × 104 cells/mL overnight and exposed to chlorinated nucleosides (4, 8, 16 μM) for 24 and 48 h at 37 °C with 5% CO2. 2 h prior to the end of the exposure period, the reagent was added to each sample. Once the 24 or 48 h time point was completed, the change in fluorescence was measured at excitation of 560 nm and emission of 590 nm using a SpectraMax M2e Microplate Reader (Molecular Devices).
Quantification of ATP and 8Cl-ATP in the nucleotide pool: HCAEC were lysed with Milli-Q water before nucleotide pool extraction twice using perchloric acid (0.4 M) and centrifuging to remove cell debris [37]. The supernatant was neutralised to pH 7 by addition of KOH (on ice) and filtered through a 0.2 μm centrifugal filter before separation using HPLC with a ZORBAX SAX column (5 μm 4.6 × 250 mm; Agilent). 50 μL injections were eluted at a flow rate of 1.5 mL/min from an initial mobile phase composition of 60% 5 mM ammonium phosphate (pH 2.8) and 40% 0.75 M ammonium phosphate (pH 3.6) to a final composition of 100% 0.75 M ammonium phosphate (pH 3.6), over 65 min. Eluents were detected with UV absorbance at 254 nm using a SPD-M20A detector. ATP was also quantified by luminescence using the ATPlite assay system (PerkinElmer). HCAEC (1 x 104) were plated overnight in black 96 well plates before exposure to 8ClA for 48 h at 37 °C with 5% CO2. Following treatment, the cells were washed and lysed using the cell lysis solution provided, before addition of the substrate solution and dark adapting the plate for 10 min as per the manufacturer's instructions. The luminescence was recorded using a SpectraMax M2e Microplate Reader (Molecular Devices).
Oxidative phosphorylation and glycolysis: Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were analysed using a Seahorse XF24 Extracellular flux analyser and the cell Mito stress test kit (Agilent). HCAEC were washed with Seahorse XF DMEM supplemented with 20 mM glucose, 40 μM l-glutamine and 80 μM sodium pyruvate after treatment with 8ClA. OCR was measured at 37 °C, in the absence and following sequential injection of inhibitors: oligomycin (1.7 μM), carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP, 0.7 μM) and rotenone/antimycin A (0.5 μM). ECAR was measured in HCAEC washed with Seahorse XF DMEM supplemented with 40 μM l-glutamine followed by incubation at 37 °C and an injection of 10 mM glucose into each sample. Each sample was normalised to protein, quantified using the DC protein assay (BioRad). To confirm the extent of glycolysis, the glycolysis cell-based assay kit (Cayman Chemical) was used to measure the secretion of l-lactate into the extracellular media.
Glucose uptake: The uptake of glucose was assessed using the Glucose Uptake Cell-Based Assay Kit (Cayman Chemical) in HCAEC exposed to 8ClA or Ad (8 μM) for 48 h. Cell culture media was then removed and replaced with glucose free HBSS with or without apigenin (50 μM) followed by incubation for 1 h. The fluorescent glucose analog 2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)-amino]-d-glucose (NBDG, 250 μM) was then added and incubated for 20 min. Supernatant was removed and replaced with Cell-Based Assay buffer. The change in fluorescence was then detected using a SpectraMax M2e Microplate Reader (Molecular Devices) at an excitation and emission of 485 and 535 nm, respectively.
Enzymatic activity of GAPDH: HCAEC were lysed in water following treatment with the nucleosides and washing with PBS to remove cell media. The lysates were added to a solution of NAD+ (20 mM) in 45 mM sodium pyrophosphate/60 mM sodium phosphate (pH 7.4) buffer. Immediately prior to analysis, glyceraldehyde 3-phosphate (180 μM) was added to each sample to start the reaction with the formation of NADH determined by absorbance at 340 nm, monitored at 37 °C over time using a SpectraMax M2e Microplate Reader (Molecular Devices) with activity normalised to protein concentration determined using the bicinchoninic acid (BCA) assay.
Quantitative real-time polymerase chain reaction (qPCR) for mRNA expression: RNA was extracted using ReliaPrep RNA Cell Miniprep System (Promega), then quantified using a Nanodrop 2000C Spectrophotometer (Thermo Scientific). RNA concentrations were normalised to 500 ng/reaction, with reverse transcription performed using the iScript cDNA synthesis kit and thermo-cycling at 25 °C for 5 min, 42 °C for 30 min, 85 °C for 5 min and infinite hold at 4 °C. cDNA was then diluted 1:3 before qPCR using iQ SYBR Green supermix (BioRad). Each sample was then heated to 95 °C for 3 min, annealed at 60 °C for 30 s extended for 30 s at 72 °C, over 40 cycles, with denaturation at 95 °C for 2 min. At the end of each cycle, a dissociation curve at 0.5 °C intervals between 65 °C and 95 °C was plotted. Each sample was compared to Ct values obtained from a standard curve from 1:10 serial dilutions of cDNA for each primer and normalised to two housekeeping genes, 18S ribosomal RNA and RPL13. Primer sequences of the housekeeping and target genes are listed in Supplementary Table 1.
Protein expression and Western blotting: HCAEC were lysed using the cell lytic reagent (Sigma-Aldrich) containing 1% (v/v) protease inhibitors, then separated into cytoplasmic and nuclear fractions using the NE-PER Nuclear and Cytosolic extraction kit (Thermo Fisher Scientific). Protein concentration was normalised using the BCA assay. Samples were loaded onto 4–12% Bis-tris gels (Life Technologies) and transferred onto iBlot 2 PVDF membrane (Thermo Fisher Scientific). The membrane was then blocked in 5% (w/v) BSA in Tris-buffered saline containing 0.1% Tween-20 (TBST; 20 mM Tris-HCl, pH 7.4, 135 mM NaCl, 0.1% (v/v) Tween) for 2 h prior to incubation with 1:1000 anti-ATF4 primary antibody or anti-TXNIP primary antibody for 2 h. A 1:2000 dilution anti-rabbit IgG in 5% (w/v) BSA in TBST (Cell Signaling Technology) was then added and incubated at 21 °C for 1 h. The membrane was imaged by chemiluminescence using Western Lighting – plus ECL (PerkinElmer). Ponceau S staining or a β-actin primary antibody was used to confirm equal protein loading across the Western blot.
Flow cytometry: For apoptosis and necrosis studies, HCAEC were resuspended in Annexin V binding buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2, pH 7.4) prior to addition of Annexin V-APC (BD Biosciences) and propidium iodide (Sigma-Aldrich). Each sample was then vortexed and incubated in the dark at 37 °C, 5% CO2 for 15 min. For calcium release into the cytosol, HCAEC were resuspended in 5 μM Fluo-4AM (Thermo Fisher Scientific) in calcium-free HBSS for 45 min in the dark at 37 °C, 5% CO2. Samples were stored on ice prior to analysis using a FACSVerse flow cytometer (BD Biosciences).
Statistical analyses: Statistical analyses were performed using GraphPad Prism software 7.0 (GraphPad Software, San Diego, USA) using one-way or two-way ANOVA with appropriate multiple comparison post-hoc tests with p < 0.05 taken as significant as outlined in the Figure Legends.
3. Results
3.1. Incorporation of chlorinated nucleosides into the cellular RNA and DNA
Initial studies were performed to determine the extent of incorporation of each chlorinated nucleoside into the RNA and DNA of HCAEC over 72 h. Exposure of HCAEC to the chlorinated ribose nucleosides 8ClA, 8ClG and 5ClC (16 μM, 0.16 pmol/cell) resulted in their incorporation into the cellular RNA in a nucleoside and time-dependent manner (Fig. 1A–C). The incorporation of 8ClA increased over 24 h, with this nucleoside found to incorporate at higher levels in the RNA compared to 8ClG and 5ClC, which were also incorporated but reached maximal levels in the RNA by 6 h of treatment (Fig. 1A–C). Treatment of HCAEC with the chlorinated deoxyribose nucleosides 8CldA, 8CldG and 5CldC (16 μM, 0.16 pmol/cell) resulted in their incorporation into the cellular DNA (Fig. 1D–F). In this case, the maximal incorporation of each nucleoside was achieved at 48 h, with 8CldG found to incorporate to a greater extent compared to 8CldA and 5CldC (Fig. 1D–F). There was no chlorination of the RNA or DNA detected in the control cells, which were incubated in the absence of each chlorinated nucleoside.
Fig. 1.
The incorporation of chlorinated nucleosides into cellular RNA and DNA. HCAEC (1 × 106 cells) were treated with A) 8ClA, B) 8ClG, C) 5ClC, D) 8CldA, E) 8CldG and F) 5CldC (16 μM; 0.16 pmol/cell) for 2–72 h (grey bars) or 24 h with washing to remove exogenous chlorinated nucleosides and re-incubation of the cells in normal cell media for 48 h (white bars, labelled 24 + 48), before extraction, hydrolysis and dephophorylation of the RNA and DNA and analysis by LC-MS. The concentration of chlorinated nucleoside was normalised to the respective parent nucleoside and is expressed as a 1:1000 ratio. Data are the mean ± SEM samples from at least three independent cell donors. *, ** and **** represent a significant (p < 0.05, 0.01 and 0.0001) difference in the level of chlorinated nucleosides compared to the respective non-treated controls, with a, b, and c showing a significant (p < 0.05) difference on comparing the 24 + 48 sample with the 24 h, 48 h or 72 h samples respectively, as determined by 1-way ANOVA with the Sidak multiple comparison post-hoc test.
In each case, there was no significant change in the concentration of the parent nucleosides, or any evidence for the incorporation of ribose nucleosides into DNA and deoxyribose nucleosides into RNA (data not shown). Further experiments were performed to examine whether the concentration of chlorinated nucleoside present in the RNA or DNA was altered in the absence of an exogenous source of 8Cl(d)A, 8Cl(d)G or 5Cl(d)C. In these experiments, the HCAEC were treated with each indicated chlorinated nucleoside for 24 h, before washing the cells, and re-culturing in the absence of the chlorinated nucleoside for a further 48 h. In general, there was no significant difference in the extent of incorporation of each chlorinated nucleoside following the washing and 48 h re-incubation, compared to that observed on analysis immediately after 24–72 h incubation (Fig. 1, white bars, labelled 24 + 48). However, exceptions to this behaviour were observed with 8ClA, where a decrease in 8ClA within the RNA was observed on re-incubation compared to the 24 h treatment (Fig. 1A), and 8CldG, where a decrease was observed compared to the 48 and 72 h treatments. A decrease, albeit non-significant, was also seen in cells exposed to 5ClC and 5CldC (Fig. 1C,F).
These data are consistent with the removal of the 8ClA (and possibly 5ClC) from the RNA, which did not appear to be the case with 8ClG. With 8CldG, the concentration of this nucleoside in the DNA was similar at 24 h to that seen at 24 h with further re-incubation in the absence of 8CldG. This suggests that there is little clearance of this nucleoside, with additional incorporation seen on longer incubation times in the presence of 8CldG, though less incorporation is apparent at 72 h compared to 48 h. There is also a marked, but non-significant, decrease in the concentration of 5CldC within the DNA following re-incubation compared to that seen at 24 h. There was some variation in the pattern and extent of chlorinated nucleoside incorporation in some cases between the individual HCAEC donors, resulting in a large error associated with these analyses.
3.2. 8ClA decreases metabolic activity and glycolysis in HCAEC
The metabolic activity of HCAEC was determined after exposure to chlorinated nucleosides (4, 8 and 16 μM) for 24 h (Fig. 2A) and 48 h (Fig. 2B) using the Prestoblue assay. An alteration in metabolic activity compared to the non-treated, control, cells was only apparent in HCAEC exposed to 8ClA, with no significant changes seen with the other chlorinated nucleosides (Fig. 2). The decrease in metabolic activity observed with 8ClA treatment was greater following 48 h (Fig. 2B) rather than 24 h (Fig. 2A), and was independent of the concentration of 8ClA used, with a similar loss in activity observed in experiments with 4, 8 and 16 μM 8ClA. These differences in metabolic activity were not observed in the analogous experiments with non-chlorinated adenosine controls (data not shown).
Fig. 2.
The effect of chlorinated nucleosides on metabolic activity. HCAEC (1 × 104 cells) were treated each chlorinated nucleoside (4, 8, 16 μM) for A) 24 h or B) 48 h before assessing metabolic activity by conversion of the resazurin-based PrestoBlue® reagent to fluorescent resorufin at λex = 560 nm and λem = 590 nm. Results represent the mean ± SEM of multiple experiments with at least 3 independent cell donors. * and ** show a significant (p < 0.05 and 0.01) change in metabolic activity compared to the non-treated control by two-way ANOVA with the Dunnett's multiple comparison post-hoc test.
It is well established that 8ClA can be metabolised to form the chlorinated analogue of ATP, 8Cl-ATP in malignant cell types, which can lead to a decrease in the production of ATP [24,26,27]. Therefore, the ratio of ATP to 8Cl-ATP was assessed by HPLC following extraction of the nucleotide pool of HCAEC treated with 8ClA (8 μM) for 24 h. Evidence was obtained for the formation of 8Cl-ATP on treatment of the HCAEC with 8ClA but not in control experiments with adenosine (Fig. 3B). The formation of 8Cl-ATP was associated with a small, but non-significant decrease in ATP (Fig. 3A). In addition, there was no significant change in the concentration of GTP, CTP or UTP on treatment of HCAEC with either 8ClA or adenosine treatment (Suppl. Fig. S1). Studies were extended to examine the concentration of ATP following exposure of the HCAEC to 8ClA for 48 h, where a more pronounced change in metabolic activity was seen. In this case, a decrease in ATP was observed in the HCAEC, which was significant in experiments with 16 μM (0.16 pmol/cell) 8ClA, whereas no change in ATP was seen on treatment with non-chlorinated adenosine (Fig. 3C).
Fig. 3.

Treatment of HCAEC with 8ClA results in 8Cl-ATP formation. (A) and (B) HCAEC (6 × 105 cells) were treated with 8ClA (8 μM) or adenosine (8 μM) for 24 h before quantifying the concentration of ATP (A) and 8Cl-ATP (B) in the cellular nucleotide pool following extraction with perchloric acid and separation by HPLC. (C) HCAEC (1 × 104 cells) were treated with 8ClA (4–16 μM) or adenosine (16 μM) for 48 h with ATP quantified using the ATPlite assay. Data represent the mean ± SEM of multiple replicates with at least 2 independent cell donors. * shows a significant (p < 0.05) change compared to the non-treated control samples by one-way ANOVA with Dunnett's multiple comparison post-hoc test.
Given 8ClA treatment of HCAEC decreased the metabolic activity of the cells and endogenous ATP concentration, studies were extended to examine mitochondrial respiration using a Seahorse analyser to measure changes in OCR following a mitochondrial stress test, where oligomycin, FCCP and rotenone/antimycin A are added in series to inhibit ATP production, uncouple the mitochondria and inhibit the electron transport chain (ETC), respectively. HCAEC were treated with 8ClA (4, 8 and 16 μM) or adenosine (16 μM) for 48 h, where a significant decrease in metabolic activity was observed (Fig. 2B). However, there were no differences in the basal respiration of the cells (no inhibitors), the ATP coupler response (with oligomycin), the ETC accelerator response (with FCCP), coupling efficiency, spare respiratory capacity or non-mitochondrial respiration (with rotenone/antimycin A) in the presence of 8ClA or adenosine compared to the control non-treated cells (Fig. 4 and Suppl. Fig. S2). Taken together, these data suggest that 8ClA treatment does not have a significant effect on the mitochondrial production of ATP.
Fig. 4.
The effect of 8ClA on mitochondrial respiration. The change in mitochondrial respiration of HCAEC (3.2 × 104 cells) treated with 8ClA (4, 8, 16 μM) or adenosine (16 μM) for 48 h was determined by measuring the OCR using a Seahorse XF Analyser with the sequential addition of oligomycin (1.7 μM), FCCP (0.7 μM) and rotenone/antimycin A (0.5 μM). The results represent the mean ± SEM of 4 biological replicates with 3 independent cell donors.
The effect of 8ClA on the glycolytic activity of the HCAEC was also examined using a Seahorse analyser to measure changes in ECAR on stimulation of the cells with glucose (10 mM). There were no differences in the baseline ECAR readings of the HCAEC under any of the treatment conditions, which were recorded in the absence of glucose (Fig. 5A). However, on injection of glucose, a marked decrease in the ECAR was seen in the HCAEC exposed to 8ClA, compared to the adenosine or non-treated cells (Fig. 5A and B). This is consistent with 8ClA treatment resulting in a decrease in the glycolytic activity of the HCAEC. These data were supported by measuring the concentration of lactate (the end-product of glycolysis) secreted into the extracellular environment, which was significantly decreased in cells treated with 8ClA (Fig. 5C).
Fig. 5.
The effect of 8ClA on glycolysis and GAPDH activity. HCAEC (3.2 × 104 cells) were treated with 8ClA (4, 8, 16 μM) or adenosine (16 μM) for 48 h before assessing: A,B) basal glycolytic rate using a Seahorse XF analyser to measure ECAR following injection of glucose (10 mM), C) the concentration of extracellular l-lactate and D) GAPDH activity. The results represent the mean ± SEM of 4 biological replicates with 3 independent cell donors. * (p < 0.05), ** (p < 0.01), *** (p < 0.001), **** (p < 0.0001) represent a significant decrease compared to control samples analysed using a one-way ANOVA with Dunnett's multiple comparison post-hoc test.
The uptake of glucose by the HCAEC in the presence and absence of 8ClA was examined using the fluorescent glucose analog NBDG. However, no changes in the glucose uptake were apparent after exposure of the HCAEC to 8ClA compared to the adenosine or non-treated cells (Suppl. Fig. S3). This was in contrast to experiments were HCAEC were treated with apigenin (50 μM), a known inhibitor of Glut-1, where a decrease in the uptake of NBDG was observed (Suppl. Fig. S3). As the decrease in glycolysis was not related to altered glucose uptake, these studies were extended to measure the activity of the glycolytic enzyme GADPH. Exposure of HCAEC to 8ClA (8 μM) for 48 h resulted in a significant (35%) decrease in the activity of GAPDH compared to that observed in the non-treated control cells (Fig. 5D). A less marked, but statistically significant decrease in GAPDH activity was also observed on treatment of the cells with adenosine (Fig. 5D). The reason for this is not certain, given that adenosine treatment does not result in significant changes in either the ECAR or lactate release to the extracellular media.
3.3. 8ClA induces ER stress, alters the cellular antioxidant responses and promotes apoptotic cell death in HCAEC
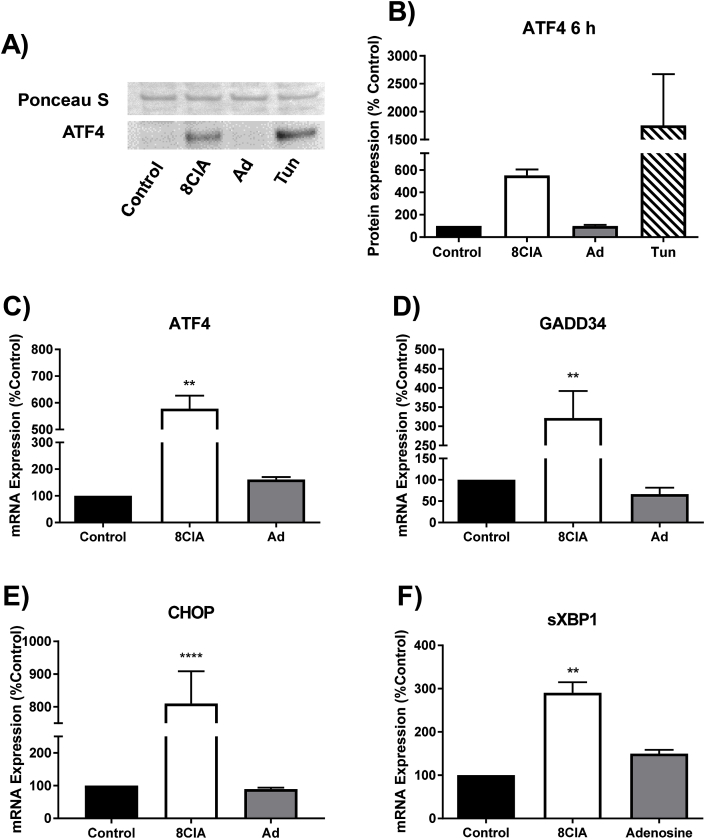
Given the marked effect of 8ClA on cellular metabolism, these studies were extended to examine alterations in the expression of genes associated with stress-related signalling pathways. Evidence was obtained for a significant increase in both the mRNA expression and nuclear translocation of ATF4 in HCAEC treated with 8ClA (Fig. 6A,B,C), which was also observed on treatment of the cells with tunicamycin, a known inducer of the UPR [30] (Fig. 6A and B). There was also a significant increase in the expression of other UPR-associated genes downstream of ATF4, including growth arrest DNA damage inducible 34 (GADD34) and CCAAT-enhancer-binding protein homologous protein (CHOP) (Fig. 6D and E). Treatment of HCAEC with 8ClA also increased the mRNA expression of spliced x-box binding protein 1 (sXBP1; Fig. 6F), which is a marker reflecting IRE1 and ATF6 signalling in response to ER stress [38]. These changes in mRNA expression were not observed in the corresponding experiments with adenosine (Fig. 6).
Fig. 6.
8ClA treatment induces ER stress and activation of the UPR. HCAEC (1 × 105 cells) were treated with 8ClA or adenosine (8 μM) for 6 h (panel A) or 24 h (panels B–E) before assessing markers of ER stress. (A,B) Western blot analysis of nuclear extracts showing translocation of ATF4 from the cytosol into the nucleus compared to treatment with tunicamycin (Tun; 2.5 μg/mL). Graphs (C–F) show the mRNA expression of ATF4, GADD34, CHOP and sXPB1 at 24 h, as analysed by qRT-PCR with normalisation to the expression of the housekeeping genes 18S and RPL13. Results represent the mean ± SEM of 3 biological replicates with 3 independent cell donors and are expressed as an increase relative to the non-treated control. ** (p < 0.01) and **** (p < 0.0001) represent a significant increase compared to control using a one-way ANOVA with Dunnett's multiple post-hoc comparisons test.
ER stress is associated with the induction of cellular antioxidant responses, particularly the Nrf2-Keap1 signalling pathway [35]. Therefore, these studies were extended to examine the effect of 8ClA treatment on the expression of a range of antioxidant genes. Incubation of HCAEC with 8ClA (8 μM) for 24 h resulted in a significant increase in the mRNA expression of NADPH dehydrogenase quinone 1 (NQO1), heme oxygenase 1 (HO-1), and glutamate-cysteine ligase catalytic subunit (GCLC), which supports activation of Nrf2 (Fig. 7A–C). Similarly, there was a significant increase in both the mRNA and protein expression of thioredoxin interacting protein (TXNIP; Fig. 7D,E,F), which lends further support to a role of 8ClA in the induction of cellular stress. However, under these experimental conditions, there were no changes in the mRNA expression of other antioxidant genes, including SOD1, SOD2, glutathione peroxidase 4 (GPx4), thioredoxin (Trx) or glutathione synthase (GS) (Suppl. Fig. S4). In each case, there was also no change in mRNA expression on treatment of the HCAEC with non-chlorinated adenosine (Fig. 6, Suppl. Fig. S4).
Fig. 7.
8ClA alters the expression of cellular antioxidant responses in HCAEC. HCAEC (1 × 105 cells) were treated with 8ClA or adenosine (8 μM) for 24 h before assessing the expression of antioxidant genes or proteins. Graphs (A–D) show the mRNA expression of NQO1, HO-1, GCLC and TXNIP at 24 h, as analysed by qRT-PCR with normalisation to the expression of the housekeeping genes 18S and RPL13. Panels E and F show the alteration in expression of TXNIP protein by Western blotting, normalised to the expression of β-actin. Results represent the mean ± SEM of 3 biological replicates with 3 independent cell donors and are expressed as an increase relative to the non-treated control. * (p < 0.05) and ** (p < 0.01) represent a significant increase compared to control using a one-way ANOVA with Dunnett's multiple post-hoc comparisons test.
Prolonged ER stress results in alterations to the intracellular calcium storage and apoptotic cell death. Therefore, the release of calcium from the ER into the cytosol was assessed in HCAEC treated with 8ClA using flow cytometry with Fluo-4AM staining. Treatment of the HCAEC with 8ClA (8 μM) for 24 h resulted in a significant increase to 1.5-fold of the non-treated control in cytosolic calcium (Fig. 8A, Suppl. Fig. S5). This increase in cytosolic calcium was not observed on pre-treatment of the HCAEC with a pharmacological inhibitor of PERK (10 μM GSK2606414), prior to exposure to 8ClA (Fig. 8B). Similarly, flow cytometry analysis with Annexin V and propidium iodide staining showed that treatment of HCAEC with 8ClA (8 μM) for 48 and 72 h resulted in a significant decrease in the population of viable cells and corresponding increase in the population of Annexin V positive cells, which was not seen with the adenosine or non-treated cells (Fig. 8C and D, Suppl. Fig. S6). With 8ClA treatment, a small, but not significant, increase in the cells containing propidium iodide was also observed, consistent with some necrotic cell death (Fig. 8C and D). Taken together, these data are consistent with the induction of HCAEC death primarily by apoptosis on prolonged exposure to 8ClA.
Fig. 8.
8ClA changes the distribution of intracellular calcium and promotes apoptotic cell death in HCAEC. HCAEC (1 × 105 cells) were treated with 8ClA or adenosine (8 μM) before assessing A,B) changes in cytosolic calcium with and without pre-treatment of the HCAEC with the PERK inhibitor GSK2606414 (10 μM) following 24 h treatment with 8ClA using Fluo-4AM. C) Cell viability following 48 h and D) 72 h treatment with 8ClA using Annexin V and propidium iodide staining, with flow cytometry analysis in each case. Results represent the mean ± SEM of 3 biological replicates with 3 independent cell donors. * (p < 0.05), ** (p < 0.01), *** (p < 0.001) and **** (p < 0.0001) represent a significant increase compared to control using a two-way ANOVA with Dunnett's multiple post-hoc comparisons test.
4. Discussion
There is increasing evidence for the formation of chlorinated nucleosides in different inflammatory pathologies [9,[13], [14], [15]]. This is attributed to the overproduction of the MPO-derived oxidant HOCl, which reacts readily with RNA and DNA, forming a range of stable chlorinated products [2]. Whether these species play a role in promoting the development of disease is less clear, though the presence of chlorinated nucleobases in DNA is mutagenic (e.g. Ref. [18]) and has been implicated as a key factor in the initiation of tumour growth in inflammatory cancers [19,20]. This study examined the effect of chlorinated nucleosides on HCAEC, as MPO is a key driver of endothelial damage in atherosclerosis [4], and there is evidence for the presence of elevated levels of chlorinated nucleobases in human atheroma [15].
The chlorinated ribose nucleotides 8ClA, 8ClG and 5ClC are incorporated into cellular RNA, whereas the chlorinated deoxyribose nucleotides 8CldA, 8CldG and 5CldC, are incorporated into DNA of HCAEC. The lack of incorporation of the chlorinated ribose nucleosides into DNA supports previous data showing that ribonucleotide reductase displays a lack of affinity for chlorinated nucleotides [14,24]. However, this may be cell type specific, as there is evidence for the incorporation of 8ClA into the DNA of human gliomal cell lines for example [39]. In general, the accumulation of each chlorinated nucleoside increased with incubation time, reaching a maximum level after 24–48 h. It has been shown previously that 5Cl(d)C and 8Cl(d)G can penetrate cells to accumulate in the nucleotide pool of the EA.hy926 endothelial cell line, though in this case, only incorporation of 5ClC was evident in the RNA [14]. This contrasts with our findings in HCAEC, and may reflect the difference between primary cells used in the present study, compared to an immortalised cell line. In support of this notion, the penetration of chlorinated nucleosides into primary epithelial prostatic cells is reported to be 40–227 times higher than in the EA.hy926 cells [14]. Thus, the incorporation of 8ClA into the cellular RNA agrees well with studies performed in various malignant cell types [24,26].
The clearance of chlorinated nucleosides from the RNA and DNA was examined by re-culturing the cells for 48 h in the absence of each modified nucleoside after an initial 24 h incubation period. Under the re-incubation conditions, a significant decrease in 8ClA in the RNA was observed, whereas there was no significant difference in the concentration of 5ClC and 8ClG. With 8ClA, this may reflect the destabilisation of the RNA, owing to this nucleoside adopting a syn-rather than anti-conformation, in accord with previous studies [24]. A decrease in RNA stability has also been reported on incorporation of 5ClC in the RNA of EA.hy926 endothelial cells [14]. With HCAEC, although there was a decrease in the concentration of 5ClC in the RNA, the change was non-significant, though this could reflect the variation in the levels of incorporation that were seen between the different cell donors tested here.
With the deoxyribose chlorinated nucleosides, a decrease in the concentration of each modified nucleoside in the DNA was seen, particularly with 8CldG and 5CldC, which were both initially incorporated into the DNA to a higher extent compared to 8CldA. This is likely to reflect some differences between the ability of the chlorinated nucleosides to either penetrate the cells or to be recognised by DNA polymerases and/or ribonuclease reductase for incorporation into the DNA, particularly in the case of 8CldG. A limitation of the current study is that the concentration of chlorinated species in the cellular nucleotide pool was not assessed, but previous reports are consistent with a greater uptake of 8CldG in both endothelial and epithelial prostatic cells [14]. Whether there are differences in the ability of DNA polymerases to recognise chlorinated nucleotides has not been examined in detail, though 5CldC is reported to be incorporated into DNA in a non-cellular, model system more readily than 8CldG [14]. Similarly, the cellular pathways responsible for the removal of chlorinated nucleosides from the DNA are not clear. The decrease in 5CldC within the DNA under the re-incubation conditions could also reflect deamination to uracil, rather than clearance, which may be mediated by cytidine deaminase or dCMP deaminase [40].
Although all the chlorinated nucleosides incorporated into RNA or DNA, a significant difference in metabolic activity was only apparent in experiments with 8ClA. The decrease in metabolic activity occurred over a similar time period to the accumulation of 8ClA within the nucleotide pool as 8Cl-ATP, and was associated with a decrease in glycolysis, rather than altered mitochondrial respiration, concomitant with apoptotic cell death. 8ClA is a potent inducer of apoptosis in multiple cancer cell lines, which has led to its use as a therapeutic agent for the treatment of chronic lymphocytic leukemia. In general, the mechanism of cell death has been proposed to be through the formation of 8Cl-ATP and depletion of ATP [22,28,41]. However, cell death in malignant cells can also result from alterations in cellular signalling, leading to the inhibition of transcription of anti-apoptotic proteins [22] or autophagy [28].
The autophagic cell death reported in breast cancer cell lines is caused by the activation of the AMP activated protein kinase (AMPK), which is induced by the depletion of endogenous ATP on exposure of the cells to 8ClA [28]. In this case, activation of AMPK attenuated mTOR signaling to result in the phosphorylation of the autophagy factor, Unc51-like kinase 1 [28]. Evidence for the activation of AMPK was also obtained in experiments with 8Cl-cAMP, where again, a marked decrease in ATP is apparent [27]. Whether AMPK is activated in HCAEC exposed to 8ClA remains to be established. However, although a marked increase in the accumulation of 8Cl-ATP was apparent in HCAEC, the concentrations of ATP were only decreased on prolonged treatment (48 h) with 8ClA. This is in contrast to the case with malignant cells, where in general, a more rapid and pronounced decrease in endogenous ATP levels are observed [25,28]. With the HCAEC, the loss of ATP may be associated with an ability of 8ClA to compromise energy production by glycolysis, shown by a decrease in ECAR, lactate production, and GAPDH activity, as there was no evidence for altered mitochondrial respiration. The perturbation of glycolysis on exposure of cells to 8ClA has been reported previously [28]. In this case, the decrease in glycolysis was attributed to altered glucose uptake, and occurred together with a decrease in basal mitochondrial respiration, which is in contrast to the current data with HCAEC.
However, treatment of HCAEC with 8ClA altered stress-related cellular signalling cascades, with strong evidence to support the induction of ER stress and the activation of the UPR, which may be responsible for the apoptotic cell death observed with these cells. The pathway responsible for triggering ER stress in this case is not clear, but may be related to the dysregulation of cellular bioenergetics observed on treating HCAEC with 8ClA, given the central role of the ER in metabolism. Activation of the UPR is intended to restore cellular homeostasis and promote cell survival, but prolonged ER stress triggers apoptosis and the activation of additional stress responses within the cells [35]. Our data are consistent with activation of both the PERK and IRE1 branches of the UPR, resulting in the nuclear translocation of ATF4, and the increased expression of GADD34 and the transcription factor CHOP, which promotes cell death via apoptosis [38].
Evidence was also obtained in support of the activation of the transcription factor Nrf2, with a significant increase in the expression of several Nrf2-associated antioxidant genes, including NQO1, HO-1 and GCLC, observed in HCAEC treated with 8ClA. This may be a consequence of the induction of ER stress, as PERK can phosphorylate Nrf2, which promotes its dissociation from Keap1 [35]. Similarly, treatment of HCAEC with 8ClA increases the expression of TXNIP, which plays a vital role in the regulation of glucose metabolism, together with regulating the activity of protein disulphide isomerases [42]. TXNIP increases the activity of ER resident protein disulphide isomerases, to promote protein folding. Therefore, the induction of ER stress has been shown in a number of cell types to elevate TXNIP expression [43,44]. The overexpression of TXNIP is also linked to pro-apoptotic activity and redox stress [43], which may contribute to the HCAEC cell death in this study.
Taken together, our results demonstrate that 8ClA is detrimental to HCAEC survival and homeostasis, by perturbing cellular metabolism and inducing ER stress and the upregulation of antioxidant and stress responses, culminating in apoptotic cell death. ER stress is a characteristic of both early and late lesions in atherosclerosis, and is a key pathway to endothelial dysfunction [32,34]. Whether 8ClA is present in sufficiently high concentration at inflammatory sites to induce this effect requires further investigation. However, it is notable that comparable concentrations of 8ClA (and other chlorinated nucleosides) are formed in cells exposed to pathologically-relevant concentrations of HOCl [11,36]. This suggests that 8ClA may be able to contribute pathologically during chronic inflammation, in addition to providing a potential biomarker for disease diagnosis. Furthermore, it is important to note that 8ClA exerts these cellular effects on HCAEC at concentrations comparable to the plasma levels achieved therapeutically in previous intervention studies [29] and clinical trials with the related analogue 8Cl-cAMP [30], which is reported to have the same mechanism of action [26]. This raises questions as to the wider implications of the use of 8ClA on the vasculature for example, in a chemotherapeutic context.
Funding sources
This work was supported by the Australian Research Council, through the Future Fellowship scheme (FT120100682) and the Novo Nordisk Foundation (Project Grant NNF17OC0028990). VT acknowledges the receipt of an Australian Government Research Training Program Scholarship from the University of Technology Sydney.
Acknowledgements
The authors would like to thank Mr Pat Pisansarakit for assistance with the tissue culture, Dr Verena Taudte for assistance with LC-MS data acquisition and analysis and Ms Anne Bredegaard for assistance with the ATP quantification studies.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101274.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Libby P. Inflammation in atherosclerosis, arterioscler. Thromb. Vasc. Biol. 2012;32(9):2045–2051. doi: 10.1161/ATVBAHA.108.179705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies M.J., Hawkins C.L., Pattison D.I., Rees M.D. Mammalian heme peroxidases: from molecular mechanisms to health implications. Antioxidants Redox Signal. 2008;10(7):1199–1234. doi: 10.1089/ars.2007.1927. [DOI] [PubMed] [Google Scholar]
- 3.Rayner B.S., Love D.T., Hawkins C.L. Comparative reactivity of myeloperoxidase-derived oxidants with mammalian cells. Free Radic. Biol. Med. 2014;71:240–255. doi: 10.1016/j.freeradbiomed.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Teng N., Maghzal G.J., Talib J., Rashid I., Lau A.K., Stocker R. The roles of myeloperoxidase in coronary artery disease and its potential implication in plaque rupture. Redox Rep. 2017;22(2):51–73. doi: 10.1080/13510002.2016.1256119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang R., Brennan M.L., Fu X., Aviles R.J., Pearce G.L., Penn M.S., Topol E.J., Sprecher D.L., Hazen S.L. Association between myeloperoxidase levels and risk of coronary artery disease. J. Am. Med. Assoc. 2001;286(17):2136–2142. doi: 10.1001/jama.286.17.2136. [DOI] [PubMed] [Google Scholar]
- 6.Vita J.A., Brennan M.-L., Gokce N., Mann S.A., Goormastic M., Shishehbor M.H., Penn M.S., Keaney J.F., Jr., Hazen S.L. Serum myeloperoxidase levels independently predict endothelial dysfunction in humans. Circulation. 2004;110:1134–1139. doi: 10.1161/01.CIR.0000140262.20831.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawkins C.L., Davies M.J. Hypochlorite-induced damage to nucleosides: formation of chloramines and nitrogen-centered radicals. Chem. Res. Toxicol. 2001;14:1071–1081. doi: 10.1021/tx010071r. [DOI] [PubMed] [Google Scholar]
- 8.Whiteman M., Jenner A., Halliwell B. 8-Chloroadenine: a novel product fromed from hypochlorous acid-induced damage to calf thymus DNA. Biomarkers. 1999;4(4):303–310. doi: 10.1080/135475099230831. [DOI] [PubMed] [Google Scholar]
- 9.Asahi T., Kondo H., Masuda M., Nishino H., Aratani Y., Naito Y., Yoshikawa T., Hisaka S., Kato Y., Osawa T. Chemical and immunochemical detection of 8-halogenated deoxyguanosines at early stage inflammation. J. Biol. Chem. 2010;285(12):9282–9291. doi: 10.1074/jbc.M109.054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byun J., Henderson J.P., Heinecke J.W. Identification and quantification of mutagenic halogenated cytosines by gas chromatography, fast atom bombardment, and electrospray ionization tandem mass spectrometry. Anal. Biochem. 2003;317(2):201–209. doi: 10.1016/s0003-2697(03)00093-9. [DOI] [PubMed] [Google Scholar]
- 11.Stanley N.R., Pattison D.I., Hawkins C.L. Ability of hypochlorous acid and N-chloramines to chlorinate DNA and its constituents. Chem. Res. Toxicol. 2010;23(7):1293–1302. doi: 10.1021/tx100188b. [DOI] [PubMed] [Google Scholar]
- 12.Noyon C., Delporte C., Dufour D., Cortese M., Rousseau A., Poelvoorde P., Neve J., Vanhamme L., Zouaoui Boudjeltia K., Roumeguere T., Van Antwerpen P. Validation of a sensitive LC/MSMS method for chloronucleoside analysis in biological matrixes and its applications. Talanta. 2016;154:322–328. doi: 10.1016/j.talanta.2016.03.087. [DOI] [PubMed] [Google Scholar]
- 13.Henderson J.P., Byun J., Takeshita J., Heinecke J.W. Phagocytes produce 5-chlorouracil and 5-bromouracil, two mutagenic products of myeloperoxidase, in human inflammatory tissue. J. Biol. Chem. 2003;278(26):23522–23528. doi: 10.1074/jbc.M303928200. [DOI] [PubMed] [Google Scholar]
- 14.Noyon C., Roumeguere T., Delporte C., Dufour D., Cortese M., Desmet J.M., Lelubre C., Rousseau A., Poelvoorde P., Neve J., Vanhamme L., Boudjeltia K.Z., Van Antwerpen P. The presence of modified nucleosides in extracellular fluids leads to the specific incorporation of 5-chlorocytidine into RNA and modulates the transcription and translation. Mol. Cell. Biochem. 2017;429(1–2):59–71. doi: 10.1007/s11010-016-2936-2. [DOI] [PubMed] [Google Scholar]
- 15.Takeshita J., Byun J., Nhan T.Q., Pritchard D.K., Pennathur S., Schwartz S.M., Chait A., Heinecke J.W. Myeloperoxidase generates 5-chlorouracil in human atherosclerotic tissue. A potential pathway for somatic mutagenesis by macrophages. J. Biol. Chem. 2006;281(6):3096–3104. doi: 10.1074/jbc.M509236200. [DOI] [PubMed] [Google Scholar]
- 16.Pal B.C., Cumming R.B., Walton M.F., Preston R.J. Environmental pollutant 5-chlorouracil is incorporated in mouse-liver and testes DNA. Mutat. Res. 1981;91(4–5):395–401. doi: 10.1016/0165-7992(81)90021-x. [DOI] [PubMed] [Google Scholar]
- 17.Morris S.M. The genetic toxicology of 5-fluoropyrimidines and 5-chlorouracil. Mutat. Res. 1993;297:39–51. doi: 10.1016/0165-1110(93)90006-9. [DOI] [PubMed] [Google Scholar]
- 18.Fedeles B.I., Freudenthal B.D., Yau E., Singh V., Chang S.C., Li D., Delaney J.C., Wilson S.H., Essigmann J.M. Intrinsic mutagenic properties of 5-chlorocytosine: a mechanistic connection between chronic inflammation and cancer. Proc. Natl. Acad. Sci. U.S.A. 2015;112(33):E4571–E4580. doi: 10.1073/pnas.1507709112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weitzman S.A., Gordon L.I. Inflammation and cancer: role of phagocyte-generated oxidants in carcinogenesis. Blood. 1990;76:655–663. [PubMed] [Google Scholar]
- 20.Weitzman S.A., Weitberg A.B., Clark E.P., Stossel T.P. Phagocytes as carcinogens; malignant transformation produced by human neutrophils. Science. 1985;227:1231–1233. doi: 10.1126/science.3975611. [DOI] [PubMed] [Google Scholar]
- 21.Hale J.T., Bigelow J.C., Mathews L.A., McCormack J.J. Analytical and pharmacokinetic studies with 5-chloro-2'-deoxycytidine. Biochem. Pharmacol. 2002;64(10):1493–1502. doi: 10.1016/s0006-2952(02)01413-2. [DOI] [PubMed] [Google Scholar]
- 22.Balakrishnan K., Stellrecht C.M., Genini D., Ayres M., Wierda W.G., Keating M.J., Leone L.M., Gandhi V. Cell death of bioenergetically compromised and transcriptionally challenged CLL lymphocytes by chlorinated ATP. Blood. 2005;105:4455–4462. doi: 10.1182/blood-2004-05-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stellrecht C.M., Ayres M., Arya R., Gandhi V. A unique RNA-directed nucleoside analog is cytotoxic to breast cancer cells and depletes cyclin E levels. Breast Canc. Res. Treat. 2010;121(2):355–364. doi: 10.1007/s10549-009-0481-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stellrecht C.M., Rodriguez C.O., Ayres M., Gandhi V. RNA-directed actions of 8-chloro-adenosine in multiple myeloma cells. Cancer Res. 2003;63(22):7968–7974. [PubMed] [Google Scholar]
- 25.Dennison J.B., Balakrishnan K., Gandhi V. Preclinical activity of 8-chloroadenosine with mantle cell lymphoma: roles of energy depletion and inhibition of DNA and RNA synthesis. Br. J. Haematol. 2009;147(3):297–307. doi: 10.1111/j.1365-2141.2009.07850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gandhi V., Ayres M., Halgren R.G., Krett N.L., Newman R.A., Rosen S.T. 8-chloro-cAMP and 8-chloro-adenosine act by the same mechanism in multiple myeloma cells. Cancer Res. 2001;61(14):5474–5479. [PubMed] [Google Scholar]
- 27.Han J.H., Ahn Y.H., Choi K.Y., Hong S.H. Involvement of AMP-activated protein kinase and p38 mitogen-activated protein kinase in 8-Cl-cAMP-induced growth inhibition. J. Cell. Physiol. 2009;218(1):104–112. doi: 10.1002/jcp.21573. [DOI] [PubMed] [Google Scholar]
- 28.Stellrecht C.M., Vangapandu H.V., Le X.F., Mao W., Shentu S. ATP directed agent, 8-chloro-adenosine, induces AMP activated protein kinase activity, leading to autophagic cell death in breast cancer cells. J. Hematol. Oncol. 2014;7:23. doi: 10.1186/1756-8722-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gandhi V., Chen W., Ayres M., Rhie J.K., Madden T.L., Newman R.A. Plasma and cellular pharmacology of 8-chloro-adenosine in mice and rats. Cancer Chemother. Pharmacol. 2002;50:85–94. doi: 10.1007/s00280-002-0456-0. [DOI] [PubMed] [Google Scholar]
- 30.Tortora G., Ciardiello F., Pepe S., Tagliaferri P., Ruggiero A., Bianco C., Guarrasi R., Miki K., Bianco A.R. Phase I clinical study with 8-chloro-cAMP and evaluation of immunological effects in cancer patients. Clin. Cancer Res. 1995;1(4):377–384. [PubMed] [Google Scholar]
- 31.Cao S.S., Kaufman R.J. Unfolded protein response. Curr. Biol. 2012;22(16):R622–R626. doi: 10.1016/j.cub.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Tabas I. The role of endoplasmic reticulum stress in the progression of atherosclerosis. Circ. Res. 2010;107(7):839–850. doi: 10.1161/CIRCRESAHA.110.224766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Civelek M., Manduchi E., Grant G.R., Stoeckert C.J., Jr., Davies P.F. Discovery approaches to UPR in athero-susceptible endothelium in vivo. Methods Enzymol. 2011;489:109–126. doi: 10.1016/B978-0-12-385116-1.00007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou A.X., Tabas I. The UPR in atherosclerosis. Semin. Immunopathol. 2013;35(3):321–332. doi: 10.1007/s00281-013-0372-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Espinosa-Diez C., Miguel V., Mennerich D., Kietzmann T., Sanchez-Perez P., Cadenas S., Lamas S. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 2015;6:183–197. doi: 10.1016/j.redox.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Badouard C., Masuda M., Nishino H., Cadet J., Favier A., Ravanat J.L. Detection of chlorinated DNA and RNA nucleosides by HPLC coupled to tandem mass spectrometry as potential biomarkers of inflammation. J. Chromatogr. B. 2005;827(1):26–31. doi: 10.1016/j.jchromb.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 37.Gandhi V., Danhauser L., Plunkett W. Separation of 1-β-d-arabinofuranosylcytosine 5′-triphosphate and 9-β-d-arabinofuranosyl-2-fluoroadenine 5′-triphosphate in human leukemia cells by high-performance liquid chromatography. J. Chromatogr. B. 1987;413:293–299. doi: 10.1016/0378-4347(87)80242-6. [DOI] [PubMed] [Google Scholar]
- 38.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 39.Langeveld C.H., Jongenelen C.A., Theeuwes J.W., Baak J.P., Heimans J.J., Stoof J.C., Peters G.J. The antiproliferative effect of 8-chloro-adenosine, an active metabolite of 8-chloro-cyclic adenosine monophosphate, and disturbances in nucleic acid synthesis and cell cycle kinetics. Biochem. Pharmacol. 1997;53(2):141–148. doi: 10.1016/s0006-2952(96)00593-x. [DOI] [PubMed] [Google Scholar]
- 40.Cummins R.R., Balinsky D. Activities of some enzymes of pyrimidine and DNA synthesis in a rat transplantable hepatoma and human primary hepatomas, in cell lines derived from these tissues, and in human fetal liver. Cancer Res. 1980;40(4):1235–1239. [PubMed] [Google Scholar]
- 41.Chen L.S., Nowak B.J., Ayres M.L., Krett N.L., Rosen S.T., Zhang S., Gandhi V. Inhibition of ATP synthase by chlorinated adenosine analogue. Biochem. Pharmacol. 2009;78(6):583–591. doi: 10.1016/j.bcp.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee S., Min Kim S., Dotimas J., Li L., Feener E.P., Baldus S., Myers R.B., Chutkow W.A., Patwari P., Yoshioka J., Lee R.T. Thioredoxin-interacting protein regulates protein disulfide isomerases and endoplasmic reticulum stress. EMBO Mol. Med. 2014;6(6):732–743. doi: 10.15252/emmm.201302561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Q., Che X., Zhang H., Fan P., Tan G., Liu L., Jiang D., Zhao J., Xiang X., Liang Y., Sun X., He Z. Thioredoxin-interacting protein links endoplasmic reticulum stress to inflammatory brain injury and apoptosis after subarachnoid haemorrhage. J. Neuroinflammation. 2017;14(1):104. doi: 10.1186/s12974-017-0878-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szpigel A., Hainault I., Carlier A., Venteclef N., Batto A.F., Hajduch E., Bernard C., Ktorza A., Gautier J.F., Ferre P., Bourron O., Foufelle F. Lipid environment induces ER stress, TXNIP expression and inflammation in immune cells of individuals with type 2 diabetes. Diabetologia. 2018;61(2):399–412. doi: 10.1007/s00125-017-4462-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.