Graphical abstract
Keywords: Interbacterial competition, Contractile phage tail-like particle, R-type tailocin, Type VI secretion system, Kosakonia, Kosakonicin
Highlights
-
•
Suggestion to simplify and unify the nomenclature of phage tail-like particles.
-
•
Discovery of kosakonicin, a new bacteriocin and tailocin.
-
•
Microscopy of kosakonicin from Kosakonia radicincitans DSM 16656.
-
•
Discovery of multiple tail fiber genes in the kosakonicin gene cluster.
-
•
Discovery of large genetic diversity in the kosakonicin tail fiber locus among ten Kosakonia strains.
Abstract
Type VI secretion systems and tailocins, two bacterial phage tail-like particles, have been reported to foster interbacterial competition. Both nanostructures enable their producer to kill other bacteria competing for the same ecological niche. Previously, type VI secretion systems and particularly R-type tailocins were considered highly specific, attacking a rather small range of competitors. Their specificity is conferred by cell surface receptors of the target bacterium and receptor-binding proteins on tailocin tail fibers and tail fiber-like appendages of T6SS. Since many R-type tailocin gene clusters contain only one tail fiber gene it was appropriate to expect small R-type tailocin target ranges. However, recently up to three tail fiber genes and broader target ranges have been reported for one plant-associated Pseudomonas strain. Here, we show that having three tail fiber genes per R-type tailocin gene cluster is a common feature of several strains of Gram-negative (often plant-associated) bacteria of the genus Kosakonia. Knowledge about the specificity of type VI secretion systems binding to target bacteria is even lower than in R-type tailocins. Although the mode of operation implicated specific binding, it was only published recently that type VI secretion systems develop tail fiber-like appendages. Here again Kosakonia, exhibiting up to three different type VI secretion systems, may provide valuable insights into the antagonistic potential of plant-associated bacteria. Current understanding of the diversity and potential of phage tail-like particles is fragmentary due to various synonyms and misleading terminology. Consistency in technical terms is a precondition for concerted and purposeful research, which precedes a comprehensive understanding of the specific interaction between bacteria producing phage tail-like particles and their targets. This knowledge is fundamental for selecting and applying tailored, and possibly engineered, producer bacteria for antagonizing plant pathogenic microorganisms.
Bacteriophages and phage tail-like particles – relationships and benefits
Recent studies have provided important insight into so-called phage tail-like particles. The nanoscale size of these structures renders microscopical research challenging, but close relationships to well-studied bacteriophages have helped to develop a clear image. This review presents important scientific discoveries over the past few years about the composition, relationship and benefits of phage tail-like particles, and attempts to sort out the inconsistent terminology that is rich in synonyms. In the second part we provide our own results on the Gram-negative genus Kosakonia to support recent ideas about the complexity of phage tail-like particle-encoding gene clusters, which may allow a previously unexpected wide range of targets.
Bacteria that contain phages or phage tail-like particles may be used for biocontrol
New research has produced evidence that plants cannot survive without bacteria, since bacteria protect plants against filamentous fungi and oomycetes [1]. However, particular strains of bacteria are themselves destructive plant pathogens and cause severe plant diseases, as well as harvest losses in agriculture. In nature the equilibrium between pathogenic, commensal, and beneficial bacteria supports the well-being of plants, whereas in agriculture (particularly in mono-cropping), yield is seriously threatened by pathogenic bacteria.
Antibiotic-resistant bacteria are emerging worldwide and careless use renders broad-spectrum antibiotics ineffective, also in agriculture [2]. Hence, tailored narrow-spectrum antibiotics should be applied in the future, but applicable agents are lacking. Phage therapy proved long ago that bacteriophages (phages) have the potential to close this gap. Phages have not only been applied successfully in humans to cure them of multidrug-resistant bacteria, but also in agriculture to protect crops against plant-pathogenic bacteria [reviewed in 3]. The sensitivity of phages to environmental stressors such as UV irradiation and soil (type, pH, moisture, organic matter content) is still an obstacle to phage therapy on plants, but promising advances have been achieved by combining phages with biocontrol bacteria [3]. Remarkably, not only phages are found in bacteria but also phage tail-like particles, which do not require isolation from the host prior to application, and may thus be employed more easily as promising biocontrol agents.
Bacteria may suffer from virulent phages and benefit from temperate phages
Phages are viruses that infect bacteria by injecting their own genome into the host cytoplasm. Phages can be virulent or temperate. A temperate phage differs from a virulent phage in its ability to integrate its genome into the chromosome of the bacterial host. The phage genome is then called a prophage. Since all phages have the potential to lyse the host cell wall it is of crucial importance whether a phage is integrated into the bacterial genome or not. Virulent phages start their lytic cycle by killing the bacterial host cell immediately after injecting their DNA into the host cytoplasm. In contrast, prophages can be vertically transmitted to bacterial daughter cells, possibly providing beneficial traits to the host, and may remain latent for many generations without causing any harm to the host. The prophage remains latent until environmental changes induce the lytic life cycle, where the prophage is excised and expressed, phage progeny produced, and the host cell destroyed. Prophages have played a major role in bacterial evolution by contributing to the diversification of the bacterial genome architecture [4].
Phage tail-like particles increase interbacterial competition
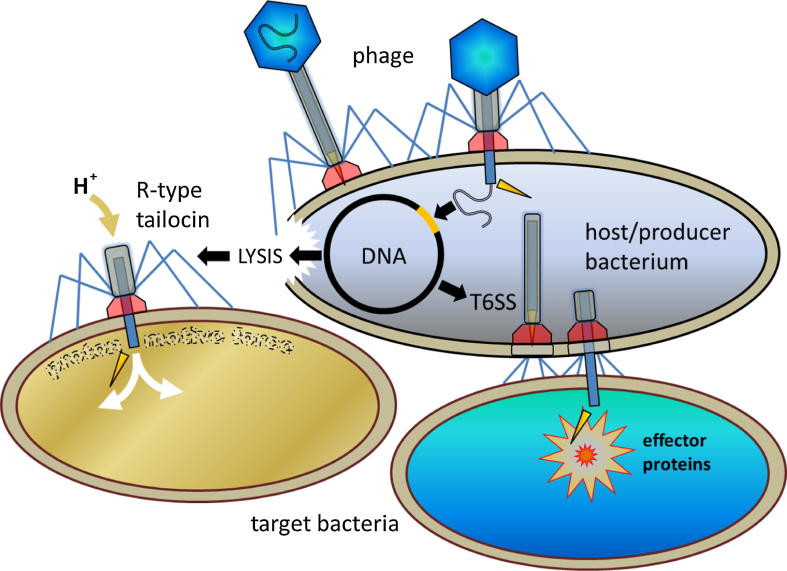
There are different modes of interbacterial competition, but remarkably, two highly efficient bacterial nanomachines have been attributed to derivatives of tailed phages (Fig. 1). In both cases subcellular bacterial components resembling fractions of phages confer exceptional qualities to bacteria. The first component is the “type VI secretion system (T6SS)”, a syringe-like injection apparatus up to several µm long that penetrates the cell wall of other bacteria (and eukaryotes) in order to inject toxic effector proteins. The second component is the much shorter tailocin. T6SS and tailocin differ from phage in lacking the capsid that carries the phage genome. The suffix “–cin” is used in many antibiotics (e.g. streptomycin) and bacteriocins (e.g. colicin) and refers to their ability to kill bacteria. There are two types of tailocins, the rigid R-type resembling the tail of myophages (contractile tail phages of the family Myoviridae), and the flexible F-type resembling the tail of siphophages. The 0.1–0.2 µm short “R-type tailocin” destroys the membrane potential of other bacteria by puncturing their cell wall, leading to collapse of their proton motive force (Fig. 1).
Fig. 1.
Schematic overview of bacteriophages and two contractile phage tail-like particles. Depicted are the type VI secretion system (T6SS) and the R-type tailocin including sites and modes of action. Note: (i) development of T6SSs and tailocins from prophages represent long-term evolutionary events (assuming prophages evolved before T6SSs and tailocins); (ii) bacterial cells carrying phages are called “host” cells, but bacterial cells exhibiting T6SSs and tailocins are called “producer” cells; (iii) the release of tailocins requires lysis of the producer cell, which implies that the same cell cannot produce functional T6SSs at the same time. Phages and contractile phage tail-like particles are not drawn to scale: T6SSs are much longer than phage tails and tailocins.
Although lacking the contractile sheath of myophages, siphophages create channels in bacterial cell walls. Involved in this process is a central tail protein, the TMP (tape measure protein). The working mechanism has not yet been determined, but there are two models: (i) DNA from the phage head forces TMP out of the tail tube, which then penetrates the cell membrane and forms the channel for DNA passage; (ii) opening of the tail at the bottom allows TMP to emerge and the DNA follows [5]. Since F-type tailocins carry neither phage heads nor DNA, model 1 can be excluded for these phage tail-like particles.
The following part of this article focuses on R-type tailocins, and although there are several synonyms for tailocins and R-type tailocins, for simplification purposes we use the superordinated terminology suggested by Gill and Young [6].
T6SSs and R-type tailocins kill target bacteria by different mechanisms
T6SS, R-type tailocin and myophage share the contractile tail (which is a central spike-carrying tube surrounded by a contractile sheath), the baseplate and the associated “tail fibers” (TFs). Only recently, T6SSs have been found to possess tail fiber-like appendages located on the bacterial outer surface, also called “extracellular bacteriophage tail fiber-like antennae”, that are presumably required for recognizing targets [7] (Fig. 1).
Due to the similarity to phage tails, T6SSs and tailocins have been termed “phage tail-like particles” [8], which is a technical expression that was coined decades ago [9]. In contrast to phages, phage tail-like particles do not bind to the outer surface of host cells, here more precisely called “producer” cells, but support producer cells in attacking cells of other bacterial strains, hereafter referred to as target cells or target bacteria. Tailocins are bacteriocins and as such they are supposed, and were repeatedly shown, to inhibit the growth of closely related bacteria. In contrast, T6SSs have been considered to affect distantly related bacteria and eukaryotes (see below). However, there are aberrant observations: recent research has revealed that T6SSs may also be applied for intraspecific [10] and tailocins for intergeneric bacterial competition (see below). Nonetheless, there are important differences between T6SSs and R-type tailocins: (i) functional T6SSs are bacterial producer cell wall-anchored appendages resembling inverted phages and functional R-type tailocins are producer-derived but extracellular particles resembling non-inverted phages; (ii) T6SSs kill bacteria by injecting toxins and R-type tailocins kill bacteria by destroying the proton motive force; (iii) a producer cell can apply T6SSs to protect itself from other bacterial strains, but has to die in order to release R-type tailocins that protect sister cells. Lysis of a few bacterial producer cells supposedly enables R-type tailocin utilization to a colony-benefiting extent, i.e. the bacterial colony sacrifices some cells for the “sake” of the population; (iv) T6SSs are much longer than R-type tailocins, since they have to bridge a longer distance: both nanomachines penetrate the cell wall of the target bacterium, but only T6SSs have to also “penetrate” the producer cell wall and bridge the interbacterial space. Thus, longer T6SSs are favored by the producer, and sheath length is limited only by producer cell diameter [11] (Fig. 1).
The T6SS is only one of several (currently nine) described types of bacterial protein secretion systems [12]. These secretion systems have in common that they enable the bacterial producer cell to transport proteins from its cytoplasm into the environment, other bacteria or eukaryotic cells. The bacterial protein secretion systems differ in composition, cargo and origin. The T6SS is the only one proposed to be of (ancient) myophage origin [13].
Phage-like protein-translocation structures represent a third group of contractile phage tail-like particles and resemble partly T6SSs and partly R-type tailocins
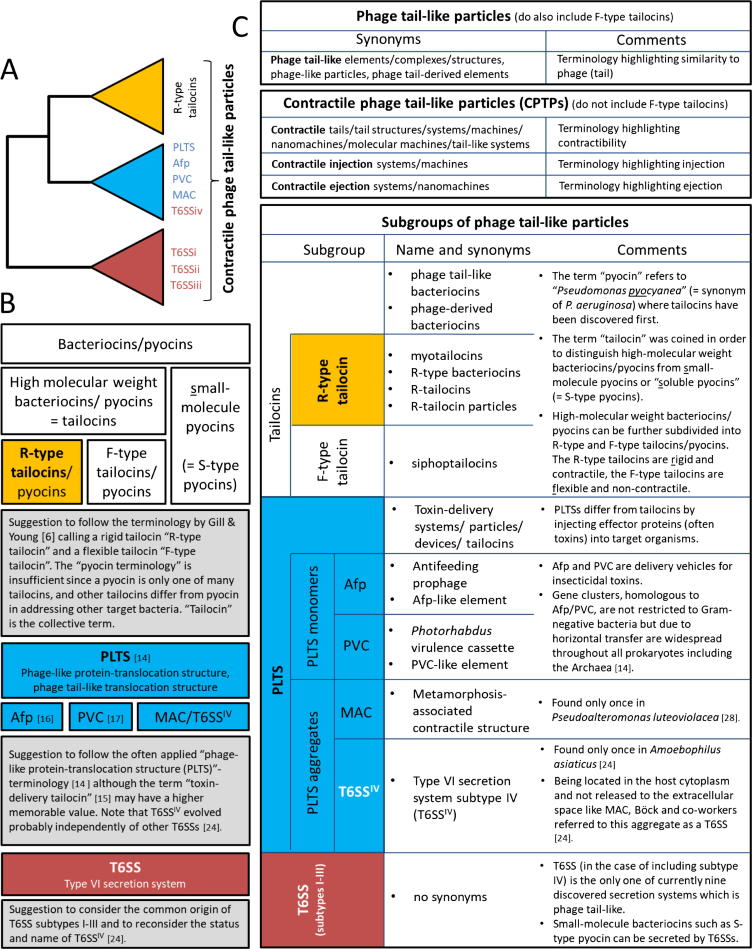
Besides T6SSs and tailocins there is a third group of headless phage tail-like particles, the so-called “phage-like protein-translocation structures” (PLTSs) [14]. T6SSs, R-type tailocins (but not F-type tailocins) and PLTSs are contractile phage tail-like particles (CPTPs). While PLTSs resemble R-type tailocins in morphology [15] and in fulfilling their task outside the producer cell, they resemble T6SSs in secreting proteins. PLTSs include “antifeeding prophages (Afp)” [16] and “Photorhabdus virulence cassettes (PVCs)” [17]. The different names describe the characteristics of PLTSs. The term “protein translocation” refers to the ability of PLTSs to secrete proteins, which is the main characteristic distinguishing PLTSs from R-type tailocins. Accordingly, Heymann and co-workers called the PLTS a “toxin-delivery tailocin” [15]. Remarkably, many proteins secreted by PLTSs are insecticides. The term “antifeeding” refers to the impact of PLTSs on herbivore insects, causing cessation of feeding activity. The term “virulence” has been adopted since PVCs provide the bacterial producer with virulence against insects.
The terminology of contractile phage tail-like particles (CPTPs) is confusing and misleading
Two synonyms of CPTPs have been coined that refer to their contractibility: (1) contractile injection systems, and (2) contractile ejection systems. The term “injection” refers to the ability of many CPTPs to inject substances into target cells; “ejection” addresses the ability to eject substances from the producer cell. All CPTPs are ejection systems, but not all of them are injection systems. R-type tailocins eject a target cell-puncturing device, the tail tube [18], but do not inject substances into the target cell [19], except probably the spike (Fig. 1), but this is supposedly not affecting the target or at least not participating as an active component of the functional (proton motive force depleting) R-type tailocin. Nonetheless, the term “contractile injection system (CIS)” or contractile injection machine has repeatedly been applied lately as a synonym for CPTPs, including R-type tailocins [20], [21], [22], [23].
Distinguishing external CIS (eCIS = R-type bacteriocins/tailocins and PLTSs) from internal CIS (T6SSs), as suggested by Böck and co-workers [24], makes the terminology even more complicated. Rather than referring to “contractile phage tail-like particles (CPTPs)” as the generic term for T6SSs, PLTSs and R-type tailocins seems more appropriate. In order to distinguish extracellularly from intracellularly functioning structures one may refer to external and internal CPTPs.
CPTPs are manifold and affect all domains of life
By attacking insects, Afps and PVCs significantly increase the target spectrum of CPTPs. Bacteria that possess all three types of CPTPs are able to affect important kingdoms in all domains of life. A single bacterial strain may have an impact on other bacteria, archaea, animals (e.g. mammals, insects and nematodes), plants and fungi by expressing CPTP-encoding gene clusters. CPTPs have been found to be involved in shaping polymicrobial communities. For instance deployment of T6SSs was shown to enable simultaneous attack on prokaryotic and fungal rivals [25]. The effect of T6SSs on fungi was not known until recently when Trunk and co-workers found that the T6SS of Serratia marcescens deploys antifungal effectors Tfe1 and Tfe2 against Saccharomyces cerevisiae and two species of the genus Candida. Interestingly, R-type tailocins from Pseudomonas also target strains of other bacterial genera, such as Campylobacter, Haemophilus, Neisseria [reviewed in 26] and Xanthomonas [27], suggesting that these bacteria share specific cell surface receptors with Pseudomonas spp.
Some CPTPs form complex aggregates
Remarkably, some bacteria tend to form ordered arrays (or aggregates) of CPTPs within or outside producer cells that are not involved in antagonizing competitors. Two case studies have been reported where a hexagonal arrangement of several CPTPs was observed: (1) the “metamorphosis-associated contractile structure (MAC)” in Pseudoalteromonas luteoviolacea [28], and (2) the “Type VI secretion system subtype IV (T6SSIV)” in Amoebophilus asiaticus [24]. The gene clusters of both aggregates are closely related to Afp and PVC, but differ significantly in their function: MACs induce the metamorphosis of marine tubeworms from free-swimming larvae into sessile juveniles, and T6SSIV are produced by intracellular bacterial symbionts of amoebae in order to evade digestion in phagosomes. Böck and co-workers hypothesized that either T6SSIV evolved from an Afp/MAC-like injection system independently of other subtypes of T6SS, or that T6SSIV represents a primordial system from which all other phage tail-like particles and phages have evolved [24]. In either case, referring to this as a completely new secretion system and not as a subtype of T6SS would have helped to avoid confusion in terminology of phage tail-like particles (BOX 1).
BOX 1.
Phage tail-like particles: Subgroups, definitions and frequent synonyms; only contractile phage tail-like particles (CPTPs) are highlighted by background colour. A. Summary of phylogenetic analyses from previous studies [8], [14], [24] depicting relationships of CPTPs. B. Overview of subgroups of bacteriocins and phage tail-like particles including suggestions for terminology. C. Frequent synonyms and comments on nomenclature of phage tail-like particles and subgroups.
T6SSs and PLTSs may have evolved only once
T6SSs (subtypes I, II and III together) and PLTSs have supposedly each evolved only once in earth history [14], [29]. However, since horizontal transfer of T6SSs [29] and PLTSs [14] is a frequent phenomenon in many prokaryotes, both types of phage tail-like particles can be found in several lineages of Gram-negative bacteria, and PLTSs even in Gram-positive bacteria and Archaea. The R-type tailocin situation appears to be complicated: the common ancestor of PLTSs is sister to a group of R-type tailocins and several myophages [8], [14] suggesting that myophages have turned into R-type tailocins repeatedly and possibly vice versa (see below).
Enterobacteriophage P2 and R-type tailocins of several Gammaproteobacteria share a common ancestry
Many temperate phages that are frequently found in Gram-negative bacteria belong to the viral family Myoviridae. A prominent member of this family is enterobacteriophage P2, which was first discovered in, and isolated from Escherichia coli. So-called P2-like prophages constitute a subgroup of Myoviridae and are widespread among enteric bacteria (=members of the family Enterobacteriaceae). Interestingly, the R-type tailocin of Pseudomonas is derived from a common ancestral origin with phage P2 [30]. Nakayama and co-workers stated that the stable gene cluster organization suggests that the R-type tailocin is not a defective phage, but a phage tail that has been evolutionarily specialized as a bacteriocin [30].
Tailocins are diverse and nested among complete phages in phylogenetic trees
Many tailocins have been described under genus-specific or even species-specific names. For instance tailocins (both R-type and F-type) of Pseudomonas have been called “pyocins”. This term was already coined in 1954 by Jacob [31] when he discovered an “antibiotic” of Pseudomonas pyocyanea (=synonym of P. aeruginosa). Tailocins appear to be common within Gammaproteobacteria [8] and have been described in many strains of the family Enterobacteriaceae. According to their enteric producers, R-type tailocins have been called xenorhabdicin (Xenorhabdus nematophila) [32], carotovoricin (Pectobacterium carotovorum) [33], enterocoliticin (Yersinia enterocolitica) [34], fonticin (Pragia fontium) and aquaticin (Budvicia aquatica) [35]. It has to be stressed that the allocation of species-specific names to R-type tailocins of closely related taxa is legitimate, since the antagonistic activity of R-type tailocins is often restricted to particular strains of a species.
Remarkably, not all R-type tailocins are derived from the same evolutionary event, i.e. the “transformation” of bacteriophages to R-type tailocins (including the loss of the capsid) has likely occurred independently in different species [36]. Phylogenies comprising both tailocin and bacteriophage genes suggest that formation of new tailocins may be a common phenomenon among bacteria. The observation that tailocins are nested among complete phages [8] appears to contradict the statement of Nakayama and co-workers that tailocins are not degenerate prophages (see above). Furthermore, in the genus Pseudomonas several tailocin gene clusters (of both R-type and F-type) have been found to occur in pairs or triplets and to be combined with intact prophages [26]. This might indicate that recombination of tailocins and intact prophages can result in new tailocins and possibly new phages. Comprehensive bioinformatics work will provide important insights into relationships of phages and phage tail-like particles.
Occurrence and use of tailocins in plant growth-promoting bacteria (PGPB) such as Kosakonia radicincitans
Kosakonia radicincitans is a bacterium of the Gram-negative family Enterobacteriaceae with plant growth-promoting properties [37]. The species has been found to be a native endophyte of a large variety of crop plants around the world [38]. Type strain DSM 16656 was repeatedly inoculated into diverse plant species and proved capable of establishing biofilms on roots [39]. Comparison of the genome of DSM 16656 to other fully sequenced genomes of K. radicincitans and closely related bacterial taxa revealed that all to date known members of K. radicincitans exhibit genomic features that are absent from other closely related bacterial species, such as a duplicated flagellar system and an additional type VI secretion system [38].
The bacterial rhizosphere community of plants is supposedly plant species-specific [40], [41], [42]. However, the environment and some microbial keystone species have a large impact on the plant and its associated microbiota [43], [44]. The inoculation of K. radicincitans DSM 16656 into tomato plants caused significant changes in the bacterial community composition of roots and shoots [38]. The fact that different bacterial strains compete with each other for plant-created niches in the rhizosphere and the phyllosphere [45] suggests that K. radicincitans might possess exceptional tools for interbacterial competition.
Kosakonia radicincitans has the potential to become a model organism for R-type tailocin and T6SS research
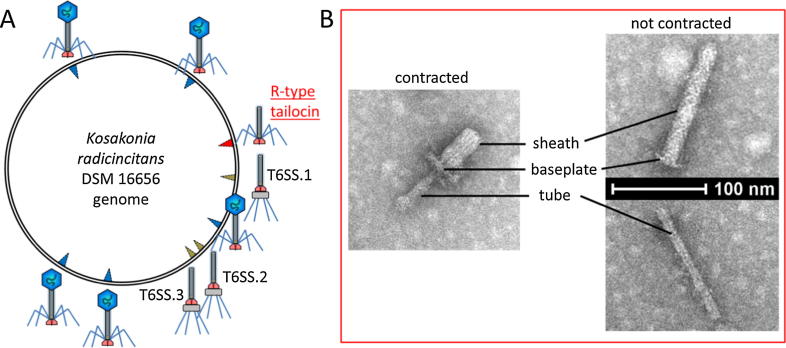
Recently it was shown that Kosakonia radicincitans DSM 16656 has six prophages, of which “phage region 2” differs from the other five phages by being highly conserved among strains of both Kosakonia subgroups [38], indicating an important function. Using PHASTER software [46] and our own bioinformatics pipeline for homologous gene detection showed that Kosakonia phage region 2 differs from the other five phages by lacking the capsid-encoding genes, suggesting it might code for a tailocin (Fig. 2A) (see Supplementary File 1 for Material and Methods).
Fig. 2.
Prophages and contractile phage tail-like particles of Kosakonia radicincitans DSM 16656. A. Schematic overview of prophage, R-type tailocin, and T6SS positions in the genome of DSM 16656. B. Transmission electron micrographs of DSM 16656 R-type tailocins.
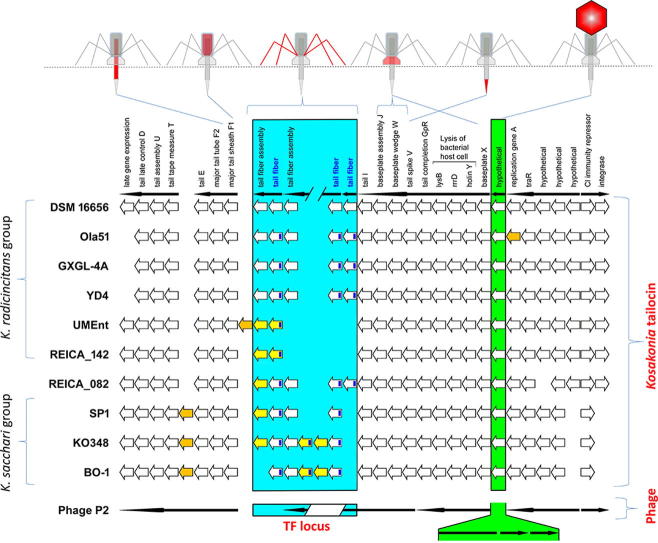
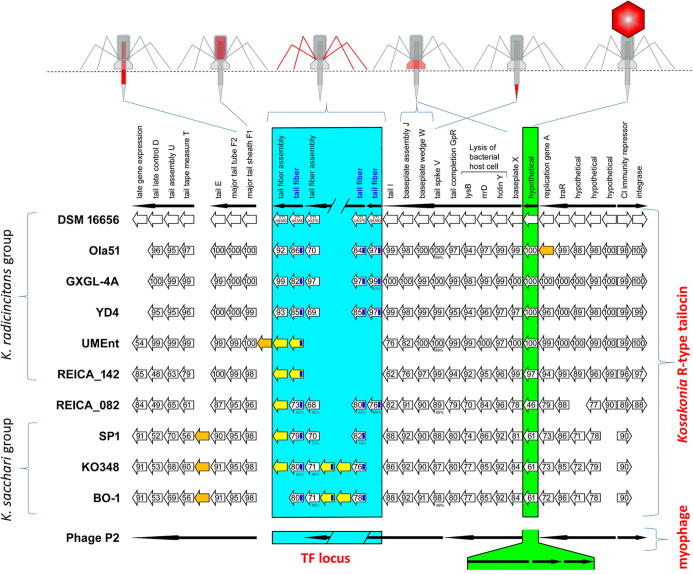
BLASTn searches [47] of Kosakonia phage region 2 showed high homology of this gene cluster to enterobacteriophage P2. Homology of phage P2 and R-type tailocins was previously shown for other Gammaproteobacteria (see above). The capsid locus of phage P2 comprises eight genes that have been replaced by a single hypothetical protein-encoding gene in all ten Kosakonia strains (Fig. 3).
Fig. 3.
R-type tailocin gene cluster of ten Kosakonia strains. Each empty white arrow represents a gene of DSM 16656; NCBI accessions are given for the five tail fiber (TF) locus genes. Black arrows above represent the operons of DSM 16656. Numbers in arrows indicate the aa sequence identity of genes from other Kosakonia strains homologous to DSM 16656; Sequence coverage is 97–100% if not indicated otherwise with lower percentages below arrows; yellow arrows represent TF locus genes (either TF genes or chaperones) missing in DSM 16656; orange arrows represent genes missing in DSM 16656 that are neither TF genes nor chaperones. The closely related phage P2 is depicted below Kosakonia strains, here black arrows show operons of phage P2; the green box indicates the position of the capsid genes including associated genes in phage P2 (together eight genes) and the gene encoding an alternative hypothetical protein in all Kosakonia strains. The blue box highlights the hyper-variable TF locus and provides the position and number of TF genes in Kosakonia strains and phage P2; in the latter one TF gene and one chaperon occur. The dark blue rectangle in arrows representing TF genes shows the position of a conserved gene fraction.
Transmission electron microscopy (TEM) analyses (see Supplementary File 1 for Material and Methods) allowed the screening of bacterial cultures of DSM 16656 for tailocins and revealed nanoscale structures in pure cultures of DSM 16656 outside bacterial cells, which resembled R-type tailocins of other bacteria [e.g. 27] in shape and size (Fig. 2B). While screening Kosakonia cultures, neither capsids nor complete phages could be detected, suggesting that Kosakonia R-type tailocins were found. The Kosakonia R-type tailocins were 130 ± 10 nm long and 20–21 nm in diameter; the inner tube was slightly shorter than the complete tailocin and had a diameter of 10–11 nm; the contracted sheath was 55 ± 10 nm short and had a diameter of 26–27 nm.
Tailocin-producing bacteria may increase bacterial target ranges by having multiple tailocin gene clusters or combining several tail fiber genes in the same tailocin gene cluster
The host range of phages and the target range of tailocins are determined by bacterial cell surface receptors and receptor-binding proteins (RBPs) on tail fibers. This applies to both phage tail fibers and tailocin tail fibers. The larger the diversity of tail fibers and RBPs, presumably the wider is the target range. Already in 1992, Haggård-Ljungquist postulated the horizontal transfer of tail fiber genes (TF genes) as one of five methods for extending the host range of phages [48], and Tétart and co-workers reported that tail fiber genes respond to strong selection pressure by homologously recombining gene fragments between unrelated phages [49]. Williams and co-workers demonstrated that chimeric tail-fibers, where the variable C-terminus of P. aeruginosa was replaced by tail fiber C-termini of different phages, showed a killing spectrum broader than the host range of the phage [50].
Two tailocin gene clusters within one genome have been reported in rhizosphere-occupying bacteria that encounter a large variety of bacterial competitors [51], [52].
Until recently it was believed that a single tailocin gene cluster usually has one TF gene and one associated chaperon (although a recombination of R-type and F-type tailocin was found in the same tailocin gene cluster, carrying TF genes of both types [26]), but three TF genes in a single tailocin gene cluster have only been reported for Pseudomonas chlororaphis by Dorosky and co-workers [52]. Generating different gene deletion mutants, the authors found distinct killing spectra for bacteria possessing different TF genes.
The discovery of Dorosky and co-workers was presumably the first and to date only report of three TF genes in a single tailocin gene cluster. However, Fig. 3 shows that several strains of the genus Kosakonia also have three TF genes. The analysis revealed that there are three different TF genes and two associated tail fiber assembly genes (chaperones) in the R-type tailocin gene cluster of Kosakonia radicincitans DSM 16656. The comparison of the DSM 16656 genome to genomes of other Kosakonia strains unveiled that some strains share the three TF genes with DSM 16656 or “replaced” one of them by another TF gene, while other strains have evolved less complex TF loci and exhibit only the minimum number of genes, i.e. one TF gene and one chaperone (Fig. 3).
Future analyses of other bacterial taxa should show whether having three TF genes is an exceptional or a more widespread phenomenon than currently known. K. radicincitans DSM 16656 carries its three TF genes on three separate operons, indicating that differential expression of the three genes is possible and potentially triggered by different niches and associated microbes. In future, combined microscopic, gene expression and gene mutation analyses will help elucidate the spectrum and limits of tailocin diversity in Kosakonia species.
Closely-related bacteria may have different tailocins pointing to different target spectra
Comparing DSM 16656 with other strains of Kosakonia revealed that closely-related strains of the same bacterial genus may all possess a highly conserved tailocin gene cluster, which only differs significantly in number and composition of TF genes. Comparison of ten different Kosakonia strains revealed a hyper-variable locus within the R-type tailocin gene cluster where highly diverse TF genes have evolved. Four strains of Kosakonia radicincitans (from Germany (DSM 16656), China (Ola 51 [53], GXGL-4A [54]), and Argentina (YD4) [55] share all three TF genes and differ only in gene sequence identity, whereas two other strains of the same species s.l. (here referred to as the K. radicincitans group) from Malaysia (UMEnt) [56] and Philippines (REICA_142) [57] lack all three TF genes but have each acquired another TF gene and another chaperone with no homology to the DSM 16656 tailocin genes. In the three considered strains of the closely-related K. sacchari group, one TF homolog of DSM 16656 is missing, but two strains acquired an additional TF gene [58], [59], [60]. This large diversity of the TF locus appears even more remarkable in the light of the highly conserved major tail sheath and major tail tube genes that share more than 95% of their gene sequence among all ten investigated Kosakonia strains. A comparison of the three TF protein sequences of Kosakonia radicincitans DSM 16656 showed that these genes are highly conserved at the N-terminus (68.6% identity within the first 153 aa of the consensus sequence), and are very variable at the C-terminal end (8.7% identity within the last 219 aa of the consensus sequence). This indicates that the three tail fiber proteins may have different target ranges. The position of the conserved fraction of the TF genes is indicated in dark blue in Fig. 3.
Are Gram-negative bacteria more competitive than other bacteria?
While T6SSs and tailocins have only been found in Gram-negative bacteria, phage-like-protein-translocation structures (PLTSs) have also been discovered in Gram-positive bacteria and even in Archaea. However, this is supposed to reflect horizontal transfer of PLTSs from Gram-negative bacteria to other prokaryotic groups [14]. Hence, regarding the modification of phages for bacterial purposes Gram-negative bacteria seem to be a few steps ahead of other prokaryotes.
Having three T6SSs and an R-type tailocin with up to three distinct TF genes, Kosakonia radicincitans is a suitable species for studying T6SSs as well as R-type tailocin evolution and function. Tailocins with more than three TF genes have not been reported yet and three or more than three type VI secretion systems (T6SSs) may occur in less than 2% of bacteria (as inferred from: “T6SSs occur in only one quarter of bacteria” [61] and “only 7% of T6SS-containing bacteria possess three or more T6SSs” [29]).
Kosakonicin appears to be an appropriate term for the Kosakonia R-type tailocin
Tailocins are “high molecular weight bacteriocins”, in which the term “bacteriocin” refers to the ability to kill closely related bacterial strains. The naming of a particular bacteriocin is not derived from the sensitive target strain but from the producer strain. This is the first report on the genus Kosakonia producing tailocins. Since all investigated Kosakonia strains exhibited the same R-type tailocin gene cluster, the term “kosakonicin” seems to suit all of them. However, since the kosakonicin gene cluster varies between the different Kosakonia taxa in the TF locus, considering species-specific names, such as “radicincicin” and “saccharicin”, might be justified.
Tailocin specificity: Why do tailocins predominantly kill closely related bacteria?
Phages bind specifically to cell surface receptors of their bacterial host. The specificity remains high over time due to co-evolution of phage and host. If a tailocin evolves by transition from a phage, the target range has to change from self to niche-competing bacteria. It has been shown for Pseudomonas that R-type tailocin-producing strains are typically resistant to the R-type tailocins they release [62]. Instead, tail fibers bind to different strains of the same species or closely related species, which have evolved from a common ancestor [49], [52]. The target recognition mechanisms of tailocins have been analyzed mainly for R-type tailocins from P. aerigunosa and Burkholderia cenocepacia: The tailocin fiber genes encode receptor-binding proteins at their C-terminus that bind to specific bacterial surface components, which are very often lipopolysaccharide (LPS) residues [63], [64].
Future studies should show whether a particular tailocin gene cluster that may have existed for millions of years (as it is probably the case in the genus Kosakonia) is still predominantly addressing closely related strains. Taking into account that (i) any new tail fiber gene may broaden the range of target bacteria, and (ii) tail fiber genes may be horizontally transferred or duplicated prior to evolving into different directions, it remains to be tested whether the Kosakonia R-type tailocin is also able to affect distantly related bacteria. In vitro studies of Kosakonia spp. and other bacteria will help unravel the antagonistic potential of this genus. Assuming that each TF gene allows binding to, and killing of another bacterial strain competing for the same niche, it follows that the more distinct TF genes a bacterium has, the more niches it can putatively occupy.
Conclusions and future perspectives
The fact that 95% of all phages carry tails despite of the metabolic cost of building these structures implies that tails must provide phages with a tremendous evolutionary advantage [5], [42]. Bacteria have supposedly taken advantage of this ideal device by transforming the phage tail into phage tail-like particles. However, whether tailed phages or phage tail-like particles evolved first remains unknown.
Since tailed phages and phage tail-like particles share the ability to penetrate bacterial cell walls, both components could be applied as pharmaceutical or biocontrol agents against pathogenic bacteria. The application of tailored or even engineered biocontrol bacteria possessing specific phage tail-like particles for antagonizing plant pathogens is a challenging goal for the future. The interaction between bacterial cell surface receptors and receptor-binding proteins on tail fibers is extremely complex [65]. If one manages to find tight correlations between tail fiber genes and cell surface receptors of target bacteria this will pave the way for selecting appropriate candidate bacteria from strain collections and molecular engineering of phage tail-like particles as a promising tool to combat pathogenic bacteria.
Conflict of interest
The authors have declared no conflict of interest.
Compliance with Ethics requirement
This article does not contain any studies with human or animal subjects.
Acknowledgements
We would like to thank Christina Maaß for excellent technical assistance. MB received funding from DIP (grant 742266/1) and BB from BMBF (grant FKZ 031A282).
Biographies

Sascha Patz is a PhD student at the University of Tuebingen in the research group “Algorithms in Bioinformatics” of Prof. Dr. Huson. During his diploma thesis at the Martin-Luther University and Leibniz Institute of Plant Biochemistry Halle (Saale), afterwards as Leonardo da Vinci fellow at the Eötvös Loránd University (Budapest), and at the Leibniz Institute of Vegetable and Ornamental Crops (Grossbeeren) he studied the effect of microbial inoculants on plants. Currently, his research topics comprise comparative genomics and metagenomics related to beneficial crop-microbe interaction and to organic wastewater bioreactor microbiomes for carbon recovery together with Prof. Dr. Ir. Angenent.

Yvonne Becker is a senior scientist at the Julius Kühn Institute, Institute for Epidemiology and Pathogen Diagnostics in Braunschweig, Germany. Her research focuses on molecular characterisation and cell biology of plant-fungus and plant-bacterial interactions as well as on fungal-bacterial interactions. She received her PhD in 2008 at the Westfälische Wilhelms-University Münster in Germany working on the Ergot fungus Claviceps purpurea – rye interaction. In 2008 she moved to Massey University, New Zealand where she did a PostDoc on the Epichloë festucae- ryegrass symbiosis. In 2015 she returned to Germany where she worked for almost two years on the PGP Bacterium Kosakonia radicincitans at the Leibniz Institute of Vegetable and Ornamental Crops.

Katja R. Richert-Pöggeler is the head of electron microscopy at the Julius Kühn Institute, Federal Research Centre for Cultivated Plants – Institute for Epidemiology and Pathogen Diagnostics. She received her Diploma and PhD for Agricultural Sciences at the Georg August University Göttingen, Germany. During her postdoctoral studies at the University of Kentucky, Lexington, USA and the Friedrich Miescher Institute in Basel, Switzerland she discovered integration and activation of pararetroviruses in the plant genome and investigated their control by the host. Her research interests are virus structure and genome organization, virus-host interactions, virus evolution and function, retroelements, biodiversity, epigenetics and genomics.

Beatrice Berger is a plant biologist. She received her PhD in 2008 at the Max Planck Institute for Chemical Ecology, Jena in Germany working on the molecular and chemical characterization of the Manduca sexta –Wild Tobacco Interaction. After finishing PhD, she extended her research interests to the field of Plant-Microbe Interactions the Leibniz Institute of Vegetable and Ornamental Crops, Grossbeeren, Germany. From 2009-2017 she focused on the impact of the PGP Bacterium Kosakonia radicincitans on crop plants including basic and applied research aspects. Since 2017 she conducts research at the Julius Kühn Institute, Institute for National and International Plant Health in Braunschweig, Germany on the field of diagnosis and control of plant pests.

Silke Ruppel is the head of Research group 2.2 at Leibniz Institute of Vegetable and Ornamental Crops, Germany. Her main research interests are: Functional interaction between the plant and its microbiome; Impact of native bacterial diversity and selected bacterial strains on plant nutrition and plant vitality; Analyses of functional interactions by complex omics techniques, from the plant and bacterial point of view, depending on the plant genetic composition and N- and P-fertilization strategies. Additionally, the risk and pros of diverse bacterial community compositions are her focus of interest in respect to potential human pathogenic bacteria and probiotic bacteria. She is also conducting advisory work for farmers applying bacterial preparations, and for politics in biodiversity.

Daniel Huson studied mathematics at Bielefeld University. He was a post-doc in phylogenetics with Tandy Warnow at the University of Pennsylvania and Princeton University. He worked as a senior staff scientist at Celera Genomics Corp. in Gene Myers group. He has been a Professor for Algorithms in Bioinformatics at the University of Tuebingen since 2002 and is currently also a Visiting Professor at the Life Sciences Institute of the National University of Singapore. He is the author of a number of widely used bioinformatics tools, such as SplitsTree, Dendroscope and MEGAN.

Matthias Becker is a botanist and microbiologist. After finishing his PhD on xerophytic african plants in 2006 he was a lecturer for evolution and biodiversity of plants at Münster University in Germany. From 2008-2015 he conducted research on alpine plants and endophytic fungi at Massey University in New Zealand. From 2015 to date he has worked for Leibniz Institute of Vegetable and Ornamental Crops and Julius Kühn Institute, where he focussed on endophytic bacteria of crop plants. His research interest is to unravel complex interactions between plants of extreme environments (desert, alpine, agriculture) and plant-associated insects, fungi and bacteria.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2019.04.003.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Durán P., Thiergart T., Garrido-Oter R., Agler M., Kemen E., Schulze-Lefert P. Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell. 2018;175(4):973–983. doi: 10.1016/j.cell.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sundin G.W., Wang N. Antibiotic resistance in plant-pathogenic bacteria. Annu Rev Phytopathol. 2018;56:161–180. doi: 10.1146/annurev-phyto-080417-045946. [DOI] [PubMed] [Google Scholar]
- 3.Buttimer C., McAuliffe O., Ross R.P., Hill C., O’Mahony J., Coffey A. Bacteriophages and bacterial plant diseases. Front Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brüssow H., Canchaya C., Hardt W.-D. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68(3):560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson A.R., Cardarelli L., Pell L.G., Radford D.R., Maxwell K.L. Long noncontractile tail machines of bacteriophages. Adv Exp Med Biol. 2012;726:115–142. doi: 10.1007/978-1-4614-0980-9_6. [DOI] [PubMed] [Google Scholar]
- 6.Gill J., Young R.F. Therapeutic applications of phage biology: history, practice and recommendations. Caister Academic Press; Norfolk: 2011. Emerging trends in antibacterial discovery: answering the call to arms; pp. 367–410. [Google Scholar]
- 7.Chang Y.-W., Rettberg L.A., Ortega D.R., Jensen G.J. In vivo structures of an intact type VI secretion system revealed by electron cryotomography. EMBO Rep. 2017;18(7):1090–1099. doi: 10.15252/embr.201744072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghequire M.G.K., De Mot R. The tailocin tale: peeling off phage tails. Trends Microbiol. 2015;23(10):587–590. doi: 10.1016/j.tim.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Ogata S., Mihara O., Ikeda Y., Hongo M. Inducible phage tail-like particles of Clostridium saccharoperbutylacetonicum and its related strains. Agric Biol Chem. 1972;36(8):1413–1421. [Google Scholar]
- 10.Speare L., Cecere A.G., Guckes K.R., Smith S., Wollenberg M.S., Mandel M.J. Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc Natl Acad Sci USA. 2018;115(36):E8528–E8537. doi: 10.1073/pnas.1808302115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vettiger A., Winter J., Lin L., Basler M. The type VI secretion system sheath assembles at the end distal from the membrane anchor. Nature Commun. 2017;8 doi: 10.1038/ncomms16088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lasica A.M., Ksiazek M., Madej M., Potempa J. The type IX secretion system (T9SS): highlights and recent insights into its structure and function. Front Cell Infect Microbiol. 2017;7:215. doi: 10.3389/fcimb.2017.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leiman P.G., Basler M., Ramagopal U.A., Bonanno J.B., Sauder J.M., Pukatzki S. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009;106(11):4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarris P.F., Ladoukakis E.D., Panopoulos N.J., Scoulica E.V. A phage tail-derived element with wide distribution among both prokaryotic domains: a comparative genomic and phylogenetic study. Genome Biol Evol. 2014;6(7):1739–1747. doi: 10.1093/gbe/evu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heymann J.B., Bartho J.D., Rybakova D., Venugopal H.P., Winkler D.C., Sen A. Three-dimensional structure of the toxin-delivery particle antifeeding prophage of Serratia entomophila*. J Biol Chem. 2013;288(35):25276–25284. doi: 10.1074/jbc.M113.456145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurst M.R.H., Glare T.R., Jackson T.A. Cloning Serratia entomophila antifeeding genes—a putative defective prophage active against the grass grub Costelytra zealandica. J Bacteriol. 2004;186(15):5116–5128. doi: 10.1128/JB.186.15.5116-5128.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang G., Dowling A.J., Gerike U., ffrench-Constant R.H., Waterfield N.R. Photorhabdus virulence cassettes confer injectable insecticidal activity against the wax moth. J Bacteriol. 2006;188(6):2254–2261. doi: 10.1128/JB.188.6.2254-2261.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bönemann G., Pietrosiuk A., Mogk A. Tubules and donuts: a type VI secretion story. Mol Microbiol. 2010;76(4):815–821. doi: 10.1111/j.1365-2958.2010.07171.x. [DOI] [PubMed] [Google Scholar]
- 19.Ge P., Scholl D., Leiman P.G., Yu X., Miller J.F., Zhou Z.H. Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states. Nat Struct Mol Biol. 2015;22(5):377–382. doi: 10.1038/nsmb.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zoued A., Durand E., Brunet Y.R., Spinelli S., Douzi B., Guzzo M. Priming and polymerization of a bacterial contractile tail structure. Nature. 2016;531(7592):59–63. doi: 10.1038/nature17182. [DOI] [PubMed] [Google Scholar]
- 21.Taylor N.M.I., Prokhorov N.S., Guerrero-Ferreira R.C., Shneider M.M., Browning C., Goldie K.N. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature. 2016;533(7603):346–352. doi: 10.1038/nature17971. [DOI] [PubMed] [Google Scholar]
- 22.Taylor N.M.I., van Raaij M.J., Leiman P.G. Contractile injection systems of bacteriophages and related systems. Mol Microbiol. 2018;108(1):6–15. doi: 10.1111/mmi.13921. [DOI] [PubMed] [Google Scholar]
- 23.Guerrero-Ferreira R.C., Hupfeld M., Nazarov S., Taylor N.M., Shneider M.M., Obbineni J.M. Structure and transformation of bacteriophage A511 baseplate and tail upon infection of Listeria cells. EMBO J. 2019;38(3) doi: 10.15252/embj.201899455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Böck D., Medeiros J.M., Tsao H.-F., Penz T., Weiss G.L., Aistleitner K. In situ architecture, function, and evolution of a contractile injection system. Science. 2017;357(6352):713–717. doi: 10.1126/science.aan7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trunk K., Peltier J., Liu Y.-C., Dill B.D., Walker L., Gow N.A.R. The type VI secretion system deploys antifungal effectors against microbial competitors. Nat Microbiol. 2018;3(8):920–931. doi: 10.1038/s41564-018-0191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghequire M.G.K., De Mot R. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol Rev. 2014;38(4):523–568. doi: 10.1111/1574-6976.12079. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez M., Godino A., Príncipe A., Morales G.M., Fischer S. Effect of a Pseudomonas fluorescens tailocin against phytopathogenic Xanthomonas observed by atomic force microscopy. J Biotechnol. 2017;256:13–20. doi: 10.1016/j.jbiotec.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Shikuma N.J., Pilhofer M., Weiss G.L., Hadfield M.G., Jensen G.J., Newman D.K. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science. 2014;343(6170):529–533. doi: 10.1126/science.1246794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernal P., Llamas M.A., Filloux A. Type VI secretion systems in plant-associated bacteria. Environ Microbiol. 2017;20(1):1–15. doi: 10.1111/1462-2920.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakayama K., Takashima K., Ishihara H., Shinomiya T., Kageyama M., Kanaya S. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol. 2000;38(2):213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 31.Jacob F. Induced biosynthesis and mode of action of a pyocine, antibiotic produced by Pseudomonas aeruginosa. Ann Inst Pasteur (Paris) 1954;86(2):149–160. [PubMed] [Google Scholar]
- 32.Morales-Soto N., Gaudriault S., Ogier J.-C., Thappeta K.R.V., Forst S. Comparative analysis of P2-type remnant prophage loci in Xenorhabdus bovienii and Xenorhabdus nematophila required for xenorhabdicin production. FEMS Microbiol Lett. 2012;333(1):69–76. doi: 10.1111/j.1574-6968.2012.02600.x. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen A.H., Tomita T., Hirota M., Sato T., Kamio Y. A simple purification method and morphology and component analyses for carotovoricin Er, a phage-tail-like bacteriocin from the plant pathogen Erwinia carotovora Er. Biosci Biotechnol Biochem. 1999;63(8):1360–1369. doi: 10.1271/bbb.63.1360. [DOI] [PubMed] [Google Scholar]
- 34.Strauch E., Kaspar H., Schaudinn C., Dersch P., Madela K., Gewinner C. Characterization of enterocoliticin, a phage tail-like bacteriocin, and its effect on pathogenic Yersinia enterocolitica strains. Appl Environ Microbiol. 2001;67(12):5634–5642. doi: 10.1128/AEM.67.12.5634-5642.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smarda J., Benada O. Phage tail-like (high-molecular-weight) bacteriocins of Budvicia aquatica and Pragia fontium (Enterobacteriaceae) Appl Environ Microbiol. 2005;71(12):8970–8973. doi: 10.1128/AEM.71.12.8970-8973.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hockett K.L., Renner T., Baltrus D.A. Independent co-option of a tailed bacteriophage into a killing complex in pseudomonas. MBio. 2015;6(4) doi: 10.1128/mBio.00452-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berger B., Patz S., Ruppel S., Dietel K., Faetke S., Junge H. Formulation and application of plant growth-promoting Kosakonia radicincitans in maize cultivation. Biomed Res Int. 2018;2018 doi: 10.1155/2018/6439481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becker M., Patz S., Becker Y., Berger B., Drungowski M., Bunk B. Comparative genomics reveal a flagellar system, a Type VI secretion system and plant growth-promoting gene clusters unique to the endophytic bacterium Kosakonia radicincitans. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.01997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witzel K., Strehmel N., Baldermann S., Neugart S., Becker Y., Becker M. Arabidopsis thaliana root and root exudate metabolism is altered by the growth-promoting bacterium Kosakonia radicincitans DSM 16656T. Plant Soil. 2017;419(1–2):557–573. [Google Scholar]
- 40.Hartmann M., Lee S., Hallam S.J., Mohn W.W. Bacterial, archaeal and eukaryal community structures throughout soil horizons of harvested and naturally disturbed forest stands. Environ Microbiol. 2009;11(12):3045–3062. doi: 10.1111/j.1462-2920.2009.02008.x. [DOI] [PubMed] [Google Scholar]
- 41.Bulgarelli D., Schlaeppi K., Spaepen S., van Loren Themaat E ver, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 42.Bulgarelli D., Garrido-Oter R., Münch P.C., Weiman A., Dröge J., Pan Y. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe. 2015;17(3):392–403. doi: 10.1016/j.chom.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Heijden M.G.A., Hartmann M. Networking in the plant microbiome. PLoS Biol. 2016;14(2) doi: 10.1371/journal.pbio.1002378. 10.1371/journal.pbio.1002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banerjee S., Schlaeppi K., Heijden van der Marcel G.A. Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol. 2018;16(9):567. doi: 10.1038/s41579-018-0024-1. [DOI] [PubMed] [Google Scholar]
- 45.Lindow S.E., Brandl M.T. Microbiology of the phyllosphere. Appl Environ Microbiol. 2003;69(4):1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44(W1):W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 48.Haggård-Ljungquist E., Halling C., Calendar R. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J Bacteriol. 1992;174(5):1462–1477. doi: 10.1128/jb.174.5.1462-1477.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tétart F., Desplats C., Krisch H.M. Genome plasticity in the distal tail fiber locus of the T-even bacteriophage: recombination between conserved motifs swaps adhesin specificity 1 1Edited by M. Yaniv. J Mol Biol. 1998;282(3):543–556. doi: 10.1006/jmbi.1998.2047. [DOI] [PubMed] [Google Scholar]
- 50.Williams S.R., Gebhart D., Martin D.W., Scholl D. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl Environ Microbiol. 2008;74(12):3868–3876. doi: 10.1128/AEM.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dorosky R.J., Yu J.M., Pierson L.S., Pierson E.A. Pseudomonas chlororaphis produces two distinct R-tailocins that contribute to bacterial competition in biofilms and on roots. Appl Environ Microbiol. 2017;83(15) doi: 10.1128/AEM.00706-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dorosky R.J., Pierson L.S., Pierson E.A. Pseudomonas chlororaphis produces multiple R-tailocin particles that broaden the killing spectrum and contribute to persistence in rhizosphere communities. Appl Environ Microbiol. 2018;84(18) doi: 10.1128/AEM.01230-18. 10.1128/AEM.01230-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng G., Zhang W., Luo H., Xie H., Lai W., Tan Z. Enterobacter oryzae sp. nov., a nitrogen-fixing bacterium isolated from the wild rice species Oryza latifolia. Int J Syst Evol Microbiol. 2009;59(Pt 7):1650–1655. doi: 10.1099/ijs.0.65484-0. [DOI] [PubMed] [Google Scholar]
- 54.Li Q.-J., Cheng J.-J., Sun S.-X., Chen Y.-P. Isolation, identification and characterization of associative nitrogen-fixing endophytic bacterium Kosakonia radicincitans GXGL-4A in maize. Microbiol China. 2016;43:2456–2463. [Google Scholar]
- 55.Bergottini V.M., Filippidou S., Junier T., Johnson S., Chain P.S., Otegui M.B. Genome sequence of Kosakonia radicincitans strain YD4, a plant growth-promoting rhizobacterium isolated from yerba mate (Ilex paraguariensis St. Hill.) Genome Announc. 2015;3(2) doi: 10.1128/genomeA.00239-15. 10.1128/genomeA.00239-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suhaimi N.S.M., Yap K.-P., Ajam N., Thong K.-L. Genome sequence of Kosakonia radicincitans UMEnt01/12, a bacterium associated with bacterial wilt diseased banana plant. FEMS Microbiol Lett. 2014;358(1):11–13. doi: 10.1111/1574-6968.12537. [DOI] [PubMed] [Google Scholar]
- 57.Hardoim P.R., van Overbeek L.S., Berg G., Pirttilä A.M., Compant S., Campisano A. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev. 2015;79(3):293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meng X., Bertani I., Abbruscato P., Piffanelli P., Licastro D., Wang C. Genome sequence of rice endophyte-associated isolate Kosakonia oryzae KO348. Genome Announc. 2015;3(3) doi: 10.1128/genomeA.00594-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu B., Zhou Q., Lin L., Hu C., Shen P., Yang L. Enterobacter sacchari sp. nov., a nitrogen-fixing bacterium associated with sugar cane (Saccharum officinarum L.) Int J Syst Evol Microbiol. 2013;63(Pt 7):2577–2582. doi: 10.1099/ijs.0.045500-0. [DOI] [PubMed] [Google Scholar]
- 60.Shinjo R., Uesaka K., Ihara K., Loshakova K., Mizuno Y., Yano K. Complete genome sequence of Kosakonia sacchari strain BO-1, an endophytic diazotroph isolated from a sweet potato. Genome Announc. 2016;4(5) doi: 10.1128/genomeA.00868-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bingle L.E., Bailey C.M., Pallen M.J. Type VI secretion: a beginner's guide. Curr Opin Microbiol. 2008;11(1):3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 62.Michel-Briand Y., Baysse C. The pyocins of Pseudomonas aeruginosa. Biochimie. 2002;84(5–6):499–510. doi: 10.1016/s0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 63.Köhler T., Donner V., van Delden C. Lipopolysaccharide as shield and receptor for R-pyocin-mediated killing in Pseudomonas aeruginosa. J Bacteriol. 2010;192(7):1921–1928. doi: 10.1128/JB.01459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buth S.A., Shneider M.M., Scholl D., Leiman P.G. Structure and analysis of R1 and R2 pyocin receptor-binding fibers. Viruses. 2018;10(8) doi: 10.3390/v10080427. 10.3390/v10080427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nobrega F.L., Vlot M., de Jonge P.A., Dreesens L.L., Beaumont H.J.E., Lavigne R. Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol. 2018;16(12):760–773. doi: 10.1038/s41579-018-0070-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.