Abstract
Purpose:
Alzheimer’s disease (AD) and dementia with Lewy bodies (DLB) are representative disorders of dementia of the elderly and the neuroimaging has contributed to early diagnosis by estimation of alterations of brain volume, blood flow and metabolism. A brain network analysis by MR imaging (MR connectome) is a recently developed technique and can estimate the dysfunction of the brain network in AD and DLB. A graph theory which is a major technique of network analysis is useful for a group study to extract the feature of disorders, but is not necessarily suitable for the disorder differentiation at the individual level. In this investigation, we propose a deep learning technique as an alternative method of the graph analysis for recognition and classification of AD and DLB at the individual subject level.
Materials and Methods:
Forty-eight brain structural connectivity data of 18 AD, 8 DLB and 22 healthy controls were applied to the machine learning consisting of a six-layer convolution neural network (CNN) model. Estimation of the deep learning model to classify AD, DLB and non-AD/DLB was performed using the 4-fold cross-validation method.
Results:
The accuracy, average precision and recall of our CNN model were 0.73, 0.78 and 0.73, and the specificity precision and recall were 0.68 and 0.79 in AD, 0.94 and 0.65 in DLB and 0.73 and 0.75 in non-AD/DLB. The triangular probability map of the MR connectome revealed the probability of AD, DLB and non-AD/DLB in each subject.
Conclusion:
Our preliminary investigation revealed the adaptation of deep learning to the MR connectome and proposed its utility in the differentiation of dementia disorders at the individual subject level.
Keywords: Alzheimer’s disease, deep learning, dementia with Lewy bodies, structural brain connectivity
Introduction
Alzheimer’s disease (AD) and dementia with Lewy bodies (DLB) are representative disorders of dementia of the elderly. The prevalence of such senile dementia has been increasing globally and AD and DLB account for 70% of senile dementia and are the important social problems worldwide.1,2
Neuroimaging in clinical medicine of dementia was previously mainly used to exclude other organic disorders, however now neuroimaging is one of the leading components of the early diagnosis of dementia disorders, because it can estimate the early structural and functional changes such as brain atrophy, hypoperfusion and hypometabolism in AD and DLB.3–7
The MR connectome is a recently developed neuroimaging technique for brain network analysis. Functional MR connectome with resting state functional MRI and a structural MR connectome based on diffusion tensor MR imaging have revealed the significant alteration of the brain network in various neurodegenerative disorders.8–10
In AD, several reports revealed that the disconnection and disability of “hub” areas in AD brain which integrating its cognitive symptoms.11–14 Further, the alteration of the brain network in AD, and DLB compared with healthy controls was also reported.15,16 A graph analysis based group analysis is useful to reveal the alteration of the brain network in dementia disorders, but it is not easy to apply the results to estimate and classify disorders at the individual subject level. In this investigation, we proposed the adaption of machine learning as an alternative method of the graph theory to solve the disadvantage of brain network analysis.
Machine learning is one of the techniques that support artificial intelligence technology and the convolution neural network (CNN) is one of the major machine learning models that are suitable for image recognition and classification.17 A multilayered neural network model including multiple convolution layers is called the “deep learning” machine learning model and has a high accuracy in image recognition and classification.
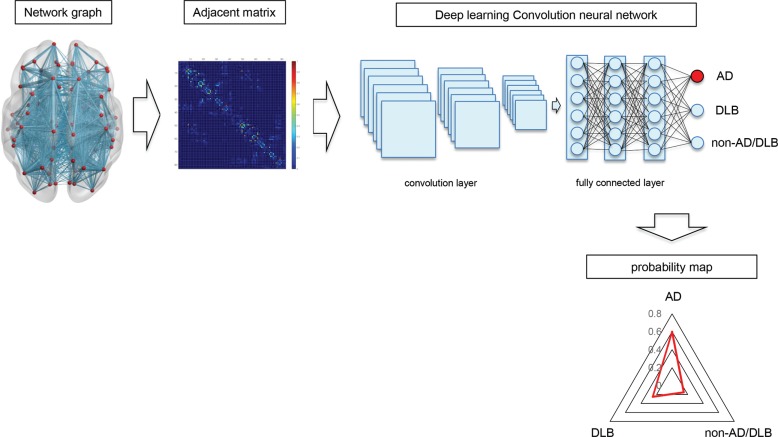
The network data handling in the graph analysis can be revealed as an adjacent matrix and its data type is similar to image data (Fig. 1). Therefore, the brain network can become a subject for machine learning and it may contribute to the recognition and classification of dementia disorders. The high accuracy of machine learning in image recognition and classification may be translocated to the network analysis by handling the adjacent matrix data of network as an image data. In this investigation, we try to adapt the machine learning to the structural MR connectome to distinguish and classify AD and DLB from healthy subjects.
Fig. 1.
The principle of the deep learning MR connectome. The connection between brain areas is expressed by a network graph consisting of nodes and edges. The “network graph” can be converted to an adjacent matrix and calculated by a graph theory. The adjacent matrix is similar to the image data and it can be an input of convolution neural network (CNN) model. The deep learning of MR connectome outputs the probability map which estimates the probability of AD, DLB and non-AD/DLB with a triangular graph. AD, Alzheimer’s disease; DLB, dementia with Lewy bodies.
Materials and Methods
Subjects
The Institutional Review Board approved this study and written informed consent was obtained from all subjects. Brain connectivity data from 48 subjects (18 AD; 7 males and 11 females average age 73.4 y.o., 8 DLB; 2 males and 6 females, 72.8 y.o. and 22 HC; healthy control subjects; 10 males and 12 females, 72.0 y.o.) were used. The diagnosis of AD was based on the criteria of DSM-4 and that of DLB was based on the clinical diagnostic criteria of DLB.1,18
MR imaging and structural network analysis
All MR studies were performed on a 3T MR unit (Achieva; Philips Medical Systems, Best, The Netherlands) with an 8-channel head coil. The structural connectivity data were constructed by a connectome mapper (http://www.connectomics.org/mapper/) based on deterministic tractography with 32 axis diffusion tensor MR images (TR/TE = 5452/70 ms, resolution = 1.75 × 1.75 mm, slice thickness (TH) = 3.0 mm, FOV = 224 × 224 mm, matrix = 128 × 128, number of excitations (NEX) = 2, b value = 0, 1000 s/mm, motion probing gradient (MPG) directions = 32, Δ/δ = 39.0/28.0 ms) and high-resolution 3D T1-weighted images (MPRAGE; TR/TE/inversion time (TI) = 15/3.54/1100 ms, resolution = 0.81 × 0.81 mm, TH = 0.86 mm, FOV = 260 × 260 mm, matrix = 320 × 320 mm).19
Machine learning and estimation
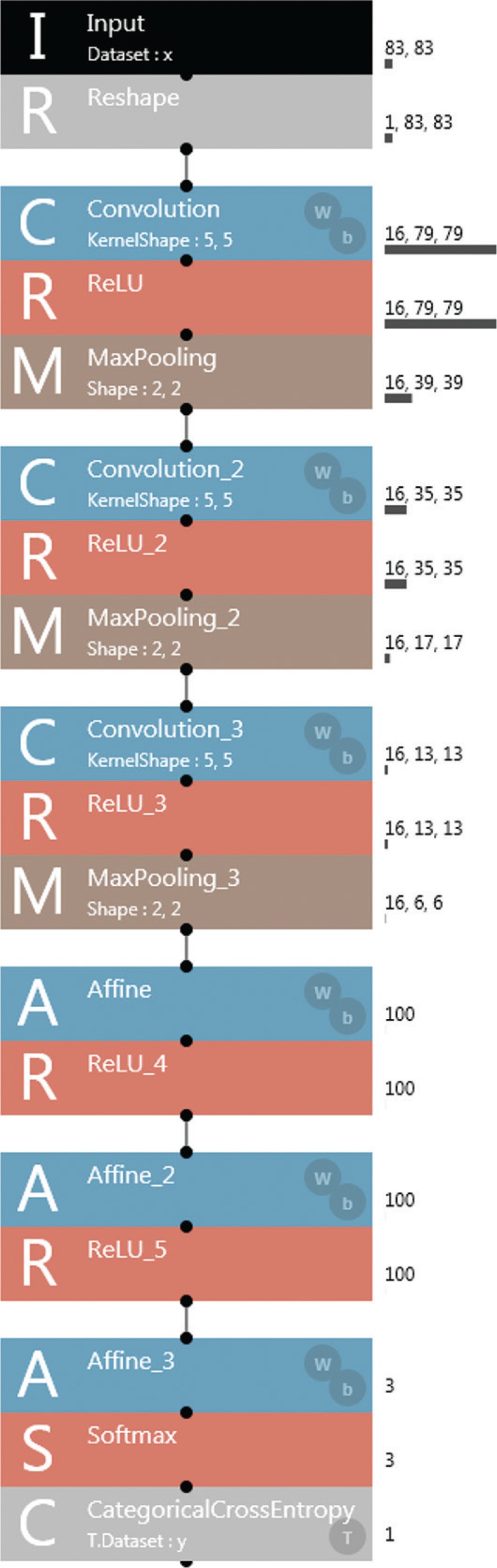
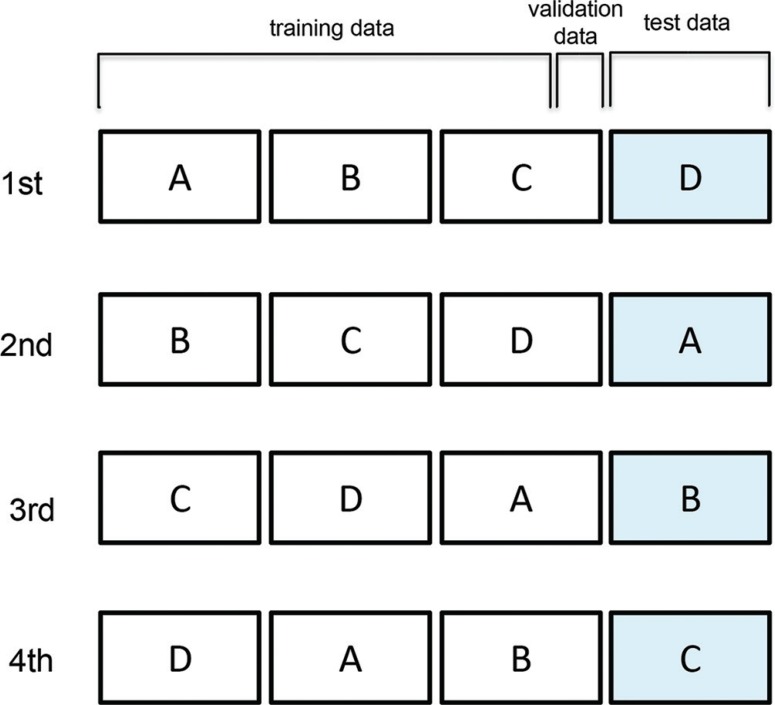
For the construction and modification of the machine learning model, Windows PC with Intel Core i5 2GHz 16GB and Neural Network Console version 1.10 (https://dl.sony.com/) was used as a deep learning integrated development environment. As a learning model, a six layer CNN model including three convolution layers and three fully connected layers was adapted (Fig. 2). The parameters of each layer are described in Fig. 2. Adam was applied as a parameter update method. The 4-fold cross-validation method was used for the training and estimation of the learning model (Fig. 3). In the 4-fold cross-validation method the data set was established with all data were divided into four groups. Three groups were used as the training data including five cases for validation and the other group was used as the test data. The learning model was trained by 100 epochs learning with the training data and the estimation was executed 10 times for the test data and the average value was recorded as an output of the data set. The average values of accuracy, precision, recall (sensitivity) and F-measure in four estimations with four data sets were recorded as the final outputs. The accuracy, precision, recall and F-measure were calculated by the following numerical formulas:
Fig. 2.
A convolution neural network (CNN) machine learning model adapted to the MR connectome. As a machine learning model, a six-layer CNN model with three convolution layers (each consisting of convolution, ReLU; Rectified Linear Unit and MaxPooling) and three fully connected layers (Affine with ReLU) was adapted. The specific elements of the layer were described on the right side of each layer.
Fig. 3.
The 4-fold cross-validation method. In the 4-fold cross-validation method, all sample data were split into four groups. One group was set as the test data and the remaining three groups were set as the training and validation data. An average of four times of investigations was estimated as the performance of the machine learning model.
Results
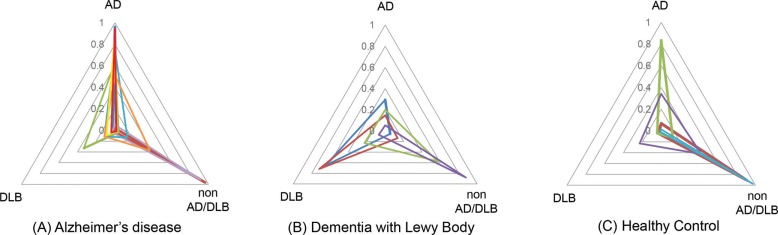
The accuracy of CNN learning model for classification of AD, DLB and HC was 0.73 and the average precision and recall were 0.78 and 0.73 in the 4-fold cross validation method (Table 1). In the confusion matrix, the specificity precision and recall were 0.68 and 0.79 in AD, 0.94 and 0.65 in DLB and 0.73 and 0.75 in non-AD/DLB. The probability map of CNN learning model in classification AD, DLB and non-AD/DLB was revealed by a radar chart. The probability map was provided by the softmax function at the output part of the deep CNN model and revealed the possibility of AD, DLB and non-AD/DLB in each subject ranging from 0.0 to 1.0 (Fig. 4).
Table 1.
Results of the estimation of the deep learning MR connectome in the classification of AD, DLB and HC
| Accuracy | 0.73 | AD | DLB | HC | |
| Avg. precision | 0.78 | Precision | 0.68 | 0.94 | 0.73 |
| Avg. recall | 0.73 | Recall | 0.79 | 0.65 | 0.75 |
| Avg. F-measure | 0.74 | F-measure | 0.72 | 0.76 | 0.73 |
AD, Alzheimer’s disease; DLB, dementia with Lewy bodies; HC, healthy control.
Fig. 4.
Probability map provided by the deep learning MR connectome of AD (A), DLB (B) and HC (C). The triangular radar graph reveals the probability of AD, DLB and non-AD/DLB in each subject ranging from 0.0 to 1.0. AD, Alzheimer’s disease; DLB, dementia with Lewy bodies; HC, healthy control.
Discussion
The human brain consists of over 10 billion neurons with a complex brain network of various sized connections from micro- to macro-level connections.20 It is difficult to recognize and estimate the micro-level brain network technically, but the macro-level scale brain network can be estimated by the structural and functional MR connectome using diffusion tensor MR tractography and function MRI.21,22
By using a graph theory, complex network can be visualized by a “graph” consisting of nodes and edges. In structural MR connectivity of the brain, the node is a brain area and the edge is the number of streams in the MR tractography between two nodes.23 The complex geometric structure of the graph can be converted to a simple adjacent matrix, and analyzed mathematically with various metrics reflecting the features of the network, such as centricity, clustering coefficients, small-worldness, global or local efficiency and characteristic path length.24–26 Group studies using the functional and structural MR connectome revealed the alteration of brain network about various brain degenerative diseases.23,27–30 Group analysis is a useful method for the estimation of brain disorders, but it is not easy to adapt its results to classify each individual subject. To classify each subject as AD, DLB or non-AD/DLB, we must extract the characteristics of AD, DLB and non-AD/DLB from many network metrics and establish a rule and equation to differentiate these conditions.
For extraction of the characteristics of the brain network, a machine learning model of image recognition and classification can be employed that may contribute to the assignment of differentiation and classification of the individual brain network. This consideration was based on the similarity of the type of network and image data and which suggested that machine learning, and especially the CNN model, was suitable for the estimation of the individual brain network.31,32 The high accuracy of the CNN model in image recognition and classification is based on three features; the convolution layer, multilayered neural network classifier part and a optimization by a back propagation method.33,34 The convolution layer extracts the features of matrix data by multiple filters and the multilayer (deep) convolution can extract the high dimensional features and its output is transmitted to following multiple layer neural network.17 Further, the back propagation is an optimization algorithm to improve the accuracy of the current deep learning machine learning model which name comes from “backward propagation of errors”. The back propagation tunes up the calculation parameters in each layer of multiple layer neural network to minimize the error between the final output and expected result.34
In this investigation, MR connectome in combination with a six-layer deep CNN model provided 0.71 accuracy in identification and classification of AD, DLB and non-AD/DLB. The 0.71 accuracy of our investigation was not beyond that of previous studies, which found 0.88 accuracy using CT, 0.97 using brain volumetry and fluorodeoxyglucose-positron emission tomography (FDG-PET), 0.95 using Cerebrospinal fluid (CSF) data and 0.76 using electroencephalogram.35–39 However, our preliminary study revealed a new possibility of the MR connectome in the estimation of dementia disorders. The MR connectome together with a deep learning technique provides the probability of dementia disorders in an individual subject. If the probability map of deep learning MR connectome is introduced into clinical practice of dementia, it may support the clinician in decision making and treatment of dementia disorders.
Our investigation has some limitations. The number of subjects is insufficient; especially the relative insufficiency of DLB subjects is a problem that should not be ignored. The relatively low accuracy of DLB classification might be induced by the small number of subjects in addition to the variety of DLB pathologies. In the construction of the structural MR connectome, the fiber tracking method was a deterministic method, not the latest probabilistic method. The adoption of the latest two-shell probabilistic method is ideally desirable.40 The accuracy of our machine learning model in the classification of dementia may be improved by solving these problems.
Conclusion
Deep CNN machine learning technique could be adapted to structural brain network analysis and a MR connectome in combination with deep learning would contribute to the differentiation of dementia disorders in individual subjects.
Acknowledgments
This work was supported by the program for Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS) from Japan Agency for Medical Research and development, AMED.
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest associated with this manuscript.
References
- 1.McKeith IG, Dickson DW, Lowe J, et al. Diagnosis and management of dementia with Lewy bodies: third report of the DLB consortium. Neurology 2005; 65:1863–1872. [DOI] [PubMed] [Google Scholar]
- 2.Alzheimer’s Association 2017 Alzheimer’s disease facts and figures. J. Alzheimers Dis 2017; 13:325–373. [Google Scholar]
- 3.Scheltens P, Fox N, Barkhof F, De Carli C. Structural magnetic resonance imaging in the practical assessment of dementia: beyond exclusion. Lancet Neurol 2002; 1:13–21. [DOI] [PubMed] [Google Scholar]
- 4.Karas GB, Burton EJ, Rombouts SA, et al. A comprehensive study of gray matter loss in patients with Alzheimer’s disease using optimized voxel-based morphometry. Neuroimage 2003; 18:895–907. [DOI] [PubMed] [Google Scholar]
- 5.Shonk TK, Moats RA, Gifford P, et al. Probable Alzheimer disease: diagnosis with proton MR spectroscopy. Radiology 1995; 195:65–72. [DOI] [PubMed] [Google Scholar]
- 6.Wolk DA, Detre JA. Arterial spin labeling MRI: an emerging biomarker for Alzheimer’s disease and other neurodegenerative conditions. Curr Opin Neurol 2012; 25:421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishii K. PET approaches for diagnosis of dementia. AJNR Am J Neuroradiol 2014; 35:2030–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie T, He Y. Mapping the Alzheimer’s brain with connectomics. Front Psychiatry 2011; 2:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu D, Li K, Terry DP, et al. Connectome-scale assessments of structural and functional connectivity in MCI. Hum Brain Mapp 2014; 35:2911–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stam CJ, Jones BF, Nolte G, Breakspear M, Scheltens P. Small-world networks and functional connectivity in Alzheimer’s disease. Cereb Cortex 2007; 17:92–99. [DOI] [PubMed] [Google Scholar]
- 11.Delbeuck X, van der Linden M, Collette F. Alzheimer’s disease as a disconnection syndrome? Neuropsychol Rev 2003; 13:79–92. [DOI] [PubMed] [Google Scholar]
- 12.Bokde AL, Ewers M, Hampel H. Assessing neuronal networks: understanding Alzheimer’s disease. Prog Neurobiol 2009; 89:125–133. [DOI] [PubMed] [Google Scholar]
- 13.Filippi M, van den Heuvel MP, Fornito A, et al. Assessment of system dysfunction in the brain through MRI-based connectomics. Lancet Neurol 2013; 12:1189–1199. [DOI] [PubMed] [Google Scholar]
- 14.Dai Z, He Y. Disrupted structural and functional brain connectomes in mild cognitive impairment and Alzheimer’s disease. Neurosci Bull 2014; 30:217–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peraza LR, Taylor JP, Kaiser M. Divergent brain functional network alterations in dementia with Lewy bodies and Alzheimer’s disease. Neurobiol Aging 2015; 36: 2458–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowther ER, O’Brien JT, Firbank MJ, Blamire AM. Lewy body compared with Alzheimer dementia is associated with decreased functional connectivity in resting state networks. Psychiatry Res 2014; 223:192–201. [DOI] [PubMed] [Google Scholar]
- 17.Fukushima K, Miyake S. Neocognitron: a new algorithm for pattern recognition tolerant of deformations and shifts in position. Pattern Recognit 1982; 15:455–469. [Google Scholar]
- 18.American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders, 4th ed Washington: Amer Psychiatric Pub Inc; 2000:1–943. [Google Scholar]
- 19.Daducci A, Gerhard S, Griffa A, et al. The connectome mapper: an open-source processing pipeline to map connectomes with MRI. PLoS One 2012; 7:e48121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Catani M, Thiebaut de Schotten M, Slater D, Dell’Acqua F. Connectomic approaches before the connectome. Neuroimage 2013; 80:2–13. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh A, Deriche R. From Diffusion MRI to Brain Connectomics. In: Cazals F, Kornprobst P, eds. Modeling in computational biology and biomedicine. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013: 193–234. [Google Scholar]
- 22.Sporns O, Tononi G, Kötter R. The human connectome: a structural description of the human brain. PLoS Comput Biol 2005; 1:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stam CJ, Reijneveld JC. Graph theoretical analysis of complex networks in the brain. Nonlinear Biomed Phys 2007; 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 2010; 52:1059–1069. [DOI] [PubMed] [Google Scholar]
- 25.Smith S. Introduction to the neuroImage special issue “Mapping the connectome”. Neuroimage 2013; 80:1. [DOI] [PubMed] [Google Scholar]
- 26.Hosseini SM, Kesler SR. Influence of choice of null network on small-world parameters of structural correlation networks. PLoS One 2013; 8:e67354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bassett DS, Bullmore ET. Human brain networks in health and disease. Curr Opin Neurol 2009; 22:340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minati L, Varotto G, D’Incerti L, Panzica F, Chan D. From brain topography to brain topology: relevance of graph theory to functional neuroscience. Neuroreport 2013; 24:536–543. [DOI] [PubMed] [Google Scholar]
- 29.Fornito A, Bullmore ET. Connectomics: a new paradigm for understanding brain disease. Eur Neuropsychopharmacol 2015; 25:733–748. [DOI] [PubMed] [Google Scholar]
- 30.van Essen DC, Ugurbil K, Auerbach E, et al. The human connectome project: a data acquisition perspective. Neuroimage 2012; 62:2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukushima S, Narita Y, Shinomiya A, et al. A case of unclassified high-grade glioma with polar spongioblastoma pattern. Neuropathology 2012; 32:604–610. [DOI] [PubMed] [Google Scholar]
- 32.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Neural Information Processing Systems 2012; doi: 10.1145/3065386. [DOI] [Google Scholar]
- 33.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521:436–444. [DOI] [PubMed] [Google Scholar]
- 34.Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature 1986; 323:533–536. [Google Scholar]
- 35.Gao XW, Hui R, Tian Z. Classification of CT brain images based on deep learning networks. Comput Methods Programs Biomed 2017; 138:49–56. [DOI] [PubMed] [Google Scholar]
- 36.Shi J, Zheng X, Li Y, Zhang Q, Ying S. Multimodal neuroimaging feature learning with multimodal stacked deep polynomial networks for diagnosis of Alzheimer’s disease. IEEE J Biomed Health Inform 2018; 22:173–183. [DOI] [PubMed] [Google Scholar]
- 37.Suk HI, Lee SW, Shen D. Deep sparse multi-task learning for feature selection in Alzheimer’s disease diagnosis. Brain Struct Funct 2016; 221:2569–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Triggiani AI, Bevilacqua V, Brunetti A, et al. Classification of healthy subjects and Alzheimer’s disease patients with dementia from cortical sources of resting state EEG rhythms: a study using artificial neural networks. Front Neurosci 2016; 10:604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaharchuk G, Gong E, Wintermark M, Rubin D, Langlotz CP. Deep learning in neuroradiology. AJNR Am J Neuroradiol 2018; 39:1776–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamagata K, Zalesky A, Hatano T, et al. Connectome analysis with diffusion MRI in idiopathic Parkinson’s disease: evaluation using multi-shell, multi-tissue, constrained spherical deconvolution. Neuroimage Clin 2018; 17:518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]