Abstract.
The methods used for digital processing of optical coherence tomography (OCT) and crosspolarization (CP) OCT images are focused on improving the contrast ratio of native structural OCT images. Such advances are particularly important for the intraoperative detection of glioma margins where the visual assessment of OCT images can be difficult and lead to errors. The aim of the study was to investigate the application of optical coefficients obtained from CP OCT data for the differentiation of glial tumorous tissue from a normal brain. Pseudocolor en-face OCT maps based on two optical coefficients (the commonly used rate of attenuation in the cochannel, and in addition, the interchannel attenuation difference) were constructed for normal rat brain coronal cross sections and for brains with a 101.8 rat glioblastoma model. It was shown that the use of optical coefficients significantly increased the available information from the OCT data in comparison with unprocessed images. As a result, this allowed contrasting of the white matter from the gray matter and tumorous tissue ex vivo, and for this purpose, the interchannel attenuation difference worked better. The interchannel attenuation difference values of white matter were at least seven and two times higher than corresponding values of the cortex and tumorous tissue, whereas the same parameter for cochannel attenuation coefficient values of white matter are about 4 and 1.4. However, quantitative analysis shows that both coefficients are suitable for the purpose of glioblastoma detection from normal brain tissue regardless of whether a necrotic component was present (in all compared groups ).
Keywords: brain imaging, crosspolarization optical coherence tomography, white and gray matter, glioblastoma model, optical coefficients, image processing
1. Introduction
Optical coherence tomography (OCT) is one of the most promising, innovative, and rapidly emerging in vivo biomedical imaging modalities.1 Due to the high resolution (2 to ) and imaging depth of up to 2 mm, OCT can provide valuable information about the inner structure of tissues. The development trends are particularly toward OCT multimodality [the combination of several modalities in one device, for example, structural OCT and Doppler effects; structural, polarization-sensitive (PS) and angiographic OCT] and the enhancement of OCT image processing.2
In recent decades, a number of studies have shown the promising results of using OCT in the assessment of health and disease in brain tissue (including glial brain tumors).3,4 Recently, OCT has been integrated into operating microscopes for neurological surgery.5,6 As for structural OCT, time-domain OCT7,8 and swept-source OCT9,10 have been used for the detection of brain tumor margins. Optical coherence microscopy (OCM) with micrometer-level resolution has been extensively studied for visualizing the cerebral cortex,11 the laminar structure of the isocortex,12 neuronal cell bodies in the brain,13,14 single myelin fibers,15 tumorous tissue,16 and for delineating fiber tracts on a reconstructed single brain image using wide-field OCM.17 For fiber tracking/quantification of the fiber orientation, polarization sensitive OCM and PS-OCT provide suitable technology because the myelinated fibers exhibit birefringence.18–21
Crosspolarization OCT (CP OCT) is a variant of PS OCT that allows the visualization of crosspolarization and copolarization scattering simultaneously. CP OCT can be used both for myelin fiber mapping and better brain tumor margin detection due to its additional sensitivity to cross-scattering together with backscattering and birefringence.22,23
Distinguishing between tumorous and nontumorous brain tissue during surgical removal of astrocytic (glial) brain tumors can be difficult due to infiltrative growth of the tumor. The detection of the tumor margin can increase both the accuracy of determining the extent of tumor resection and the survival of the patient.24 For this reason, several studies based on data from experimental glial tumor models,23 ex vivo human brain samples,9 and from live surgery7 have been carried out. The differences between tumorous and nontumorous brain tissues were revealed using two approaches: qualitative and quantitative assessment of the OCT data. Qualitative or visual analysis of two-dimensional (2-D) cross-sectional OCT images allows the separation of tissue types by using several criteria such as signal intensity (white matter is characterized by high scattering due to the presence of myelinated fibers and tumor tissue by its low scattering)22,25 or homogeneity (white matter and cortex have well-ordered structures, and therefore, the corresponding OCT images look more homogeneous in comparison to images of tumorous tissue).16,26 Structural variations in the different tissue types, especially meningioma and astrocytoma with microcysts, can already be seen in the native OCT images,16 but are not acceptable as discriminatory criteria. Qualitative OCT assessment is familiar to surgeons through their “visual” analysis of clinical imaging data (e.g., ultrasonography). However, it is also relatively subjective and depends on the surgeon’s experience with OCT imaging, as well as on the properties of the OCT image itself, such as its resolution and sampling density.
Quantitative assessment of the OCT data can be based on the calculation of optical coefficients, for example, the attenuation coefficient7–10,27 or machine-learning (classification) methods28,29 and seems to be more objective. In patients, Böhringer et al.8 used an attenuation coefficient calculated from the log scale attenuation curves of areas with a homogeneous signal in the cross-section images, thus describing the linear part of the attenuation curve, but no data relating to sensitivity or specificity were demonstrated. Kut et al.9 have demonstrated the high diagnostic value of the attenuation coefficient in distinguishing between glioma tissue and white matter: for high-grade patients, the specificity and sensitivity were 100% and 92%, respectively; for low-grade patients: 80% and 100%. This group suggested a combined approach of qualitative and quantitative assessment of the three-dimensional (3-D) OCT images by building en-face color-coded maps using three colors: green and red colors to denote the white matter and glioma tissue, respectively; and yellow color to show the infiltrative zone. The attenuation threshold was established on the basis of studies of ex vivo human brain tissues. By applying the same attenuation threshold, the possibility of reliably distinguishing cancer from surrounding noncancerous areas in vivo during surgery in a rodent model of human brain cancer was demonstrated. This combination realizes the advantages of each approach: visibility and objectivity. In the paper,30 texture analysis was used to distinguish between healthy brain and meningioma tissue on ex vivo OCT images.
CP OCT can provide supplementary information about tissue structure, which also allows the calculation of optical coefficients, characterizing polarization-related processes in the tissue in addition to attenuation. This paper performs a method for calculating two optical coefficients based on the detection of light scattered in two orthogonal polarizations and by building color-coded maps of tumorous and nontumorous brain tissues for potential future intraoperative use. In this study, CP OCT images of whole cross sections of the rat brain were obtained to image fresh ex vivo samples (without any additional preparation). This was done to study both the tissue types (gray matter, white matter, and tumor), and the structures they form (e.g., cortex, hippocampus, and corpus callosum) as well as the tumor margins. Quite large tissue volumes were analyzed to reveal clearly the optical difference between tumorous and normal brain tissues and to achieve reliable and accurate results.
2. Materials and Methods
2.1. Animals
The study was performed on 13 female Wistar rats (five rats with healthy brains and eight with 101.8 rat glioblastoma), with weights of . All animals were delivered from the Laboratory of Neuromorphology of the Research Institute of Human Morphology (Moscow, Russian Federation). The animals were maintained, two per cage, in a temperature-, humidity-, and light-controlled room, with free access to water and food. Homogenized 101.8 glioblastoma ( tumor cells) had been inoculated into the ventricle of the right hemisphere of the brain of each of the eight relevant rats using a standard procedure.31
Glioblastoma 101.8 is a primary chemically induced brain tumor (grade IV), but after cultivation, it takes on morphological properties similar to human glioblastoma (high cellular density, cell polymorphism, presence of necrotic and hemorrhage areas, and infiltrative growth). CP OCT imaging was performed in four rats on days 7 to 9 after the glioma injection, when the tumor had matured but it did not have multiple necrosis; four rats were studied between 12 to 15 days, when extensive hemorrhages and necrosis appeared in the tumor. The study was approved by the Ethics Committee of the Nizhny Novgorod State Medical Academy (protocol no. 14 from December 10, 2013).
2.2. CP OCT Device
A spectral domain CP OCT device (Institute of Applied Physics of the Russian Academy of Sciences, Nizhny Novgorod, Russia) with cross-polarization detection was used in the study.32,33 The central wavelength of this device is 1310 nm with a radiation power of 20 mW. The axial resolution is and the lateral resolution is in air. The probing beam uses circular polarization. The device has a 20,000 A-scans/s scanning rate and performs 2-D lateral scanning within an area of to obtain the 3-D distribution of backscattered light in the polarization with the same and reversed rotations of the electric-field vector.31 Scanning was performed in contactless mode. An advantage of the CP OCT device used is its real-time image formation. Structural 2-D (cross-sections and en-face images) in co- and crosspolarization and a 3-D image in copolarization are displayed on the personal computer monitor in the process of scanning, thereby facilitating assessment of the image quality.
2.3. Study Design
This study describes an approach for the calculation of CP OCT optical coefficients from different brain areas including the cortex, white matter, and glioma tissue and the construction of color-coded maps based on them.
3-D CP OCT images of the brain were acquired ex vivo immediately after brain excision and crosscutting in the coronal section. The plane of the brain crosscutting for obtaining the CP OCT scans of various brain structures was fixed at a distance of behind the bregma, allowing comparison of analogous structures in healthy brains with those in the brains with tumors. Sectioning was performed at two sites in this plane: (1) through the center of the tumor to visualize tumor margins with white matter and cerebral cortex and (2) closer to the edge of the tumor for detecting infiltration of the white matter (corpus callosum) by the tumor in greater detail.
A sequential scanning of the entire brain surface was performed row by row with overlapping images (from the cerebral cortex, corpus callosum, and hippocampus to the amygdala and hypothalamus) with further digital processing: building individual color-coded maps—an en face distribution of the optical coefficients values following their reconstruction into a single, complete picture of the rat brain. Between 32 and 46 CP OCT data sets were collected from each rat brain. Each imaging data set consisted of A-scans forming the OCT imaging volume. For each collected A-scan set, the optical coefficients were calculated according to the method described in the following section (see Sec. 2.4). After CP OCT scanning, histological evaluations of all the samples were performed.
To estimate whether the optical coefficients would increase the OCT contrast, the color-coded maps were compared with initial structural en-face OCT images in co- and crosspolarization and with the histological data. Unprocessed en-face OCT images were saved from the 3-D volume at a depth of below the tissue surface to avoid image artifacts like glare from the surface (especially from the white matter), while preserving the contrast between the tissues, which rapidly decreases with depth. A-scan realignment was performed to ensure coefficient calculation from the same depth range.
2.4. Calculation of Optical Coefficients
Given that brain tissue is optically homogeneous, the OCT signal in both polarization channels is attenuated exponentially with depth:
where and are the signals in the co- and crosspolarization channels, respectively,
Note that , () are the attenuation coefficients in the corresponding channels and is the depth coordinate. Cochannel attenuation coefficient and interchannel attenuation difference were used. The choice of the interchannel attenuation difference instead of crosschannel attenuation was made to highlight possible situations in which the crosschannel signal decays more rapidly than the cochannel signal.
The co- and crosschannel attenuation coefficients can be obtained by linear fitting of the logarithms of the corresponding signals and used for quantitative analyses of the CP OCT data. For each collected A-scan set, the optical coefficients were calculated in the same predefined depth range starting from below the tissue surface (pixel #10) to a depth of below the tissue surface (pixel #50). In addition to the conventionally used cochannel attenuation coefficient of the samples, the difference in attenuation between the two orthogonal polarization channels was also investigated as a potential predictor of the state of the brain tissue. This coefficient was referenced in this study as the interchannel attenuation difference (). The A-scans of a white matter sample in co- and crosspolarization on a logarithmic scale and with corresponding linear fits are shown in Fig. 1. En-face color-coded maps of different tissue types were created based on the calculated optical coefficients (cochannel attenuation and interchannel attenuation difference).
Fig. 1.

A-scan examples and corresponding values of cochannel attenuation () and interchannel attenuation difference () for various types of brain tissues. (a) Cortex with negative interchannel attenuation difference; (b) cortex with positive interchannel attenuation difference; (c) white matter; and (d) glioblastoma 101.8. Dashed vertical line represents the tissue surface. Coefficients’ calculations were made in the range of 70 to .
Optical coefficients were recorded for the 3-D CP OCT images in four groups: group 1, normal gray matter (); group 2, normal white matter (); group 3, glioblastoma 101.8 without necrosis (), and group 4, glioblastoma 101.8 with necrosis (). 3-D CP OCT images of normal tissues were obtained from the five rats with healthy brains (two sets of CP OCT images from the right and left hemispheres were scanned from each of two brain subjects after brain incision and coronal cutting). To quantify the tumor tissue, CP OCT data sets containing only tumor tissue were taken (images including tumor margins with gray or white matter were excluded). These images were obtained from the eight rats with inoculated glioblastoma 101.8 (two sets of CP OCT images from the right hemisphere were scanned from each of two brain subjects after brain incision and coronal cutting).
2.5. Histology
After the OCT scanning, brain samples were fixed in 10% formalin for 48 h and a series of histological sections were made. Thus, the en-face plane of the CP OCT images coincided with the plane of the histological sections. The histological sections were stained with hematoxylin and eosin (H&E) and Luxol fast blue stain with crezyl violet (to identify both myelinated fibers and the neuronal tissue structures) and evaluated by two independent histopathologists using light microscopy (Leica DM2500, Germany). Agreement between the specialists was revealed in 98% of cases.
2.6. Statistical Analysis
The median value among all values of the optical coefficients calculated for each A-scan of a 3-D CP OCT image was used. The results are expressed as the Me [Q1; Q3] where Me is the median of the coefficient and [Q1; Q3] are the 25th and 75th percentile values, respectively. To compare the tissue type using each coefficient, we used multiple comparisons with the Bonferroni correction. The statistical data processing was done in MS Excel 2010 with a public domain software plugin for statistical analysis STATISTICA 10 (StatSoft, Inc., Tulsa, Oklahoma) and the Prism v6 software (GraphPad Software, La Jolla, California).
3. Results
3.1. Unprocessed CP OCT Images versus Color-Coded Maps of Normal Brain
The first part of the study was aimed at illustrating the different sensitivities of the optical coefficients in the visualization of healthy gray and white matter. Due to the structural complexity of the brain, a sequential scanning of the whole brain slice was performed to identify the various brain structures in comparison with the histological data and a stereotaxic rat atlas.34 This provided high-precision identification of the brain areas. Then, to reveal the most sensitive/specific coefficients for distinguishing different brain tissue types, several regions of interest were chosen.
Representative wide-field CP OCT images of a healthy rat brain are shown in Fig. 2. Unprocessed (native) OCT images in co- [Fig. 2(b)] and crosspolarization [Fig. 2(d)] show evident optical differences between white matter structures (such as the corpus callosum, internal capsule, striatum, and optic tract) and gray matter (such as the cerebral cortex, hippocampus, and thalamus). The color-coded maps [Figs. 2(c) and 2(e)] provide much better visualization and differentiation of the white and gray matter that are schematically shown in Fig. 2(a). The cochannel attenuation coefficient [Fig. 2(c)] is seen to be more sensitive to the density of the scattering structures, which is why the highest values (red color) correspond to the white matter tracts (densely packed myelinated fibers); medium values (green and yellow colors) correspond to areas of accumulation of myelinated fibers or to individual, spreading ones and to gray matter with high neuronal density; the remaining structures (bodies of neurons and glia) have low values of cochannel attenuation coefficient (dark blue color). The interchannel attenuation difference [Fig. 2(e)] is more sensitive to elongated scattering structures, providing excellent visualization of white matter areas.
Fig. 2.
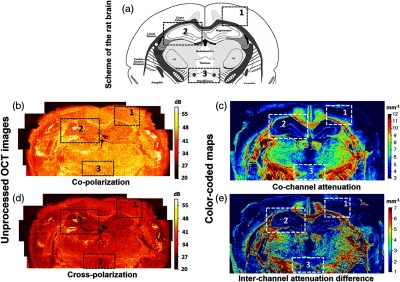
Wide-field unprocessed CP OCT images and color-coded maps of healthy brain (coronal cross section). (a) Schematic view of the rat brain showing the relationship between gray matter (white color) and white matter (gray color).34 The dark gray color marks brain structures with densely packed myelinated fibers and tracts, while the bright gray color indicates areas with thin myelinated fibers or single bundles. (b)–(e) Wide-field brain images reconstructed from unprocessed images in (b) co-, and (d) crosspolarization; (c), (e) the corresponding color-coded maps: (c) cochannel attenuation coefficient, (e) interchannel attenuation difference. Color-coded maps based on the calculated optical coefficients provide better contrast for clear visualization of the different brain structures. Black dotted boxes (1, 2, 3) indicate regions of interest for detailed analysis (see comments in the text).
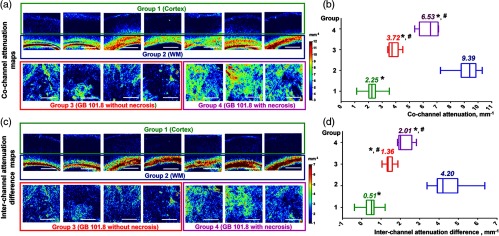
We chose three different regions of interest in Fig. 2 to demonstrate the best correspondence of the interchannel attenuation difference (red-color) for visualizing the white matter in comparison with the unprocessed images and even the cochannel attenuation maps. Box #1 shows the margin between the cerebral cortex (stratified gray matter) and, located directly below it, white matter (corpus callosum) with strongly oriented fibers. This region is of vital neurosurgical interest because the predominant localization of glial tumors in humans is subcortical with a tendency to extensive growth toward the cerebral cortex and germination through it. Variations in the CP OCT color-coded maps of normal cortex and white matter are shown in Figs. 5(a) and 5(c), upper row. Results of the quantitative analysis of these tissue types are shown in Figs. 5(b) and 5(d) and in Table 1. The cortex was well differentiated from the white matter using both cochannel attenuation and the interchannel attenuation difference ( and versus and , ).
Fig. 5.
Variability of color-coded maps of (a) cochannel attenuation and (c) interchannel attenuation difference in four groups of tissues: normal cortex (group 1), normal white matter (group 2), glioblastoma without necrosis (group 3), and glioblastoma with necrosis (group 4). The distribution of (b) cochannel attenuation and (d) interchannel attenuation difference coefficient values between different tissue types. Data are presented as medians with 25th and 75th percentile values. *Significant differences between white matter and other groups (); #significant differences between the cortex and groups with glioblastoma (). Scale bars 1 mm.
Table 1.
Optical coefficients’ difference between tumorous and nontumorous brain tissue types.
| Tissue type | Cochannel attenuation (), | Interchannel attenuation difference (), | ||||
|---|---|---|---|---|---|---|
| Me a | Ratio to WM | Ratio to cortex | Me a | Ratio to WM | Ratio to cortex | |
| 1. Normal cortex () | 2.25 [2.03; 2.51]* | 4.17 | — | 0.51 [0.24; 0.63]* | 7.92 | — |
| 2. Normal white matter () | 9.39 [8.77; 9.85] | — | 4.17 | 4.2 [3.91; 4.97] | — | 8.24 |
| 3. Glioblastoma 101.8 without necrosis () | 3.72 [3.51; 4.17]*,** | 2.52 | 1.65 | 1.36 [1.32; 1.58]*,** | 3.09 | 2.67 |
| 4. Glioblastoma 101.8 with necrosis () | 6.53 [5.72; 7.10]*,** | 1.44 | 2.69 | 2.01 [1.92; 2.59]*,** | 2.09 | 3.94 |
Me : Me—median; [Q1;Q3]—25th and 75th percentiles values, respectively.
Statistically significant difference compared to white matter (multiple comparisons with the Bonferroni correction, ).
Statistically significant difference compared to cortex (multiple comparisons with the Bonferroni correction, ).
Two other regions of interest (boxes #2 and #3) were chosen to stress the high level of contrast between structures that contain only gray matter or only white matter and that these can be clearly identified using the optical coefficients. In box #2, the hippocampus, consisting of gray matter (low values of optical coefficients, blue color), is clearly delineated because of being surrounded by white matter having higher values of the optical coefficients (yellow-green and red colors). Box #3 demonstrates that using the cross-scattering coefficient, isolated white matter bundles (left and right crus of the fornix) passing through the gray matter can be detected.
In summary, comparison of the color-coded maps and unprocessed CP OCT images is indicative of the benefit of using optical coefficient calculation (postprocessing) for higher-contrast imaging of the different brain structures. The interchannel attenuation difference has high specificity for the visualization of white matter containing myelin fibers when compared to the cochannel attenuation coefficient.
3.2. Unprocessed CP OCT Images Versus Color-Coded Maps of the Brain in the Glioblastoma Model
The next step was to determine the differences in optical properties between nontumorous and tumorous brain tissue using CP OCT color-coded maps. For this purpose, the tumor core tissue and its margin with infiltration into surrounding white and gray matter were analyzed. Identification of glioma infiltration is quite challenging, but is a very important task during glioma surgery.
Wide-field structural CP OCT images and color-coded maps of the brain samples with 101.8 glioblastoma were reconstructed and compared carefully with histological data. Several regions of interest, including the tumor core, tumor margin, and surrounding brain tissue, were identified by a pathologist and then analyzed in detail in the CP OCT data.
Figure 3 shows the potential of CP OCT for the tumor core detection and its margin with the cortex. Morphologically, tumor tissue has a heterogeneous structure mainly due to the presence of multiple small or large areas of hemorrhages and necrosis [Figs. 3(e) and 3(f)]. In unprocessed CP OCT images, the tumor tissue appears homogeneous and darker compared to the gray matter [Figs. 3(a) and 3(b)]. By contrast, variegated OCT color-coded maps clearly demonstrate tumor heterogeneity [Figs. 3(b) and 3(d), bright-green spots on dark blue background] compared with the more or less homogeneous OCT signal from normal gray matter [Figs. 2(c) and 2(e)], Fig. 5(a), upper row, dark blue color] and white matter [Figs. 2(c) and 2(e)], Fig. 5(a), upper row, bright stripe with colors from green to red]. It should be noted that by visual assessment of the color-coded maps, the cochannel attenuation coefficient is more sensitive for the detection of tumor cell clusters, blood, and necrotic areas than is the interchannel attenuation difference. Therefore, cochannel attenuation maps appear to be better for the visualization of tumor tissue.
Fig. 3.
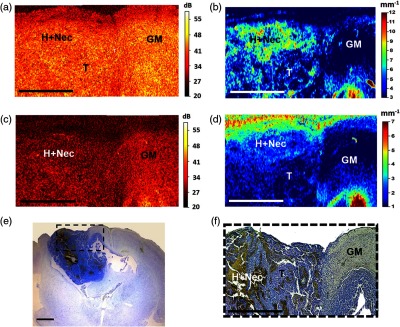
Ex vivo CP OCT identification of glioblastoma with necrotic areas and its margin with the cerebral cortex. Unprocessed OCT images (a) in co- and (c) in crosspolarization. Color-coded maps: (b) the cochannel attenuation coefficient; (d) interchannel attenuation difference. (e), (f) Histological images with Luxol blue and crezyl violet staining. The area of interest in (e) is presented under high magnification in (f). T, tumor; GM, gray matter; H + Nec, hemorrhage and necrosis. For comparison with the corresponding histology, en-face (same orientation as the color-coded maps) has been provided. Scale bars 1 mm.
In the case of extensive infiltration of the normal brain tissue with glioblastoma cells [Fig. 4(e)], the myelin fibers are destroyed [Fig. 4(f)]. In the interchannel attenuation difference maps, the OCT signal of the area involving fragmented white matter is significantly reduced [Fig. 4(d), infiltrated white matter—infWM] compared to the white matter of the peritumoral area and healthy brain tissue [Fig. 2(e), Fig. 5(a), upper row, colors from green to red]. In the cochannel attenuation maps, the OCT signal from the white matter can indicate high values close to those of healthy tissues and the outer contour of the white matter area can be observed [Fig. 4(b), infWM].
Fig. 4.
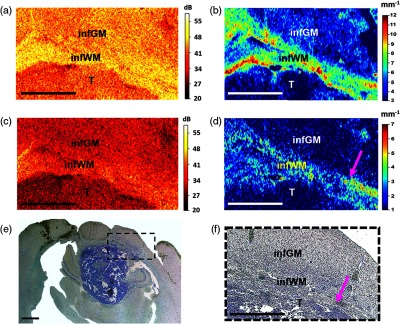
Improving OCT contrast by building color-coded maps based on optical coefficients in the detection of brain tissue infiltration by glioblastoma cells. Unprocessed OCT images (a) in co- and (c) in crosspolarization. Color-coded maps: (b) the cochannel attenuation coefficient; (d) interchannel attenuation difference. (e), (f) Histological images with Luxol blue and crezyl violet staining. The area of interest in (e) is presented under high magnification in (f). Pink arrow indicates the transition point of the most destroyed and preserved myelinated fibers. T, tumor; infGM, infiltrated gray matter; infWM, infiltrated white matter. For comparison with the corresponding histology, en-face (same orientation as the color-coded maps) has been provided. Scale bars 1 mm.
The infiltration of gray matter leads to significantly increased cellular density and due to this, the scattering properties of the cortex are locally increased. This is manifested in the cochannel attenuation maps by localized increases of the OCT signal values and makes the cortex appear more heterogeneous [Fig. 4(b), infiltrated gray matter, infGM] compared to a healthy equivalent [Fig. 2(c), Fig. 5(a), upper row, dark blue color]. Conversely, in the interchannel attenuation difference maps, the cortex infiltration is not obvious [Fig. 3(d), infGM].
The cochannel attenuation and interchannel attenuation difference values for glioblastoma 101.8 without necrosis were 3.72 and 1.36 , respectively; for glioblastoma 101.8 with necrosis 6.53 and 2.01 , respectively (Table 1). The results show that by using both coefficients, tumor tissue can be well distinguished from the white matter () and from the cortex () [see Figs. 5(b) and 5(d)], but the ratio of the interchannel attenuation difference of the white matter and other tissue groups is higher than for the cochannel attenuation coefficient (Table 1), indicating the higher specificity of this coefficient for white matter in differentiating from other tissue types in comparison with the cochannel attenuation coefficient.
Interchannel attenuation difference detects the same tendency for differences of other tissue groups from the cerebral cortex. Therefore, this coefficient can serve as a very promising tool for intraoperative normal brain tissue detection in glioma surgery, especially if the glioblastoma has extensive necrotic areas.
4. Discussion
The development of methods of intraoperative optical imaging is a priority in neurosurgery, as it allows for increased accuracy of surgical procedures. Recent trends in optical bioimaging lie in moving from a qualitative (visual) assessment to quantitative image processing. Various approaches for the quantitative assessment of OCT images for distinguishing gliomas and healthy tissue have been actively developed,8–10,30 but still need further studies. Kut et al.9 demonstrated that the cochannel attenuation coefficient estimated from 3-D OCT images could be used to differentiate tumorous and nontumorous tissues and that it is promising to use during brain tumor resection. A color-coded map denoting regions of high and low cochannel attenuation, which correspond to white matter and to tumor tissue, respectively, was generated to provide visual feedback to the surgeon. The system’s algorithm was developed using ex vivo human tissues and tested during in vivo surgery on a mouse glioma model. However, the coefficient values achieved from the ex vivo human specimen study need to be clarified for intraoperative use during glioma resection.
The crosspolarization modality in OCT widens its functionality and provides benefits for quantitative OCT data processing by adding the interchannel attenuation difference. Using crosspolarization offers increased OCT sensitivity to the destruction of myelinated fibers and, therefore, improves the visualization of tumor infiltration zones. Nevertheless, quantitative analysis shows that both coefficients are suitable for the purpose of identification of tumorous tissue.
Some observations from this study deserve particular mention: (1) wide-field CP OCT imaging can provide comprehensive information about brain morphology and (2) quantitative processing of CP OCT data and the building of color-coded maps based on various optical coefficients can provide significant additional detail from the images obtained.
Using optical coefficients, the cerebral cortex, corpus callosum, hippocampus, and other regions can be clearly visualized and identified in brain cross sections due to the increased image contrast. Although initial structural CP OCT images with knowledge of the anatomy also enables such identification, contrast increasing is important in relation to the translation of the research to the clinic since it makes it much easier for a user who is unfamiliar with the OCT imaging to interpret images and to make a decision. The cochannel attenuation and interchannel attenuation differences have different sensitivities to different tissue types. The cochannel attenuation rate is higher the greater the concentration of the scattering elements, whereas interchannel attenuation difference depends on the presence and density of fiber-like structures (myelinated fibers or densely packed cells in ribbon structures such as are found with the neurons in the hippocampus).
In our study, cochannel attenuation coefficient for normal gray matter was 2.25 ; for normal white matter 9.39 ; for the rat glioblastoma model 3.72 in regions without necrosis and 6.53 in necrotic areas. These absolute values differ from Kut et al.9 results. Attenuation coefficient values estimated using ex vivo human samples and the nonpolarization OCT method in the research9 were: for normal white matter ; for high-grade gliomas (cancer core) ; cortex was not evaluated, but the infiltration zone has a attenuation rate. The differences in the obtained data in these two studies could be explained by different objects as well as the individual specifications of OCT devices and different approaches to signal processing. However, the tendency of statistically higher values of the white matter in cochannel attenuation coefficient in comparison to tumor tissue is present in both studies (). Moreover, in our previous study,27 it was shown that the presence of particular morphological patterns can lead to dramatic changes in coefficient values and that the cortex shows attenuation coefficients in the range of that of different tumor types.
It should be noted that these calculations of the coefficients were made while disregarding the confocal factor of the OCT setup. Since OCT imaging was performed in a noncontact mode, the position of the focal plane relative to the object surface was not fixed, which made compensation of the confocal factor cumbersome. Thus, it was ignored during the calculations, and its effect on the obtained coefficients was estimated. First, the OCT signal with respect to the confocal factor can be written as
where is the focal plane position, is the refractive index of the medium, and is the Rayleigh length. Since the co- and crosschannel attenuation coefficients are estimated from the logarithm of the OCT signal, the logarithm of the confocal factor acts as a depth-dependent bias on the linear part, depending on the attenuation coefficient:
The effect of such a bias was modeled numerically taking into account that the position of the focal plane was uniformly distributed in the depth range 200 to below the surface. Depending on the exact location of the focal plane, the bias was distributed in the range from to with a mean of and a standard deviation of . Thus, due to ignoring the confocal parameter, the cochannel attenuation coefficient was systematically underestimated, but the error relative to the lowest values of the cochannel attenuation coefficient did not exceed 10%. Note that since the attenuation coefficients in both polarization channels had the same bias, the estimation of interchannel attenuation is unbiased.
To summarize existing data relative to OCT imaging of tumorous and nontumorous brain tissue for clinical use,9,10,27 it is suitable to build color-coded maps with a reduced number of colors to make the visual assessment of the scanning area convenient and simplify the information for the surgeon. This could be done by determining the cut-off values of each coefficient for distinguishing tumorous and nontumorous tissues. In addition to using OCT for determination of tissue type during surgery for gliomas, OCT could be helpful in stereotactic procedures such as deep brain stimulation or lesioning and brain tumor biopsy. During biopsy, OCT can be used for both: (1) direct viewing of the place for sampling (detecting tumorous tissue) and (2) avoidance of intraoperative bleeding by detecting blood vessels. The clear differences of OCT data between deep seated gray matter (hippocampus, thalamus) demonstrated in this study support using OCT for targeting basal nuclei (e.g., subthalamic nucleus) during deep brain stimulation and also during damage procedures for the size of destruction.
In fact, the calculation of the cochannel attenuation and interchannel attenuation difference in two orthogonal polarization channels can be used to widen the received OCT data in different clinical fields where tissue evaluation with the cochannel attenuation coefficient has already proven to be useful. Examples of such applications include the evaluation of atherosclerotic plaques,35 of oral mucosae,36 and of nanocontrast of the optical properties.37
5. Conclusion
A method for 3-D CP OCT data quantification using co- and cross-scattering images was developed and applied to the visualization of healthy and tumorous brain tissues. Building color-coded maps based on the offered optical coefficients allows for revealing more detail from OCT images compared with unprocessed OCT images. An additional optical coefficient—the interchannel attenuation difference—was proposed, which, as it turned out, has a high sensitivity for white matter detection. It can be extremely useful for the determination of regions of white matter infiltrated by a tumor. Further studies will allow adaptation of the presented approach for neurological surgery, in particular for the rapid and accurate intraoperative detection of glioma margins.
Acknowledgments
The experimental part of the study and selection of optimal quantitative characteristics for numerical image processing were supported by Russian Foundation for Basic Research; Award No. 18-29-01049_mk. The method for optical coefficients calculation and tissue quantitative analysis was supported by Russian Foundation for Basic Research; Award no. 16-32-60178 mol_a_dk. Color map construction and statistical analysis were supported by the Russian President Grant for Young Scientists; Award no. MK-6634.2018.7.
Biographies
Elena B. Kiseleva has a PhD in biology (biophysics) and is a senior researcher of Laboratory of Optical Coherence Tomography (OCT) at the the Institute of Biomedical Technologies, PRMU, Russia. Her interests and focus are on the optical imaging of biotissues, in particular, crosspolarization (CP) OCT, polarization, and nonlinear microscopy. She has contributed to the development of the numerical processing of CP OCT images for differential diagnostics of mucosal and brain diseases and to studies of myelinated fiber changes due to glioma invasion.
Konstantin S. Yashin has a PhD in medicine and is a neurosurgeon and neurooncologist at the University Clinic, PRMU, Russia. His crossdisciplinary research has focused on introducing advance optical technologies (OCT, PDT, and fluorescence) in the surgery of glial brain tumors.
Alexander A. Moiseev has a PhD in physics and mathematics and is a senior researcher at the Laboratory of Highly Sensitive Optical Measurements of the Institute of Applied Physics at the RAS, Nizhny Novgorod, Russia. His fields of scientific interest include multimodal OCT, especially quantitative analysis of OCT data (structural, polarization, and angiographic images) and subdiffraction fluorescence microscopy. He is the author of 23 research publications.
Lidia B. Timofeeva has a PhD in biology and is an assistant at the Department of Histology of PRMU. Her fields of scientific interest include investigation of biological tissue morphology by light microscopy and immunohistochemistry. She specializes in mechanisms of reparative processes in the peripheral and central nervous system, and oncomorphology, including brain tumors. She is the author of 18 research publications.
Vera V. Kudelkina has an MD in biology (biochemical and bionanotechnology) and is a researcher of the Laboratory of Neuromorphology at the Research Institute of Human Morphology, Russia. She is working on modeling of tumors of the nervous system, studying the effectiveness of anticancer drugs on the tumor models of the central nervous system. Her background includes over four years in experimental tumor chemotherapy and over ten years in clinical tumor morphology.
Anna I. Alekseeva has an MSc degree in biophysics and is a researcher of Laboratory of Neuromorphology in the Research Institute of Human Morphology, in Russia. Her interests and focus are on experimental tumor chemotherapy, in particular, study of the effect of physical and chemical parameters of nanoparticles in the treatment of nervous tumors, and the study of antitumor activity. Her background includes over five years in experimental tumor chemotherapy with nanoparticle usage.
Svetlana V. Meshkova is a member of the Radiology Department at PRMU, Russia. Her research areas include commercially adoptable technologies that use multimodal optical imaging systems and advanced image processing methods to improve the contrast of optical images. She conducts experiments with glioma models and on human samples with brain tumors.
Anastasia V. Polozova received her MSc degree in biology. She works at the Histology Laboratory of the Institute of Biomedical Technologies in PRMU, Russia, where she specializes in preparing samples of tissue for standard and immunohistochemical staining. She is interested in studies of structural changes in tissues under various pathological conditions of the body using light and fluorescent microscopy methods.
Grigory V. Gelikonov has a DSc degree in physics and mathematics and is head of the Laboratory of Highly Sensitive Optical Measurements of the Institute of Applied Physics of the RAS Nizhny Novgorod, Russia. He has been a laureate of the State Prize in Science and Technology for Research on OCT since 1999. His research interests are related to the development of time- and spectral-domain OCT, polarization-sensitive measurements, optical coherence angiography and elastography, and super-resolution optical imaging.
Elena V. Zagaynova has a DSc degree in medicine and is a professor at the RAS and a director of the Institute of Biomedical Technologies PRMU, Russia. She is the author of more than 90 research publications on OCT, which are pioneering works on the interpretation of OCT images and the evaluation of clinical possibilities of OCT in endoscopy, oncology, and urology. Currently, her research interests involve the development of new methods for cancer diagnosis and treatment, stem cells research, and regenerative medicine.
Natalia D. Gladkova, MD, has a DSc degree in medicine and is a professor and head of the Laboratory of OCT, Institute of Biomedical Technologies, PRMU, Russia. Her scientific efforts are focused on organization and carrying out of multidisciplinary and interdisciplinary basic and applied scientific research in the development of innovative approaches for diagnosis and treatment of diseases. She is interested in studying the possibilities of optical bioimaging methods, especially OCT, to evaluate the structure and microcirculation of normal and pathological biotissues.
Disclosures
No conflicts of interest, financial or otherwise, are declared by the authors.
References
- 1.de Boer J., Leitgeb R., Wojtkowski M. “Twenty-five years of optical coherence tomography: the paradigm shift in sensitivity and speed provided by Fourier domain OCT,” Biomed. Opt. Express 8(7), 3248–3280 (2017). 10.1364/BOE.8.003248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drexler W., et al. , “Optical coherence tomography today: speed, contrast, and multimodality,” J. Biomed. Opt. 19(7), 071412 (2014). 10.1117/1.JBO.19.7.071412 [DOI] [PubMed] [Google Scholar]
- 3.Men J., et al. , “Optical coherence tomography for brain imaging and developmental biology,” IEEE J. Sel. Top. Quantum Electron. 22(4), 1–13 (2016). 10.1109/JSTQE.2015.2513667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan Y., et al. , “Optical coherence tomography for precision brain imaging, neurosurgical guidance and minimally invasive theranostics,” BioSci. Trends 12(1), 12–23 (2018). 10.5582/bst.2017.01258 [DOI] [PubMed] [Google Scholar]
- 5.Lankenau E., et al. , “Combining optical coherence tomography (OCT) with an operating microscope,” Adv. Med. Eng. 114, 343–348 (2007). 10.1007/978-3-540-68764-1 [DOI] [Google Scholar]
- 6.El-Haddad M. T., Tao Y. K., “Advances in intraoperative optical coherence tomography for surgical guidance,” Curr. Opin. Biomed. Eng. 3, 37–48 (2017). 10.1016/j.cobme.2017.09.007 [DOI] [Google Scholar]
- 7.Giese A., et al. , “Non-invasive intraoperative optical coherence tomography of the resection cavity during surgery of intrinsic brain tumors,” Proc. SPIE 6078, 60782Z (2006). 10.1117/12.674436 [DOI] [Google Scholar]
- 8.Bohringer H. J., et al. , “Imaging of human brain tumor tissue by near-infrared laser coherence tomography,” Acta Neurochir. 151(5), 507–517 (2009). 10.1007/s00701-009-0248-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kut C., et al. , “Detection of human brain cancer infiltration ex vivo and in vivo using quantitative optical coherence tomography,” Sci. Trans. Med. 7(292), 292ra100 (2015). 10.1126/scitranslmed.3010611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan W., et al. , “Robust and fast characterization of OCT-based optical attenuation using a novel frequency-domain algorithm for brain cancer detection,” Sci. Rep. 7, 44909 (2017). 10.1038/srep44909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srinivasan V. J., Radhakrishnan H., “Optical coherence tomography angiography reveals laminar microvascular hemodynamics in the rat somatosensory cortex during activation,” Neuroimage 102, 393–406 (2014). 10.1016/j.neuroimage.2014.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magnain C., et al. , “Blockface histology with optical coherence tomography: a comparison with Nissl staining,” Neuroimage 84, 524–533 (2014). 10.1016/j.neuroimage.2013.08.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F., et al. , “Nondestructive evaluation of progressive neuronal changes in organotypic rat hippocampal slice cultures using ultrahigh-resolution optical coherence microscopy,” Neurophotonics 1(2), 025002 (2014). 10.1117/1.NPh.1.2.025002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magnain C., et al. , “Optical coherence tomography visualizes neurons in human entorhinal cortex,” Neurophotonics 2(1), 015004 (2015). 10.1117/1.NPh.2.1.015004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arous J. B., et al. , “Single myelin fiber imaging in living rodents without labeling by deep optical coherence microscopy,” J. Biomed. Opt. 16(11), 116012 (2011). 10.1117/1.3650770 [DOI] [PubMed] [Google Scholar]
- 16.Assayag O., et al. , “Imaging of non-tumorous and tumorous human brain tissues with full-field optical coherence tomography,” Neuroimage Clin. 2, 549–557 (2013). 10.1016/j.nicl.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamborski S., et al. , “Extended-focus optical coherence microscopy for high-resolution imaging of the murine brain,” Biomed. Opt. Express 7(11), 4400–4414 (2016). 10.1364/BOE.7.004400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H., Lenglet C., Akkin T., “Structure tensor analysis of serial optical coherence scanner images for mapping fiber orientations and tractography in the brain,” J. Biomed. Opt. 20(3), 036003 (2015). 10.1117/1.JBO.20.3.036003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H., et al. , “Polarization sensitive optical coherence microscopy for brain imaging,” Opt. Lett. 41(10), 2213–2216 (2016). 10.1364/OL.41.002213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C. J., et al. , “Visualizing and mapping the cerebellum with serial optical coherence scanner,” Neurophotonics 4(1), 011006 (2017). 10.1117/1.NPh.4.1.011006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boas D., et al. , “Polarization-sensitive optical coherence tomography of the human brain connectome,” SPIE Newsroom 1–4 (2017). 10.1117/2.1201701.006834 [DOI] [Google Scholar]
- 22.Yashin S., et al. , “Multimodal optical coherence tomography in visualization of brain tissue structure at glioblastoma (experimental study),” Sovrem. Technol. Med. 8(1), 73–81 (2016). 10.17691/stm2016.8.1.10 [DOI] [Google Scholar]
- 23.Kiseleva B., et al. , “Quantitative cross-polarization OCT detection of infiltrative tumor margin in rat glioma model: pilot study,” Sovrem. Technol. Med. 10(1), 6–14 (2018). 10.17691/stm2018.10.1.01 [DOI] [Google Scholar]
- 24.Sanai N., Berger M. S., “Extent of resection influences outcomes for patients with gliomas,” Rev. Neurol. 167(10), 648–654 (2011). 10.1016/j.neurol.2011.07.004 [DOI] [PubMed] [Google Scholar]
- 25.Yashin S., et al. , “Ex vivo visualization of human gliomas by cross-polarization optical coherence tomography: pilot study,” Sovrem. Technol. Med. 8(4), 14–22 (2016). 10.17691/stm2016.8.4.02 [DOI] [Google Scholar]
- 26.Bohringer H. J., et al. , “Time-domain and spectral-domain optical coherence tomography in the analysis of brain tumor tissue,” Lasers Surg. Med. 38, 588–597 (2006). 10.1002/(ISSN)1096-9101 [DOI] [PubMed] [Google Scholar]
- 27.Yashin K. S., et al. , “Quantitative nontumorous and tumorous human brain tissue assessment using microstructural co- and cross-polarized optical coherence tomography,” Sci. Rep. 9, 2024 (2019). 10.1038/s41598-019-38493-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moiseev A., et al. , “Pixel classification method in optical coherence tomography for tumor segmentation and its complementary usage with OCT microangiography,” J. Biophotonics 11(4), e201700072 (2018). 10.1002/jbio.2018.11.issue-4 [DOI] [PubMed] [Google Scholar]
- 29.Marvdashti T., et al. , “Classification of basal cell carcinoma in human skin using machine learning and quantitative features captured by polarization sensitive optical coherence tomography,” Biomed. Opt. Express 7(9), 3721–3735 (2016). 10.1364/BOE.7.003721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenz M., et al. , “Automated differentiation between meningioma and healthy brain tissue based on optical coherence tomography ex vivo images using texture features,” J. Biomed. Opt. 23(7), 071205 (2018). 10.1117/1.JBO.23.7.071205 [DOI] [PubMed] [Google Scholar]
- 31.Baklaushev V. P., et al. , “Treatment of glioma by cisplatin-loaded nanogels conjugated with monoclonal antibodies against Cx43 and BSAT1,” Drug Delivery 22(3), 276–285 (2015). 10.3109/10717544.2013.876460 [DOI] [PubMed] [Google Scholar]
- 32.Moiseev A. A., et al. , “Noniterative method of reconstruction optical coherence tomography images with improved lateral resolution in semitransparent media,” Laser Phys. Lett. 10(12), 125601 (2013). 10.1088/1612-2011/10/12/125601 [DOI] [Google Scholar]
- 33.Gelikonov V. M., et al. , “Cross-polarization optical coherence tomography with active maintenance of the circular polarization of a sounding wave in a common path system,” Radiophys. Quantum Electron. 60(11), 897–911 (2018). 10.1007/s11141-018-9856-9 [DOI] [Google Scholar]
- 34.Paxinos G., Watson C., The Rat Brain in Stereotaxic Coordinates, 4th ed., Elsevier, Academic Press, New York: (1998). [Google Scholar]
- 35.Xu C., et al. , “Characterization of atherosclerosis plaques by measuring both backscattering and attenuation coefficients in optical coherence tomography,” J. Biomed. Opt. 13(3), 034003 (2008). 10.1117/1.2927464 [DOI] [PubMed] [Google Scholar]
- 36.Tomlins P. H., et al. , “Scattering attenuation microscopy of oral epithelial dysplasia,” J. Biomed. Opt. 15(6), 066003 (2010). 10.1117/1.3505019 [DOI] [PubMed] [Google Scholar]
- 37.Xi J., Chen Y., Li X. D., “Characterizing optical properties of nano contrast agents by using cross-referencing OCT imaging,” Biomed. Opt. Express 4(6), 842–851 (2013). 10.1364/BOE.4.000842 [DOI] [PMC free article] [PubMed] [Google Scholar]