Highlights
-
•
MRI-CT deformable image registration was not superior to rigid registration.
-
•
Dice similarity coefficients were 0.65, 0.62, and 0.63 for deformable registrations.
-
•
Dice similarity coefficient was 0.63 for rigid registration.
-
•
Registration quality was superior in muscle and gland compared to bone and vessel.
Abbreviations: RT, radiation therapy; MRI, magnetic resonance imaging; MRIgRT, MRI-guided radiotherapy planning; CT, computed tomography; OAR, organ(s) at risk; MRL, MRI linear accelerator; DIR, deformable image registration; RIR, rigid image registration; HNC, head and neck cancer; IMRT, intensity-modulated radiation therapy; DICOM, digital imaging and communications in medicine; DSC, dice similarity coefficient; HD max, Hausdorff maximum distance; HD mean, Hausdorff mean distance; HPV, human papillomavirus; sCT, synthetic computed tomography; MAE, mean absolute error; HU, Hounsfield units
Keywords: MRI-guided radiotherapy, CT-MRI image registration, Deformable image registration, Rigid image registration, Quality assessment
Abstract
Background
MRI-guided radiotherapy planning (MRIgRT) may be superior to CT-guided planning in some instances owing to its improved soft tissue contrast. However, MR images do not communicate tissue electron density information necessary for dose calculation and therefore must either be co-registered to CT or algorithmically converted to synthetic CT. No robust quality assessment of commercially available MR-CT registration algorithms is yet available; thus we sought to quantify MR-CT registration formally.
Methods
Head and neck non-contrast CT and T2 MRI scans acquired with standard treatment immobilization techniques were prospectively acquired from 15 patients. Per scan, 35 anatomic regions of interest (ROIs) were manually segmented. MRIs were registered to CT rigidly (RIR) and by three commercially available deformable registration algorithms (DIR). Dice similarity coefficient (DSC), Hausdorff distance mean (HD mean) and Hausdorff distance max (HD max) metrics were calculated to assess concordance between MRI and CT segmentations. Each DIR algorithm was compared to DIR using the nonparametric Steel test with control for individual ROIs (n = 105 tests) and for all ROIs in aggregate (n = 3 tests). The influence of tissue type on registration fidelity was assessed using nonparametric Wilcoxon pairwise tests between ROIs grouped by tissue type (n = 12 tests). Bonferroni corrections were applied for multiple comparisons.
Results
No DIR algorithm improved the segmentation quality over RIR for any ROI nor all ROIs in aggregate (all p values >0.05). Muscle and gland ROIs were significantly more concordant than vessel and bone, but DIR remained non-different from RIR.
Conclusions
For MR-CT co-registration, our results question the utility and applicability of commercially available DIR over RIR alone. The poor overall performance also questions the feasibility of translating tissue electron density information to MRI by CT registration, rather than addressing this need with synthetic CT generation or bulk-density assignment.
1. Introduction
MRI-guided radiotherapy planning (MRgRT) has attractive advantages over CT for its potential to deliver more personalized treatment. First, MRI has superior visualization of soft tissue anatomy for more accurate definition of target volumes and organs at risk (OARs). Second, emerging novel devices such as MR-Linear accelerators (MRL) create an opportunity to use MRI rather than CT as a tool for online target and OAR visualization and for adaptive re-planning during treatment course [1]. Third, functional MRI sequences may leverage tissue-specific contrast agents, as well as uncover biomarkers that may predict treatment outcomes [2], [3].
MRgRT has been established in many cancer subsites, including brain, spine, liver and pancreas, prostate, and nasopharyngeal cancers [2], [3]. This landscape is expanding. For example, an MRgRT system (ViewRay, 0.35 Tesla, OH, USA) [4] was operational in 2014 at the Washington University in St. Louis, and in the intervening four-and-a-half years has changed their practice for abdominal and breast cancers [1], [5]. There is reason to hope that these developments herald new indications for MRI, especially as improved systems with more precise targeting and steeper gradient drop-offs are grandfathered into practice. At our institution, we successfully treated the first patient in the United States using a recently FDA-approved high-field Elekta-Philips Unity MRL (Elekta AB, 1.5 T, Stockholm, Sweden; Philips, Amsterdam, Netherlands) [6], [7].
However, the integration of MRI with radiotherapy planning on a broader landscape faces many challenges including those related to MRI acquisition, geometric distortion, and onerous MR-CT registration techniques [2], [8], [9]. At our institution, we already established that an MRI acquisition technique for simulation (i.e. in the treatment position) using RT planning immobilization devices improved performance over diagnostic (i.e. non-immobilized) MR acquisition for radiotherapy planning. This method demonstrated superior image quality, decreased geometric distortion, and better MR-CT co-registration [10], [11]. Prior work to ours has also confirmed the feasibility of MR acquisition in the treatment position [12]. Nevertheless, we still lack a rigorous quantification of the quality of deformable and rigid MR-CT co-registration techniques and their anatomic fidelity when both MR and CT are acquired in the treatment position. MR-CT co-registration is potentially useful since MRI does not provide tissue electron-density information needed to calculate radiation dose. Prior studies suggest that deformable image registration (DIR) techniques may offer an advantage over conventional rigid image registration (RIR) for head and neck cancer (HNC) patients [13], [14], [15], [16], [17]. Nevertheless, not all these studies evaluated the quality of DIR across a robust number of anatomic regions of interest (ROIs), and some used MRIs acquired using standard diagnostic acquisition techniques. To this end, we assessed the quality of MR-CT co-registration using commercially available image registration software for images acquired using strict RT immobilization techniques for both MRIs and CTs in HNC cancer patients receiving definitive IMRT. The specific aims are: 1) Assess whether the use of DIR software provides an advantage over the conventional RIR, and 2) Determine the quality of MR-CT registration tools for HNC using overlap and surface distance metrics.
2. Methods
2.1. Study population
Eligible patients for this study had biopsy-proven intact squamous cell carcinoma of the head and neck, treatment with curative-intent intensity-modulated radiation therapy, no chemotherapy between CT and MRI scans, and not greater than 4 weeks between CT and MRI acquisition to avoid conflating our results with anatomic changes attributable to disease progression or treatment. For patients that met the criteria, nonenhanced simulation CT and nonenhanced simulation T2-weighted (T2w) MRI Digital Imaging and Communications in Medicine (DICOM) scans were prospectively acquired from the University of Texas MD Anderson Cancer Center clinical databases in agreement with an IRB protocol designed to evaluate imaging parameters for MR simulation. Anonymized scans are made available online at https://figshare.com/s/a5e09113f5c07b3047df.
2.2. Immobilization technique
For both CT and MRI, patients were immobilized in the treatment position using a customized head, neck, and shoulder thermoplastic masks, mold head support, and dental stent. For further details we refer the reader to the MRI immobilization protocol we have previously validated [11].
2.3. Imaging properties
Non-contrast enhanced simulation CT scans were acquired on one of four machine models (LightSpeed RT16, GE Medical Systems, Waukesha, WI, USA; Brilliance 64 or Brilliance Big Bore, Philips Healthcare, Cambridge, MA, USA; SOMATOM Definition Edge, Siemens, Washington D.C., USA) using x-ray tube currents between 208 and 434 mA at 120 kVp. All scans had slice thicknesses of either 2.5 or 3 mm, with 2.5 or 3 mm of space between slices. Sections were displayed in 512 × 512 pixel matrices and reconstructed in diameters between 500 and 600 mm. Non-contrast enhanced simulation T2w MRI scans were acquired in one of two machine models (Discovery MR750, GE Medical Systems, Waukesha, WI, USA; Ingenia, Philips Healthcare, Cambridge, MA, USA) in 3 Tesla magnetic fields with echo times between 97.72 and 101.82 ms. All scans were acquired with slice thicknesses of 2.5 mm, with 2.5 or 4 mm of space between slices. Sections were displayed in 512 × 512 pixel matrices and reconstructed in 256 mm diameters. The median time between CT and MRI acquisition was 8 days (range 1–16 days).
2.4. Manual segmentation of ROIs
Anatomic ROIs included the C1-C4 vertebrae, mandible, spinal cord, thyroid cartilage, genioglossus, geniohyoid, hyoglossus, soft palate, tongue, and bilateral mastoids, submandibular and parotid glands, anterior digastrics, myelohyoids, masseters, medial and lateral pterygoids, sternocleidomastoids, internal jugular veins and common carotid arteries. These were contoured on CT and T2-MRI images independently by four radiation oncologists (M.A.M, M.K.J., S.P.N., H.E.) with at least five years of experience in head and neck radiotherapy (including residency) and approved by the principal investigator (C.D.F). All CT and MRI scans were acquired between 11/21/2013 and 06/09/2016. Each patient’s CT and MRI pair was contoured by the same radiation oncologist to evade interobserver variability per patient.
2.5. Image registration
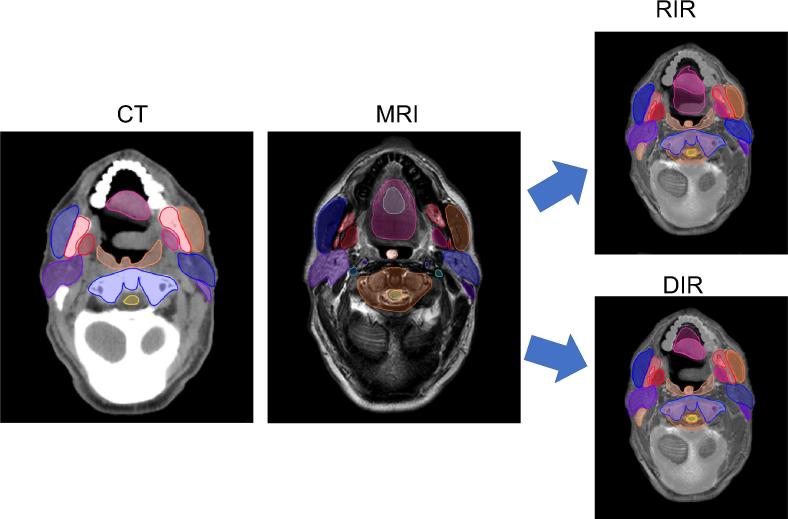
Three DIR algorithms (Admire version 1.13.5, a commercial research tool, Elekta, Stockholm, Sweden; Pinnacle v. 9.1, Philips Radiation Oncology Systems, Madison, WI, USA; and Velocity-deformable, Velocity AI, version 3.0.1, Atlanta, GA, USA) and one RIR algorithm (Velocity-rigid, Velocity AI, version 3.0.1, Atlanta, GA, USA) were used in this study. RT planning CT scan for each patient was used as the primary (i.e. fixed) image set and the T2-MRI was the secondary (i.e. mobile) image. Following the co-registration, we evaluated overlap and surface distance metrics for the 35 ROIs in all patients. Fig. 1 summarizes the study workflow.
Fig. 1.
Illustration of study workflow. T2-weighted MRI and non-contrast simulation CT scans were both acquired in the standard RT position and thirty-five ROIs were contoured on each scan by an expert radiation oncologist. MRI was then registered to CT by one of three DIR algorithms or by RIR, and performance metrics were calculated for the alignment of MRI contours and CT contours.
2.6. Statistical analysis and visualization
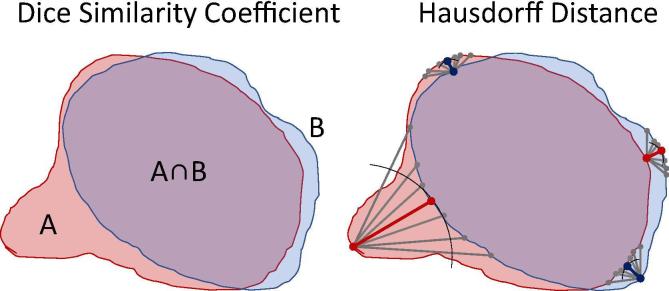
Statistical analyses were performed in JMP Pro version 14.0 software (SAS institute, Cary, NC). Three performance metrics were calculated on 525 ROIs (1575 total measurements): Dice similarity coefficient (DSC) [18], Hausdorff maximum distance (HD max), and Hausdorff mean distance (HD mean) [19]. The DSC is calculated as two times the overlap volume (or area) between two contour volumes (or areas) A and B divided by the sum A and B. DSC values range from 0 to 1 where 0 indicates no overlap and 1 indicates perfect overlap (Fig. 2). Hausdorff distances are calculated in the following way: the distances between every point on contour A and its nearest point on contour B, and between every point on B and its nearest point on A, are noted. The HD max and mean are respectively the maximum and mean of these distances. Smaller HD max and mean values signify more similar contours. The results were assessed visually in histograms and were tested with Shapiro-Wilks [20] to determine that the null hypotheses (that the distribution is normal) were in every case rejected (p values <0.0001). Therefore, nonparametric statistical tests were employed since they make no assumptions about the normality of their underlying distributions. Median metrics for each DIR algorithm were pitted against DIR for all ROIs using the Steel method with control (n = 105 tests) [21]. On a per-metric basis, DIR algorithms were compared to RIR for all ROIs in aggregate (n = 3 tests). Data were aggregated by tissue type (muscle, bone, vessel, spinal cord, and gland), and median metrics per tissue type were evaluated against one another using the Wilcoxon method for pairwise comparisons (n = 12 tests) [22]. The alpha threshold was lowered by a Bonferroni correction to account for multiple comparisons [23]. Results were represented graphically in the Python programming language version 3.6.7 (Python Software Foundation, https://www.python.org) leveraging code housed in the Pandas version 0.23.4 [24], Matplotlib version 3.0.1 [25] and Seaborn version 0.9.0 (Michael Waskom, http://seaborn.pydata.org/) libraries.
Fig. 2.
Illustration of metrics. The DSC is computed as and may take values from 0 to 1. A larger number indicates better volume overlap. Hausdorff Distances are the minimum distances from every point in A to any point in B (e.g. red lines) and from B to A (e.g. blue lines). The maximum and mean of these distances are the HD max and HD mean. Smaller numbers indicate less extreme deviations between contour surfaces. HD max is more sensitive to projection-like deviations than DSC. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.7. Interobserver variability
To exclude the influence of interobserver variability, we statistically analyzed our results after stratifying by radiation oncologist. DIR algorithms were compared to RIR with the Steel Method using the DSC, HD max, and HD mean metrics on a per-radiation oncologist, per-tissue type basis (n = 162 comparisons) with an exception: the “cord” tissue type results (which consists of only one ROI, the spinal cord) could not be subjected to this test for two radiation oncologists because these two radiation oncologists each contoured the scans of only one patient.
3. Results
3.1. Patient characteristics
Fifteen patients were identified who met study inclusion criteria. The median patient age was 57 years (range 45–81) and 80% were males. Primary tumor sites included the tongue base and tonsils ranging from T1–T4, and 87% had nodal involvement. In all but one patient the tumor was found to be HPV positive. Fifty-three percent of patients were treated with photon-based radiotherapy modalities, and 47% of patients were treated with protons. The median final dose was 70 Gy (range 67.75–70 Gy) and all treatments were delivered in 33 fractions. Additionally, 93% of patients underwent concurrent chemotherapy with either cisplatin or cetuximab, although chemotherapeutic treatment during the interval between CT and MRI was an exclusion criterion. Patient characteristics are summarized in Table 1.
Table 1.
Summary of patient characteristics.
| Median age | ||
| 57 (range 45–81) | ||
| Gender | ||
| Male | 12 | |
| Female | 3 | |
| Ethnicity | ||
| White | 12 | |
| Hispanic | 2 | |
| Black | 1 | |
| Tumor site | ||
| Base of tongue | 7 | |
| Glossopharyngeal sulcus | 2 | |
| Tonsil | 6 | |
| Tumor stage | ||
| T1 | 2 | |
| T2 | 8 | |
| T3 | 3 | |
| T4 | 2 | |
| Nodal stage | ||
| N0 | 2 | |
| N1 | 1 | |
| N2 | 12 | |
| Median dose | ||
| 70 Gy (range 67.75–70) | ||
| Number of fractions | ||
| 33 | ||
| Treatment modality | ||
| IMRT | 7 | |
| VMAT | 1 | |
| IMPT | 7 | |
| Concurrent chemo | ||
| Cetuximab | 5 | |
| Cisplatin | 9 | |
| None | 1 | |
3.2. Comparison of DIR with RIR
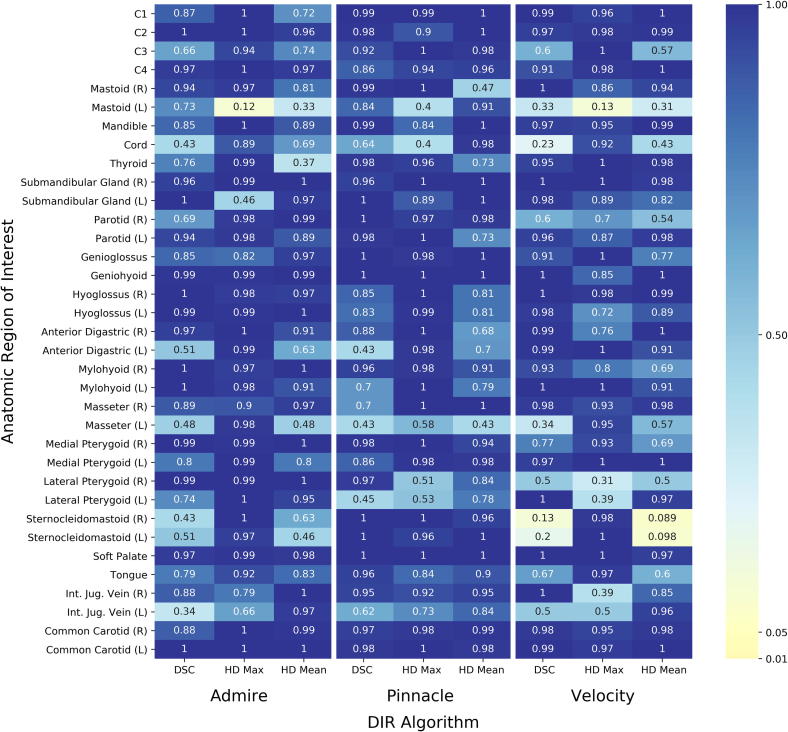
Our results could not demonstrate a statistically significant improvement in any deformable algorithm’s performance over a rigid registration alone, regardless of ROI assessed or metric used to make the assessment (Fig. 3). Likewise, no improvement could be demonstrated between DIRs and RIR when all ROIs were grouped in one aggregate (p values all >0.05).
Fig. 3.
Nonparametric Steel tests with control failed to demonstrate superiority of DIR to RIR. Steel tests were calculated comparing each DIR algorithm to RIR by individual ROI and performance metric. Resulting p-values are plotted on a heat map. No statistically significant difference exists between DIR and RIR for any ROI.
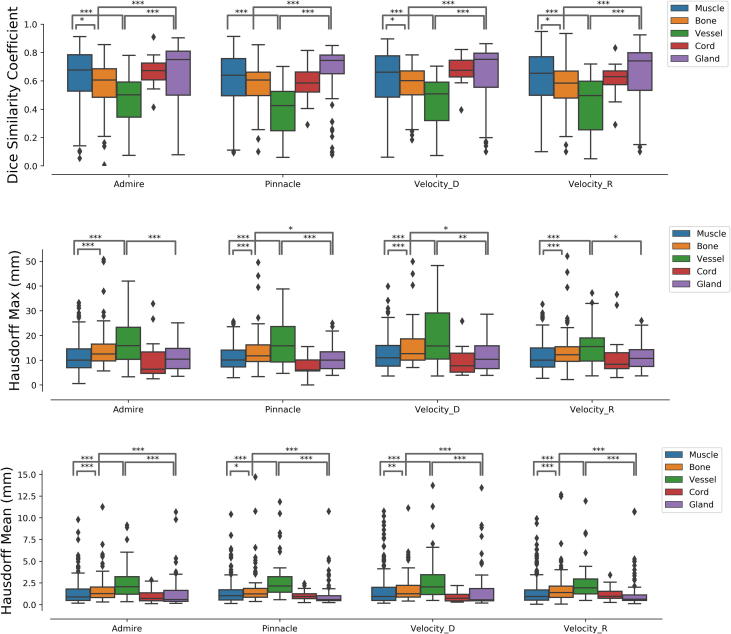
3.3. Registration performance by tissue type
Soft tissue ROIs consistently demonstrated superior concordance across all registration methods, including RIR. Selected statistically significant differences are shown in Fig. 4. Median DSC and HD metrics are enumerated for all tissue types in Table 2. The apparent trend is that muscle gland ROIs were superior to vessel and bone. For example, the median DSC was higher for muscle than for bone (0.66 vs 0.61, p value <0.0001), and the median Hausdorff max and mean were lower (respectively 10.23 vs. 12.18, p value <0.005; 0.94 vs. 1.29, p value <0.0001), indicating superior performance in muscle than bone. This statistically significant pattern was also observed between gland and bone. Vessel ROIs (internal jugular veins and common carotid arteries) suffered the least conformal registration independent of DIR or RIR method, as well as the most volatile surface distance metrics.
Fig. 4.
Nonparametric pairwise comparisons using the Wilcoxon method demonstrated improved registration fidelity for soft tissues compared to other tissue types. Selected statistically significant relationships are shown illustrating superior DSC, Hausdorff max, and Hausdorff mean results for muscle and gland ROIs compared to bone, vessel, and spinal cord ROIs. This trend is generally consistent for all registration methods. Vessel ROIs were the least conformal and exhibited the greatest variance in surface distance metrics.
Table 2.
Median DSC and HD metrics per tissue type, stratified by registration algorithm and in aggregate.
| Median |
||||||
|---|---|---|---|---|---|---|
| Bone | Cord | Gland | Muscle | Vessel | ||
| DSC | DIR 1 | 0.61 | 0.67 | 0.75 | 0.68 | 0.50 |
| DIR 2 | 0.60 | 0.59 | 0.74 | 0.64 | 0.43 | |
| DIR 3 | 0.60 | 0.67 | 0.75 | 0.78 | 0.60 | |
| RIR | 0.58 | 0.63 | 0.74 | 0.65 | 0.50 | |
| In aggregate | 0.61 | 0.65 | 0.75 | 0.66 | 0.47 | |
| HD max (mm) | DIR 1 | 12.52 | 6.37 | 10.42 | 10.04 | 15.9 |
| DIR 2 | 11.78 | 6.10 | 10.05 | 10.12 | 15.86 | |
| DIR 3 | 12.68 | 7.75 | 10.38 | 10.97 | 15.78 | |
| RIR | 12.24 | 8.39 | 10.74 | 10.08 | 15.53 | |
| In aggregate | 12.18 | 7.56 | 10.42 | 10.23 | 15.77 | |
| HD mean (mm) | DIR 1 | 1.27 | 0.71 | 0.59 | 0.88 | 2.06 |
| DIR 2 | 1.23 | 0.93 | 0.60 | 1.03 | 2.16 | |
| DIR 3 | 1.26 | 0.74 | 0.55 | 0.93 | 2.05 | |
| RIR | 1.39 | 0.96 | 0.63 | 0.94 | 1.93 | |
| In aggregate | 1.29 | 0.84 | 0.60 | 0.94 | 2.05 | |
3.4. Comparison between radiation oncologists
In all comparisons of results between radiation oncologists there was no statistically significant difference between any DIR algorithm and RIR, with one exception for one radiation oncologist in one tissue type (+0.07 median DSC conformality for the muscle tissue type with Admire DIR over RIR, p = 0.027 after Bonferroni correction). This single result notwithstanding, these results demonstrate that our data are not confounded by interobserver variability.
4. Discussion
This study did not demonstrate that commercially-available DIR algorithms confer a better registration than RIR alone for simulation T2-weighted MRI registration to simulation CT in the context of HNC radiotherapy workflow. Since our primary objective of this study was to evaluate the quality of registration methods for MRI applications in radiotherapy planning, treatment, and post-treatment surveillance, this result is informative. We used rigorous, nonparametric statistical methods to compare between DIR and RIR, but the statistical rigor may be moot because none of the registration algorithms – deformable or rigid – achieved robust, clinically satisfactory fidelity to manually segmented anatomy. Across all ROIs, the median DSCs were only 0.65, 0.62, 0.63, and 0.63 for Admire, Pinnacle, Velocity-deformable, and Velocity-rigid registrations, respectively. Notably, our conclusions do not nominally accord with Fortunati et al, who report improvement in DIR compared to RIR for MR-CT registration (acquired with patient-specific RT immobilization methods) over 12 segmented ROIs in 12 patients [13]. However, per ROI, Fortunati et al. report differences in DSC and HD measurements between DIR and RIR that were practically indistinguishable even when statistically distinguishable (refer to Fig. 3 in their paper). The left and right eye lens ROIs are exceptions, which do show meaningful improvements with DIR of approximately +0.2 DSC, −0.5 mm modified Hausdorff surface distance, and −2 mm 3-dimensional Hausdorff surface distance, but this may be explained by that fact that the eyes are not immobilized, as the authors discuss. Therefore, while statistically and nominally different, we believe that Fortunati et al.’s results concord with ours.
Ancillary to the discussion above, we did find that all registrations methods demonstrated superior concordance in soft tissue ROIs, consistent with the detailed soft tissue visualization intrinsic to MRI. Vessel ROIs were, as a group, the least concordant tissue type. This may be due to the difficulty of distinguishing vessels from surrounding tissue without the aid of contrast. Vessels are small and serpentine and can be difficult to delineate even for experienced clinicians. This suggests that for T2-weighted MRI to non-contrast CT registration, vessels may be the most difficult anatomic structures to faithfully represent for DIR or RIR. These findings suggest that registration algorithms may need the greatest improvement in head and neck anatomic regions that are not well visualized in MRI or may benefit from contrast.
In contrast to CT to CT co-registration where several studies showed the superiority of DIR over RIR in the context of diagnostic and simulation CTs co-registration [26], [27], [28], [29], our CT to MR co-registration results did not demonstrate the same trend of DIR superiority. The significant improvement in DIR for diagnostic CT to simulation CT is mainly attributable to the poor performance of RIR since the diagnostic CTs are not acquired under any structured positioning. For example, a previous study done by our group for CT-CT co-registration quality assurance showed markedly inferior RIR performance compared to the present study, in which both the CT and MRI were acquired according to RT immobilization protocols. Judged by DSC and HD metrics, MRI-CT RIR performance is nearly as good as the best CT-CT DIR for muscle and gland tissue types in that study, but not for other tissue types, particularly bone. In sum, DIR currently appears capable of achieving better overall concordance for diagnostic CT to simulation CT co-registrations than simulation CT to MRI co-registrations.
To further the integration of MRI in radiotherapy applications, our results allow for following one of three paths: 1) manage MRI-CT registration using rigid methods alone, or 2) improve the performance of existing DIR algorithms, or 3) approach MR-guided radiotherapy from MR-only workflows, relying on synthetic CT (sCT) generation and eliminating MR-CT registration altogether. The first path may be unattractive because of the mediocre performance of RIR already discussed. The second path may be a preferable alternative to the first, but currently the lack of an immediately viable DIR stalls the integration of MRI. Moreover, there is no guarantee when nor if an improved DIR algorithm will become available. Therefore, MR-only workflow using sCT may be the best path continuing MRI integration with radiotherapy applications.
Significant challenges to MR-only radiotherapy exist. Principally, the best metrics and parameters for quality assessment of sCT have not yet been established nor standardized, nor have the optimal methods for generating sCTs been established in large clinical trials [8]. Multiple methods have been developed for generating sCTs, most of which fall into three groups: 1) voxel-based, 2) atlas-based, or 3) bulk-override [8], [9]. The plurality of methods are voxel-based and generate sCTs that model dose deposition with no more than 2.5% error from standard CT. Synthetic CTs have mean absolute errors (MAE) reported between 40 and 200 Hounsfield Units (HU), depending on body organ [9]. Atlas-based methods have reported similar efficacies, but can be more computationally intensive and are challenged by atypical anatomy [8]. Atlas-based sCT generation requires at least one image registration (sometimes more), but to an MRI atlas rather than to a CT. Although the acquisition and evaluation of sCTs lack standardization, the metrics in studies to date (ΔDose%, DSC, and HU MAE) seem to sustain the notion that clinically acceptable plans can be generated from standard MRI sequences, even though ultra-short echo time sequences improve the delineation of bone and air [8], [9]. At our institution a high field (1.5 Tesla) MRL treated the first patient in North America in January 2019, following suit behind University Medical Center Utrecht in the Netherlands, which pioneered the first treatment in late 2017 [7].
In summary, we present results evaluating the performance of DIR and RIR algorithms for T2w simulation MRI to simulation CT registration. We found the median DSC to be mediocre even for tissue types that best lend themselves to registration, and that DIR conferred no improvement in registration fidelity over RIR when images were acquired with standard immobilization protocols. Together, these results either usher the timely improvement of existing co-registration methods or else herald the use of sCT over MRI-CT registration for MR-guided dose deposition modeling in HNC, thereby signaling the need for standardization of sCT acquisition methods and evaluation metrics in order to scale MR-guided radiotherapy applications.
5. Declarations of interest
S.P.N. reports funding from the Australian Postgraduate Award. K.K.B. reports research funding from the NIH and RaySearch Laboratories and has a licensing agreement with RaySearch Laboratories. C.D.F. reports research funding from the NIH, the Netherlands Organisation for Scientific Research, and Elekta AB.
Sources of funding
C.D. F. has received funding from the National Institute for Dental and Craniofacial Research Award (1R01DE025248-01/R56DE025248) and Academic-Industrial Partnership Award (R01 DE028290) , the National Science Foundation (NSF), Division of Mathematical Sciences, Joint NIH/NSF Initiative on Quantitative Approaches to Biomedical Big Data (QuBBD) Grant (NSF 1557679), the NIH Big Data to Knowledge (BD2K) Program of the National Cancer Institute (NCI) Early Stage Development of Technologies in Biomedical Computing, Informatics, and Big Data Science Award (1R01CA214825), the NCI Early Phase Clinical Trials in Imaging and Image-Guided Interventions Program (1R01CA218148), the NIH/NCI Cancer Center Support Grant (CCSG) Pilot Research Program Award from the UT MD Anderson CCSG Radiation Oncology and Cancer Imaging Program (P30CA016672), the NIH/NCI Head and Neck Specialized Programs of Research Excellence (SPORE) Developmental Research Program Award (P50 CA097007) and the National Institute of Biomedical Imaging and Bioengineering (NIBIB) Research Education Program (R25EB025787). C.D.F has also received direct industry grant support, speaking honoraria and travel funding from Elekta AB.
Conflict of interest statement
None.
Contributor Information
Kendall Kiser, Email: Kendall.j.kiser@uth.tmc.edu.
Abdallah S.R. Mohamed, Email: asmohamed@mdanderson.org.
Mona Kamal, Email: mkjomaa@mdanderson.org.
Sweet Ping Ng, Email: sweet.ng@petermac.org.
Hesham Elhalawani, Email: hmelhalawani@mdanderson.org.
Amit Jethanandani, Email: ajethan1@uthsc.edu.
Renjie He, Email: rhe1@mdanderson.org.
Yao Ding, Email: yding1@mdanderson.org.
Houda Bahig, Email: hbahig@mdanderson.org.
Adam Garden, Email: agarden@mdanderson.org.
Stephen Lai, Email: sylai@mdanderson.org.
Jack Phan, Email: jphan@mdanderson.org.
Gary B. Gunn, Email: gbgunn@mdanderson.org.
David Rosenthal, Email: dirosenthal@mdanderson.org.
Steven Frank, Email: sjfrank@mdanderson.org.
Kristy K. Brock, Email: kkbrock@mdanderson.org.
Jihong Wang, Email: jihong.wang@mdanderson.org.
Clifton D. Fuller, Email: cdfuller@mdanderson.org.
References
- 1.Henke L.E., Contreras J.A., Green O.L. Magnetic resonance image-guided radiotherapy (MRIgRT): A 4.5-year clinical experience. Clin Oncol. 2018;30:720–727. doi: 10.1016/j.clon.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandarana H., Wang H., Tijssen R.H.N. Emerging role of MRI in radiation therapy. J Magn Reson Imaging. 2018;48:1468–1478. doi: 10.1002/jmri.26271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McWilliam A., Rowland B., Van Herk M. The challenges of using MRI during radiotherapy. Clin Oncol. 2018;30:680–685. doi: 10.1016/j.clon.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Mutic S., Dempsey J.F. The ViewRay system: Magnetic resonance–guided and controlled radiotherapy. Semin Radiat Oncol. 2014;24:196–199. doi: 10.1016/j.semradonc.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Henke L., Kashani R., Robinson C. Phase I trial of stereotactic MR-guided online adaptive radiation therapy (SMART) for the treatment of oligometastatic or unresectable primary malignancies of the abdomen. Radiother Oncol. 2018;126:519–526. doi: 10.1016/j.radonc.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 6.Kerkmeijer L.G.W., Fuller C.D., Verkooijen H.M. The MRI-linear accelerator consortium: Evidence-based clinical introduction of an innovation in radiation oncology connecting researchers, methodology, data collection, quality assurance, and technical development. Front Oncol. 2016;6 doi: 10.3389/fonc.2016.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raaymakers B.W., Jürgenliemk-Schulz I.M., Bol G.H. First patients treated with a 1.5 T MRI-LINAC: Clinical proof of concept of a high-precision, high-field MRI guided radiotherapy treatment. Phys Med Biol. 2017;62:L41–L50. doi: 10.1088/1361-6560/aa9517. [DOI] [PubMed] [Google Scholar]
- 8.Johnstone E., Wyatt J.J., Henry A.M. Systematic review of synthetic computed tomography generation methodologies for use in magnetic resonance imaging–only radiation therapy. Int J Radiat Oncol Biol Phys. 2018;100:199–217. doi: 10.1016/j.ijrobp.2017.08.043. [DOI] [PubMed] [Google Scholar]
- 9.Edmund J.M., Nyholm T. A review of substitute CT generation for MRI-only radiation therapy. Radiat Oncol. 2017;12 doi: 10.1186/s13014-016-0747-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohamed A.S.R., Hansen C., Weygand J. Prospective analysis of in vivo landmark point-based MRI geometric distortion in head and neck cancer patients scanned in immobilized radiation treatment position: Results of a prospective quality assurance protocol. Clin Transl Radiat Oncol. 2017;7:13–19. doi: 10.1016/j.ctro.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding Y., Mohamed A.S.R., Yang J. Prospective observer and software-based assessment of magnetic resonance imaging quality in head and neck cancer: Should standard positioning and immobilization be required for radiation therapy applications? Pract Radiat Oncol. 2015;5:e299–e308. doi: 10.1016/j.prro.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verduijn G.M., Bartels L.W., Raaijmakers C.P. Magnetic resonance imaging protocol optimization for delineation of gross tumor volume in hypopharyngeal and laryngeal tumors. Int J Radiat Oncol Biol Phys. 2009;74:630–636. doi: 10.1016/j.ijrobp.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Fortunati V., Verhaart R.F., Angeloni F. Feasibility of multimodal deformable registration for head and neck tumor treatment planning. Int J Radiat Oncol Biol Phys. 2014;90:85–93. doi: 10.1016/j.ijrobp.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 14.Wang X., Li L., Hu C. A comparative study of three CT and MRI registration algorithms in nasopharyngeal carcinoma. J Appl Clin Med Phys. 2009;10 doi: 10.1120/jacmp.v10i2.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webster G.J., Kilgallon J.E., Ho K.F. A novel imaging technique for fusion of high-quality immobilised MR images of the head and neck with CT scans for radiotherapy target delineation. Br J Radiol. 2009;82:497–503. doi: 10.1259/bjr/50709041. [DOI] [PubMed] [Google Scholar]
- 16.Du Bois D’Aische A, De Craene M., Geets X. Estimation of the deformations induced by articulated bodies: Registration of the spinal column. Biomed Signal Process Control. 2007;2:16–24. [Google Scholar]
- 17.Leibfarth S., Mönnich D., Welz S. A strategy for multimodal deformable image registration to integrate PET/MR into radiotherapy treatment planning. Acta Oncol. 2013;52:1353–1359. doi: 10.3109/0284186X.2013.813964. [DOI] [PubMed] [Google Scholar]
- 18.Dice L.R. Measures of the amount of ecologic association between species. Ecology. 1945;26:297. [Google Scholar]
- 19.Huttenlocher D.P., Klanderman G.A., Rucklidge W.J. Comparing images using the Hausdorff distance. IEEE Trans Pattern Anal Mach Intell. 1993;15:850–863. [Google Scholar]
- 20.Wilk M.B., Shapiro S.S. An analysis of variance test for normality (complete samples) Biometrika. 1965;52:591–611. [Google Scholar]
- 21.Steel R.G. A multiple comparison rank sum test: treatments versus control. Biometrics. 1959;15:560–572. [Google Scholar]
- 22.Wilcoxon F. Individual comparisons of grouped data by ranking methods. J Econ Entomol. 1946;39:269–270. doi: 10.1093/jee/39.2.269. [DOI] [PubMed] [Google Scholar]
- 23.Dunn O.J. Multiple comparisons among means. J Am Stat Assoc. 1961;56:52. [Google Scholar]
- 24.McKinney W. Data structures for statistical computing in Python. Proceedings of the 9th Python in Science Conference (SCIPY 2010) 2010;445:51-56..
- 25.Hunter J.D. Matplotlib: A 2d graphics environment. Comput Sci Eng. 2007;9:90–95. [Google Scholar]
- 26.Mohamed A.S., Ruangskul M.N., Awan M.J. Quality assurance assessment of diagnostic and radiation therapy-simulation CT image registration for head and neck radiation therapy: anatomic region of interest-based comparison of rigid and deformable algorithms. Radiology. 2015;274:752–763. doi: 10.1148/radiol.14132871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guy C.L., Weiss E., Che S. Evaluation of image registration accuracy for tumor and organs at risk in the thorax for compliance with TG 132 recommendations. Adv Radiat Oncol. 2019;4:177–185. doi: 10.1016/j.adro.2018.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motegi K., Tachibana H., Motegi A. Usefulness of hybrid deformable image registration algorithms in prostate radiation therapy. J Appl Clin Med Phys. 2019;20:229–236. doi: 10.1002/acm2.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarudis S., Karlsson A., Bibac D. Evaluation of deformable image registration accuracy for CT images of the thorax region. Phys Med. 2019;57:191–199. doi: 10.1016/j.ejmp.2018.12.030. [DOI] [PubMed] [Google Scholar]