Abstract
Retroviruses infect almost all vertebrates, from humans to domestic and farm animals, from primates to wild animals, where they cause severe diseases, including immunodeficiencies, neurological disorders, and cancer. Nonhuman retroviruses have also been recently associated with human diseases. To date, no effective treatments are available; therefore, finding retrovirus-specific therapeutic targets is becoming an impelling issue. G-Quadruplexes are four-stranded nucleic acid structures that form in guanine-rich regions. Highly conserved G-quadruplexes located in the long-terminal-repeat (LTR) promoter of HIV-1 were shown to modulate the virus transcription machinery; moreover, the astonishingly high degree of conservation of G-quadruplex sequences in all primate lentiviruses corroborates the idea that these noncanonical nucleic acid structures are crucial elements in the lentiviral biology and thus have been selected for during evolution. In this work, we aimed at investigating the presence and conservation of G-quadruplexes in the Retroviridae family. Genomewide bioinformatics analysis showed that, despite their documented high genetic variability, most retroviruses contain highly conserved putative G-quadruplex-forming sequences in their promoter regions. Biophysical and biomolecular assays proved that these sequences actually fold into G-quadruplexes in physiological concentrations of relevant cations and that they are further stabilized by ligands. These results validate the relevance of G-quadruplexes in retroviruses and endorse the employment of G-quadruplex ligands as innovative antiretroviral drugs. This study indicates new possible pathways in the management of retroviral infections in humans and animal species. Moreover, it may shed light on the mechanism and functions of retrovirus genomes and derived transposable elements in the human genome.
Keywords: retroviruses, G-quadruplex, genome structure, LTR promoter, conservation
Retroviruses (RVs) are the most ancient known viruses: their origin dates back to more than 450 million years ago.1 They are multifaceted viruses: they infect almost all vertebrates, ranging from humans to small animals (e.g., domestic cats and mice), farm animals (e.g., poultry, cattle, and goats), different primates, and other animals (e.g., horses and fishes). In all these organisms, RVs cause severe diseases, including immunodeficiencies, neurological disorders, and different types of cancer, representing a major threat for all species; to date, no specific and effective treatments are available.2 In addition, nonhuman RVs have been recently associated with human diseases by accidental infection, such as sporadic human breast cancer,3 or by ingestion of RV-infected meat (cattle and poultry), especially in immunocompromised individuals.4 Therefore, finding targets for therapeutic treatment of RVs is becoming an impellent issue.
The distinctive feature of RVs is retrotranscription of the two positive, single-stranded RNA genome filaments by the viral reverse-transcriptase (RT) enzyme; the generated double-stranded DNA is integrated into the host DNA to form the provirus (Figure 1A). The proviral genome is next transcribed and translated to form new virions.5 When viral-genome integration occurs in somatic cells, RVs are classified as exogenous (XRVs); conversely, after occasional integration into the host germline and concurrent disruption of key viral genes, RVs may become endogenous (ERVs). XRVs are mainly organized into two subfamilies, Orthoretrovirinae and Spumavirinae, which differ in retrotranscription timing: the first includes six genera, namely alpha-, beta-, delta-, gamma-, and epsilon-RVs and lentiviruses, whereas the second comprises the spumavirus genus.2
Figure 1.
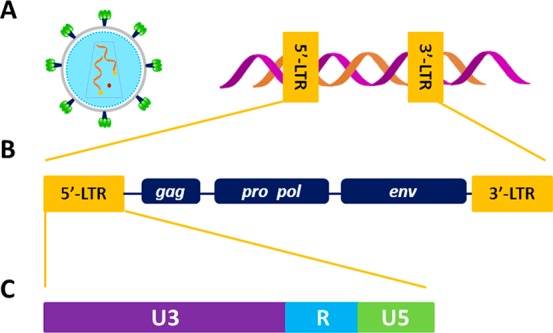
RV structure and genome organization. (A) Simplified model of an RV virion (left) and of the integrated provirus (right). (B) RV-provirus organization. (C) Regions of the 5′-LTR promoter.
The basic provirus organization is made of four coding genes, gag, pro, pol, and env, flanked by two identical untranslated regions, the long terminal repeats (LTRs, Figure 1B). Complex RVs also contain additional genes encoding for accessory proteins. The 5′-LTR is the control center for retroviral gene expression, consisting of three sections, U3, R, and U5 (Figure 1C). The U3 region, which includes binding sites for transcription factors, represents the RV-unique promoter.6 In the human immunodeficiency virus type 1 (HIV-1), we demonstrated that the LTR-U3 guanine (G)-rich region adopts noncanonical secondary structures, namely, G-quadruplexes (G4s).7 G4s may form within G-rich strands of nucleic acids when four Gs are linked together through Hoogsteen-type hydrogen bonds to assemble in self-stacked G-tetrads coordinated by monovalent cations.8 In HIV-1, the fine-tuning of G4 structures due to cellular proteins has been directly correlated to the regulation of viral transcription: stabilization and unfolding of G4s silence and promote transcription, respectively.9,10 Moreover, G4 ligands strongly reduce virus propagation.11,12 Interestingly, despite the typical great variability of the RV genomes, G-clusters in the LTR are highly conserved in all primate lentiviruses.13 We observed that the presence of G4s has been selected throughout evolution, suggesting an active and central role in lentivirus biology. G4 correlation with transcription-factor binding sites suggests exploitation of structural conserved elements as mechanosensors in the regulation of key viral steps.13 In general, bioinformatics studies traced putative G4-forming sequences (PQSs) in almost all human viruses: most of these viral PQSs are characterized by high degrees of conservation and statistically significant distributions, implying essential biological roles.14 Altogether, these findings show that despite the large mutation rates of viruses, G4s represent key elements in the viral life cycle and consequently are interesting targets in the development of innovative drugs.
In this context, with the purpose of examining the presence and role of G4s in the retroviral machinery and of ultimately identifying new targets for antiretroviral therapy, here we sought to investigate the G4 distribution and conservation in the whole Retroviridae family, and we present a comprehensive analysis of G4s within the RV genomes. Using genomewide bioinformatic analysis, we show that all RV genera contain PQSs. PQSs in the 5′-LTR promoter were focused on and investigated for their ability to actually fold into G4s. We demonstrate that, despite plentiful differences among RVs, G4s in regulatory regions represent a feature common to all genera.
Results
Putative Quadruplex-Forming Sequences (PQSs) in the LTR-Promoter Regions of Most RVs
We initially investigated the presence of PQSs in the full-length genomes of all RVs, with the exception of lentiviruses as that genus had been previously examined for the presence of G4s.13 Analysis was performed using the QuadBase2 web server,15 which allows flexible customization of loop length and inclusion of bulges, as some G4s have been reported to form even in the presence of noncontinuous Gs within G-runs.16,17 We searched for sequences located in both the forward and reverse strands of the RV integrated genomes characterized by (i) at least 3 Gs in each run, (ii) continuous or 1-nucleotide-bulged G-runs, and (iii) 1 to 12 nucleotide-long loops (G3L1–12). All the viruses investigated in this study are listed in Table S1.
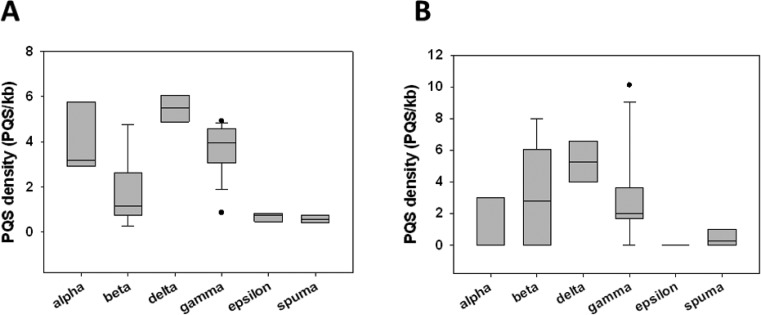
PQSs were observed in all RV genera, for a total of 1050 sequences over 48 analyzed viruses (Figure 2A). The average number of observed PQSs per genus ranged from 7 to 48. Delta-RVs were particularly enriched in PQSs, with very low variability among viruses; conversely, epsilon- and spuma-RVs showed 7- and 5-fold lower PQS amounts, respectively. Alpha-, beta-, and gamma-RV genera displayed great variability among the different viruses, with average PQSs-per-virus values of 20, 15, and 26, respectively.
Figure 2.
Box plots showing average PQS densities (PQS/Kb) in full-length genomes (A) and LTR regions (B) of RVs.
We previously observed that G4s in the LTR of the HIV-1 provirus act as regulators of viral transcription.7 The presence and pattern of G4-forming sequences is extremely conserved in all primate lentiviruses,13 thereby pointing toward a key regulatory role of LTR G4s in the whole lentivirus genus. Consequently, we here focused our analysis on the LTR region of RVs: LTR PQSs were found in all RV genera, except for the epsilon-RVs, for a total of 65 PQSs over 48 analyzed viruses; delta-RVs were confirmed to be the most enriched in PQSs among all genera (Figure 2B). About 80% of the PQSs (50 out of 65) were located in the reverse strand. All found sequences are reported in Table S2.
We also observed that the majority of PQSs (∼70%) were located in the U3 region, just upstream of the transcription start site (Figure 3). The U3 region plays a crucial role in the induction of viral transcription, as it comprises the unique promoter and transcription-factor binding sites: in this regard, we have proved that G4 sequences significantly overlap with Sp1 binding sites in the HIV-1 and primate lentiviruses.13
Figure 3.
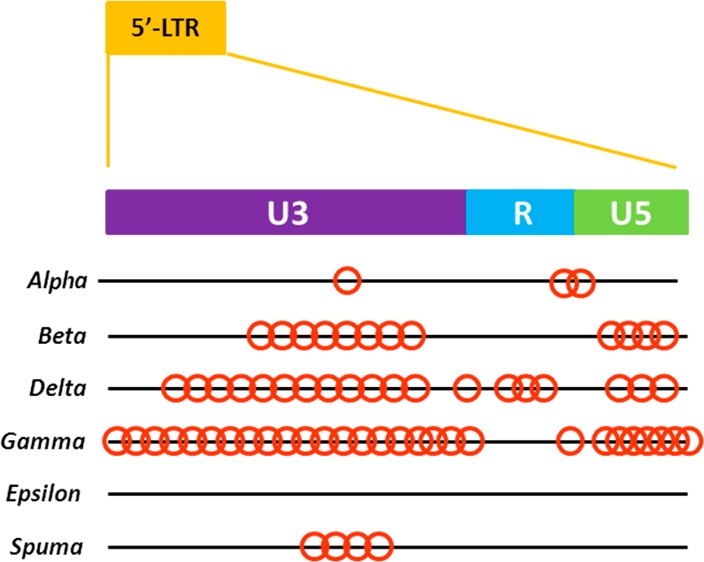
PQS distribution along the LTR regions of RVs. Each red circle indicates one PQS.
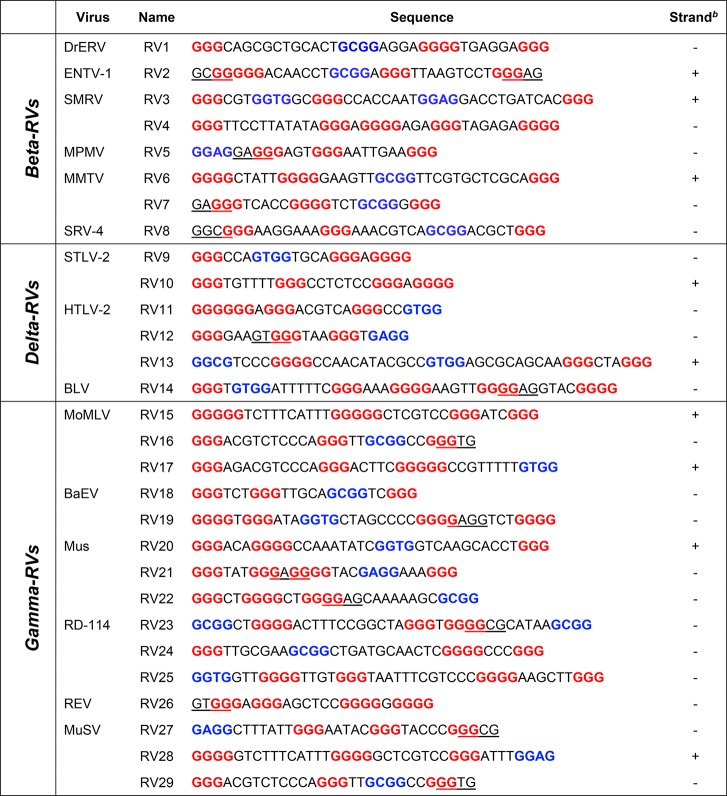
From this first screening, sequences containing more than one bulged G-tract were excluded, as the presence of too many bulged G-tracts has been reported to reduce G4 stability and even prevent their formation.18 Consequently, 29 sequences were obtained, distributed as follows: 8 in the beta-RV genus, 6 in the delta-RV genus, and 15 in the gamma-RV genus (Table 1). The observed sequences greatly varied in terms of length (22–44 nucleotides) and number of G-tracts (4–6). However, similarities were found in Mo-MLV and MuSV RVs, where the RV16 and RV29 sequences had the same base composition, and RV15 and RV28 differed by just three nucleotides in the last G-tract. Six sequences comprised continuous G-tracts, whereas the remaining 23 contained a bulged G-tract. Moreover, loop composition was quite mixed, as the sequences included very short loops (L ≤ 5 nt, in RV4, RV7, RV9, RV12, RV18, and RV21) and very long ones (9 < L > 12 nt in RV15 and RV25), whereas the remaining presented miscellaneous loop organization.
Table 1. PQS Analysis Performed with QuadBase2 within the LTR Regions of RVsa.
G3 tracts are shown in red and bold, nonoverlapping bulged G3 tracts (e.g., GGXG) are shown in blue and bold, and overlapping bulged G3 tracts (e.g., GXGGG) are underlined.
PQS location: “+” indicates the forward strand, and “–” indicates the reverse strand.
Highly Conserved PQSs in RV LTRs
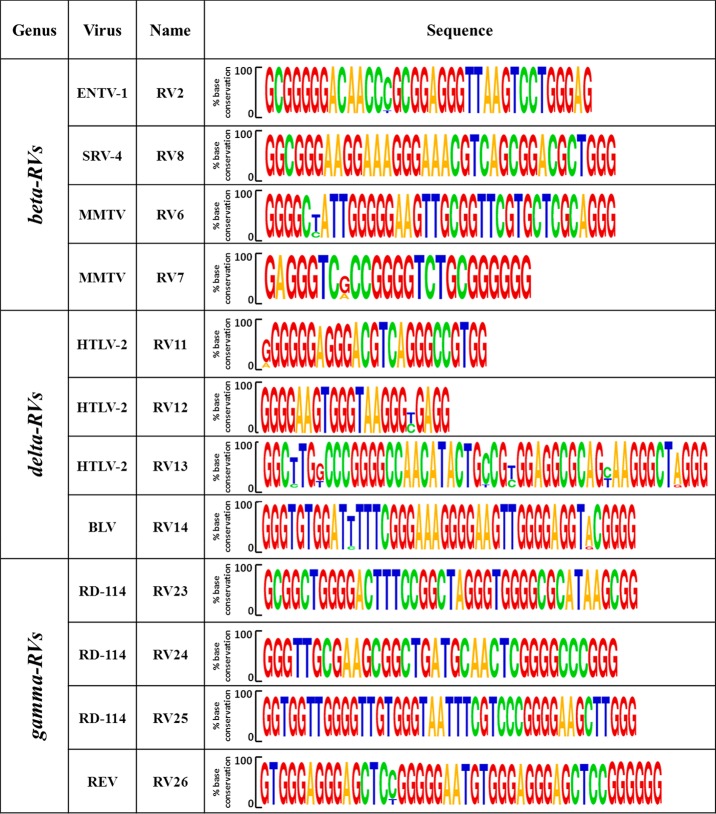
To assess the relevance of PQSs, we performed base-conservation analysis. Generally, RVs show high genetic variability, mainly as a result of error-prone proviral-genome synthesis and recombination between the two RNA copies during retrotranscription.19 Nonetheless, conservation analysis, conducted on all RVs for which five or more complete LTR sequences were available (Table S3), showed an extremely high degree of G-base conservation, especially within G-tracts that are likely involved in G4 formation (Figure 4). These results corroborate the data obtained for lentiviruses13 and herpesviruses,20−22 further suggesting that G4s are key elements in the viral cycle and therefore have been selected for during viral-genome evolution.
Figure 4.
Base conservation of putative G4-forming sequences within strains of each RV species. Consensus sequences were obtained by alignment of at least five sequences.
RV-LTR-PQS Folding into G4
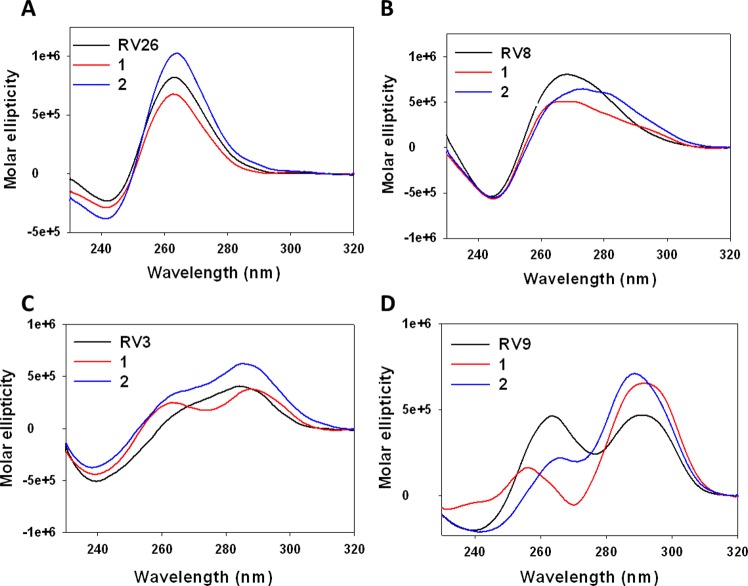
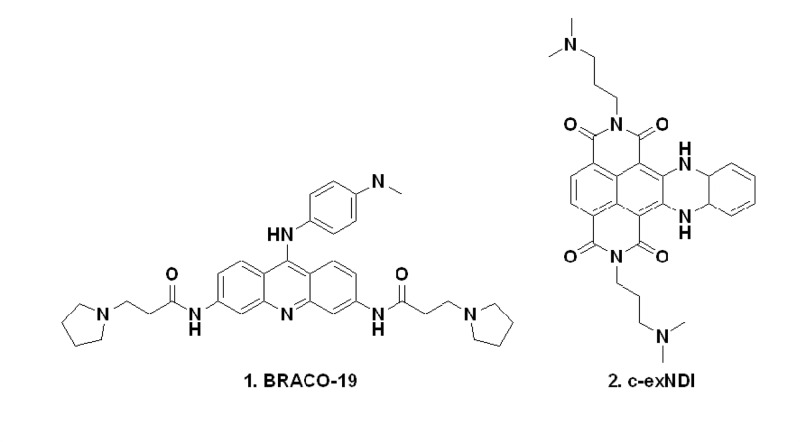
The actual ability of PQSs to fold into G4s was initially ascertained by circular-dichroism (CD) spectroscopy, as signature CD spectra are available for G4s.23 Representative CD spectra showing a G4 RV, a non-G4 RV and two different mixed G4 RVs are shown in Figure 5; CD spectra of all the analyzed sequences and their melting profiles are reported, organized by genus, in Figures S1–S4. Most of the examined oligonucleotides displayed clear-cut G4 signatures, such as RV26 (Figure 5A) and RV5 and RV7 (Figure S1). The majority of the sequences, however, were characterized by complex CD profiles (Figures 5C,D and S1–S4), likely indicating the coexistence of multiple conformations, corroborating the high dynamism and polymorphism reported for G4 DNA structures. RV3, for instance, showed two different transitions at 260 and 290 nm (Figure 5C), which may indicate the contribution of a parallel and an antiparallel conformation, respectively.23 Five sequences, RV2, RV6, RV8, RV13, and RV27, displayed a broad peak in the 260–280 nm wavelength range, indicating a prevalent non-G4 conformation (Figures 5B, S1, S2, and S4).24 We also evaluated the effects of two different compounds, BRACO-19 (B19, compound 1, Figure 6) and a core-extended naphtalenediimide (c-exNDI, compound 2, Figure 6), on RV G4 topology. Both molecules have been employed as G4 ligands in viruses:251 has been reported to inhibit HIV-1 both in lytic and latent infections,11,26 and 2 has been shown to preferentially bind and stabilize viral G4s over cellular ones.12,27 CD experiments were conducted in the presence of 4 equiv of compounds and showed diverse effects: in the case of the RV3 sequence, for example, 1 strongly increased the molar ellipticity at 260 nm, suggesting the preferential binding for one of the possible conformations. In contrast, 2 enhanced the peak at 290 nm, providing a different CD spectrum (Figure 5C). Peculiar effects were also observed for other sequences: for example, in RV9, in which the peaks at 260 and 290 nm display similar intensities, 1 totally abolished the peak at 260 nm, whereas 2 enhanced both transitions (Figure 5D). Such structure-related behaviors imply that the two compounds may exert their G4 stabilizing activities through different binding modes.
Figure 5.
Representative CD spectra of RV G4 sequences in the absence (black line) or presence of G4 ligands 1 (red line) and 2 (blue line). (A) G4 CD spectrum, characterized by a maximum peak at λ = 260 nm and a minimum one at λ = 240 nm, which define a parallel conformation. (B) Non-G4 CD spectrum, characterized by a broad signal at 260 < λ < 280 nm. (C–D) Two different mixed-G4 CD profiles.
Figure 6.
Chemical structures of the G4 ligands B19 (1) and c-exNDI (2) employed in this study.
To evaluate the stability of the RV G4s, we next performed CD thermal-denaturation experiments in the temperature (T) range of 20–95 °C. RV26 was the most stable G4, with a melting temperature (Tm) of 74.3 °C, whereas the least stable was RV24 (Tm = 41 °C). Moreover, plotting of the molar ellipticity versus T revealed two major melting transitions for hybrid G4s, at λ = 260 and 290 nm, the Tm of which are reported in Table 2. The occurrence of multiple melting transitions confirms the coexistence of different conformations in solution, each characterized by different Tm values. In some cases, such as with the RV9 sequence, two very clear transitions and thus Tm values were obtained, whereas in the other case, such as with RV3, the presence of different species was so complex that it precluded the determination of single Tm values. In general, all G4-forming sequences displayed Tm > 37 °C, suggesting that RV G4s can stably fold in conditions that are close to the physiological ones. CD melting analysis in the presence of compounds showed a general stabilization effect on G4s, the Tm values of which were generally enhanced after G4-ligand treatment (Table 2). The different effects induced by the two compounds on the different RV G4s suggest the existence of different G4-binding mechanisms.
Table 2. CD Tm Values of RV G4s in the Absence and Presence of G4 Ligands 1 and 2a.
|
Tm (°C) |
ΔTm (°C) |
|||||
|---|---|---|---|---|---|---|
| — | 1 | 2 | 1 | 2 | ||
| beta-RVs | RV1 | 48.1 ± 0.9 | 68.9 ± 0.2 | 60.6 ± 0.8 | 20.8 | 12.5 |
| RV2 | ND | ND | ND | |||
| RV3 | ND | ND | ND | |||
| RV4 | 67.1 ± 1.2 | >90 | >90 | >22.9 | >22.9 | |
| RV5 | 48.0 ± 1.9 | 68.9 ± 3.1 | 85.9 ± 1.2 | 20.9 | 37.9 | |
| ND | ND | 62.3 ± 1.1 | ND | ND | ||
| RV6 | ND | ND | ND | |||
| RV7 | 63.9 ± 0.8 | 75.8 ± 0.9 | >90 | 11.9 | >26.1 | |
| RV8 | ND | ND | ND | |||
| delta-RVs | RV9 | 65.1 ± 0.3 | >90 | >90 | >24.9 | >24.9 |
| 64.9 ± 0.3 | >90 | >90 | >25.1 | >25.1 | ||
| RV10 | 66.4 ± 1.3 | 83.8 ± 2.1 | >90 | 17.4 | >20.6 | |
| 48.9 ± 0.8 | 72.1 ± 0.9 | 70.3 ± 2.5 | 23.2 | 24.4 | ||
| RV11 | 61.4 ± 0.3 | 79.2 ± 0.7 | ND | 14.6 | ND | |
| 56.6 ± 2.1 | 69.0 ± 3.8 | 63.4 ± 0.3 | 12.4 | 6.8 | ||
| RV12 | 63.1 ± 0.4 | ND | ND | ND | ND | |
| 63.3 ± 0.4 | 66.3 ± 0.1 | 66.9 ± 0.8 | 3.2 | 3.8 | ||
| RV13 | ND | ND | ND | |||
| RV14 | 65.5 ± 0.8 | >90 | >90 | >24.5 | >24.5 | |
| gamma-RVs | RV15 | 55.4 ± 0.1 | >90 | >90 | >34.6 | >34.6 |
| ND | 67.0 ± 0.1 | 62.1 ± 2.6 | ND | ND | ||
| RV16 | ND | ND | ND | ND | ND | |
| 53.3 ± 1.4 | 63.4 ± 1.0 | 68.5 ± 2.3 | 10.1 | 15.2 | ||
| RV17 | 52.3 ± 0.8 | 86.7 ± 1.0 | 57.0 ± 3.4 | 33.7 | 4.7 | |
| RV18 | 59.9 ± 0.4 | 76.5 ± 0.1 | 70.6 ± 1.0 | 16.6 | 10.7 | |
| RV19 | 66.8 ± 0.1 | 77.6 ± 0.6 | 85.1 ± 0.1 | 10.8 | 18.3 | |
| RV20 | ND | ND | ND | |||
| RV21 | 56.8 ± 0.1 | ND | 65.6 ± 1.8 | ND | 8.8 | |
| 56.1 ± 0.1 | 60.0 ± 2.4 | 69.6 ± 2.0 | 3.9 | 13.5 | ||
| RV22 | 54.2 ± 0.8 | ND | ND | ND | ND | |
| 54.7 ± 0.6 | 64.9 ± 2.9 | 75.9 ± 3.9 | 10.2 | 21.2 | ||
| RV23 | 56.9 ± 2.7 | 83.8 ± 3.9 | 81.4 ± 2.2 | 25 | 22.6 | |
| RV24 | 41.2 ± 0.2 | 50.8 ± 0.1 | 43.9 ± 3.0 | 9.6 | 2.7 | |
| RV25 | 53.4 ± 0.1 | >90 | >90 | >35.6 | >35.6 | |
| RV26 | 73.6 ± 0.6 | >90 | >90 | >16.4 | >16.4 | |
| RV27 | ND | ND | ND | |||
| RV28 | >90 | >90 | >90 | ND | ND | |
| ND | 58.5 ± 4.5 | 71.0 ± 0.5 | ||||
| RV29 | ND | ND | ND | ND | ND | |
| 53.3 ± 1.4 | 63.4 ± 1.0 | 68.5 ± 2.3 | 10.1 | 15.2 | ||
Data are reported as mean values ± SD from at least two independent experiments. In cases of double transitions, Tm values calculated at λ = 260 nm (first value) and 290 nm (second value) are shown.
Dimethylsulfate (DMS)-footprinting analysis was next carried out to evaluate the G bases involved in G4 formation. We selected seven representative sequences, according to the folding characteristics observed in CD analysis: RV26, RV7, and RV5 for the parallel conformation; RV18 for a predominant antiparallel topology; and RV9, RV22, and RV12 for mixed arrangements. Oligonucleotides were folded in the presence and absence of KCl and treated with DMS to analyze the G residues protected from DMS-induced methylation. In the absence of K+ ions, cleavage to all Gs was observed, suggesting an unstructured oligonucleotide form. On the other hand, in the presence of KCl, all analyzed sequences showed protection of three Gs in each G-tract, indicating their involvement in G4 formation. On the basis of the DMS-footprinting pattern, we propose that each analyzed RV G4 consists of three planar tetrads formed by four contiguous or bulged G-runs (Figure S5). Deeper investigation into the secondary arrangement could allow the design of specific ligands able to selectively bind the single RV G4s.
Stalling of Polymerase Progression by RV-LTR G4s
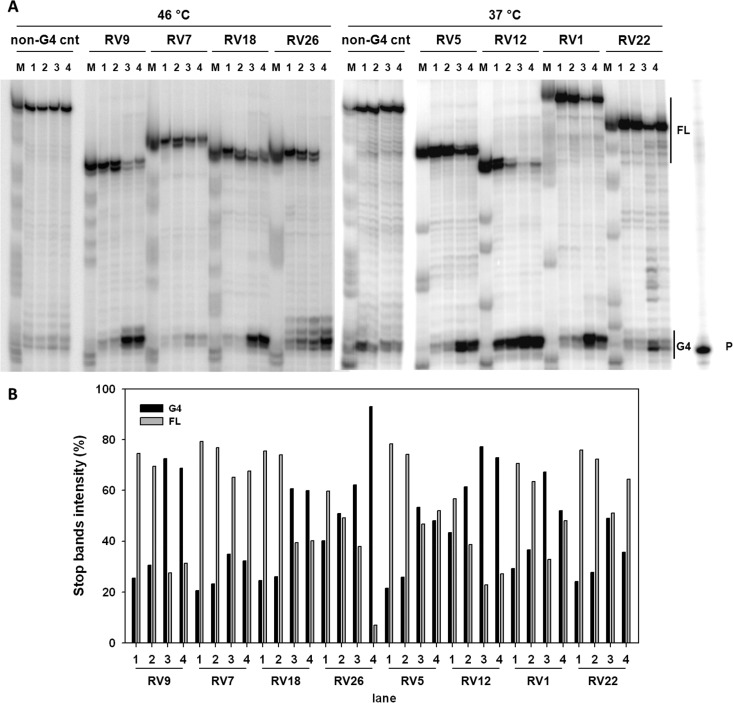
To investigate whether the identified RV G4s were able to stall polymerase progression, a Taq-polymerase stop assay was performed. Eight RV G4-forming sequences, belonging to different genera and characterized by different G4 folding topologies and stability, were selected as reported above. Extended RV G4 templates (Table S4), containing primer-annealing sequences at the 3′-ends, were annealed to the primer (Table S4) and incubated with Taq polymerase for 30 min at the indicated temperature. The chosen sequences were investigated in the absence and presence of K+ to establish G4 formation and in the presence of G4 ligands to assess ligand-induced G4 stabilization. The two investigated ligands were used at different concentrations (1 at 100 μM and 2 at 100 nM), according to their previously observed activity.11,12 In the presence of 100 mM K+ (Figure 7A, lane 2), all RV G4 templates stopped the polymerase at the most 3′-G-tract involved in G4 formation, indicating that K+ stimulates G4 folding, which in turn blocks polymerase progression. Upon addition of G4 ligands, the intensity of the G4 stop bands highly increased in all instances (Figure 7A, lanes 3 and 4), along with considerable reduction of the full-length amplicons, thus corroborating effective stabilization of the RV G4s by both compounds. In contrast, both ligands had no effect on a DNA template unable to fold into G4 (Figure 7A, non-G4 cnt, lanes 3 and 4), indicating that the observed polymerase inhibition was G4-dependent. Quantification of the stop sites corresponding to G4s and of the full-length products is shown in Figure 7B. Overall, these data are in line with those obtained by CD analysis and confirm the ability of the chosen sequences to fold into G4 and get stabilized by G4 ligands.
Figure 7.
Representative Taq-polymerase stop assay of RV G4 sequences. (A) Templates amplified by Taq polymerase at the indicated temperature in the absence (lane 1) or presence of 100 mM K+ alone (lane 2) or with G4 ligand 1 (lane 3) or 2 (lane 4). A template sequence (non-G4 cnt) made of a scrambled sequence unable to fold into a G4 was also used as an internal control. Lane P: unreacted labeled primer. Lane M: ladder of markers obtained by the Maxam and Gilbert sequencing protocol carried out on the amplified strand complementary to the template strand. Vertical bars indicate G4-specific Taq-polymerase stop sites. (B) Quantification of lanes shown in panel (A). Quantification of stop bands corresponding to G4 and of the full-length amplification product (FL) is shown.
Discussion
In the past few years, interest in the characterization of G4 structures and their role within viral genomes has greatly increased, providing new directions in the management of viral infections. In this context, our group previously demonstrated that the HIV-1 transcription machinery is modulated by the tuned folding and unfolding of G4s located in the U3 region of LTR promoter. We proved that the G4 folding pattern is highly conserved not only among almost 1000 HIV-1 strains7 but also among all primate lentiviruses,13 indicating G4s are crucial elements in viral evolution.
In this work, we investigated the presence of G4 structures in the whole Retroviridae family. In line with previously collected data on lentiviruses,13 we found PQSs in the LTRs of all RVs except for the epsilon-RVs. This last genus is the least represented, including only three virus species: it is tempting to speculate that the absence of G4s has impacted the evolution of this genus. As for the other RVs (the G4-containing RVs), we demonstrated that their PQSs (i) are well conserved, (ii) can actually adopt stable G4 arrangements, and (iii) are able to stall the polymerase enzyme.
In retrovirology, base-conservation analysis represents a critical issue, considering the high mutation rates of RVs. The limited availability of deposited sequences for most RVs hampers comprehensive conservation analysis; however, our data collected in this and previous works13 clearly indicate that G4-forming sequences are conserved elements within each RV LTR, thus representing essential elements for the virus life-cycle. Moreover, considering that all RV LTRs are characterized by the presence of PQSs, it may be hypothesized that, although LTRs greatly differ in terms of primary sequences and length, their shared functional homology could be ascribed to structural conserved elements like G4s.
The LTR is responsible for the expression of viral genes and ultimately for virus replication; it has been widely demonstrated that sequence variation in LTRs affects the binding of transcription factors, thus altering transcription.28 Therefore, targeting the LTR may be effective in the treatment of infections, and to this end, the employment of G4 ligands represents a valuable approach. In this study, we demonstrated that all RV-LTR G4s are stabilized in vitro by G4 binders and that two different molecules stabilized a third of the selected sequences by over 20 °C. Furthermore, the Taq-polymerase stop assay revealed that this significant stabilization deeply impacts polymerase progression. Notably, compounds 1 and 2 exerted comparable in vitro effects on the HIV-1 sequences and, when tested in vivo, were able to greatly reduce virus propagation.11,12 These data support the investigation of G4 ligands as promising candidates of innovative antiretroviral drugs.
It is worth noting that development of anti-RV compounds is currently limited to HIV. However, human-health-threatening RVs are not restricted to lentiviruses; besides human viruses like HTLV or HFV, there is an increasing body of evidence that correlates nonhuman RVs with human diseases. For example, the insurgence of sporadic human breast cancer has been associated with MMTV infections;3 in addition, immunocompromised people could be exposed to nutrition-related RVs, like BLV or REV, which infect cattle and poultry, respectively.4,29 The identification of structurally conserved elements like G4s in RV genomes and the consequent possibility to target them with specific compounds may thus represent a turning point in the management of the widest range of retroviral infections in humans and also in animal species of interest, such as farm animals and pets.
An additional point of interest is that characterization of LTR G4s has implications in genetics because 8% of the human genome consists of LTR-transposable elements (TE), including ERVs and single LTR segments, which have become effective parts of the mammalian genome. A recent study reported that G4s enrich the LTRs of plant TEs and human ERVs, regulating transcription.30,31 The authors intriguingly suggest that TEs could be the vehicles by which PQSs have spread into the human genome.32 Considering that (i) LTRs contain the majority of PQSs found in TEs,32 and (ii) LTR elements in the human genome are derived from ancient RV infections, RVs could represent the primordial organisms that first developed G4 structures.
Our present work expands on the theme and substantiates the consistent presence of G4s in LTR elements.
Conclusions
The work proposed here provides a comprehensive overview of the presence of G4s in RV-LTR-promoter regions. It adds to the boosting recognition of G4s as widespread elements in the broadest range of organisms, from higher to lower eukaryotes and from plants to microorganisms.33−37 It follows that research on G4s in viral LTRs has two implications: first, the possibility to manage RV infections by developing innovative drugs and, second, the opportunity to unravel the ancestral mechanisms that regulate life as we know it today.
Experimental Section
Oligonucleotides and Compounds
All the oligonucleotides used in this work were purchased from Sigma-Aldrich (Milan, Italy) and are listed in Tables 1 and S4. B19 was obtained from ENDOTHERM (Saarbruecken, Germany), c-exNDI was synthesized and kindly provided by Professor Filippo Doria and Professor Mauro Freccero (University of Pavia).
G4 Analysis of RV Genomes
Prediction of G4-forming sequences on RV genomes and LTR regions was performed using the QuadBase2 web server.15 The search was restricted to G-tracts formed by 3 Gs (continuous or including 1 nucleotide bulge) and loops from 1 to 12 nucleotides.
Base-Conservation Analysis of Predicted G4-Forming Sequences
Predicted G4-forming sequences were analyzed in terms of base conservation by aligning sequences from PubMed. Accession numbers of the whole set of sequences are reported in Table S3. Conservation analysis was performed on RVs with five or more sequences available in databases. LOGO representation of base conservation was obtained by the WebLogo software.38
Circular-Dichroism Analysis
All the oligonucleotides used in this study (Table 1) were diluted to final concentrations of 3 μM in lithium cacodylate buffer (10 mM, pH 7.4) and KCl 100 mM. Samples were heated at 95 °C for 5 min and then slowly cooled to room temperature. Where indicated, compounds were added in 4 equiv, 4 h after denaturation. CD spectra were recorded on a Chirascan-Plus (Applied Photophysics, Leatherhead, U.K.) equipped with a Peltier temperature controller using a quartz cell with a 5 mm optical-path length. Thermal-unfolding experiments were recorded from 230 to 320 nm over a temperature range of 20–90 °C. Acquired spectra were baseline-corrected for signal contribution from the buffer, and the observed ellipticities were converted to mean residue ellipticity according to θ = degree × cm2 × dmol–1 (molar ellipticity). Tm values were calculated according to the van’t Hoff equation applied for a two-state transition from a folded state to an unfolded state
DMS-Footprinting Assay
Oligonucleotides were 5′-end-labeled with [γ-32P]ATP by T4 polynucleotide kinase (Thermo Scientific, Milan, Italy) at 37 °C for 30 min and purified using MicroSpin G-25 columns (GE Healthcare Europe, Milan, Italy). They were next resuspended in lithium cacodylate buffer (10 mM, pH 7.4) in the absence or presence of 100 mM KCl, heat-denatured, and cooled to room temperature. Samples were then treated with dimethylsulfate (DMS, 0.5% in ethanol) for 5 min at room temperature, and the reaction was stopped by the addition of 10% glycerol and β-mercaptoethanol before the samples were loaded onto a 15% native polyacrylamide gel. DNA bands were localized via autoradiography, excised, and eluted in water overnight. The supernatants were recovered, ethanol-precipitated, and treated with piperidine 10% solution for 30 min at 90 °C. Reaction products were analyzed on 20% denaturing polyacrylamide gels, visualized by phosphorimaging analysis, and quantified by ImageQuant TL software (GE Healthcare Europe, Milan, Italy).
Taq-Polymerase Stop Assay
The Taq-polymerase stop assay was performed according to previously described procedures.7 The labeled primer (final concentration of 72 nM) was annealed to the template (final concentration of 36 nM, Table S4) in lithium cacodylate buffer (10 mM, pH 7.4) in the presence or absence of KCl 100 mM by heating at 95 °C for 5 min. After gradual cooling to room temperature, the samples were incubated, where indicated, with 1 (1 μM) or 2 (100 nM) at room temperature overnight. For primer extension, AmpliTaq Gold DNA polymerase (2U per reaction; Applied Biosystems, Carlsbad, CA) was employed at the indicated temperature for 30 min. Reactions were stopped by ethanol precipitation, and primer-extension products were separated on a 16% denaturing gel and finally visualized by phosphorimaging (Typhoon FLA 9000, GE Healthcare, Milan, Italy). Markers were prepared on the basis of the Maxam and Gilbert sequencing protocol.39
Acknowledgments
This work was supported by grants to S.N.R. from the European Research Council (ERC Consolidator grant number 615879) and the Bill and Melinda Gates Foundation (grant numbers OPP1035881 and OPP1097238).
Glossary
Abbreviations Used
- RV
retrovirus
- RT
reverse transcriptase
- XRV
exogenous retrovirus
- ERV
endogenous retrovirus
- LTR
long terminal repeat
- G4
G-quadruplex
- PQS
putative G-quadruplex-forming sequence
- CD
circular dichroism
- DMS
dimethyl sulfate
- TE
transposable element
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.9b00011.
Analyzed RVs, obtained sequences, accession numbers of all RVs, oligonucleotide sequences used in the biophysic assays, CD spectra, and DMS-footprinting analysis (PDF)
Author Contributions
E.R., M.T., R.P., and M.N. performed the experiments; S.N.R. conceived the work; and E.R. and S.N.R. wrote the manuscript. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Hayward A. (2017) Origin of the retroviruses: when, where, and how?. Curr. Opin. Virol. 25, 23–27. 10.1016/j.coviro.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood A. D.; Ishida Y.; O’Brien S. P.; Roca A. L.; Eiden M. V. (2018) Transmission, Evolution, and Endogenization: Lessons Learned from Recent Retroviral Invasions. Microbiol. Mol. Biol. Rev. 82, e00044-17 10.1128/MMBR.00044-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braitbard O.; Roniger M.; Bar-Sinai A.; Rajchman D.; Gross T.; Abramovitch H.; La Ferla M.; Franceschi S.; Lessi F.; Naccarato A. G.; Mazzanti C. M.; Bevilacqua G.; Hochman J. (2016) A new immunization and treatment strategy for mouse mammary tumor virus (MMTV) associated cancers. Oncotarget 7, 21168–21180. 10.18632/oncotarget.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaya-Galan N. N.; Corredor-Figueroa A. P.; Guzman-Garzon T. C.; Rios-Hernandez K. S.; Salas-Cardenas S. P.; Patarroyo M. A.; Gutierrez M. F. (2017) Bovine leukaemia virus DNA in fresh milk and raw beef for human consumption. Epidemiol. Infect. 145, 3125–3130. 10.1017/S0950268817002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P.; Coffin J. M. (2008) Effects of Retroviruses on Host Genome Function. Annu. Rev. Genet. 42, 709–732. 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- Wu Y. (2004) HIV-1 gene expression: lessons from provirus and non-integrated DNA. Retrovirology 1, 13. 10.1186/1742-4690-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone R.; Nadai M.; Frasson I.; Poe J. A.; Butovskaya E.; Smithgall T. E.; Palumbo M.; Palu G.; Richter S. N. (2013) A dynamic G-quadruplex region regulates the HIV-1 long terminal repeat promoter. J. Med. Chem. 56, 6521–6530. 10.1021/jm400914r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D.; Lipps H. J. (2015) G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 43, 8627–8637. 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosoni E.; Frasson I.; Scalabrin M.; Perrone R.; Butovskaya E.; Nadai M.; Palu G.; Fabris D.; Richter S. N. (2015) Nucleolin stabilizes G-quadruplex structures folded by the LTR promoter and silences HIV-1 viral transcription. Nucleic Acids Res. 43, 8884–8897. 10.1093/nar/gkv897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalabrin M.; Frasson I.; Ruggiero E.; Perrone R.; Tosoni E.; Lago S.; Tassinari M.; Palù G.; Richter S. N. (2017) The cellular protein hnRNP A2/B1 enhances HIV-1 transcription by unfolding LTR promoter G-quadruplexes. Sci. Rep. 7, 45244. 10.1038/srep45244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone R.; Butovskaya E.; Daelemans D.; Palu G.; Pannecouque C.; Richter S. N. (2014) Anti-HIV-1 activity of the G-quadruplex ligand BRACO-19. J. Antimicrob. Chemother. 69, 3248–3258. 10.1093/jac/dku280. [DOI] [PubMed] [Google Scholar]
- Perrone R.; Doria F.; Butovskaya E.; Frasson I.; Botti S.; Scalabrin M.; Lago S.; Grande V.; Nadai M.; Freccero M.; Richter S. N. (2015) Synthesis, Binding and Antiviral Properties of Potent Core-Extended Naphthalene Diimides Targeting the HIV-1 Long Terminal Repeat Promoter G-Quadruplexes. J. Med. Chem. 58, 9639–9652. 10.1021/acs.jmedchem.5b01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone R.; Lavezzo E.; Palù G.; Richter S. N. (2017) Conserved presence of G-quadruplex forming sequences in the Long Terminal Repeat Promoter of Lentiviruses. Sci. Rep. 7, 2018. 10.1038/s41598-017-02291-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavezzo E.; Berselli M.; Frasson I.; Perrone R.; Palù G.; Brazzale A. R.; Richter S. N.; Toppo S. (2018) G-quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide. PLOS Comput. Biol. 14, e1006675 10.1371/journal.pcbi.1006675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhapola P.; Chowdhury S. (2016) QuadBase2: web server for multiplexed guanine quadruplex mining and visualization. Nucleic Acids Res. 44, W277–W283. 10.1093/nar/gkw425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier M.; Moya-Torres A.; Krahn N. J.; McDougall M. D.; Orriss G. L.; McRae E. K. S.; Booy E. P.; McEleney K.; Patel T. R.; McKenna S. A.; Stetefeld J. (2018) Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge. Nucleic Acids Res. 46, 5319–5331. 10.1093/nar/gky307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nicola B.; Lech C. J.; Heddi B.; Regmi S.; Frasson I.; Perrone R.; Richter S. N.; Phan A. T. (2016) Structure and possible function of a G-quadruplex in the long terminal repeat of the proviral HIV-1 genome. Nucleic Acids Res. 44, 6442–6451. 10.1093/nar/gkw432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukundan V. T.; Phan A. T. (2013) Bulges in G-Quadruplexes: Broadening the Definition of G-Quadruplex-Forming Sequences. J. Am. Chem. Soc. 135, 5017–5028. 10.1021/ja310251r. [DOI] [PubMed] [Google Scholar]
- Rethwilm A.; Bodem J. (2013) Evolution of Foamy Viruses: The Most Ancient of All Retroviruses. Viruses 5, 2349–2374. 10.3390/v5102349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas B.; Kandpal M.; Jauhari U. K.; Vivekanandan P. (2016) Genome-wide analysis of G-quadruplexes in herpesvirus genomes. BMC Genomics 17, 949. 10.1186/s12864-016-3282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artusi S.; Nadai M.; Perrone R.; Biasolo M. A.; Palu G.; Flamand L.; Calistri A.; Richter S. N. (2015) The Herpes Simplex Virus-1 genome contains multiple clusters of repeated G-quadruplex: Implications for the antiviral activity of a G-quadruplex ligand. Antiviral Res. 118, 123–131. 10.1016/j.antiviral.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas B.; Kumari P.; Vivekanandan P. (2018) Pac1 Signals of Human Herpesviruses Contain a Highly Conserved G-Quadruplex Motif. ACS Infect. Dis. 4, 744–751. 10.1021/acsinfecdis.7b00279. [DOI] [PubMed] [Google Scholar]
- Vorlíčková M.; Kejnovská I.; Sagi J.; Renčiuk D.; Bednářová K.; Motlová J.; Kypr J. (2012) Circular dichroism and guanine quadruplexes. Methods 57, 64–75. 10.1016/j.ymeth.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Kypr J.; Kejnovska I.; Renciuk D.; Vorlickova M. (2009) Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 37, 1713–1725. 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero E.; Richter S. N. (2018) G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy. Nucleic Acids Res. 46, 3270–3283. 10.1093/nar/gky187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna-Przybylska D.; Sharma G.; Maggirwar S. B.; Bambara R. A. (2017) Deficiency in DNA damage response, a new characteristic of cells infected with latent HIV-1. Cell Cycle 16, 968–978. 10.1080/15384101.2017.1312225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegaro S.; Perrone R.; Scalabrin M.; Doria F.; Palu G.; Richter S. N. (2017) A core extended naphtalene diimide G-quadruplex ligand potently inhibits herpes simplex virus 1 replication. Sci. Rep. 7, 2341. 10.1038/s41598-017-02667-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs F. C.; Mehrens D.; Pomeroy S.; Goodenow M. M.; Wigdahl B. (1998) Human Immunodeficiency Virus Type 1 Long Terminal Repeat Quasispecies Differ in Basal Transcription and Nuclear Factor Recruitment in Human Glial Cells and Lymphocytes. J. Biomed. Sci. 5, 31–44. 10.1007/BF02253354. [DOI] [PubMed] [Google Scholar]
- Gyles C. (2016) Should we be more concerned about bovine leukemia virus?. Can. Vet. J. 57, 115–116. [PMC free article] [PubMed] [Google Scholar]
- Kejnovsky E.; Lexa M. (2014) Quadruplex-forming DNA sequences spread by retrotransposons may serve as genome regulators. Mob. Genet. Elements 4, e28084 10.4161/mge.28084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexa M.; Kejnovsky E.; Steflova P.; Konvalinova H.; Vorlickova M.; Vyskot B. (2014) Quadruplex-forming sequences occupy discrete regions inside plant LTR retrotransposons. Nucleic Acids Res. 42, 968–978. 10.1093/nar/gkt893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kejnovsky E.; Tokan V.; Lexa M. (2015) Transposable elements and G-quadruplexes. Chromosome Res. 23, 615–623. 10.1007/s10577-015-9491-7. [DOI] [PubMed] [Google Scholar]
- Griffin B. D.; Bass H. W. (2018) Review: Plant G-quadruplex (G4) motifs in DNA and RNA; abundant, intriguing sequences of unknown function. Plant Sci. 269, 143–147. 10.1016/j.plantsci.2018.01.011. [DOI] [PubMed] [Google Scholar]
- Vinyard W. A.; Fleming A. M.; Ma J.; Burrows C. J. (2018) Characterization of G-Quadruplexes in Chlamydomonas reinhardtii and the Effects of Polyamine and Magnesium Cations on Structure and Stability. Biochemistry 57, 6551–6561. 10.1021/acs.biochem.8b00749. [DOI] [PubMed] [Google Scholar]
- Harris L. M.; Monsell K. R.; Noulin F.; Famodimu M. T.; Smargiasso N.; Damblon C.; Horrocks P.; Merrick C. J. (2018) G-Quadruplex DNA Motifs in the Malaria Parasite Plasmodium falciparum and Their Potential as Novel Antimalarial Drug Targets. Antimicrob. Agents Chemother. 62, e01828-17 10.1128/AAC.01828-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guédin A.; Lin L. Y.; Armane S.; Lacroix L.; Mergny J.-L.; Thore S.; Yatsunyk L. A. (2018) Quadruplexes in “Dicty”: crystal structure of a four-quartet G-quadruplex formed by G-rich motif found in the Dictyostelium discoideum genome. Nucleic Acids Res. 46, 5297–5307. 10.1093/nar/gky290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turturici G.; La Fiora V.; Terenzi A.; Barone G.; Cavalieri V. (2018) Perturbation of Developmental Regulatory Gene Expression by a G-Quadruplex DNA Inducer in the Sea Urchin Embryo. Biochemistry 57, 4391–4394. 10.1021/acs.biochem.8b00551. [DOI] [PubMed] [Google Scholar]
- Crooks G. E.; Hon G.; Chandonia J.-M.; Brenner S. E. (2004) WebLogo: A Sequence Logo Generator. Genome Res. 14, 1188–1190. 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M.; Gilbert W. (1980) [57] Sequencing End-Labeled DNA with Base-Specific Chemical Cleavages. Methods Enzymol. 65, 499–560. 10.1016/S0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.