Abstract
There is a need to develop diagnostic and analytical tools that allow noninvasive monitoring of bacterial growth and dissemination in vivo. For such cell-tracking studies to hold translational value to controlled human infections, in which volunteers are experimentally colonized, they should not require genetic modification, and they should allow tracking over a number of replication cycles. To gauge if an antimicrobial peptide tracer, 99mTc-UBI29–41-Cy5, which contains both a fluorescent and a radioactive moiety, could be used for such in vivo bacterial tracking, we performed longitudinal imaging of a thigh-muscle infection with 99mTc-UBI29–41-Cy5-labeled Staphylococcus aureus. Mice were imaged using SPECT and fluorescence-imaging modalities at various intervals during a 28 h period. Biodistribution analyses were performed to quantitate radioactivity in the abscess and other tissues. SPECT and fluorescence imaging in mice showed clear retention of the 99mTc-UBI29–41-Cy5-labeled bacteria following inoculation in the thigh muscle. Despite bacterial replication, the signal intensity in the abscess only modestly decreased within a 28 h period: 52% of the total injected radioactivity per gram of tissue (%ID/g) at 4 h postinfection (pi) versus 44%ID/g at 28 h pi (15% decrease). After inoculation, a portion of the bacteria disseminated from the abscess, and S. aureus cultures were obtained from radioactive urine samples. Bacterial staining with 99mTc-UBI29–41-Cy5 allowed noninvasive bacterial-cell tracking during a 28 h period. Given the versatility of the presented bacterial-tracking method, we believe that this concept could pave the way for precise imaging capabilities during controlled-human-infection studies.
Keywords: bacterial infection, cell-tracking, ubiquicidin, multimodal, SPECT, fluorescence
To contribute to the development of innovative antimicrobial drugs and vaccines, controlled-human-infection (CHI) trials are being initiated to find solutions for monitoring bacterial persistence and for better understanding the immune parameters involved.1,2 In such studies, volunteers are purposefully colonized with a pathogen (e.g., bacteria) in order to gain mechanistic insights into the potential of these pathogens to colonize tissue and to study the interaction of specific bacteria with the existing microbiome and the native immune system. These studies have led to remarkable breakthroughs with regards to understanding local microbiome interactions (for example, experimental Neisseria lactamica colonization in healthy students protecting against Neisseria meningitis carriage) or interactions between different pathogens (live-attenuated-influenza vaccines increasing the likelihood of pneumococcal carriage).3,4 Longitudinal molecular imaging of these experimental bacterial infections could help provide better insights into the dynamics of colonization and metastatic-abscess formation and allow monitoring during antimicrobial interventions in animal models and potentially in future CHIs. Such investigation of bacterial colonization in humans would benefit from methods for tracking bacteria using noninvasive imaging modalities.5 Thus far, dedicated imaging of bacterial infections in preclinical studies mostly relies on optical imaging of genetically modified bioluminescent strains that express plasmid-integrated luciferase genes6 or ultrasound imaging of strains that express acoustic reporter genes.7 Although a useful preclinical tool, the use of genetically modified organisms (GMOs) purely for clinical-imaging purposes is not considered to be acceptable for safety reasons. An alternative approach involves using molecular-imaging techniques8,9 for labeling of bacteria employing tracers with fluorescent,10−12 magnetic,13 or radioactive imaging labels.14 In these cases, the ability to label bacteria with tracers provides a non-GMO approach for tracking bacteria in preclinical models after in vitro labeling of bacteria.
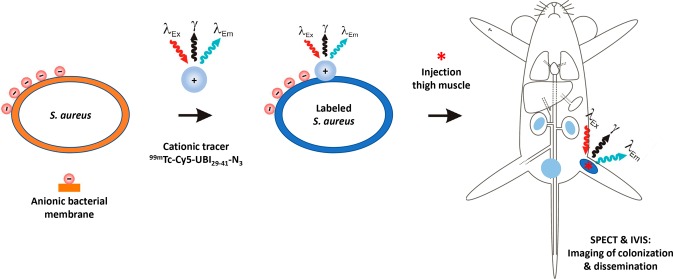
In this study, we used Staphylococcus aureus as a model system for studying experimental bacterial colonization, as these bacteria are well-known for their potential to disseminate and cause metastatic abscesses.15,16 The UBI29–41 peptide was selected because of its proven ability to label bacteria in vitro and in vivo.10,17,18 To support longitudinal in vivo cell tracking during a 28 h period, we labeled the bacteria with a derivative of the previously reported hybrid ubiquicidin peptide 99mTc-UBI29–41-Cy5-N3, which provides a new addition to the hybrid-UBI family.19 We assessed the effect of labeling on bacterial viability and the strengths of both the radioactive and fluorescent signals after various replication cycles in vitro. Subsequently, using our S. aureus model, we monitored the biodistribution and bacterial dissemination of labeled S. aureus after inoculation into a thigh muscle.
Results and Discussion
Both radioactive and fluorescence imaging can be used to monitor the progression of a Staphylococcus aureus thigh-muscle infection in a mouse model using wild-type bacteria. The strength of such a multimodal bacterial-cell-tracking concept lies in the fact that establishment of infection and subsequent dissemination of bacteria can be monitored in a noninvasive manner using the radioactive signature. Fluorescence imaging complements radioactive findings with cellular-imaging resolution and optical inspection of infected sites.
Radiolabeling of S. aureus with UBI29–41-Cy5-N3
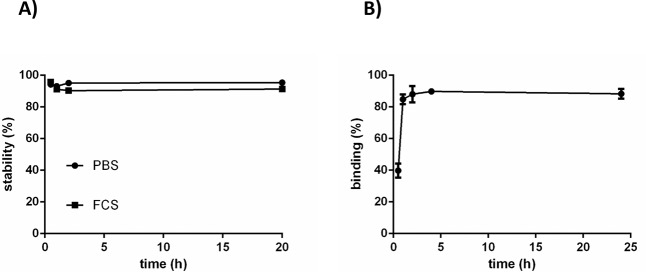
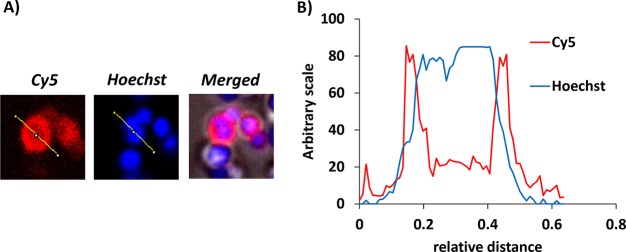
A new UBI analogue, UBI29–41-Cy5-N3, was synthesized (see the Supporting Information). Tracer stability was determined in PBS and FCS; after incubation for 20 h at 37 °C, a less than 10% release of radioactivity was observed (Figure 1A). As shown in Figure 1B, the labeling of S. aureus with 99mTc-UBI29–41-Cy5-N3 was optimally achieved (85–90%) after 1 h of incubation, and over the course of 20 h, 99mTc-UBI29–41-Cy5-N3 did not reveal dissociation from the bacteria. Fluorescence microscopy of a detailed section containing aggregated bacteria demonstrated that 99mTc-UBI29–41-Cy5-N3 accumulated in the bacterial membrane (Figure 2A,B).
Figure 1.
(A) Stability of 99mTc-UBI29–41-Cy5-N3 in PBS and FCS at 37 °C. (B) In vitro binding of 99mTc-UBI29–41-Cy5-N3 to S. aureus (99mTc-SA) at various time intervals. Data are expressed as the percent of radioactivity bound to bacteria in a pellet after two centrifugation steps. Error bars represent standard deviations.
Figure 2.
(A) Fluorescent staining with UBI29–41-Cy5 and Hoechst enabling live confocal-microscopy imaging of bacteria. UBI29–41-Cy5 demonstrates high signal-to-noise for staining of bacterial membranes (red), whereas Hoechst stains the cytoplasm (blue). (B) Analysis of a plotted section of a single bacterium with two plot profiles (red, Cy5; blue, Hoechst) across the same bacterium at 1000× magnification.
Effect of Bacterial Replication on Signal Strength
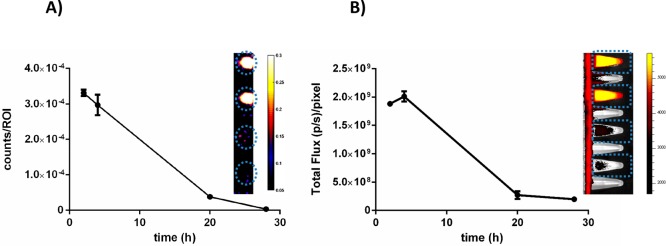
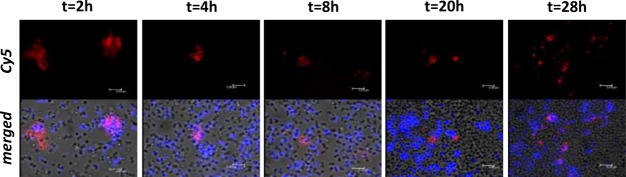
After labeling of the parental bacteria, their replication over time could cause dilution of the signal intensity. We tried to map the effect of such a dilution over a 28 h period; in vitro, this is expected to present four to five bacterial-replication cycles. After correcting for the number of viable bacteria, we observed a reduction of about 10 times in terms of both radioactivity- and fluorescence-signal strength per bacterium (Figure 3A,B). Remarkably, qualitative assessment via fluorescence microscopy at 28 h revealed a clustering of Cy5 signal around the aggregated bacteria, which could indicate there is biased tracer allotment to the daughter cells remaining in the aggregates versus those daughter cells “budding off” (Figure 4).
Figure 3.
(A) Dilution of radioactivity of 99mTc-SA during replication in BHI medium. Data are expressed as the radioactivity counts per ROI after correction for the number of viable bacteria per milliliter at the same interval. (B) Dilution of fluorescence of 99mTc-SA during replication in BHI medium. Data are expressed as the total flux ((photons/s)/pixel) per ROI after correction for the number of viable bacteria per milliliter at the same interval. The inserts on both graphs are representative images of (A) SPECT- and (B) IVIS-imaging Eppendorf tubes containing the various dilutions.
Figure 4.
Fluorescence-microscopy imaging of UBI29–41-Cy5 (red)- and Hoechst (blue in merged images)-stained S. aureus during replication, demonstrating clusters of Cy5 fluorescent signals, which remain without dilution in new cells during the replication cycles.
SPECT Imaging and Biodistribution of 99mTc-UBI29–41-Cy5-N3-Labeled Bacteria in Mice
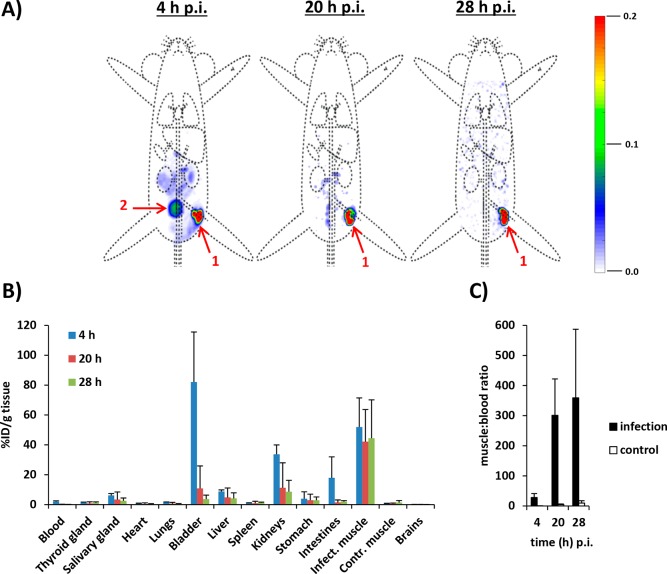
Because of the 6 h half-life of 99mTc, the current setup limited monitoring to the initial 28 h time span following infection. This time span reflects approximately five cycles of bacterial replication20,21 and a theoretical 32-fold dilution of signal per bacterium. SPECT imaging revealed a strong signal at the site of bacterial deposition (Figure 5A). Longitudinal biodistribution analysis yielded relatively constant focal radioactivity values of 51.9 ± 19.5% of the total injected radioactivity per gram of tissue (%ID/g; 4 h postinfection, pi), 42.1 ± 21.5%ID/g (20 h pi), and 44.4 ± 25.7%ID/g (28 h pi, Figure 5B and Table S1), and differences were not significant. Over time, the muscle-to-blood ratios increased significantly (p < 0.05) from 28.0 ± 13.2 (4 h pi) to 301.2 ± 120.8 (20 h pi) and eventually to 359.2 ± 227.8 (28 h pi, Figure 5C), indicating rapid blood clearance of initial residual bacteria-associated radioactivity (biological half-life t1/2 = 35.3 ± 4.0 min, R2 = 0.990). This, together with the high amounts of radioactivity observed in the kidneys and bladder at 4 h pi (33.7 ± 6.3 and 82.1 ± 33.6%ID/g, respectively), suggested early renal dissemination of S. aureus upon establishment. The absence of significant uptake of “free” 99mTc in known background tissues, such as the salivary gland, thyroid, and stomach, or metabolites in urine, confirmed the in vivo stability of the 99mTc-conjugation of UBI29–41-Cy5-N3 in mice.22,23 Interestingly, the signal distribution over bacteria that was observed in vitro (see above) did not adversely affect signal intensities during imaging. In vivo, only a 15%ID/g decrease of radioactive-signal intensity was observed at the inoculation site (Table S1). We consider this persistent signal intensity a major asset of the labeling technology, as it will also potentially allow for the investigation of cellular immune responses. With confocal-microscopy imaging of the phagocytosis of UBI29–41-Cy5-N3-labeled S. aureus by human macrophages, we demonstrated that within the measured time frame, the Cy5 signal remained with the bacteria (Figure S9A–C), and dead bacteria remained stained as well (Figure S9D). This, however, means that during our imaging studies of the abscess, we could not differentiate between viable bacteria and bacteria taken up by the immune cells.
Figure 5.
(A) Time-related (4, 20, and 28 h pi) isotope SPECT imaging of 99mTc-SA in vivo after inoculation in a thigh muscle. Organs are marked as (1) inoculation site and (2) bladder. The scale bars indicate the intensities of radioactivity expressed as arbitrary units. (B) Uptake of 99mTc-SA in various tissues at various intervals after inoculation in a thigh muscle, as determined by radioactivity calculations in excised tissues. Data are expressed as the mean percentages (±SD) of total injected radioactivity per gram of tissue (%ID/g) of three observations. Error bars represent the standard deviations. (C) Muscle-to-blood ratios at various intervals of radioactivity counts obtained from excised tissues.
Use of other, more stable radioisotopes could considerably lengthen the window of time during which bacterial colonization and dissemination can be monitored. For instance, as the half-life of 111In is 2.8 days, the use of, for example, 111In-labeled DTPA-Cy5-UBI29–41 would facilitate an extension of bacterial tracking to 1–2 week window.19 The ability to precisely image bacteria for such a period of time could provide valuable information about the early stages of infection with bacteria and pathogens and will help researchers decide if such strategies are amenable to early interventions.24
Fluorescence Imaging and Biodistribution of S. aureus in Mice
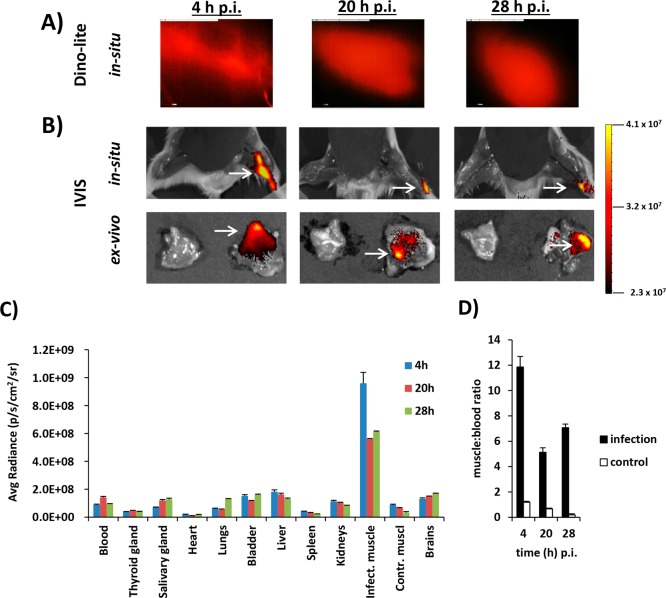
In contrast to the collateral noninfected thigh muscle, a clear fluorescent Cy5 signal could be detected in the infected thigh muscle (Figure 6A–C). However, the accuracy of bacterial-infection monitoring via whole-body fluorescence imaging was somewhat limited because of tissue absorption, scattering, and autofluorescence.25 This effect was also reflected by the fact that muscle-to-blood ratios with fluorescence imaging were much lower as compared with radioactivity measurements, calculated at 4 h (11.0 ± 1.4), 20 h (8.2 ± 0.3), and 28 h (8.9 ± 0.6, Figure 6D).
Figure 6.
(A,B) Time-related (4, 20, and 28 h pi) fluorescence imaging with either (A) Dino-lite (infected muscle in situ only) or (B) IVIS (infected and noninfected muscles in situ and ex vivo) at equal settings. The infected muscle is indicated with a white arrow. The scale bar indicates the intensity of fluorescence expressed as photons/s/cm2. (C) Time-dependent uptake of 99mTc-SA in various tissues after inoculation in a thigh muscle, as determined by assessment of the fluorescent signal in excised tissues. Data are expressed as the average radiance (photons/s/cm2/sr). As the stomach and intestines are highly autofluorescent, these organs were excluded from the analysis. Error bars represent the standard deviations. (D) Muscle-to-blood ratios of the fluorescent signals obtained from ex vivo imaging of excised thigh muscles.
Renal Clearance of 99mTc-UBI29–41-Cy5-N3-Labeled Bacteria
We also measured the outgrowth of renally excreted bacteria recovered from urine samples and found viable bacteria in urine samples of infected as opposed to uninfected mice (Figure S8B). Although the colony-forming units in urine remained quite constant throughout the 28 h infection period, there was a substantial decline in both radioactive and fluorescent signals over time (Figure S8C). As most of the radioactive and fluorescent signals were found in the bacterial pellet, we assumed that the release of 99mTc-UBI29–41-Cy5-N3 was minimal and not responsible for these observations. However, notwithstanding the robustness of bacterial labeling that we determined in vitro, release of 99mTc-UBI29–41-Cy5-N3 from the bacteria in mice cannot be fully excluded as accounting for these observations. 99mTc likely binds to UBI29–41-Cy5-N3 through multiple sites, and the relatively weak coordination of 99mTc may explain why the dissociation of the radiolabel is much faster than the disappearance of the fluorescent signal, which relies on covalent conjugation. Renal clearance of S. aureus is a frequent but unexplained phenomenon in patients with staphylococcal bacteremia,26−28 and in preclinical studies, colonization of the kidneys by S. aureus has been observed as well.29−31 Thus, we concluded that S. aureus bacteria deposited in the tight muscle persisted in the kidneys, the urinary bladder, or both.
Because UBI29–41 binds to a broad spectrum of microorganisms,32−34 the presented UBI29–41-based bacterial tracking provides a platform technology for colonization studies with a wide range of Gram-positive and -negative bacteria.9 Additionally, the ability of the UBI29–41 peptide to bind metabolically inactive bacteria could enable insightful colonization studies that aim to dissect responses to such metabolically inactive bacteria (rather than metabolically active bacteria), as they have been shown to play a role in chronic and therapy-resistant bacterial infections.35 Given the successful clinical precedent of UBI29–41 use,36,37 such applications may soon be amenable to translation into experimental human-colonization models.24
Conclusions
In conclusion, we have validated the feasibility of using a multimodal-platform technology for in vivo tracking of wild-type bacteria such as S. aureus. By labeling the bacteria with 99mTc-UBI29–41-Cy5-N3, we were able to combine in vivo microscopic imaging with ex vivo evaluation of the infection site strengthened by quantitative biodistribution studies. We believe that this hybrid imaging technology could pave the way for the development of diagnostic and analytical options in fundamental and translational research on infectious diseases in CHI.
Materials and Methods
General
All chemicals and solvents were obtained from commercial sources and used without further purification. NMR spectra were obtained using a Bruker DPX 300 spectrometer (300 MHz, 1H NMR). All spectra were referenced to residual solvent signal or TMS. HPLC was performed on a Waters system by using a 1525EF pump and a 2489 UV detector. For preparative HPLC, a Dr. Maisch GmbH, Reprosil-Pur 120 C18-AQ 10 μm column (250 × 20 mm) was used, and a gradient of 0.1% TFA in H2O/CH3CN (95:5) to 0.1% TFA in H2O/CH3CN (5:95) in 40 min was employed. For analytical HPLC, a Dr. Maisch GmbH, Reprosil-Pur C18-AQ 5 μm (250 × 4.6 mm) column was used, and a gradient of 0.1% TFA in H2O/CH3CN (95:5) to 0.1% TFA in H2O/CH3CN (5:95) in 40 min was employed. Mass spectrometry was performed on a Bruker microflex MALDI-TOF using α-cyano-4-hydroxycinnamic acid as the matrix with granuliberin R as the internal standard.
Synthesis
The scheme of the chemical-synthesis pathway for 99mTc-UBI29–41-Cy5-N3 is depicted in Figure S1 of the Supporting Information.
Synthesis of 1-(5-Carboxypentyl)-2,3,3-trimethyl-3H-indol-1-ium
2,3,3-Trimethylindolenine (1.0 mL, 6.3 mmol) and 6-bromohexanoic acid (2.5 g, 12.6 mmol) were heated at 60 °C in CH3CN (10 mL) for 72 h. Precipitation in ethyl acetate (100 mL) yielded a white precipitate, which was filtered using a P3 glass sintered filter and washed with ethyl acetate. After desiccation, 447 mg (26%) of an off-white, fluffy solid was obtained, which was used without further purification.
Synthesis of 1-Azido-4-bromobutane
1,4-Dibromobutane (14.3 mL, 120 mmol), and sodium azide (3.9 g, 60 mmol) were stirred in DMF (100 mL) at 50 °C under a N2 atmosphere. After 48 h, diethyl ether (100 mL) and 1 M NaOH (25 mL) were added. The aqueous layer was extracted twice with diethyl ether; the organic layer was dried using Na2SO4 and filtered, and the solvent was removed in vacuo. After purification by column chromatography using n-hexane, 3.2 g (30%) of solid was obtained.
Synthesis of In1-(4-azidobutyl)-2,3,3-trimethyl-3H-indol-1-iumdole-N3
2,3,3-Trimethylindolenine (1.6 mL, 10 mmol) and 1-azido-4-bromobutane (1.8 g, 10 mmol) were stirred in CH3CN at 55 °C. After 7 days, the solvent was removed in vacuo, and n-hexane was added. An oily residue was obtained after decanting. After stirring in n-hexanes two more times, stirring in diethyl ether yielded a blue precipitate (1.2 g, 47%).
Synthesis of 1-(4-Azidobutyl)-2-((1E,3E)-5-((E)-1-(5-carboxypentyl)-3,3-dimethylindolin-2-ylidene)penta-1,3-dien-1-yl)-3,3-dimethyl-3H-indol-1-ium (Cy5-N3)
Malonaldehyde dianilide hydrochloride (100 mg, 388 μmol), DiPEA (135 μL, 775 μmol), and acetic anhydride (44 μL, 465 μmol) were stirred in cold DCM (10 mL) for 2 h. After removal of the solvent, 1-(4-azidobutyl)-2,3,3-trimethyl-3H-indol-1-ium (97 mg, 379 μmol) and sodium acetate (311 mg, 3788 μmol) in ethanol (15 mL) were added, and the mixture was stirred at room temperature (rt). After 19 h, 1-(5-carboxypentyl)-2,3,3-trimethyl-3H-indol-1-ium (104 mg, 379 μmol) was added, and the mixture was stirred overnight at 60 °C. Additional 1-(4-azidobutyl)-2,3,3-trimethyl-3H-indol-1-ium (49 mg, 190 μmol) and 1-(5-carboxypentyl)-2,3,3-trimethyl-3H-indol-1-ium (52 mg, 190 μmol) were added, and the solution was refluxed to complete the reaction. After removal of the solvent in vacuo, the crude product was purified in portions by HPLC, yielding the product as a blue solid after lyophilization in an estimated yield of 8% (24.5 mg originating from 300 mg of purified crude product). MALDI-TOF calcd 566.8, found 566.3 (Figure S2).
Synthesis of 1-(4-Azidobutyl)-2-((1E,3E)-5-((E)-1-(6-((2,5-dioxopyrrolidin-1-yl)oxy)-6-oxohexyl)-3,3-dimethylindolin-2-ylidene)penta-1,3-dien-1-yl)-3,3-dimethyl-3H-indol-1-ium
1-(4-Azidobutyl)-2-((1E,3E)-5-((E)-1-(5-carboxypentyl)-3,3-dimethylindolin-2-ylidene)penta-1,3-dien-1-yl)-3,3-dimethyl-3H-indol-1-ium (24.5 mg, 43 μmol), HSPyU (89 mg, 216 μmol), and DiPEA (75.3 μL, 433 μmol) were dissolved in DMSO (800 μL, dried on molecular sieves) and stirred overnight at rt. Ethyl acetate/diethyl ether (1:1) was added, and the resulting suspension was transferred to a falcon tube and centrifuged. After being washed with diethyl ether (2 × 50 mL), the precipitate was dissolved in CH3CN and concentrated in vacuo. Purification by HPLC and subsequent lyophilization of the correct fractions yielded a blue solid (22.4 mg, 78%). MALDI-TOF calcd 663.8, found 663.3 (Figure S3). 1H NMR (300 MHz, CDCl3): δ 7.95 (t, J = 13.0 Hz, 2H, CH–CH–CH–CH–CH), 7.44–7.30 (m, 4H, Ar–H), 7.24 (t, J = 7.4 Hz, 2H, Ar–H), 7.10 (dd, J = 7.8, 3.9 Hz, 2H, Ar–H), 6.70–6.59 (t, J = 12.5 Hz, 1H, CH–CH–CH–CH–CH), 6.23 (dd, 17.3, 13.7 Hz, 2H, CH–CH–CH–CH–CH), 4.17–3.93 (m, 4H, 2× N–CH2), 3.41 (t, J = 6.3 Hz, 2H, CH2–N3), 2.85 (s, 4H, 2× CH2 from OSu) 2.65 (t, J = 7.1 Hz, 2H, −CH2–COOSu), 2.01–1.53 (m, 22H, 4× C–CH3, −CH2–CH2–CH2–CH2–COOH, −CH2–CH2–CH2–N3). For the spectra, see Figure S4A,B.
Synthesis of UBI29–41-Cy5-N3
The UBI29–41 peptide (H-TGRAKRRMQYNRR-NH2) was synthesized by standard Fmoc solid-phase peptide synthesis (SPPS) and dissolved in H2O to yield a 1 mM solution. To 300 μL of this solution (0.5 mg, 0.3 μmol), 0.1 M phosphate buffer (pH 8.5, 300 μL) and 1-(4-azidobutyl)-2-((1E,3E)-5-((E)-1-(5-carboxypentyl)-3,3-dimethylindolin-2-ylidene)penta-1,3-dien-1-yl)-3,3-dimethyl-3H-indol-1-ium (0.6 mg, 0.9 μmol) dissolved in DMSO (100 μL) were added, and the mixture was stirred overnight at rt. The solution was filtered, and the blue filtrate was purified using HPLC (Figure S5). The product-containing fraction was collected and lyophilized (0.25% yield). The mixture of N-terminal-labeled and lysine-labeled products was dissolved in PBS (50 μL, 15 μM) and used for subsequent radiolabeling and experiments. MALDI-TOF calcd 2241.7, found 2241.5 (Figure S6). The compound was dissolved in PBS (50 μL, 15 μM) and stored at −20 °C.
Radiolabeling of UBI29–41-Cy5-N3 and Stability
Radiolabeling of UBI29–41-Cy5-N3 with 99mTc was performed according to a previous procedure.32 In short, to 5 μL of UBI29–41-Cy5-N3 (15 μM in PBS), 4 μL of SnCl2·2H2O (0.44 mg/mL saline, Technescan PYP, Mallinckrodt Medical B.V. Petten), 4 μL of NaOH (0.1 M), and 100 μL of a freshly eluted 99mTc-Na-pertechnetate solution (1000 MBq/mL, Mallinckrodt Medical B.V.) were added, and the mixture was gently stirred in a shaking water bath for 1 h at 37 °C.32 After 1 h, the reaction mixture was purified by size-exclusion chromatography with sterile PBS as the mobile phase using Sephadex G-25 desalting columns (PD-10, GE Healthcare Europe GmbH). Fractions containing 99mTc-UBI29–41-Cy5-N3, counted for radioactivity in a dose-calibrator, were collected and directly used for labeling of bacteria. The stability of 99mTc-labeling was determined after incubation of 10 μL of tracer at rt in 1 mL of PBS or fetal-calf serum (20%, v/v, FCS, Life Technologies Inc.), and samples were analyzed at various intervals until 24 h. The release of radioactivity was determined by instant thin-layer chromatography (ITLC) on 1 × 7 cm ITLC-SG paper strips (Agilent Technologies) with PBS as the mobile phase. A schematic depiction of 99mTc-UBI29–41-Cy5-N3 is shown in Figure S7.
Labeling of S. aureus with 99mTc-UBI29–41-Cy5-N3 (99mTc-SA)
To allow monitoring of the inoculation site and dissemination of bacteria in mice they were labeled with a hybrid reporter compound consisting of a bacteria-binding peptide (UBI29–41), which was functionalized with a 99mTc radioisotope and a Cy5 dye. S. aureus was obtained from the American Type Culture Collections (ATCC 25922). Bacteria cultured for 24 h yielded samples with approximately 3 × 109 viable bacteria per milliliter of brain–heart-infusion broth (37 g/L, BHI medium, Sigma-Aldrich). These samples were stored in Eppendorf tubes in BHI/glycerol (80:20, v/v) at −20 °C until further use. For labeling, a stock vial (3 × 109 cfu/mL) with S. aureus was defrosted, washed three times (4 min × 3500 rpm) in 25 mM Na–NH4–acetate (pH 5) and diluted to 1 × 108 cfu/mL. Next, 100 μL of 99mTc-UBI29–41-Cy5-N3 (99mTc-SA, 0.3 μM in 25 mM Na–NH4–acetate, pH 5; 100 MBq) was added to 1 mL of a bacterial suspension containing 2 × 108 cfu of S. aureus, and the mixture was gently stirred at rt. The efficiency of the rate of bacterial labeling was measured at different intervals within a 24 h time frame. Before the determination of the tracer uptake at each interval, 0.1 mL of the bacteria in the labeling suspension was washed two times with phosphate-buffered saline (PBS) and centrifuged (4 min × 3500 rpm). The amount of radioactivity associated with the bacteria 99mTc-SA was determined at various intervals. The efficiency of the bacterial labeling was expressed as the percentage of the total added amount of radioactivity: (MBqadded/MBqbacteria-associated) × 100. The amount of fluorescence associated with bacteria was measured in a fluorescence spectrophotometer (PerkinElmer LS 55 fluorescence spectrometer equipped with a red-sensitive PMT).
Confocal Microscopy
Confocal microscopy was used to visually confirm bacterial staining with Cy5 after labeling.19 To visualize the interaction of UBI29–41-Cy5 with bacterial membranes, 1 mL of a solution containing S. aureus (2 × 108 cfu/mL; 25 mM Na–NH4–acetate, pH 5) was stained with 10 μL of UBI29–41-Cy5-N3 (15 μM in PBS). For counterstaining of the bacterial cytoplasm, 25 μL of Hoechst 33342 (1 μg/mL) was added with gently rotation for 1 h at rt in the dark. Thereafter, the excess of unbound dyes was washed away after three centrifugation steps with PBS as described above, and then the cells were resuspended in 1 mL of PBS. Then, 10 μL of stained bacteria was pipetted onto culture dishes with glass inserts (35 mm diameter glass-bottom dishes, No. 15, poly-d-lysine coated, γ-irradiated, MatTek Corporation). Images were acquired in a single field of view using a Leica SP5 WLL confocal microscope (λex = 633 nm, λem = 650–700 nm) under 100× magnification using Leica Application Suite software. Pictures were loaded in ImageJ software (ImageJ 1.44p, NIH) to draw a profile line over a single representative stained bacterium in the Cy5 and Hoechst spectra to estimate the distributions of both dyes in the bacterial membrane and cytoplasm.
Effect of Bacterial Replication on Signal Strength
After inoculation of the labeled bacteria in mice, it can be expected that within dividing bacteria, the signal strengths of both the radioactivity and fluorescence become diluted, which may affect the imaging resolution of bacteria during dissemination. To study this effect on the intensity of the signal of radiolabeled bacteria during bacterial replication, 1 mL of BHI containing 5 × 107 cfu of 99mTc-SA was pipetted into 1 mL of BHI medium and incubated in the dark at 37 °C for 28 h in a shaking water bath. At various intervals, samples were taken and pipetted into 0.2 mL vials to determine both the number of viable bacteria and the amount of radioactivity in a three-headed U-SPECT-2 (MILabs) as described before.19 For each interval, the radioactivity per region of interest (ROI) was determined from reconstructed images using AMIDE’s Medical Image Data Examiner (http://amide.sourceforge.net). At each interval, the radioactivity counts were corrected for background activity and radioactive decay and data were expressed as the radioactivity per colony-forming unit. Subsequently, the same vials were analyzed for intensity of fluorescence using a preclinical IVIS Spectrum Imaging System (Caliper Life Science). Images of the Cy5 dye were acquired following excitation at 640 nm, and light was collected at >680 nm (acquisition time of 5 s). Quantitative analysis of the fluorescence in the tissues (photons/sec/cm2) was performed using the Living Image software from Xenogeny v 3.2 (Caliper LS) at equal image-adjustment settings. A similar setup was made for fluorescence microscopy to image Cy5-labeled S. aureus (without radioactivity) to obtain visual information about the dilution of optical signal in dividing bacteria. Therefore, after washing with BHI, 200 μL was pipetted from a shaking culture maintained at 37 °C onto culture dishes, and microscopic imaging was performed at various intervals until 28 h using an inverted, widefield Leica AF6000 fluorescence microscope (λex = 633 nm, λem = 650–700 nm) under 100× magnification with Leica Application Suite software.
Bacterial Viability
Following labeling of bacteria with 99mTc-UBI29–41-Cy5-N3, we determined the bacterial viability, because UBI and UBI-derived peptide analogues have therapeutic effects, although the amount of peptide (0.3 μM) used in this study for labeling of bacteria with 99mTc-UBI29–41-Cy5-N3 was about 100× below the required dose for UBI29–41 to cause an in vitro antimicrobial effect of killing 50% of the bacteria (IC50 = 20 μM).38 To determine the effect that 99mTc-UBI29–41-Cy5-N3 exerts on S. aureus, viability was assessed 1 h after labeling of bacteria. For this purpose, samples of 99mTc-SA (2 × 108 cfu/mL in PBS) were centrifuged three times and washed with PBS as described above, and 10 mL drops from serial dilutions (range of 101 to 108) of bacteria were pipetted onto BHI agar plates and cultured overnight; the viability, expressed as the number of colony-forming units per milliliter, was compared with that of unlabeled S. aureus.
Controlled-Mouse-Infection Model
A thigh-muscle-infection model was generated using 2–3 month old Swiss mice (20–25 g, Crl:OF1 strain, Charles River Laboratories). The mice were kept under specific pathogen-free conditions in the animal-housing facility of the Leiden University Medical Center (LUMC). Food and water were given ad libitum. Animals were inoculated in the right thigh muscle with 0.1 mL of PBS containing (2.0 ± 0.2) × 107 viable 99mTc-SA (10 MBq).19 The in vivo outgrowth of S. aureus after 28 h (about five replication cycles) was assessed to be (8.0 ± 4.2) × 108. All animal studies were approved by the institutional Animal Ethics Committee (DEC permit 12160) of LUMC.
Single-Photon-Emission-Computed-Tomography (SPECT) Imaging
SPECT imaging was performed as described before.19 Briefly, at various intervals after injection of 99mTc-SA, mice were placed and fixed onto a dedicated positioned bed of a three-headed U-SPECT-2 under continuous 1–2% isoflurane anesthesia. Radioactivity counts from total body scans were acquired for 20 min, and images were reconstructed accordingly. Imaging occurred 4, 20, and 28 h pi with n = 3 mice at each interval, and the same mice were euthanized for each time interval by an intraperitoneal injection of 0.25 mL of Euthasol (ASTfarma). This was done because of the follow-up fluorescence imaging using two imaging modalities, and the biodistribution assays of the same mice at each interval (n = 3) needed to be performed at the same interval as with SPECT imaging.
Fluorescence Imaging
To allow local inspection of thigh-muscle infections and reduce reflection of light using fluorescence imaging, the fur and skin of the hind legs of the euthanized mice were removed. Fluorescence imaging was performed using a Dino-lite hand-held digital microscope dedicated to Cy5 detection (Dino-Lite Edge AM4115T-DFRW, AnMo Electronics Corporation). Acquisition was performed at λex,max = 620 nm with λem,max = 650 nm and in a magnification range of 20–100×. Images were captured using DinoCapture 2.0 software (AnMo Electronics Corporation). Next, in situ imaging of the exposed infected muscle and ex vivo fluorescence imaging of excised tissues was carried out using the IVIS Spectrum Imaging System (Caliper Life Science).19 During acquisition, the abdomen of the mice was shielded against overexposure by autofluorescence signals originating from food contents in the stomach and intestines.39
Biodistribution
After SPECT and fluorescence imaging, organs and tissues were weighed and counted for their radioactive content using a gamma counter (2470 automatic gamma counter, PerkinElmer). Counts per minute were converted into megabecquerels (MBq) and corrected for physical decay of technetium-99m (t1/2 = 6 h) at the time point of injection (t = 0), and the percentage of the injected dose per gram of tissue (%ID/g) was calculated as follows: [(MBqtissue/MBqinjected) × 100]/grams of tissue.
Culturing of Urine Samples
Because accumulation of radioactivity was observed in the urinary bladder in early SPECT imaging in mice, urine samples were collected and centrifuged, and the pellets containing the bacteria were washed twice in PBS as described above. Serial dilutions of the pellets were cultured overnight on BHI-agar plates to determine the number of viable S. aureus that could be recovered from urine. As a control for the presence of native flora, urine collected from noninfected mice were cultured as well. No other tissues were processed for microbial culturing.
Statistical Analysis
All data are presented as mean values ± standard deviations of three independent series of imaging and biodistribution. Statistical analysis for differences between groups in the animal studies were performed by Student’s two-tailed t test. Significance was defined as p < 0.05. All analyses and calculations were performed using GraphPad Prism version 5.01 for Windows (GraphPad Software).
Acknowledgments
We acknowledge Matthias N. van Oosterom for his assistance with the reconstruction of the SPECT images. The research leading to these results was funded with grants from The Netherlands Organization for Scientific Research (VIDI-grant STW BGT11272). M.R. was supported by a VENI grant from ZONMW and a Gisela Thier fellowship from LUMC.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.9b00015.
Biodistribution of 99mTc-UBI29–41-Cy5-N3 in mice, chemical-synthesis pathway of UBI29–41-Cy5-N3, MALDI-TOF spectrum of 1-(4-azidobutyl)-2-((1E,3E)-5-((E)-1-(5-carboxypentyl)- 3,3-dimethylindolin-2-ylidene)penta-1,3-dien-1-yl)-3,3-dimethyl-3H-indol-1-ium, MALDI-TOF and 1H-NMR spectra of 1-(4-azidobutyl)-2-((1E,3E)-5-((E)-1-(6-((2,5-dioxopyrrolidin-1-yl)oxy)-6-oxohexyl)-3,3-dimethylindolin-2-ylidene)penta-1,3-dien-1-yl)-3,3-dimethyl-3H-indol-1-ium, reverse-phase-HPLC purification and characterization of UBI29–41-Cy5-N3, MALDI-TOF spectrum of HPLC-purified UBI29–41-Cy5-N3, schematic depiction of 99mTc-labeled UBI29–41-Cy5-N3, BHI-agar plates containing serial dilutions of S. aureus from urine samples, assessment of viable bacteria in urine, radioactivity (99mTc) and fluorescence (Cy5) measurements, and confocal-microscopy imaging of phagocytosis of UBI29–41-Cy5-N3-labeled S. aureus by human macrophages (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Roestenberg M.; Hoogerwerf M.-A.; Ferreira D. M.; Mordmüller B.; Yazdanbakhsh M. (2018) Experimental infection of human volunteers. Lancet Infect. Dis. 18, e312–322. 10.1016/S1473-3099(18)30177-4. [DOI] [PubMed] [Google Scholar]
- Balasingam S.; Horby P.; Wilder-Smith A. (2014) The potential for a controlled human infection platform in Singapore. Singapore Med. J. 55 (9), 456–461. 10.11622/smedj.2014114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deasy A. M.; Guccione E.; Dale A. P.; Andrews N.; Evans C. M.; Bennett J. S.; Bratcher H. B.; Maiden M. C.; Gorringe A. R.; Read R. C. (2015) Nasal inoculation of the commensal Neisseria lactamica inhibits carriage of Neisseria meningitidis by young adults: A controlled human infection study. Clin. Infect. Dis. 60 (10), 1512–1520. 10.1093/cid/civ098. [DOI] [PubMed] [Google Scholar]
- Rylance J.; de Steenhuijsen Piters W. A. A.; Mina M. J.; Bogaert D.; French N.; Ferreira D. M. (2019) Two randomized trials of effect of live attenuated influenza vaccine on pneumococcal colonization. Am. J. Respir. Crit. Care Med. 10.1164/rccm.201811-2081LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsen K.; Hertlein T. (2018) Towards clinical application of non-invasive imaging to detect bacterial infections. Virulence 9 (1), 943–945. 10.1080/21505594.2018.1425072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocan T. M.; Panchal R. G.; Bavari S. (2015) Applications of in vivo imaging in the evaluation of the pathophysiology of viral and bacterial infections and in development of countermeasures to BSL3/4 pathogens. Mol. Imagin. Biol. 17 (1), 4–17. 10.1007/s11307-014-0759-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sole R.; Conde-Pueyo N. (2018) Ultrasound approach tracks gut microbes. Nature 553 (7686), 36–37. 10.1038/d41586-017-08911-0. [DOI] [PubMed] [Google Scholar]
- Gemmel F.; Van den Wyngaert H.; Love C.; Welling M. M.; Gemmel P.; Palestro C. J. (2012) Prosthetic joint infections: radionuclide state-of-the-art imaging. Eur. J. Nucl. Med. Mol. Imaging 39 (5), 892–909. 10.1007/s00259-012-2062-7. [DOI] [PubMed] [Google Scholar]
- Bunschoten A.; Welling M. M.; Termaat M. F.; Sathekge M.; van Leeuwen F. W. B. (2013) Development and prospects of dedicated tracers for the molecular imaging of bacterial infections. Bioconjugate Chem. 24 (12), 1971–1989. 10.1021/bc4003037. [DOI] [PubMed] [Google Scholar]
- Akram A. R.; Avlonitis N.; Lilienkampf A.; Perez-Lopez A. M.; McDonald N.; Chankeshwara S. V.; Scholefield E.; Haslett C.; Bradley M.; Dhaliwal K. (2015) A labelled-ubiquicidin antimicrobial peptide for immediate in situ optical detection of live bacteria in human alveolar lung tissue. Chem. Sci. 6, 6971–6979. 10.1039/C5SC00960J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geva-Zatorsky N.; Alvarez D.; Hudak J. E.; Reading N. C.; Erturk-Hasdemir D.; Dasgupta S.; von Andrian U. H.; Kasper D. L. (2015) In vivo imaging and tracking of host–microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nat. Med. 21, 1091. 10.1038/nm.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Zhu Y.; Chen X. (2017) Selective imaging of gram-negative and gram-positive microbiotas in the mouse gut. Biochemistry 56 (30), 3889–3893. 10.1021/acs.biochem.7b00539. [DOI] [PubMed] [Google Scholar]
- Hoerr V.; Tuchscherr L.; Hüve J.; Nippe N.; Loser K.; Glyvuk N.; Tsytsyura Y.; Holtkamp M.; Sunderkötter C.; Karst U.; Klingauf J.; Peters G.; Löffler B.; Faber C. (2013) Bacteria tracking by in vivo magnetic resonance imaging. BMC Biol. 11, 63. 10.1186/1741-7007-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushtaq S.; Choi M. H.; Yang J. E.; Shim H. E.; Song L.; Song H. Y.; Choi Y. J.; Jeon J. (2018) Technetium-99m-based simple and convenient radiolabeling of Escherichia coli for in vivo tracking of microorganisms. J. Radioanal. Nucl. Chem. 317 (2), 997–1003. 10.1007/s10967-018-5941-y. [DOI] [Google Scholar]
- Cheng A. G.; DeDent A. C.; Schneewind O.; Missiakas D. (2011) A play in four acts: Staphylococcus aureus abscess formation. Trends Microbiol. 19 (5), 225–232. 10.1016/j.tim.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomer L.; Schneewind O.; Missiakas D. (2016) Pathogenesis of Staphylococcus aureus bloodstream infections. Annu. Rev. Pathol.: Mech. Dis. 11, 343–364. 10.1146/annurev-pathol-012615-044351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welling M. M.; Bunschoten A.; Kuil J.; Nelissen R.; Beekman F. J.; Buckle T.; van Leeuwen F. W. B. (2015) Development of a hybrid tracer for SPECT and optical imaging of bacterial infections. Bioconjugate Chem. 26 (5), 839–849. 10.1021/acs.bioconjchem.5b00062. [DOI] [PubMed] [Google Scholar]
- Welling M. M.; Hensbergen A. W.; Bunschoten A.; Velders A. H.; Roestenberg M.; van Leeuwen F. W. B. (2019) An update on radiotracer development for molecular imaging of bacterial infections. Clin. Transl. Imag. 7, 105–124. 10.1007/s40336-019-00317-4. [DOI] [Google Scholar]
- Welling M. M.; Bunschoten A.; Kuil J.; Nelissen R. G. H. H.; Beekman F. J.; Buckle T.; van Leeuwen F. W. B. (2015) Development of a hybrid tracer for SPECT and optical imaging of bacterial infections. Bioconjugate Chem. 26 (5), 839–849. 10.1021/acs.bioconjchem.5b00062. [DOI] [PubMed] [Google Scholar]
- Calame W.; van der Waals R.; Mattie H.; van Furth R. (1989) Influence of etoposide and cyclophosphamide on the efficacy of cloxacillin and erythromycin in an experimental staphylococcal infection. Antimicrob. Agents Chemother. 33 (6), 980–982. 10.1128/AAC.33.6.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calame W.; Feitsma H. I. J.; Ensing G. J.; Goedemans W. T.; Camps J. A. J.; van Furth R.; Pauwels E. K. J. (1991) Detection of a local staphylococcal infection in mice with technetium-99m-labeled polyclonal human immunoglobulin. J. Nucl. Med. 32 (3), 468–474. [PubMed] [Google Scholar]
- International Commission on Radiological Protection . (1980) Limits for intakes of radionuclides by workers, Pergamon Press, Oxford. [PubMed] [Google Scholar]
- Leggett R.; Giussani A. (2015) A biokinetic model for systemic technetium in adult humans. J. Radiol. Prot. 35 (2), 297. 10.1088/0952-4746/35/2/297. [DOI] [PubMed] [Google Scholar]
- Roestenberg M.; Mo A.; Kremsner P. G.; Yazdanbakhsh M. (2017) Controlled human infections: A report from the controlled human infection models workshop, Leiden University Medical Centre 4–6 May 2016. Vaccine 35 (51), 7070–7076. 10.1016/j.vaccine.2017.10.092. [DOI] [PubMed] [Google Scholar]
- Leblond F.; Davis S. C.; Valdes P. A.; Pogue B. W. (2010) Pre-clinical whole-body fluorescence imaging: Review of instruments, methods and applications. J. Photochem. Photobiol., B 98 (1), 77–94. 10.1016/j.jphotobiol.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muder R. R.; Brennen C.; Rihs J. D.; Wagener M. M.; Obman A.; Obman A.; Stout J. E.; Yu V. L. (2006) Isolation of Staphylococcus aureus from the urinary tract: association of isolation with symptomatic urinary tract infection and subsequent staphylococcal bacteremia. Clin. Infect. Dis. 42 (1), 46–50. 10.1086/498518. [DOI] [PubMed] [Google Scholar]
- Chihara S.; Popovich K. J.; Weinstein R. A.; Hota B. (2010) Staphylococcus aureus bacteriuria as a prognosticator for outcome of Staphylococcus aureus bacteremia: a case-control study. BMC Infect. Dis. 10, 225. 10.1186/1471-2334-10-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. K.; Crossley K.; Gerding D. N. (1978) The association between Staphylococcus aureus bacteremia and bacteriuria. Am. J. Med. 65 (2), 303–306. 10.1016/0002-9343(78)90824-0. [DOI] [PubMed] [Google Scholar]
- Sawai T.; Tomono K.; Yanagihara K.; Yamamoto Y.; Kaku M.; Hirakata Y.; Koga H.; Tashiro T.; Kohno S. (1997) Role of coagulase in a murine model of hematogenous pulmonary infection induced by intravenous injection of Staphylococcus aureus enmeshed in agar beads. Infect. Immun. 65 (2), 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacconi M.; Haag A. F.; Chiarot E.; Donato P.; Bagnoli F.; Delany I.; Bensi G. (2017) In vivo analysis of Staphylococcus aureus-infected mice reveals differential temporal and spatial expression patterns of fhuD2. Infect. Immun. 85 (10), e00270-17 10.1128/IAI.00270-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Vyver H.; Bovenkamp P. R.; Hoerr V.; Schwegmann K.; Tuchscherr L.; Niemann S.; Kursawe L.; Grosse C.; Moter A.; Hansen U.; Neugebauer U.; Kuhlmann M. T.; Peters G.; Hermann S.; Löffler B. (2017) A novel mouse model of Staphylococcus aureus vascular graft infection: noninvasive imaging of biofilm development in vivo. Am. J. Pathol. 187 (2), 268–279. 10.1016/j.ajpath.2016.10.005. [DOI] [PubMed] [Google Scholar]
- Welling M. M.; Paulusma-Annema A.; Balter H. S.; Pauwels E. K. J.; Nibbering P. H. (2000) Technetium-99m labelled antimicrobial peptides discriminate between bacterial infections and sterile inflammations. Eur. J. Nucl. Med. Mol. Imaging 27 (3), 292–301. 10.1007/s002590050036. [DOI] [PubMed] [Google Scholar]
- Lupetti A.; Welling M. M.; Mazzi U.; Nibbering P. H.; Pauwels E. K. J. (2002) Technetium-99m labelled fluconazole and antimicrobial peptides for imaging of Candida albicans and Aspergillus fumigatus infections. Eur. J. Nucl. Med. Mol. Imaging 29 (5), 674–679. 10.1007/s00259-001-0760-7. [DOI] [PubMed] [Google Scholar]
- Ebenhan T.; Chadwick N.; Sathekge M. M.; Govender P.; Govender T.; Kruger H. G.; Marjanovic-Painter B.; Zeevaart J. R. (2014) Peptide synthesis, characterization and Ga-68-radiolabeling of NOTA-conjugated ubiquicidin fragments for prospective infection imaging with PET/CT. Nucl. Med. Biol. 41 (5), 390–400. 10.1016/j.nucmedbio.2014.02.001. [DOI] [PubMed] [Google Scholar]
- Garzoni C.; Kelley W. L. (2009) Staphylococcus aureus: new evidence for intracellular persistence. Trends Microbiol. 17 (2), 59–65. 10.1016/j.tim.2008.11.005. [DOI] [PubMed] [Google Scholar]
- de Murphy C. A.; Gemmel F.; Balter J. (2010) Clinical trial of specific imaging of infections. Nucl. Med. Commun. 31 (8), 726–733. 10.1097/MNM.0b013e32833a3d7f. [DOI] [PubMed] [Google Scholar]
- Ostovar A.; Assadi M.; Vahdat K.; Nabipour I.; Javadi H.; Eftekhari M.; Assadi M. (2013) A pooled analysis of diagnostic value of Tc-99m-ubiquicidin (ubi) scintigraphy in detection of an infectious process. Clin. Nucl. Med. 38 (6), 413–416. 10.1097/RLU.0b013e3182867d56. [DOI] [PubMed] [Google Scholar]
- Brouwer C. P. J. M.; Bogaards S. J. P.; Wulferink M.; Velders M. P.; Welling M. M. (2006) Synthetic peptides derived from human antimicrobial peptide ubiquicidin accumulate at sites of infections and eradicate (multi-drug resistant) Staphylococcus aureus in mice. Peptides 27 (11), 2585–2591. 10.1016/j.peptides.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Inoue Y.; Izawa K.; Kiryu S.; Tojo A.; Ohtomo K. (2008) Diet and abdominal autofluorescence detected by in vivo fluorescence imaging of living mice. Mol. Imaging 7 (1), 21–27. 10.2310/7290.2008.0003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.