Abstract
Despite a wealth of knowledge on Salmonella phages worldwide, little is known about poultry-associated Salmonella phages from Thailand. Here, we isolated 108 phages from Thai poultry farms that infect Salmonella enterica serovar Typhimurium. Phages STm101 and STm118 were identified as temperate Siphoviridae phages. Genome sequencing and analyses revealed these phages share approximately 96% nucleotide sequence similarity to phage SPN19, a member of the Chi-like virus genus. PCR amplification of the gene encoding capsid protein E of the Chi-like phage was positive for 50% of phage isolates, suggesting a predominance of this phage type among the sampled poultry farms. In addition to the flagella, two phages required the lipopolysaccharide to infect and lyse Salmonella. Furthermore, phylogenomic analysis demonstrated that phages STm101 and STm118 formed a monophyletic clade with phages isolated from Western countries, but not from closer isolated phages from Korea. However, further investigation and more phage isolates are required to investigate possible causes for this geographic distribution.
Keywords: bacteriophage, Chi-like virus, flagellotropic, Salmonella, siphovirus, poultry farm, prevalent, phylogenomic tree
1. Introduction
Bacteriophages (phages) are viruses that infect bacteria, and play important roles in regulating abundance, diversity, and composition of bacterial communities [1]. A significant number of Salmonella phages have been isolated and characterized from diverse environments. This includes sequencing of complete genomes from a wide variety of dsDNA, tailed phages of the order Caudovirales [2,3]. Salmonella phages are exploited in various lab-based applications including strain construction via transduction [4,5,6] and phage typing for epidemiological purposes [7,8]. The inherent Salmonella-specificity of certain phages (and their component proteins) has also been exploited to develop highly specific bio-probes for the detection of foodborne Salmonella [9,10,11,12] and as antibiotic alternatives for eliminating Salmonella in food production [13,14,15,16].
The Siphoviridae Salmonella phage χ (Chi) was first isolated by Sertic and Boulgakov in 1936 [17]. It is a flagellotropic phage and attaches to the flagella of motile bacteria as its primary receptor [17]. Due to the relatively low likelihood of encounters between phages and bacteria in certain environments, it has been proposed that phage Chi, as well as others [18,19,20], recognize motile flagella and/or pili to increase the probability of interacting with a metabolically active host suitable for infection [19]. The genomic sequence of phage Chi is approximately 59 kb long with 56.5% GC content and 75 open reading frames (ORFs) [21,22]. Related Chi-like phages with similar genome sizes, gene contents, and gene orders to phage Chi include Salmonella phages FSLSP030, FSL_SP-039, FSLSP088, FSL_SP-124, and SPN19 [21,23], Enterobacter cancerogenus phage Enc34 [24], and Providencia stuartii phage RedJac [25]. Additionally, non-Chi-like flagellotropic phages have been identified such as Caulobacter crescentus phages ϕCb13 and ϕCbK [19], the Gram-positive targeting Bacillus subtilis phage PBS1 and Bacillus pumilus phage PBP1 [26], and more recently the Agrobacterium phage 7-7-1 [27]. The broad host ranges typically observed for flagellotropic phages (e.g., phage Chi spans Salmonella and Escherichia coli hosts [28,29]) makes them attractive candidates for development into tools for pathogen detection and remediation.
Here, we describe the isolation of 108 Salmonella phages from chicken fecal and boot swap samples of Thai poultry farms. Over 50% of the phages were identified as members of the Chi-like genus, suggesting a predominance of this phage type within Thai poultry farms. Two phages from the same farm were randomly chosen for further characterization using transmission electron microscopy (TEM) and whole genome sequencing. The phylogenomic relationship among these newly isolated Thai Chi-like phages and other available reported Chi-like phages was also investigated. Finally, using S. Typhimurium (S. Tm) outer membrane mutants, we identified that in addition to the flagella, these Chi-like phages required the lipopolysaccharide (LPS) for efficient host attachment and infection.
2. Materials and Methods
2.1. Bacterial Strains and Media
All strains used for host range analysis and mutant studies are listed in Table 1, Table 3, and Supplementary Table S1. S. Tm MA8508 was used for Salmonella phage isolation and propagation from fecal and boot swab samples. Cells were grown at 37 °C in trypticase soy broth (TSB) or agar (TSA) (Titan Biotech, Delhi, India). Saline magnesium (SM) buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 8.0 mM MgSO4, and 0.01% gelatin) was used for phage storage at 4 °C.
Table 1.
Host range infection of Salmonella phages STm101 and STm118.
| Salmonella Serovar (s) | No. of Isolates | Spot Lysis (% Infectivity) of Phages | |
|---|---|---|---|
| STm101 | STm118 | ||
| S. Enteritidis | 32 | 3 (9.4%) | 21 (65.6%) |
| S. Typhimurium | 27 | 20 (74.1%) | 17 (63%) |
| S. Virchow | 15 | 7 (46.7%) | 4 (26.7%) |
| S. Hadar | 11 | 3 (27.3%) | 2 (18.2%) |
| S. Choleraesuis | 13 | 10 (76.9%) | 6 (46.2%) |
| S. Agona | 3 | 3 (100%) | 3 (100%) |
| S. Saintpaul | 1 | 1 (100%) | 1 (100%) |
| S. Kentucky | 1 | 0 (0%) | 1 (100%) |
| S. Albany | 3 | 0 (0%) | 0 (0%) |
| S. I 4,[5],12:i:- | 4 | 0 (0%) | 0 (0%) |
| S. Altona | 1 | 0 (0%) | 0 (0%) |
| S. Corvallis | 2 | 0 (0%) | 0 (0%) |
| S. Give | 3 | 0 (0%) | 0 (0%) |
| S. Schwarzengrund | 1 | 0 (0%) | 0 (0%) |
| S. Singapore | 1 | 0 (0%) | 0 (0%) |
| Total | 118 | 47 (39.8%) | 55 (46.6%) |
| Control strains | |||
| Listeria monocytogenes | 13 | 0 (0%) | 0 (0%) |
| Vibrio spp. | 8 | 0 (0%) | 0 (0%) |
| Escherichia coli | 5 | 0 (0%) | 0 (0%) |
| Staphylococcus aureus | 5 | 0 (0%) | 0 (0%) |
| Total | 31 | 0 (0%) | 0 (0%) |
2.2. Isolation, Purification, and High Titer Phage Stock Preparation
With the owners’ authorization, 252 fecal samples were obtained from four free-range chicken farms dispersed across the Lamphun province, Northern Thailand, and 169 boot swab samples were collected from staff working at a commercial farm in central Thailand. Each sample was screened for the presence of phages using spot assays as described [30]. Briefly, 2 g of fecal sample was suspended in SM buffer then centrifuged at 4500× g for 30 min while boot swab sample in buffered peptone water (Merck, Branchburg, New Jersey, USA) was centrifuged at 4500× g for 10 min. Then the centrifuged supernatant was filtered through a 0.45 µm membrane filter (GE Healthcare Life Sciences, Marlborough, MA, USA). Ten microliters of the filtrated suspension were spotted onto a bacterial lawn containing 100 μL of a mid-log phase S. Tm MA8508 culture mixed with 3 mL soft agar, and incubated overnight at 37 °C. Samples producing plaques (zones of localized lysis) were confirmed to contain phages using the double agar overlay plaque technique as previously described [31]. Single plaques were then picked and purified a minimum of five times by repeated double agar overlays.
For high titer phage stock production, double agar overlays were used to produce ten semi- confluent lysis plates (diameter 150 mm) from a single plaque of an isolated phage. Plates were gently agitated at room temperature with SM buffer for 5 hours to produce a crude phage lysate. The lysate was centrifuged at 6000× g for 45 min at 4 °C, and the supernatant filtered through a 0.45 µm membrane filter, before a final centrifugation at 18,000× g for 30 min at 4 °C. The supernatant was gently removed, and 2 mL of SM buffer added to the pellet and incubated overnight at 4 °C. On the next day, the phage suspension was centrifuged at 1800× g, for 15 min at 4 °C. For each phage, a high titer of approximately 1011 pfu/mL (plaque forming units/mL) was recovered and stored at 4 °C.
2.3. Host Range Analysis of Isolated Phages
Spot assays were also used to determine phage host range. Briefly, 10 μL of phage solution taken from a serial dilution was spotted onto a bacterial lawn containing 100 μL of mid-log phase bacterial culture mixed with 3 mL of 0.35% TSA supplemented with 5 mM CaCl2 (soft agar), and incubated overnight at 37 °C. The presence of a lytic zone was considered evidence of phage susceptibility or lysis from without due to a high multiplicity of infection (MOI) from direct spotting of undiluted phage solutions; no lysis was considered evidence of phage resistance. All negative results were confirmed again using double agar overlay plaque assay [30].
2.4. Amplification of Salmonella Chi-Like Phage Capsid Protein E DNA
The gene encoding the major capsid protein E of the Chi-like Salmonella phage SPN19 (GenBank accession number NC_019417.1) was used as a template to design two PCR primers: forward (5’-TTCAGACCCACGGATGGTTG-3’) and reverse (5’-AGAAAGCGGCTACAACACGA-3’), corresponding to nucleotide positions 41,881–41,830 and 42,302–42,321, respectively, on the complete genome. The PCR cycle composed of a hot start at 94 °C for 3 min, followed by 35 cycles of 94 °C for 1 min, 60 °C for 30 s, and 72 °C for 30 s, before a final extension of 72 °C for 5 min. The amplified DNA fragment was 511 bp in length and was detected after 1% agarose gel (Vivantis Technologies, Selangor Darul Ehsan, Malaysia) electrophoresis and stained with GelRed® nucleic acid gel stain (Biotium, Fremont, CA, USA).
2.5. One-Step Phage Growth Curve Analysis
One-step growth curves were performed as described [30]. Briefly, a culture of S. Tm MA8508 was grown to OD600 of 0.2–0.5 and mixed with individual phage suspensions at a multiplicity of infection (MOI) of 0.01. Phage adsorption was performed for 15 min at 37 °C, after which the suspension was filtered through a 0.45 μm membrane filter. The membrane filter was placed in 50 mL of TSB supplemented with 5 mM CaCl2 (Merck, New Jersey, USA) and incubated at 37 °C. Every 5 min, 2 × 500 µL of suspension was collected and aliquoted into separate tubes. Using double agar overlay plaque assay, the first aliquot was used for pfu/mL determination of “free phages” (after removal by centrifugation of bacterial cells) and the second aliquot was chloroform treated to release intracellular phages for the “total phage” count. Briefly, 10 μL of each aliquot was serially diluted, mixed with 100 μL of mid-log phase S. Tm MA8508, 3 mL of 0.35% TSA, and poured onto a TSA supplemented with 5 mM CaCl2. After overnight incubation at 37 °C, total pfu/mL was determined. Combining these numbers, the latent period, eclipse period, and burst size were calculated as a one-step growth curve.
2.6. Transmission Electron Microscopy
Transmission electron microscopy was performed as previously described [30]. Briefly, a high titer (>109 pfu/mL) phage solution was dropped onto a 100-mesh copper grid coated with Formvar (Ted Pella, Redding, CA, USA) then negative stained with 2% potassium phosphotungstate (Sigma-Aldrich, St. Louis, MO, USA), pH 7.2. The grid was examined using a TEM-1230 transmission electron microscope (JEOL, Tokyo, Japan) equipped with a Dual vision digital camera (Gatan, Pleasanton, CA, USA). Phage morphology was classified based on order and family according to Ackermann [32,33].
2.7. Next-Generation Sequencing of Phage Genome and Sequences Analysis
Phage DNA was extracted by a phenol/chloroform extraction method as described [34]. Genome sequencing of phages were performed using the Ion Torrent PGM with Ion 314 V2 chips, following the manufacturer’s protocol for 200 bp genomic DNA fragment library preparation (Ion Xpress Plus gDNA fragment library system), template preparation (Ion OneTouch system), and sequencing (Ion PGM 200 sequencing kit) (Thermo Fisher Scientific, Waltham, MA, USA). De novo assembly of the reads was performed via succinct de Bruijn graph using MEGAHIT software version 1.2.x [35,36]. CAP3 Assembly Program was used to assemble the final phage contig and to remove the overlapping sequences [31]. To annotate the contigs, the genome sequences in FASTA format were annotated using Prokka version 1.2 [37]. The outputs of each phage were compared and manually curated against the reference Salmonella phage SPN19 (NC_019417.1) to obtain the final set of genes in each phage genome. Easyfig software version 2.2.3 was used to generate the phage alignment figure [38]. The completed genome sequences of Salmonella phages STm101 and STm118 are available from GenBank under accession numbers KX765862 and KX765863, respectively.
2.8. Comparison, Clustering, and Phylogenomic Analysis of Phage Genomes
Comparative genomics was performed using CLC Genomics Workbench (Version 12.0, QIAGEN) using Salmonella phage SPN19 (NC_019417.1) as a reference strain. Phage genome sequences were mapped to the reference using default parameters and the mapped reads were further analyzed for genome variation with the CLC Genomics Workbench. Only reads that have minimum coverage of 30X and frequency of more than 70% were included for the analyses. The 139 Salmonella phage genomes were downloaded from the GenBank database [39] and screened for completeness before being added to the analysis, as shown in Supplementary File 1. In addition, geographic areas of the selected Salmonella phages were summarized in Supplementary Table S2. Coding sequences were identified and translated from all genomes. Protein sequences from each genome were stored separately. Those protein sequences were compared all-versus-all with standalone blastp via a Perl script (“get_homologues.pl”) with the following command and options: get_homologues.pl -A -t 0 -e -z -G [40]. In brief, the Perl script used a “cluster of orthologous group” (COG) algorithm to identify and cluster sets of homologous protein sequences using an E-value threshold of 10−5. The in-paralogs found in the process were omitted. An all-versus-all matrix containing the average amino acid identity (AAI) of all protein coding sequences within each genome was generated. Pairwise distances from a correlation matrix built from all pairwise AAI values were hierarchically clustered using R-script [41] to identify phage genomes with significant similarity to Salmonella phage genomes. The command used was: hclust (as.dist (1 – cor (t (aai.matrix), use = “pairwise.complete.obs”)), method = “average”).
Using COG analysis, a cluster of 14 related phages, including STm101 and STm118 were identified as sharing a core set of 28 orthologs. Protein multiple sequence alignments (MSAs) from these orthologs were built with default settings in MUSCLE v3.8.31 [42] and used to build codon-aware multiple alignments of the nucleotide sequences corresponding to each protein coding sequence with another Perl script (“pal2nal.pl”) [43]. The codon-aligned nucleotide sequences for the 28 orthologs in each phage genome were concatenated and used to build an unrooted phylogenomic tree in FastTree Version 2.1.10 [44] using the generalized time reversible nucleotide substitution model. The reliability of splits in the tree is estimated by local support values that are tested on 1000 resamples using the Shimodaira–Hasegawa method [45] on nearest-neighbor interchanges at each split.
3. Results
3.1. Host Range Analysis of the Two Thai Phages
In total, 252 chicken droppings from four farms and 169 boot swab samples from one farm were tested for phages that infect S. Tm MA8508. Overall, 108 samples produced lysis and formed single plaques, indicating phage activity. A single plaque (representing an individual phage) was picked from each of the 108 plaque-positive samples, passaged a minimum of five times, and then plaque morphology assessed. Three different phage morphologies were observed: (i) clear (29.6%); (ii) turbid (16.7%); and (iii) halo-forming plaques (53.7%), with halo-forming plaques accounting for over half of phages isolated. Seventeen phage isolates were randomly selected from the five farms for host range analysis using 44 different Salmonella strains, as shown in Supplementary Table S3. All phages presented different host ranges, including phages isolated from the same farm. The two clear plaque-forming phages isolated from the same farm named STm101 and STm118 were selected for further investigation.
The host ranges of the two phages were tested against a broader 118 strains of Salmonella, including 98 strains from the five serovars S. Enteritidis, S. Typhimurium, S. Virchow, S. Choleraesuis, and S. Hadar, as they are commonly associated with Salmonella infections of poultry plus 20 strains of other Salmonella serovars, as shown in Table 1. Overall, phages STm101 and STm118 were capable of infecting 39.8% (47/118) and 46.6% (55/118) of the tested strains, respectively. Phages STm101 and STm118 could lyse 9.4% (3/32) and 65.6% (21/32) of tested S. Enteritidis strains, respectively. None of the total 15 tested strains of S. Albany, S. I 4,[5],12:i:-, S. Altona, S. Corvallis, S. Give, S. Schwarzengrund, and S. Singapore could be infected by phages STm101 and STm118. Phage STm118, but not STm101, could infect a single S. Kentucky strain. None of the phages infected any of the other Gram-negative strains tested, which involved five E. coli strains and eight Vibrio spp. strains, or Gram-positive strains of Staphylococcus aureus and Listeria monocytogenes, as shown in Table 1.
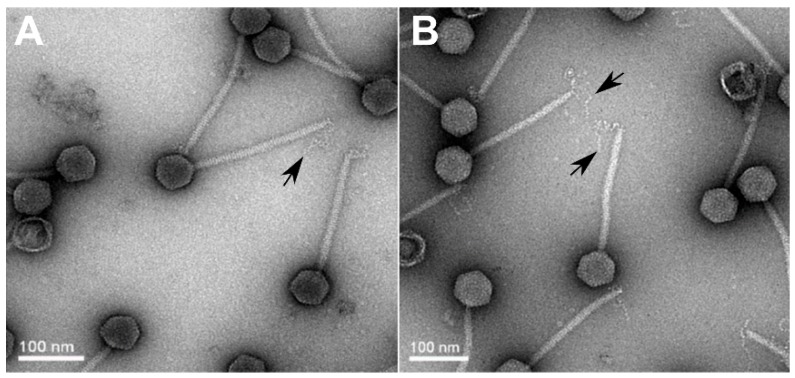
TEM analysis revealed that the two phages belong to the Siphoviridae family, featuring isometric capsids (62 ± 5 nm) with long, non-contractile tails (220 ± 12 nm), as shown in Figure 1. Attached at the tip of each phage tail is the specialized receptor-binding protein (RBP)—a single, kinked tail fiber. The single tail fiber is a common feature among Chi-like phages and is used for the initial attachment to the flagella of host bacteria [20,21,22,28].
Figure 1.
TEM-based designation of the Salmonella phages as Chi-like Siphoviruses. According to the isometric capsid (62 ± 5 nm) and non-contractile tail (220 ± 12 nm) morphologies, phages STm101 (A) and STm118 (B) are Siphoviruses. Black arrows designate the single, baseplate-attached tail fiber used by phage Chi and Chi-like phages for host attachment via interaction with the bacterial flagella [20,28,47].
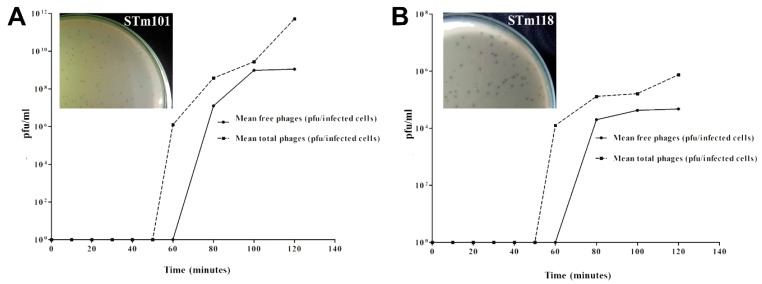
Data from one-step phage growth curves performed at MOI = 0.01 showed that phages STm101 and STm118 have similar growth patterns when using S. Tm MA8508 as a host, as shown in Figure 2. At ~60 min, the latent periods of both phages were extended more than the typical latent periods of 30 min observed for most Salmonella phages. STm118 also produced a reduced burst size (48 pfu/infected cell) compared to STm101 (112 pfu/infected cell) and other characterized Salmonella phages, which typically produce 100–230 pfu/infected cell [28,46]. However, these values depend heavily on the host and may change if tested using a different Salmonella host strain. Nevertheless, from the differences observed in their host ranges and plaque morphologies, we classified STm101 and STm118 as different phages.
Figure 2.
One-step growth curves of the Salmonella phages using a multiplicity of infection of 0.01. The mean free phages (solid line) and mean total phages liberated using chloroform (dotted line) are calculated as plaque forming units (pfu) for phages STm101 (A) and STm118 (B) infecting S. Tm MA8508. Shown as insets are pictures of the clear plaque morphologies for the two phages, STm101 and STm118 (A and B).
3.2. Genomic Analysis Identifies Thai Phage Isolates as Chi-Like Viruses
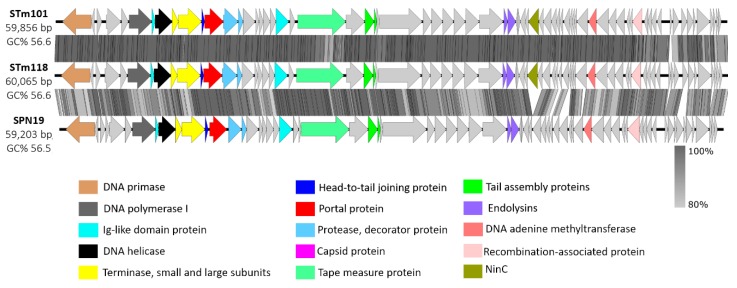
The genomes of phages STm101 and STm118 are 59,856 and 60,065 bp, respectively, and share 98.8% nucleotide sequence similarity. Overall, the genomes of these phages are composed of 71 (STm101) and 72 (STm118) ORFs with a GC content of 56.6%, with no evident tRNAs, as shown in Supplementary Tables S4 and S5. These phages have approximately 93.6% to 96.2% nucleotide sequence identity to phage Chi (KM458633) and the Chi-like SPN19 (NC_019417), respectively, confirming both as members of the Chi-like virus genus. The phages all contain an integrase (int; gp59) with homology to integrases found in other Chi-like phages, e.g., gp14 in SPN19, suggesting a lysogenic lifestyle of these Chi-like phages. Moreover, during our studies we observed the formation of turbid plaques (a common trait of lysogenic phages) for STm101 and STm118 upon infection of S. Tm strain DB7155, providing evidence towards a lysogenic nature of these phages. As revealed in the phage Chi genome, both STm101 and STm118 feature the same cos-site (5′-GGTGCGCAGAGC-3’) at their genome termini, located 570 bps away from the first gene encoding a putative DNA primase [21]. As expected from their 98.8% sequence similarity, STm101 and STm118 present identical genomic organizations, with strong similarity to the Chi-like SPN19 phage (NC_019417.1), as shown in Figure 3. In brief, all three genomes can be aligned from the terminal cos-site follow by genes related to replication, i.e., those encoding putative DNA primase, polymerase, helicase, and the terminase subunits. However, as sequence variation between STm101 and STm118 was present across their entire genomes, determining a genetic link to their variable host ranges will require further investigation.
Figure 3.
Alignment of Salmonella STm101, STm118, and the reference Chi-like SPN19 phage genomes based upon nucleotide sequence similarity, and aligned relative to their shared, terminal cos-sites at the left end. Predicted open reading frames (ORFs) are shown as arrows pointing in the direction of transcription and are color-coded according to their predicted functions.
3.3. Phylogenomic Tree Inference of the Chi-Like Phage Cluster
Table 2 provides a comparison of our two phages to the 12 other members of the Chi-like cluster of Salmonella-infecting phages. The Chi-like cluster was identified among all Salmonella phages by hierarchically clustering all phage genomes according to their average amino acid identity, as shown in Supplementary Figure S1 and Supplementary Files 1 and 2. Of these, phages 35 and 37 encode the most genes, as shown in Table 2, despite being smaller than or the same size as the other phage genomes. These statistics are reflected in the average gene size for phage 35 and 37, which is 561 bp and 520 bp, respectively, compared to more than 700 bp for other genomes within the cluster. Furthermore, the genomes of phages 35 and 37 share the fewest genes with the rest of the genomes, with an average orthologous cluster size of 7.0 and 6.3, respectively, compared to 11.0–13.0 for the other phages. The gene content from genomes of the newly identified Salmonella phages STm101 and STm118 is relatively well conserved among genomes from the Chi-like phage cluster, as indicated by their large average cluster of orthologous group (COG) size. In addition, all genomes from the Chi-like phage cluster encode 28 orthologs, which can be used to infer phylogeny for the cluster, as shown in Supplementary File 3.
Table 2.
Genome characteristics of Salmonella phages in the Chi-like phage cluster. CDSs: coding sequences; COG: cluster of orthologous group.
| Phage | Accession no. | Size (bp) | GC (%) | CDSs | Avg CDS Size (bp) | tRNAs | Avg COG Size | Family | Bacterial Host | Source | Country | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Salmonella phage STm101 | KX765862 | 59,856 | 56.6 | 71 | 716 | 0 | 16.8 | Siphoviridae | S. Typhimurium | Chicken feces | Thailand | This study |
| Salmonella phage STm118 | KX765863 | 60,065 | 56.6 | 72 | 624 | 0 | 16.6 | Siphoviridae | S. Typhimurium | Chicken feces | Thailand | This study |
| Salmonella phage STm374 | KX765864 | 59,934 | 56.6 | 72 | 502 | 0 | 16.2 | Siphoviridae | S. Typhimurium | Chicken feces | Thailand | - |
| Salmonella phage 118970_sal1 | NC_031930.1 | 59,518 | 56.5 | 71 | 782 | 0 | 11.5 | Siphoviridae | S. Enteritidis | Water buffalo feces | Italy | [48] |
| Salmonella phage 35 | KR296689.1 | 55,391 | 56.8 | 91 | 561 | 0 | 7 | Podoviridae | S. Gallinarum | Sewage | India | [49] |
| Salmonella phage 37 | NC_029045.1 | 60,216 | 56.5 | 105 | 520 | 0 | 6.3 | Siphoviridae | S. Gallinarum | Sewage | India | [49] |
| Salmonella phage BP12C | NC_031228.1 | 60,606 | 56.4 | 76 | 757 | 0 | 11.7 | Siphoviridae | S. Hadar | Sewage | USA | - |
| Salmonella phage Chi | JX094499.1; KM458633.1 | 59,407 | 56.5 | 75 | 749 | 0 | 11.9 | Siphoviridae | Salmonella enterica, E. coli | - | France | [21,22] |
| Salmonella phage FSL SP-039 | KC139514 | 59,815 | 56.6 | 71 | 776 | 0 | 12.3 | Siphoviridae | S. Cerro | Dairy farms | USA | [23] |
| Salmonella phage FSL SP-124 | KC139515 | 59,245 | 56.5 | 71 | 765 | 0 | 12.2 | Siphoviridae | S. Cerro | Dairy farms | USA | [23] |
| Salmonella phage FSL SP-030 | NC_021779.1 | 59,746 | 56.6 | 71 | 776 | 0 | 12.3 | Siphoviridae | S. Dublin | Dairy farms | USA | [23] |
| Salmonella phage FSL SP-088 | NC_021780.1 | 59,454 | 56.4 | 70 | 779 | 0 | 12.3 | Siphoviridae | S. Typhimurium | Dairy farms | USA | [23] |
| Salmonella phage iEPS5 | NC_021783.1 | 59,254 | 56.3 | 73 | 765 | 0 | 12 | Siphoviridae | S. Typhimurium | Sewage | South Korea | [28] |
| Salmonella phage SPN19 | NC_019417.1 | 59,203 | 56.5 | 72 | 770 | 0 | 12.1 | Siphoviridae | S. Typhimurium | Sewage | South Korea | - |
Phage genomes are subjected to frequent horizontal gene transfer events that may obscure distant evolutionary relationships between them [50,51,52,53,54]. Additionally, they lack a universally conserved marker gene, which makes it difficult or impossible to determine phage phylogenies that extend deeply into evolutionary history, even with an abundance of phage genomes [52]. However, among small clusters of similar phage genomes, a core set of orthologs can allow us to infer the phylogenomic relationship among those phages. We identified 28 such orthologs for the Chi-like phages and inferred their phylogeny from the concatenated codon-aligned nucleotide sequences for these orthologs, as shown in Supplementary File 4. Internal nodes in the tree all have local support values (based on 1000 resamples of nearest-neighbor interchanges using the Shimodaira–Hasegawa test) of 100%, except for the node separating Salmonella phage FSL SP-030 and Salmonella phage FSL SP-088, which has a local support of 90.9%, as shown in Figure 4. Large local support values for every internal node suggested that this tree accurately represents the phylogeny of the Chi-like phages and that there is indeed a strong phylogenomic signal for the evolutionary history of this phage cluster. We did not root the tree due to the weak phylogenomic signal between the Chi-like cluster and other phage clusters. In agreement with the genome statistics in Table 2, the phylogenomic tree shows that phages 35 and 37 are the most distantly related to the rest of the cluster, meanwhile phages STm101 and STm118 form a distinct subclade with phages 118970 sal1, 35, and 37. The other major subclade is composed of phages FSL SP-039, FSL SP-124, SPN19, iEPS5, Chi, FSL SP-030, and FSL SP-088, while phage BP12C seems to be intermediately related to both major subclades, as shown in Figure 4.
Figure 4.
Phylogenomic tree representing the inferred evolutionary relationships of the Salmonella Chi-like phages. Local support values are calculated based on 1000 resamples using the Shimodaira–Hasegawa test on nearest-neighbor interchanges at each split. The tree was built from a concatenated codon-aware nucleotide alignment of 28 orthologs shared by the phages. The tree was built with FastTree (Version 2.1.10) using the generalized time reversible nucleotide substitution model.
3.4. Chi-Like Salmonella Phages Are Predominant among the Sampled Thai Poultry Farms
Genome sequence analysis identified the two phage isolates to be members of the Chi-like virus family. The quantification of Chi-like viruses across our 108 phage isolates was performed using PCR analysis with primers specific to the gene encoding virus capsid protein E [21,22]. Among 108 phages, 54 isolates, including STm101 and STm118, were positive as indicated by the presence of a 511 bp amplicon, as shown in Supplementary Figure S2. In addition, the percentage of Chi-like phage distribution between farms was not significantly different. Since all of these phages were isolated from chicken feces of four free-range farms in Northern Thailand and one commercial poultry farm in Central Thailand, we speculate that the Chi-like virus genus could be a predominant type among poultry farms in Thailand; however, wider sampling and further genetic analysis of Salmonella phage isolates across Thailand will be required to test this hypothesis.
3.5. Flagella and Lipopolysaccharide Are Required for Efficient Infection by the Two Thai Phages
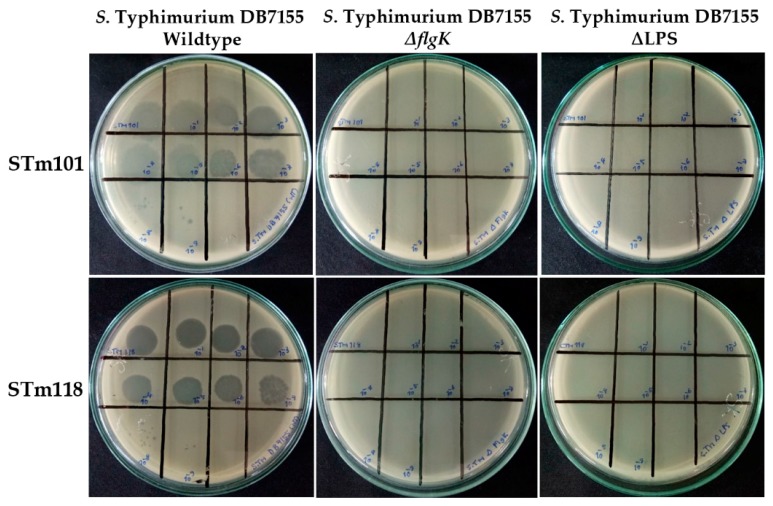
Flagellotropic phages first infect motile bacterial hosts by attaching to the rotating flagella [20,26,28]. After flagella attachment, phages move down the flagella filament towards the body of the host where they bind irreversibly to secondary receptor(s) prior to DNA ejection into the host [55]. For example, phages ϕCb13 and ϕCbK use a pilus protein PilA as the secondary receptor for host attachment, with deletion of pilA (ΔpilA) providing complete resistance to the bacteria from phage infection [19]. Recently, it was also shown that phage 7-7-1, a flagellotropic Myoviridae, uses the LPS of its host Agrobacterium sp. as a secondary receptor for infection [27]. To the best of our knowledge, only the flagella has been identified for Chi-like phages as a receptor, with little known regarding potential secondary receptors. Thus, to identify other secondary receptors used by STm101 and STm118, we tested the ability of the phages to infect and lyse (assessed by plaque formation) nine strains of S. Tm deficient for various cell wall components proposed as phage receptors, as shown in Table 3.
Table 3.
Results of spot lysis assays for STm101 and STm118 phage stocks were tested. The number of plaques formed are reported as being equivalent to wildtype (++), significantly reduced to wildtype (+), or no plaques formed (−).
| S. Typhimurium | Other Designations and Features | Source | STm101 | STm118 |
|---|---|---|---|---|
| Wildtype | DB7155 | [56] | ++ | ++ |
| ΔompA | DB7155, ΔompA::Kanr; outer membrane protein A | [56] | ++ | ++ |
| ΔompC | DB7155, ΔompC::Kanr; outer membrane protein C | [56] | ++ | ++ |
| ΔompX | DB7155, ΔompX::Kanr; outer membrane protein X | [56] | ++ | ++ |
| ΔbtuB | DB7155, ΔbtuB::Kanr; TonB-dependent vitamin B12 transporter | [56] | ++ | ++ |
| ΔfadL | DB7155, ΔfadL::Kanr; long-chain fatty acids transporter | [56] | ++ | ++ |
| Δtsx | DB7155, Δtsx::Kanr; nucleoside-specific outer membrane channel | [56] | ++ | ++ |
| ΔtonB | DB7155, ΔtonB::Kanr; active transport regulator protein TonB | [56] | ++ | ++ |
| ΔflgK | SL1344_CH502, ΔflgK::Kanr; hook-filament junction protein | [28] | - | - |
| ΔLPS | DB7155 ΔLPS::Cmr; all LPS synthesis genes for regions distal to KDO residues deleted. | [56] | - | - |
As expected for flagellotropic phages, plaque formation was inhibited for both phages against S. Tm ΔflgK (flagella hook protein deletion) compared to the wildtype strain, as shown in Figure 5. FlgK is a major component of the flagella, and therefore its deletion inhibits complete formation of this critical, primary receptor [28]. Plaque formation was not affected upon deletion of outer membrane porins (e.g., ΔOmpC) or molecular transporters (e.g., ΔFadL) compared to S. Tm wildtype and these were thus determined to not be receptors or necessary for host infection and lysis. Interestingly, both phages failed to produce plaques against S. Tm deficient in lipopolysaccharide (ΔLPS), as shown in Figure 5. This rough strain of Salmonella is completely devoid of carbohydrate substitutions to its KDO (2-keto-deoxy-d-octanoate) residues that form the core of the LPS [56]. While the LPS is a well-characterized receptor for many Gram-negative targeting phages [56,57,58,59,60], including the flagellotropic Agrobacterium phage 7-7-1 [27], this is the first study to suggest a role of the LPS for host infection by a member of the Chi-like phage genus.
Figure 5.
Spot tests of Salmonella phages STm101 and STm118 in serial dilution (10 μL of 1011–102 pfu/mL) against lawns of S. Typhimurium wildtype, ΔflgK, and ΔLPS grown on trypticase soy agar (TSA). Clear zones of lysis and single plaque formation were only observed for both phages against S. Tm wildtype.
4. Discussion
Rapid detection and remediation of Salmonella is essential for reducing the global burden associated with non-typhoidal Salmonella infections. Combined with increased emergence of multidrug resistant (MDR) Salmonella, innovative solutions are required to tackle the overuse of antibiotics in agriculture and food production [61,62], especially as these agents are often applied instead of proper hygiene measures. Phages have already been proven successful in reducing Salmonella contaminations in poultry [13,16], and for detection of food- and water-borne Salmonella [9,11,12].
Salmonella enterica is a highly diverse species of more than 2600 serovars [63]. In congruence to their highly diverse host, almost 250 Salmonella phages have so far been isolated and observed using electron microscopy [32]. In contrast, a smaller number of Salmonella phage genomes have been sequenced, meaning the study of Salmonella phage genomic diversity has remained rather limited. In this study, we isolated and characterized the biological and genomic properties of a diverse assortment of Salmonella phages isolated from poultry farms in Thailand. Host range analysis demonstrated that phages STm101 and STm118 could lyse approximately 50% of the 118 Salmonella tested strains. Unfortunately, the lysogenic lifestyle of this and other Chi-like phages is a major disadvantage for phage therapy, as virulence-associated genes could easily be spread among bacterial pathogens by phage integration [64]. However, genome-engineering strategies are rapidly advancing, and being applied to produce strictly lytic phages with adaptable host ranges or defined payloads [65] for clinical and diagnostic applications [66] including phage therapy in humans [67].
To investigate the genomic properties and diversity of the isolated Salmonella phages, the complete genomes of these phages were sequenced to identify them as members of the Chi-like virus genus. This also provides the first genomes of Salmonella Chi-like phages from Thailand, as shown in Table 2. PCR amplification of the Chi-like virus capsid protein E identified half of all phages isolated in this study as similar members of the Chi-like virus genus, suggesting that Chi-like phages could be a predominant phage family across these poultry farms. To explore the relationship between our isolated phages and previously reported Chi-like phages, we performed a phylogenomic analysis. Interestingly, phages STm101 and STm118 formed a monophyletic clade with phages isolated from Western countries, as shown in Figure 4; these included phage BP12C, isolated in USA (GenBank accession no. NC_031228) and phage 118970_sal1, isolated in Italy (GenBank accession no. NC_031930.1) [48], but not from closer isolated phages, such as phages SPN19 and iEPS5, isolated in Korea (GenBank accession no. NC_019417) [28]. However, further investigation and more phage isolates are required to investigate possible causes for this geographic distribution. Notably, the constructed phylogenomic tree was based on using the 28 core genes analysis and not the whole genome sequences. It may not reflect the relationships between the entire phages. Mavrich and Hatfull (2017) [68] reported that a set of core genes can be represented a phylogenomic relationship between phages only in the high-gene flux mode.
Initial recognition of the flagella by tail fibers of Chi-like phages is proposed as a mechanism to increase the likelihood of the phage identifying a motile and therefore metabolically active host that is suitable for progeny production [19,20]. Once the phage has navigated the length of the flagella to the cell body, it then attaches irreversibly to secondary receptors on the cell surface prior to triggered tail penetration and DNA ejection. Using S. Tm cells deficient in cell wall components, we identified the LPS as an essential component for efficient infection and lysis by phages STm101 and STm118. The LPS is composed of three distinct components—inner and outer cores and a hypervariable O-antigen—all of which are previously identified as receptors for phage adsorption. For example, Salmonella Podoviruses P22 and 9NA specifically recognize the repeating units of the O-antigen [10], meanwhile Salmonella phage S16 and E. coli phage T4 bind to the core LPS residues using two sets of tail fibers, which also triggers DNA ejection [56,69,70,71,72]. Identification of the specific LPS component recognized by the Chi-like phages, and how these are bound by the phage’s receptor-binding proteins, will be necessary to further our understanding of the infection process used by these unusual phages.
Acknowledgments
We kindly thank Lionello Bossi (Université Paris-Sud, France) for providing S. Tm MA8508, for phage propagation and S. Ryu (Seoul National University, Korea) for providing parental and ΔflgK strains of S. Tm [35].
Abbreviations
| bp | base pair |
| gp | gene product |
| kbp | kilo-base pair |
| LPS | lipopolysaccharide |
| OM | outer membrane |
| ORFs | open reading frames |
| SM buffer | saline magnesium buffer |
| S. Tm | S. Typhimurium |
| PCR | polymerase chain reaction |
| COG | cluster of orthologous group |
| AAI | average amino acid identity |
| TEM | transmission electron microscopy |
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4915/11/6/520/s1, Figure S1: A hierarchically clustering all Salmonella phage genomes according to their average amino acid identity, Figure S2: 50% of all Salmonella phage isolates from Thai poultry farms are putative members of the Chi-like virus genus, Table S1: Complete list of Salmonella strains used for phage host range analysis title, Table S2: List of all identified Salmonella phages with the country of phage isolation, Table S3: Infectivity of other Chi-like phages against 4 serovars of Salmonella enterica, Table S4: List of Salmonella phage STm101 annotated ORFs and gene products, Table S5: List of Salmonella phage STm118 annotated ORFs and gene products, Supplementary file 1 (CSV file):Video Identified Salmonella phages with accession numbersS1: title, Supplementary File 2 (TSV file): Average amino acid identities of Salmonella phages, Supplementary file 3 (TSV file): A shared core set of 28 orthologs from the sequences of 14 Chi-like phages, Supplementary file 4 (FNA file): The concatenated codon-aligned nucleotide sequences of the 28 orthologs.
Author Contributions
Conceptualization, S.K.; Funding acquisition, S.K.; Investigation, P.P., M.D., R.S., C.O., J.L., O.P., P.A., J.K., N.B., and M.I.; Resources, R.T., M.V., and R.K.; Supervision, M.D., M.R.J.C., E.E.G., and S.K.; Validation, P.P. and M.D.; Writing – original draft, P.P. and R.S.; Writing – review & editing, M.D., J.K., E.E.G., and S.K.
Funding
The National Science and Technology Development Agency (NSTDA), Thailand, and the Betagro Science Center Co. Ltd., Thailand supported this work. Additionally, Phothaworn P. was supported by the Royal Thai Golden Jubilee studentship (PHD/0137/2556).
Ethics Approval and Consent to Participate
Not applicable.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- 1.Koskella B., Brockhurst M.A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 2014;38:916–931. doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chibani-Chennoufi S., Bruttin A., Dillmann M.L., Brussow H. Phage-host interaction: An ecological perspective. J. Bacteriol. 2004;186:3677–3686. doi: 10.1128/JB.186.12.3677-3686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glazko G., Makarenkov V., Liu J., Mushegian A. Evolutionary history of bacteriophages with double-stranded DNA genomes. Biol. Direct. 2007;2:36. doi: 10.1186/1745-6150-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang H.Y., Dozois C.M., Tinge S.A., Lee T.H., Curtiss R., 3rd Transduction-mediated transfer of unmarked deletion and point mutations through use of counterselectable suicide vectors. J. Bacteriol. 2002;184:307–312. doi: 10.1128/JB.184.1.307-312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neal B.L., Brown P.K., Reeves P.R. Use of Salmonella phage P22 for transduction in Escherichia coli. J. Bacteriol. 1993;175:7115–7118. doi: 10.1128/jb.175.21.7115-7118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mann B.A., Slauch J.M. Transduction of low-copy number plasmids by bacteriophage P22. Genetics. 1997;146:447–456. doi: 10.1093/genetics/146.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabsch W. Salmonella Typhimurium phage typing for pathogens. Methods Mol. Biol. 2007;394:177–211. doi: 10.1007/978-1-59745-512-1_10. [DOI] [PubMed] [Google Scholar]
- 8.Jeoffreys N.J., James G.S., Chiew R., Gilbert G.L. Practical evaluation of molecular subtyping and phage typing in outbreaks of infection due to Salmonella enterica serotype Typhimurium. Pathology. 2001;33:66–72. doi: 10.1080/00313020124110. [DOI] [PubMed] [Google Scholar]
- 9.Dunne M., Loessner M.J. Modified bacteriophage tail fiber proteins for labeling, immobilization, capture, and detection of bacteria. Methods Mol. Biol. 2019;1918:67–86. doi: 10.1007/978-1-4939-9000-9_6. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt A., Rabsch W., Broeker N.K., Barbirz S. Bacteriophage tailspike protein based assay to monitor phase variable glucosylations in Salmonella O-antigens. BMC Microbiol. 2016;16:207. doi: 10.1186/s12866-016-0826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh A., Poshtiban S., Evoy S. Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors. 2013;13:1763–1786. doi: 10.3390/s130201763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen I.H., Horikawa S., Bryant K., Riggs R., Chin B.A., Barbaree J.M. Bacterial assessment of phage magnetoelastic sensors for Salmonella enterica Typhimurium detection in chicken meat. Food Control. 2017;71:273–278. doi: 10.1016/j.foodcont.2016.07.003. [DOI] [Google Scholar]
- 13.Hungaro H.M., Mendonca R.C.S., Gouvea D.M., Vanetti M.C.D., Pinto C.L.D. Use of bacteriophages to reduce Salmonella in chicken skin in comparison with chemical agents. Food Res. Int. 2013;52:75–81. doi: 10.1016/j.foodres.2013.02.032. [DOI] [Google Scholar]
- 14.Zinno P., Devirgiliis C., Ercolini D., Ongeng D., Mauriello G. Bacteriophage P22 to challenge Salmonella in foods. Int. J. Food Microbiol. 2014;191:69–74. doi: 10.1016/j.ijfoodmicro.2014.08.037. [DOI] [PubMed] [Google Scholar]
- 15.Bao H.D., Zhang P.Y., Zhang H., Zhou Y., Zhang L.L., Wang R. Bio-control of Salmonella Enteritidis in foods using bacteriophages. Viruses. 2015;7:4836–4853. doi: 10.3390/v7082847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waseh S., Hanifi-Moghaddam P., Coleman R., Masotti M., Ryan S., Foss M., MacKenzie R., Henry M., Szymanski C.M., Tanha J., et al. Orally administered P22 phage tailspike protein reduces Salmonella colonization in chickens: Prospects of a novel therapy against bacterial infections. PLoS ONE. 2010;5:e13904. doi: 10.1371/journal.pone.0013904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sertic V., Boulgakov N.A. Bactériophages spécifiques pour des variétés bactériennes flagellées. C. R. Soc. Biol. 1936;123:887–888. [Google Scholar]
- 18.Schmidt J.M., Stanier R.Y. Isolation and characterization of bacteriophages active against stalked bacteria. J. Gen. Microbiol. 1965;39:95–107. doi: 10.1099/00221287-39-1-95. [DOI] [PubMed] [Google Scholar]
- 19.Guerrero-Ferreira R.C., Viollier P.H., Ely B., Poindexter J.S., Georgieva M., Jensen G.J., Wright E.R. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc. Natl. Acad. Sci. USA. 2011;108:9963–9968. doi: 10.1073/pnas.1012388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meynell E.W. A phage, phi chi, which attacks motile bacteria. J. Gen. Microbiol. 1961;25:253–290. doi: 10.1099/00221287-25-2-253. [DOI] [PubMed] [Google Scholar]
- 21.Hendrix R.W., Ko C.C., Jacobs-Sera D., Hatfull G.F., Erhardt M., Hughes K.T., Casjens S.R. Genome sequence of Salmonella phage chi. Genome Announc. 2015;3 doi: 10.1128/genomeA.01229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J.H., Shin H., Choi Y., Ryu S. Complete genome sequence analysis of bacterial-flagellum-targeting bacteriophage chi. Arch. Virol. 2013;158:2179–2183. doi: 10.1007/s00705-013-1700-0. [DOI] [PubMed] [Google Scholar]
- 23.Switt A.I.M., Orsi R.H., den Bakker H.C., Vongkamjan K., Altier C., Wiedmann M. Genomic characterization provides new insight into Salmonella phage diversity. BMC Genom. 2013;14:481. doi: 10.1186/1471-2164-14-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kazaks A., Dislers A., Lipowsky G., Nikolajeva V., Tars K. Complete genome sequence of the Enterobacter cancerogenus bacteriophage Enc34. J. Virol. 2012;86:11403–11404. doi: 10.1128/JVI.01954-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Onmus-Leone F., Hang J., Clifford R.J., Yang Y., Riley M.C., Kuschner R.A., Waterman P.E., Lesho E.P. Enhanced de novo assembly of high throughput pyrosequencing data using whole genome mapping. PLoS ONE. 2013;8:e61762. doi: 10.1371/journal.pone.0061762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joys T.M. Correlation between susceptibility to bacteriophage PBS1 and motility in Bacillus subtilis. J. Bacteriol. 1965;90:1575–1577. doi: 10.1128/jb.90.6.1575-1577.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez F., Helm R.F., Broadway K.M., Scharf B.E. More than rotating flagella: lipopolysaccharide as a secondary receptor for flagellotropic phage 7-7-1. J. Bacteriol. 2018;200 doi: 10.1128/JB.00363-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi Y., Shin H., Lee J.H., Ryu S. Identification and characterization of a novel flagellum-dependent Salmonella-infecting bacteriophage, iEPS5. Appl. Environ. Microbiol. 2013;79:4829–4837. doi: 10.1128/AEM.00706-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards S., Meynell G.G. The widespred occurrence of enteric flagellar phages. J. Gen. Virol. 1968;2:443–444. doi: 10.1099/0022-1317-2-3-443. [DOI] [PubMed] [Google Scholar]
- 30.Clokie M.R.J., Kropinski A.M. Bacteriophages: Methods and Protocols. 1st ed. Humana Press; New York, NY, USA: 2009. [Google Scholar]
- 31.Huang X., Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ackermann H.W. 5500 Phages examined in the electron microscope. Arch. Virol. 2007;152:227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- 33.Ackermann H.W. Phage classification and characterization. Methods Mol. Biol. 2009;501:127–140. doi: 10.1007/978-1-60327-164-6_13. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press; New York, NY, USA: 2001. [Google Scholar]
- 35.Li D., Liu C.M., Luo R., Sadakane K., Lam T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 36.Li D., Luo R., Liu C.M., Leung C.M., Ting H.F., Sadakane K., Yamashita H., Lam T.W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 37.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 38.Sullivan M.J., Petty N.K., Beatson S.A. Easyfig: A genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coordinators N.R. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkw1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Contreras-Moreira B., Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 2013;79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Core Team. R: A Language and Environment for Statistical Computing. (2013) [(accessed on 17 May 2017)]; Available online: https://www.R-project.org/
- 42.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suyama M., Torrents D., Bork P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price M.N., Dehal P.S., Arkin A.P. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimodaira H., Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999;16:1114–1116. doi: 10.1093/oxfordjournals.molbev.a026201. [DOI] [Google Scholar]
- 46.McLaughlin M.R., King R.A. Characterization of Salmonella bacteriophages isolated from swine lagoon effluent. Curr. Microbiol. 2008;56:208–213. doi: 10.1007/s00284-007-9057-9. [DOI] [PubMed] [Google Scholar]
- 47.Schade S.Z., Adler J., Ris H. How bacteriophage chi attacks motile bacteria. J. Virol. 1967;1:599–609. doi: 10.1128/jvi.1.3.599-609.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paradiso R., Lombardi S., Iodice M.G., Riccardi M.G., Orsini M., Bolletti Censi S., Galiero G., Borriello G. Complete genome sequences of three Siphoviridae bacteriophages infecting Salmonella enterica serovar Enteritidis. Genome Announc. 2016;4:e00939-16. doi: 10.1128/genomeA.00939-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karpe Y.A., Kanade G.D., Pingale K.D., Arankalle V.A., Banerjee K. Genomic characterization of Salmonella bacteriophages isolated from India. Virus Genes. 2016;52:117–126. doi: 10.1007/s11262-015-1269-7. [DOI] [PubMed] [Google Scholar]
- 50.Jiang S.C., Paul J.H. Gene transfer by transduction in the marine environment. Appl. Environ. Microbiol. 1998;64:2780–2787. doi: 10.1128/aem.64.8.2780-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paul J.H. Microbial gene transfer: an ecological perspective. J. Mol. Microbiol. Biotechnol. 1999;1:45–50. [PubMed] [Google Scholar]
- 52.Rohwer F., Edwards R. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 2002;184:4529–4535. doi: 10.1128/JB.184.16.4529-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindell D., Sullivan M.B., Johnson Z.I., Tolonen A.C., Rohwer F., Chisholm S.W. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. USA. 2004;101:11013–11018. doi: 10.1073/pnas.0401526101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grose J.H., Casjens S.R. Understanding the enormous diversity of bacteriophages: The tailed phages that infect the bacterial family Enterobacteriaceae. Virology. 2014;468–470:421–443. doi: 10.1016/j.virol.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bender R.A., Refson C.M., O’Neill E.A. Role of the flagellum in cell-cycle-dependent expression of bacteriophage receptor activity in Caulobacter crescentus. J. Bacteriol. 1989;171:1035–1040. doi: 10.1128/jb.171.2.1035-1040.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marti R., Zurfluh K., Hagens S., Pianezzi J., Klumpp J., Loessner M.J. Long tail fibres of the novel broad-host-range T-even bacteriophage S16 specifically recognize Salmonella OmpC. Mol. Microbiol. 2013;87:818–834. doi: 10.1111/mmi.12134. [DOI] [PubMed] [Google Scholar]
- 57.Andres D., Baxa U., Hanke C., Seckler R., Barbirz S. Carbohydrate binding of Salmonella phage P22 tailspike protein and its role during host cell infection. Biochem. Soc. Trans. 2010;38:1386–1389. doi: 10.1042/BST0381386. [DOI] [PubMed] [Google Scholar]
- 58.Broeker N.K., Barbirz S. Not a barrier but a key: How bacteriophages exploit host’s O-antigen as an essential receptor to initiate infection. Mol. Microbiol. 2017;105:353–357. doi: 10.1111/mmi.13729. [DOI] [PubMed] [Google Scholar]
- 59.Washizaki A., Yonesaki T., Otsuka Y. Characterization of the interactions between Escherichia coli receptors, LPS and OmpC, and bacteriophage T4 long tail fibers. Microbiologyopen. 2016;5:1003–1015. doi: 10.1002/mbo3.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trojet S.N., Caumont-Sarcos A., Perrody E., Comeau A.M., Krisch H.M. The gp38 adhesins of the T4 superfamily: A complex modular determinant of the phage’s host specificity. Genome Biol. Evol. 2011;3:674–686. doi: 10.1093/gbe/evr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin D.M., Koskella B., Lin H.C. Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017;8:162–173. doi: 10.4292/wjgpt.v8.i3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loessner M.J., Hagens S., Slijkhuis A.J.H., Klumpp J.A., Marti R. Bacteriophage for biocontrol of Salmonella and in the manufacturing or processing of foods. Application No. 14/399,383. Patent. 2015 May 7;
- 63.Majowicz S.E., Musto J., Scallan E., Angulo F.J., Kirk M., O’Brien S.J., Jones T.F., Fazil A., Hoekstra R.M., Burd I.C.E.D., et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 64.Nilsson A.S. Phage therapy-constraints and possibilities. Ups. J. Med. Sci. 2014;119:192–198. doi: 10.3109/03009734.2014.902878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kilcher S., Loessner M.J. Engineering bacteriophages as versatile biologics. Trends Microbiol. 2019;27:355–367. doi: 10.1016/j.tim.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 66.Pires D.P., Cleto S., Sillankorva S., Azeredo J., Lu T.K. Genetically engineered phages: A review of advances over the last decade. Microbiol. Mol. Biol. Rev. 2016;80:523–543. doi: 10.1128/MMBR.00069-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dedrick R.M., Guerrero-Bustamante C.A., Garlena R.A., Russell D.A., Ford K., Harris K., Gilmour K.C., Soothill J., Jacobs-Sera D., Schooley R.T., et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019;25:730–733. doi: 10.1038/s41591-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mavrich T.N., Hatfull G.F. Bacteriophage evolution differs by host, lifestyle and genome. Nat. Microbiol. 2017;2:17112. doi: 10.1038/nmicrobiol.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leiman P.G., Arisaka F., van Raaij M.J., Kostyuchenko V.A., Aksyuk A.A., Kanamaru S., Rossmann M.G. Morphogenesis of the T4 tail and tail fibers. Virol. J. 2010;7 doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dunne M., Denyes J.M., Arndt H., Loessner M.J., Leiman P.G., Klumpp J. Salmonella phage S16 tail fiber adhesin features a rare polyglycine rich domain for host recognition. Structure. 2018;26:1573–1582. doi: 10.1016/j.str.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 71.Taylor N.M., Prokhorov N.S., Guerrero-Ferreira R.C., Shneider M.M., Browning C., Goldie K.N., Stahlberg H., Leiman P.G. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature. 2016;533:346–352. doi: 10.1038/nature17971. [DOI] [PubMed] [Google Scholar]
- 72.Thomassen E., Gielen G., Schutz M., Schoehn G., Abrahams J.P., Miller S., van Raaij M.J. The structure of the receptor-binding domain of the bacteriophage T4 short tail fibre reveals a knitted trimeric metal-binding fold. J. Mol. Biol. 2003;331:361–373. doi: 10.1016/S0022-2836(03)00755-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.